Abstract
The virological response (VR) to a tipranavir-ritonavir (TPV-RTV)-based regimen had been shown to be associated with a number of mutations in the protease gene, the use of enfuvirtide (T20), and the TPV phenotypic inhibitory quotient (IQ). The role of the TPV genotypic IQ (gIQ) has not yet been fully investigated. The aim of our study was to evaluate the relationship between the TPV gIQ and the VR at 48 weeks to TPV-based salvage regimens. Patients placed on regimens containing two nucleoside reverse transcriptase inhibitors plus TPV-RTV 500/200 mg twice a day with or without T20 were prospectively studied. Regular follow-up was performed over the study period. VR, considered a viral load (VL) decrease of ≥1 log unit and/or the achievement of <50 copies/ml with no VL rebound of >0.5 log unit compared to the maximal VL decrease at week 48, was assessed. Thirty-eight patients who had received multiple drugs were included. At week 48 the VL decrease was −1.48 (interquartile range [IQR], −2.88 to −0.48), 15 patients (39.5%) had VLs of <50 copies/ml, and the CD4+ cell count increase was 37 cells/mm3 (IQR, −30 to +175). Twenty subjects (52.6%) achieved VRs. The TPV gIQ and optimized background score (OBS) were independently associated with higher VL decreases. The TPV gIQ and OBS were also independent predictors of a VR at week 48. TPV gIQ and OBS cutoff values of 14,500 and 2, respectively, were associated with a higher rate of VR. The TPV gIQ was shown to be able to predict the VR at 48 weeks to TPV-containing salvage regimens better than the TPV trough concentration or TPV-associated mutations alone. A possible TPV gIQ cutoff value of 14,500 for reaching a VR at week 48 was suggested. Further studies are needed in order to evaluate the calculation of TPV gIQ as a new tool for the optimization of TPV-based salvage therapy.
Tipranavir (TPV) is a nonpeptidic protease inhibitor with potent in vitro activity against most human immunodeficiency virus (HIV) type 1 (HIV-1) strains resistant to other protease inhibitors (PIs) (1, 14, 15). In vitro data have shown that resistance to TPV develops slowly (5). When TPV is coadministered with ritonavir (RTV) as a booster, TPV has been shown to have potent antiviral activity in multidrug-experienced patients (3, 6, 9, 12). In the RESIST-1 and the RESIST-2 studies, the efficacy and safety of TPV-RTV (500 mg/200 mg twice daily) in 1,509 highly treatment-experienced HIV-1-positive patients were assessed. Analysis at 48 weeks showed that the TPV-RTV-containing regimens significantly improved the immune and virological responses (VRs) compared to the responses to an RTV-boosted comparator PI plus an optimized background (OB) regimen (3, 6, 8).
Different factors have been found to be associated with the virological and immunological responses: a lower viral load (VL) at the baseline, the use of enfuvirtide (T20) as a part of the OB regimen, the presence of two or more active drugs in the OB regimen (OB score [OBS], ≥2) (3, 6, 8), and the baseline numbers of specific TPV-associated resistance mutations (TPV RMs) (2). Moreover, the TPV trough concentration (Ctrough) and phenotypic inhibitory quotient (IQ) have also been shown to be associated with the VR at week 24 (12a, 16).
The genotypic IQ (gIQ), which is the ratio between the PI Ctrough and the number of PI-associated mutations, is simpler to derive than the IQ in the clinical setting. The gIQ has previously been shown to be a predictor of the therapeutic response to PI-based salvage regimens, e.g., lopinavir or fosamprenavir (7, 10, 11, 13). Preliminary data showed that TPV-gIQ correlated with the early and the middle VRs (2a, 2b). However, no data are yet available on the predictive value of the TPV gIQ on the long-term efficacy of TPV-based regimens. Therefore, the aim of our study was to perform a pharmacokinetic/pharmacodynamic evaluation of the predictors of the VR at week 48 to salvage TPV-containing regimens in the clinical setting.
MATERIALS AND METHODS
Study design.
Patients enrolled in the TPV Expanded Access Program study and administered regimens containing two nucleoside reverse transcriptase inhibitors (NRTIs) plus TPV-RTV 500/200 mg twice a day with or without T20 were prospectively evaluated. The criteria for the inclusion of the patient data in the final analysis were a baseline plasma HIV RNA load of >1,000 copies/ml, determination of the HIV genotype and the virtual phenotype in the 6 months before the initiation of the TPV-based regimen, regular follow-up through week 48, the availability of at least one TPV Ctrough measurement, and a self-reported treatment adherence rate of more than 90% in the last 7 days before each visit. The HIV RNA load and CD4+ cell count were assayed by reverse transcription-PCR (Cobas Amplicor HIV-1 Monitor test, version 1.5; Roche Molecular Systems, Switzerland) and flow cytometry, respectively, at the baseline and at weeks 12, 24, and 48 as indicators of the early, mid-, and long-term responses, respectively.
Study end points.
A VR was considered an HIV RNA load decrease of ≥1 log unit and/or achievement of an HIV RNA load of <50 copies with no HIV RNA load increase of >0.5 log unit compared to the maximal VL decrease. The intention-to-treat last-observation-carried-forward method was used. In this intention-to-treat approach, for subjects who discontinued TPV before week 48 for reasons other than virological failure (VF), the last HIV RNA load and CD4+ cell count recorded before TPV withdrawal were considered for further analysis.
Calculation of OBS.
OBS was calculated by relying on the genotypic sensitivity score. It was calculated by using the Virtual Phenotype program, version 3.6 (Virco). The drugs included in the background regimen to which the virus was reported to have full or partial susceptibility by use of the Virtual Phenotype program were scored as 1, while the drugs reported to be inactive by use of the Virtual Phenotype program were scored as 0. OBS was defined as the sum of the genotypic sensitivity scores of all drugs included in the regimen. In subjects administered T20, this drug was considered inactive if it had previously been administered to the subject and was discontinued after VF.
Genotypic resistance test.
A genotypic resistance test was performed at the baseline for subjects with VLs of >1,000 copies/ml by using the ViroSeq HIV-1 genotyping system (Celera Diagnostics, LCC, Alameda, CA) and an automatic sequencer (ABI Prism 3100; PE Biosystems, Foster City, CA). Genotype interpretation was made by use of the mutation score proposed recently (2) and was used in the combined analysis of the data from the 48-week RESIST 1 and RESIST 2 trials (8). According to this score, amino acid changes within the protease gene at positions L10V, I13V, K20M/R/V, L33F, E35G, M36I, K43T, M46L, I47V, I54A/M/V, Q58E, H69K, T74P, V82L/T, N83D, and I84V were considered TPV RMs.
Pharmacokinetic analysis.
Blood samples were collected and placed into lithium heparin-containing tubes before the morning dose; and the plasma was separated by centrifugation at 5,000 rpm, refrigerated at 4°C for 10 min, and then stored at −70°C until analysis. At the time of blood sampling, the patients were asked about the time of their last TPV dose intake. Only plasma samples obtained between 10 and 14 h postdosing were considered for Ctrough analysis. TPV concentrations were determined by using a validated high-performance liquid chromatography method with UV detection, which was linear over the range of 1,000 to 180,000 ng/ml. The intraday and interday precisions (coefficients of variation) ranged from 0.94 to 2.55% and 3.07 to 4.24%, respectively. The limit of quantification and the limit of detection were 90 ng/ml and 35 ng/ml, respectively (4). In subjects for whom more than one pharmacokinetic measurement was available, the mean value of all available Ctroughs throughout the study period was considered. The gIQ was calculated for each patient as the ratio between the mean TPV Ctrough and the baseline number of TPV RMs. The gIQ was expressed as ng/ml/mutation.
Statistical analyses.
Linear and logistic regression analyses were used to investigate the factors associated with higher HIV RNA load decreases and VRs. Variables showing P values of <0.05 by univariate analysis were considered for the multivariate analysis by the forward conditional method.
Receive operator characteristic (ROC) curve analysis was used to explore possible cutoff values for variables predictive by logistic regression analysis. The χ2 test was used to analyze the association between categorical variables. Statistical significance was considered a P value of <0.05. Statistical analysis was performed with SPSS software (2004, version 13.0; SPSS Inc., Chicago, IL).
RESULTS
Study population characteristics.
Thirty-eight subjects (84.2% male) were included in the study. Eight (21%) patients were coinfected with hepatitis C virus. The median numbers of previous PIs and NRTIs received were five (interquartile range [IQR], four to six) and five (IQR, five to six), respectively. The subjects had previous failures to a median of five (IQR, five to six) PI-based regimens.
The OB regimen included a median of three drugs (range, two to four drugs). Twenty of 38 subjects were administered with T20, and 6 of these 20 subjects were T20 experienced and had had previous VFs while they were on a regimen that included this drug. The median OBS, based on virtual phenotype interpretation, was two (range, zero to three). Complete study population characteristics are reported in Table 1.
TABLE 1.
Demographics and baseline characteristics of study population
| Characteristic | Result for total population |
|---|---|
| Total no. of patients | 38 |
| No. (%) of male subjects | 32 (84.2) |
| Median (IQR) age (yr) | 45 (39-49) |
| Median (IQR) wt (kg) | 69 (60-76) |
| Median (IQR) ht (cm) | 174 (170-180) |
| No. (%) of subjects with hepatitis C virus coinfection | 8 (21) |
| No. (%) of subjects with clinical status ofa: | |
| A | 14 (36.8) |
| B | 13 (34.2) |
| C | 11 (28.9) |
| Pharmacological history | |
| Median (range) no. of previous PIs | 5 (1-7) |
| Median (range) no. of PIs with VF | 4 (1-7) |
| Median (range) no. of previous NRTIs | 5 (2-6) |
| Median (range) no. of NRTIs with VF | 5 (2-6) |
| No. (%) of subjects previously treated with TDFb | 33 (86.8) |
| No. (%) of subjects previously treated with TDF and with VF | 31 (81.6) |
| Median (range) no. of previous NNRTIsc | 1 (0-2) |
| Median (range) of NNRTIs with VF | 1 (0-2) |
| No. (%) of subjects previously treated with T20 | 8 (21) |
| No. (%) of subjects previously treated with T20 and with VF | 6 (15) |
| OB regimen | |
| Median (range) no. of drugs in OB regimen | 3 (2-4) |
| No. (%) of subjects with OB regimen with T20 | 20 (52.6) |
| No. of subjects previously treated with T20 with VF/total no. of subjects receiving T20 (%) | 6/20 (30) |
| Median (range) OBS (no. of drugs in OB regimen considered active) | 2 (0-3) |
| No. of subjects with the following OBS: | |
| 0 | 3 (7.9) |
| 1 | 7 (18.4) |
| 2 | 21 (55.3) |
| 3 | 7 (18.4) |
| No. (%) of subjects with the following no. of TPV RMs: | |
| 0 | 3 (7.9) |
| 1 | 2 (5.2) |
| 2 | 6 (15.8) |
| 3 | 11 (28.9) |
| 4 | 9 (81.8) |
| 5 | 6 (15.8) |
| 6 | 1 (2.6) |
| Baseline immunovirology | |
| Median (IQR) log HIV RNA load | 4.75 (4.19-5.14) |
| Median (IQR) no. of CD4+ cells/ml | 241 (100-399) |
| Median (IQR) CD4+ cell % | 14.5 (10.9-20.3) |
Clinical status according to the 1993 CDC classification.
TDF, tenofovir disoproxil fumarate.
NNRTIs, nonucleoside reverse transcriptase inhibitors.
Virological and immunological outcomes.
The median CD4+ cell count increases were 24 cells/ml (IQR, −29 to 125 cells/ml), 40 cells/ml (IQR, −14 to 161 cells/ml), and 37 cells/ml (IQR, −30 to 175 cells/ml) at 12, 24, and 48 weeks, respectively. The median decays in the plasma HIV RNA load were −2.08 log units (IQR, −3.3 to −0.5 log units), −2.1 log units (IQR, −2.9 to −0.7 log units), and −1.48 log units (IQR, −2.88 to −0.48 log units) at 12, 24, and 48 weeks, respectively. VRs were observed in 23 (60.5%), 25 (65.8%), and 20 (52.6%) subjects at 12, 24, and 48 weeks, respectively; and the VLs were <50 copies/ml in 16 subjects (42.1%), 14 subjects (36.8%), and 15 subjects (39.5%) at 12, 24, and 48 weeks, respectively. The immunovirological outcomes are reported in Table 2.
TABLE 2.
Evolution of immunovirological parameters and proportion of VRs through week 48
| Time of analysis | Median (IQR) HIV RNA load (log) | Median (IQR) HIV RNA load variation (log) from baseline | No. (%) of subjects with VRa | No. (%) of subjects with <50 HIV RNA copies/ml | Median (IQR) CD4+ cell count (cells/mm3) | Median (IQR) CD4+ variation (cells/mm3) from baseline |
|---|---|---|---|---|---|---|
| Baseline | 4.75 (4.19-5.14) | 0 (0) | 241 (100-399) | |||
| Wk 12 | 2.07 (<1.7-4.31) | −2.08 (−3.3, −0.5) | 23 (60.5) | 16 (42.1) | 262 (185-402) | +24 (−29, +125) |
| Wk 24 | 2.14 (<1.7-4.25) | 2.1 (−2.9, −0.7) | 25 (65.8) | 14 (36.8) | 297 (184-449) | +40 (−14, +161) |
| Wk 48 | 2.36 (<1.7-4.45) | −1.48 (−2.88, −0.48) | 20 (52.6) | 15 (39.5) | 323 (181-459) | +37 (−30, +175) |
VR was considered an HIV RNA load decrease of >1 and/or <50 copies/ml, with no HIV RNA increase >0.5 log compared to the maximal viral load decrease.
Pharmacokinetic and genotypic analyses.
A total of 190 plasma samples from 38 subjects were collected for pharmacokinetic analysis. A median of five samples (IQR, four to six samples) were obtained from each subject. The overall mean of all the available TPV Ctrough measurements through 48 weeks was 31,937 ng/ml (standard deviation, ±14,106 ng/ml) (Table 3).
TABLE 3.
Overall pharmacokinetic analysisa
| Time of analysis | Mean (SD) TPV Ctrough (ng/ml) |
|---|---|
| Wk 2 (n = 27) | 39,664 (17,477) |
| Wk 4 (n = 25) | 32,266 (19,323) |
| Wk 8 (n = 28) | 30,310 (24,374) |
| Wk 12 (n = 31) | 27,169 (19,423) |
| Wk 24 (n = 29) | 31,980 (18,800) |
| Wk 36 (n = 25) | 30,569 (21,226) |
| Wk 48 (n = 25) | 32,293 (18,078) |
| Overall mean (n = 36) | 31,406 (14,526) |
The mean TPV Ctrough was calculated as the mean of all available Ctrough measurements. Plasma samples obtained between 10 and 14 h postdosing were considered.
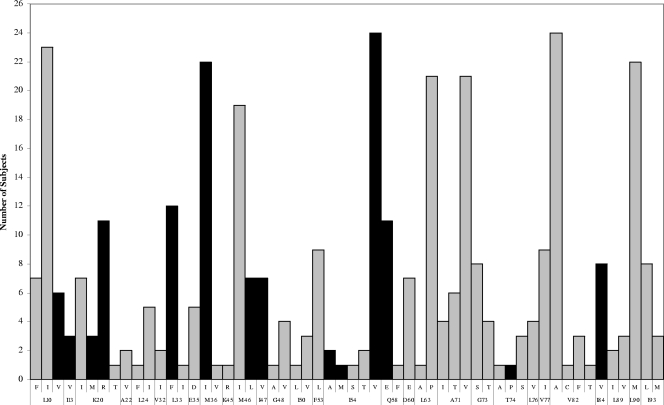
The median number of TPV RMs was three (IQR, two to four). No TPV RMs were detected in three subjects. The specific amino acid changes at specific protease codons are reported in Fig. 1. The overall median TPV gIQ was 9,688 ng/ml/mutation (IQR, 6,379 to 15,739 ng/ml/mutation).
FIG. 1.
Frequency of specific amino acid changes at specific codons in the protease gene. Black bars represent amino acid changes considered to be TPV RMs.
Pharmacological determinants of VR.
By univariate linear regression analysis, a higher VL decrease at week 48 was associated with a higher OBS (R = −0.387; P = 0.016) and a higher gIQ (R = −0.391; P = 0.015), while an association of a higher VL decrease with a lower number of TPV RMs showed a trend toward significance (R = 0.302; P = 0.066). By multivariate analysis, only OBS (R = −0.343; P = 0.025) and gIQ (R = −0.347; P = 0.023) were confirmed to be independent predictors of a higher VL decrease.
By univariate logistic regression analysis, OBS (P = 0.035), gIQ (P = 0.046), and the number of TPV RMs (P = 0.045) were shown to be predictors of a VR. By multivariate analysis, although OBS was the only factor that independently predicted a VR (P = 0.035), gIQ (P = 0.055) was also included in the final model, providing a better overall prediction of a VR than that observed in the other models tested. The results of these analyses are reported in Tables 4 and 5.
TABLE 4.
Summary results of linear regression analysisa
| Variable | Univariate analysis
|
Multivariate analysis
|
||
|---|---|---|---|---|
| R | P value | R | P value | |
| Baseline log VL | −0.147 | 0.378 | ||
| Baseline CD4+ cell count | −0.047 | 0.77 | ||
| OBS | −0.441 | 0.016 | −0.343 | 0.025 |
| TPV RM score | 0.302 | 0.06 | ||
| TPV Ctrough | 0.007 | 0.96 | ||
| gIQ3 | −0.391 | 0.015 | −0.347 | 0.023 |
| T20 coadministration | 0.039 | 0.81 | ||
The VL decrease from the baseline to week 48 was considered the dependent variable, whereas the variables listed in the first column were tested as independent variables. Boldface data indicate statistically significant results.
TABLE 5.
Summary results of logistic regression analysisa
| Variable |
P value
|
|
|---|---|---|
| Univariate analysis | Multivariate analysis | |
| Baseline log VL | 0.211 | |
| Baseline CD4+ cell count | 0.191 | |
| OBS | 0.025 | 0.035 |
| TPV RMs score | 0.045 | 0.38 |
| TPV Ctrough | 0.86 | |
| gIQ | 0.036 | 0.055 |
| T20 coadministration | 0.73 | |
A VR at 48 weeks was considered the dependent variable, whereas the variables listed in the first column were tested as independent variables. Boldface data indicate statistically significant results.
The OBS cutoff value for VR was calculated to be 2 by ROC curve analysis. It had a 90% sensitivity and a 44% specificity in predicting a VR at 48 weeks. At this time point, 18/28 (64.2%) subjects with an OBS of ≥2 showed a VR, whereas it was observed in only 2/10 (20%) subjects with an OBS <2 (χ2 = 5.79; P = 0.016). In the same way, a TPV gIQ cutoff value of 14,500 ng/ml/mutation was calculated by using ROC curve analysis. This provided a sensitivity of 50% and a specificity of 89% in predicting a VR at 48 weeks. Ten of 12 (83.3%) subjects with a TPV gIQ of >14,500 ng/ml/mutation achieved a VR, whereas the latter was recorded in only 10/26 (38.4%) subjects with a gIQ of ≤14,500 ng/ml/mutation (χ2 = 6.6; P = 0.015).
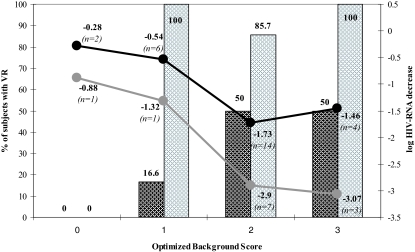
In order to evaluate the interplay between OBS and gIQ, both the VL decrease and the VR at 48 weeks were stratified according to these two variables. All values are represented graphically in Fig. 2. In the subgroup of subjects with a TPV gIQ of ≤14,500 ng/ml/mutation (n = 26), the plasma HIV RNA load decrease was significant only in case of an optimal OBS (≥2). In subjects with a TPV gIQ of >14,500 ng/ml/mutation (n = 12), a maximal VL decrease was similarly reached when the OBS was ≥2. However, differences in the magnitude of the VL decrease between the two groups could be observed. Moreover, among the subjects with a TPV gIQ of ≤14,500 ng/ml/mutation, the proportion of those with a VR gradually increased according to the OBS, achieving the maximal value (50%) when the latter was ≥2. Nevertheless, in subjects with a TPV gIQ of >14,500 ng/ml/mutation, the proportion of those with a VR was already maximal when the OBS was 1. Unfortunately, the limited sample size of each subgroup makes a statistical comparison impossible.
FIG. 2.
Mean plasma HIV RNA level decrease (lines) and proportion of the VR at week 48 (bars) stratified according to the gIQ and the OBS. Data from subjects with a TPV gIQ of ≤14,500 are in black, and those from subjects with a TPV gIQ of >14,500 are in gray.
DISCUSSION
Our study confirmed that TPV-based regimens can be effective salvage options in more than 50% of heavily pretreated subjects. Moreover, 39% of subjects administered TPV-RTV had less than 50 HIV RNA copies/ml at week 48. These results are consistent with previously published results of the data from the RESIST 1 and the RESIST 2 studies analyzed in combination. In these large trials, 58.5% of subjects reached a VR (defined as a more than 1-log-unit decrease in the plasmatic HIV RNA load), and 30.4% of patients achieved HIV RNA loads of <400 copies/ml at 48 weeks (8).
In our population, OBS and the TPV gIQ were shown to be factors associated with a VL decrease and a VR. The former was a confirmation of the results from the RESIST trials. In multidrug-experienced patients who were administered TPV and who had limited therapeutic options due to several failures of previous regimens, the choice of the optimal OB regimen with the highest number of drugs with residual activity is a crucial challenge. In our patients, in fact, OBS was an independent predictor of VR by both multivariate linear and logistic regression analyses. Moreover, subjects with two or more active drugs in the OB regimen were more likely to achieve a VR than those with less active drugs, confirming the possible OBS cutoff suggested previously (3, 6, 8).
Our study, however, is the first to analyze the long-term response according to the TPV gIQ. This parameter, which integrates pharmacokinetic and pharmacodynamic variables, was shown to be a better predictor of both a higher VL decrease and a VR at 48 weeks than the TPV Ctrough value or the number of TPV RMs considered separately. Although the number of TPV RMs, in fact, was shown to be associated with a higher VL decrease by univariate linear regression analysis, this association was not confirmed by multivariate analyses (Table 4 and Table 5). In previous reports of the findings from RESIST trials, the magnitude of the TPV plasma exposure was shown to be crucial for achievement of the effective inhibition of HIV strains, according to the individual phenotypic IQ. The TPV-RTV standard dosing could result in different plasma exposures, due to the interindividual variability of the TPV pharmacokinetics. As a consequence, the achievement of an adequate exposure in a single patient could be unpredictable, especially against strains carrying a higher number of TPV RMs. The phenotypic resistance test is complex and cannot feasibly be performed in laboratory practice; therefore, the phenotypic IQ is not useful in the clinical setting. The gIQ calculation, however, is more practical and affordable in such a context. Moreover, in our study a gIQ cutoff value of 14,500 ng/ml/mutation for a VR at 48 weeks was suggested. In other words, a TPV Ctrough of 14,500 ng/ml per each TPV RM is suggested to achieve a high probability of a VR. In this way, early TPV gIQ calculation and possible dose individualization could be options that might be explored for use in the difficult setting of deep salvage therapy.
Combined analysis of both predictive variables (TPV gIQ and OBS), shown in Fig. 2, suggested further clinical considerations. A TPV gIQ value above 14,500 ng/ml/mutation was associated with a higher virological efficacy than a TPV gIQ value below this cutoff and showed an additional VL decrease between 1.2 and 1.5 log units even in association with a high OBS (equal to or greater than 2). In a similar way, the proportion of patients with a VR also increased from 50% in subjects with a lower TPV gIQ to 87 to 100% in patients showing TPV gIQ values of >14,500 ng/ml/mutation in association with a OBS of ≥2. Moreover, although it is of anecdotal value, VR was also achieved in the only subjects with an OBS of 1 and a TPV gIQ above 14,500 ng/ml/mutation, whereas in this OBS stratum, this was true for only 16% of subjects with a lower TPV gIQ. From a clinical viewpoint, these findings suggest that optimization of the TPV gIQ should also be done in order to increase the probability of a VR in patients, with the expectation that the residual activity of drugs in association with TPV-RTV would be good.
In our study, the use of T20 as an active drug was considered in the calculation of the OBS, whereas it did not result per se as an independent predictor of VR, as it was in the RESIST trials (3, 6, 8). This could be due to the limited sample size of our study and/or to a possible imbalance in the clinical stages of the patients selected for evaluation of this association. Subjects administered T20, in fact, showed a slightly higher number of TPV RMs than the other patients, although this difference did not reach statistical significance (data not shown).
The sample size was a main limit of our analysis. Although the number of subjects included allowed univariate regression and multivariate regression analyses with three or fewer independent variables, some other variables potentially analyzed in the model remained at borderline significance, such as the number of TPV RMs. Moreover, in the calculation of the TPV gIQ, all TPV RMs were equally weighted as a unitary value, while they are supposed to affect drug susceptibility to different degrees. However, the lack of a consensual weighted score for such mutations allowed the easy and fast interpretation of the genotypic results in the clinical setting. Another possible bias could be the survival effect, due to the early discontinuation of treatment in subjects failing or intolerant of the therapy. However, the first discontinuation due to intolerance was after 85 days, and many patients showing VF were maintained on the TPV-containing regimen until the availability of a new salvage drug. Therefore, the survival effect should not significantly affect the analysis of the 48-week efficacy. Moreover, the survival effect was also not considered in the RESIST trials analysis due to study design considerations.
In conclusion, our findings suggest that TPV gIQ is an independent predictor of a long-term VR to TPV-based regimens. Therefore, the TPV gIQ cutoff value proposed warrants further evaluation in prospective therapeutic drug monitoring-guided dose modification clinical trials.
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Back, N. K., A. van Wijk, D. Remmerswaal, M. van Monfort, M. Nijhuis, R. Schuurman, and C. A. Boucher. 2000. In-vitro tipranavir susceptibility of HIV-1 isolates with reduced susceptibility to other protease inhibitors. AIDS 14:101-102. [DOI] [PubMed] [Google Scholar]
- 2.Baxter, J., J. Schapiro, C. Boucher, V. Kohlbrenner, D. Hall, J. Scherer, and D. Mayers. 2006. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J. Virol. 80:10794-10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Bonora, S., D. Gonzalez De Requena, A. Calcagno, M. G. Milia, A. D'Avolio, M. Sciandra, S. Garazzino, M. Siccardi, A. Sinicco, and G. Di Perri. 2006. Abstr. 13th Conf. Retrovir. Opportunistic Infect., abstr. 577.
- 2b.Bonora, S., D. Gonzalez de Requena, A. Calcagno, M. G. Milia, A. D'Avolio, M. Sciandra, S. Garazzino, M. Siccardi, A. Sinicco, and G. Di Perri. 2006. Abstr. 7th Int. Workshop Clin. Pharmacol. HIV Ther., abstr. 83.
- 3.Cahn, P., J. Villacian, A. Lazzarin, C. Katlama, B. Grinsztejn, K. Arasteh, P. Lopez, N. Clumeck, J. Gerstoft, N. Stavrianeas, S. Moreno, F. Antunes, D. Neubacher, and D. Mayers. 2006. Ritonavir-boosted tipranavir demonstrates superior efficacy to ritonavir-boosted protease inhibitors in treatment-experienced HIV-infected patients: 24-week results of the RESIST-2 trial. Clin. Infect. Dis. 43:1347-1356. [DOI] [PubMed] [Google Scholar]
- 4.D'Avolio, A., M. Sciandra, M. Siccardi, L. Baietto, D. Gonzalez de Requena, S. Bonora, and G. Di Perri. 2006. A simple and sensitive assay for determining plasma tipranavir concentration in the clinical setting by new HPLC method. J. Chromatogr. B 848:374-378. [DOI] [PubMed] [Google Scholar]
- 5.Doyon, L., S. Tremblay, L. Bourgon, E. Wardrop, and M. G. Cordingley. 2005. Selection and characterization of HIV-1 showing reduced susceptibility to the non-peptidic protease inhibitor tipranavir. Antivir. Res. 68:27-35. [DOI] [PubMed] [Google Scholar]
- 6.Gathe, J., D. A. Cooper, C. Farthing, D. Jayaweera, D. Norris, G. Pierone, Jr., C. R. Steinhart, B. Trottier, S. L. Walmsley, C. Workman, G. Mukwaya, V. Kohlbrenner, C. Dohnanyi, S. McCallister, D. Mayers, and the RESIST-1 Study Group. 2006. Efficacy of the protease inhibitors tipranavir plus ritonavir in treatment-experienced patients: 24-week analysis from the RESIST-1 trial. Clin. Infect. Dis. 43:1337-1346. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez de Requena, D., O. Gallego, L. Valer, I. Jimenez-Nacher, and V. Soriano. 2004. Prediction of virological response to lopinavir/ritonavir using the genotypic inhibitory quotient. AIDS Res. Hum. Retrovir. 20:275-278. [DOI] [PubMed] [Google Scholar]
- 8.Hicks, C. B., P. Cahn, D. A. Cooper, S. L. Walmsley, C. Katlama, B. Clotet, A. Lazzarin, M. A. Johnson, D. Neubacher, D. Mayers, H. Valdez, and the RESIST Investigator Group. 2006. Durable efficacy of tipranavir-ritonavir in combination with an optimised background regimen of antiretroviral drugs for treatment-experienced HIV-1-infected patients at 48 weeks in the Randomized Evaluation of Strategic Intervention in multi-drug reSistant patients with Tipranavir (RESIST) studies: an analysis of combined data from two randomised open-label trials. Lancet 368:466-475. [DOI] [PubMed] [Google Scholar]
- 9.MacGregor, T. R., J. P. Sabo, S. H. Norris, P. Johnson, L. Galitz, and S. McCallister. 2004. Pharmacokinetic characterization of different dose combinations of coadministered tipranavir and ritonavir in healthy volunteers. HIV Clin. Trials 5:371-382. [DOI] [PubMed] [Google Scholar]
- 10.Marcelin, A. G., C. Dalban, G. Peytavin, C. Lamotte, R. Agher, C. Delaugerre, M. Wirden, F. Conan, S. Dantin, C. Katlama, D. Costagliola, and V. Calvez. 2004. Clinically relevant interpretation of genotype and relationship to plasma drug concentrations for resistance to saquinavir-ritonavir in human immunodeficiency virus type 1 protease inhibitor-experienced patients. Antimicrob. Agents Chemother. 48:4687-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcelin, A. G., C. Lamotte, C. Delaugerre, N. Ktorza, H. Ait Mohand, R. Cacace, M. Bonmarchand, M. Wirden, A. Simon, P. Bossi, F. Bricaire, D. Costagliola, C. Katlama, G. Peytavin, V. Calvez, and the Genophar Study Group. 2003. Genotypic inhibitory quotient as predictor of virological response to ritonavir-amprenavir in human immunodeficiency virus type 1 protease inhibitor-experienced patients. Antimicrob. Agents Chemother. 47:594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCallister, S., H. Valdez, K. Curry, T. MacGregor, M. Borin, W. Freimuth, Y. Wang, and D. L. Mayers. 2004. A 14-day dose-response study of the efficacy, safety, and pharmacokinetics of the non peptidic protease inhibitor tipranavir in treatment-naive HIV-1-infected patients. J. Acquir. Immune Defic. Syndr. 35:376-382. [DOI] [PubMed] [Google Scholar]
- 12a.Naeger, L., J. Zheng, and K. Struble. 2006. Abstr. 13th Conf. Retrovir. Opportunistic Infect., abstr. 639a.
- 13.Pellegrin, I., D. Breilh, J. M. Ragnaud, S. Boucher, D. Neau, H. Fleury, M. H. Schrive, M. C. Saux, J. L. Pellegrin, E. Lazaro, and M. Vray. 2006. Virological responses to atazanavir-ritonavir-based regimens: resistance-substitutions score and pharmacokinetic parameters (Reyaphar study). Antivir. Ther. 11:421-429. [PubMed] [Google Scholar]
- 14.Poppe, S. M., D. E. Slade, K. T. Chong, R. R. Hinshaw, P. J. Pagano, M. Markowitz, Ho, D. D., Mo, H., R. R. Gorman III, T. J. Dueweke, S. Thaisrivongs, and W. G. Tarpley. 1997. Antiviral activity of the dihydropyrone PNU-140690, a new nonpeptidic human immunodeficiency virus protease inhibitor. Antimicrob. Agents Chemother. 41:1058-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner, S. R., J. W. Strohbach, R. A. Tommasi, P. A. Aristoff, P. D. Johnson, H. I. Skulnick, L. A. Dolak, E. P. Seest, P. K. Tomich, M. J. Bohanon, M. M. Horng, J. C. Lynn, K. T. Chong, R. R. Hinshaw, K. D. Watenpaugh, M. N. Janakiraman, and S. Thaisrivongs. 1998. Tipranavir (PNU-140690): a potent, orally bioavailable nonpeptidic HIV protease inhibitor of the 5,6-dihydro-4-hydroxy-2-pyrone sulfonamide class. J. Med. Chem. 41:3467-3476. [DOI] [PubMed] [Google Scholar]
- 16.Valdez, H., S. McCallister, V. Kohlbrenner, and D. Mayers. 2005. Abstr. 3rd IAS Conf. HIV Pathogenesis Treatment, abstr. WeOa0205.