Abstract
Isogenic L1 and L2 gene knockout mutants of Stenotrophomonas maltophilia KJ (KJΔL1 and KJΔL2, respectively) were constructed by xylE gene replacement. Induction kinetics of the L1 and L2 genes were evaluated by testing catechol 2,3-dioxygenase activity in the mutants. The results suggested that the induction of the L1 and L2 genes was differentially regulated.
Stenotrophomonas maltophilia produces two β-lactamases, known as L1 and L2 (16, 17). The expression of L1 and L2 is inducible, and β-lactams are inducers. The induction mechanism first proposed by Mett et al. (11) implies a common regulation system for both genes. Recently a differential regulation has been proposed (2). The present study investigated the induction properties of the L1 and L2 genes.
S. maltophilia KJ is a clinical isolate. Its β-lactamase extract was prepared from the periplasmic contents (9) using ampicillin (100 μg/ml) as an inducer and subjected to an isoelectric focusing (IEF) assay (10). Two β-lactamases were revealed by IEF, of pIs 6.0 and 8.2, which can be assigned as the pIs of L1 and L2, respectively (5, 15).
PCR primers L1P-F (5′-ACATTGCCTACTACACCTCC-3′), L1P-R (5′-GCTCTTTACAGAGTCGAGCC-3′), AmpRL2-F (5′-AAGCCGCCTGGATGGAAC-3′), and AmpRL2-R (5′-ATGCCGATGATGCCGAAC-3′) were designed to amplify the two β-lactamase genes based on the released genome sequences of S. maltophilia K279a (www.sanger.ac.uk/projects/S_maltophilia). The 1.6-kb and 2.2-kb PCR amplicons obtained by PCR using the L1P-F/L1P-R and AmpRL2-F/AmpRL2-R pairs of primers, respectively, were ligated into T-vector (Yeastern Biotech Co.) and sequenced. The 1.6-kb PCR amplicon contained the L1 gene and a partial Ton B-dependent receptor gene separated by a 95-bp intercistronic region (IG). The 2.2-kb PCR amplicon contained two genes, blaL2 and ampR, divergently oriented and separated by a 175-bp IG. AmpR is a typical LysR transcriptional regulator protein (19) and has been shown to be a regulator for the expression of flanking β-lactamase genes in other genera (21, 24). However, the role of ampR in S. maltophilia is still unknown. For S. maltophilia KJ, a conserved LysR motif (TCCTAACGCTTCA) (6) was found in the IG between ampR and the L2 gene, implying that AmpR could be a regulator for the expression of the L2 gene. Neither a putative transcriptional regulator gene nor any conserved LysR motif is present in the region upstream of the L1 gene. The L1 and L2 proteins encoded by the isolate KJ were 87 to 98% and 69 to 100% identical, respectively, to other L1 and L2 enzymes in the literature (1, 18, 22, 23, 25) (www.sanger.ac.uk/projects/s_maltophilia; http://www.jgi.doe.gov).
Isogenic L1 and L2 gene knockout mutants, KJΔL1 and KJΔL2, were obtained by a gene replacement strategy. The 1.6-kb and 2.2-kb PCR amplicons were subcloned into the plasmid pEX18Tc, and a catechol 2,3-dioxygenase (C23O) gene (xylE) (20) was inserted into either the SacI site of the L1 gene or the StuI site of the L2 gene to generate transcriptional fusions in the same orientation as that of the interrupted genes. The KJΔL1 and KJΔL2 mutants were obtained by conjugation, followed by two-step antibiotics/10% sucrose selection as described previously (25). The authenticity of mutants was verified by both PCR sequencing and IEF.
MICs of strains KJ, KJΔL1, and KJΔL2 were determined in triplicate according to a standard twofold serial agar dilution method (12) (Table 1). The MICs for cefepime, imipenem, and meropenem were quantified using Etest strips (AB Biodisk, Solna, Sweden). Results of susceptibility testing with the two mutants were overall consistent with the substrate profiles of the two enzymes reported previously (4, 22).
TABLE 1.
MICs of β-lactam antibiotics for S. maltophilia KJ, KJΔL1, and KJΔL2
| Antibiotic | MIC (μg/ml) of antibiotic for strain:
|
||
|---|---|---|---|
| KJ | KJΔL1 | KJΔL2 | |
| Penicillins | |||
| Ampicillin | >2,048 | >2,048 | >2,048 |
| Piperacillin | 1,024 | 256 | 512 |
| Carbenicillin | 1,024 | 1,024 | 64 |
| Cephalosporins | |||
| Cefuroxime | 2,048 | 1,024 | 1,024 |
| Cefoxitin | 1,024 | 128 | 1,024 |
| Cefoperazone | 128 | 32 | 64 |
| Cefotaxime | 256 | 128 | 128 |
| Ceftriaxone | 256 | 128 | 128 |
| Cefepime | 64 | 64 | 4 |
| Carbapenems | |||
| Imipenem | >32 | 8 | >32 |
| Meropenem | >32 | 0.5 | >32 |
| Monobactam | |||
| Aztreonam | >2,048 | >2,048 | 16 |
The inducibility of L1 and L2 enzymes was further confirmed. Strains KJ, KIΔL1, and KJΔL2 were treated with and without cefuroxime (50 μg/ml), and then levels of the expressed β-lactamase were comparatively checked with IEF and activity assay using CENTA (3) as the substrate (data not shown).
Induction experiments were performed to monitor the C23O activity of KJΔL2 and KJΔL1. Overnight cultures were diluted to an optical density at 450 nm of 0.15 and subsequently grown at 37°C for 0.5 h. Unless otherwise stated, induction was carried out using 50 μg/ml of inducer for 2.5 h. The C23O activities in intact cells were determined (8); meanwhile, the optical density of cell suspension at 450 nm was recorded. One unit of enzyme activity was defined as the amount of enzyme that converted 1 nmol substrate per minute. The specific activity of the enzyme was defined in terms of units per A450 units (one A450 unit corresponds to approximately 3.6 × 108 cells/ml). Each experiment was repeated at least three times.
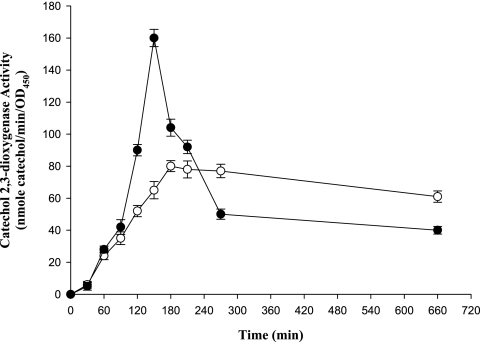
An induction time course experiment was designed to monitor the C23O activities of KJΔL1 and KJΔL2 at an interval of 30 min, using cefuroxime as an inducer (Fig. 1). Without the addition of an inducer, the C23O activity was below the level of detection for both mutants. In the presence of the inducer, the C23O activity was detectable starting with the first sampling, without any apparent lag period. This is consistent with studies with other S. maltophilia strains (14-16) and with Enterobacter cloacae (7) but different from that with Pseudomonas aeruginosa, for which a long and concentration-dependent lag phase was observed (13). Maximum C23O activities for KJΔL1 and KJΔL2 were obtained at 3 h and 2.5 h after induction, respectively. Thereafter, the C23O activity of KJΔL2 decreased at a rate significantly faster than that of KJΔL1 up to 4.5 h of induction.
FIG. 1.
Induction of C23O activity in S. maltophilia KJΔL1 and KJΔL2. The error bars indicate standard deviations (n = 3). Symbols: ○, mutant KJΔL1; •, mutant KJΔL2. OD450, optical density at 450 nm.
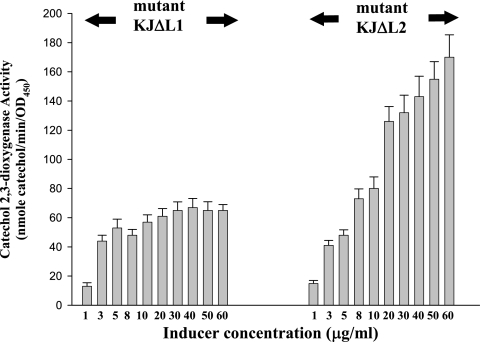
Figure 2 shows the C23O activities of KJΔL1 and KJΔL2 as a function of the cefuroxime concentration. The expression of xylE of KJΔL1 and KJΔL2 was readily induced by a small amount of cefuroxime (1 μg/ml). Apparently, the level of L1 induction was less dependent on the cefuroxime concentration than that of L2 induction.
FIG. 2.
Induction of C23O activity in S. maltophilia KJΔL1 and KJΔL2 as a function of the inducer concentration. The error bars indicate standard deviations (n = 3). OD450, optical density at 450 nm.
Rosta and Mett found that total induced β-lactamase activity in S. maltophilia decreases at a rate of 50% per generation after the maximal peak (15). In the present study, a difference between the induction of L1 and that of L2 was apparent. The difference in enzyme decline after maximum induction (Fig. 1) between KJΔL1 and KJΔL2 could be explained by the decrease in the inducer concentration in the assay system owing to its hydrolysis by the induced β-lactamase, which has a greater effect on induction of L2 (Fig. 2). In addition, the different activities of L1 and L2 with the inducer cefuroxime might also contribute to this difference.
The induced β-lactamase activity has been shown to be linearly correlated to the inducer concentration for Pseudomonas aeruginosa and S. maltophilia (13, 15). In the present study, a roughly linear correlation was observed with KJΔL2 but not with KJΔL1. This lower dependence of the induction of L1 on the inducer concentration is a phenomenon that has not yet been reported in β-lactamase induction of gram-negative bacteria (13, 15).
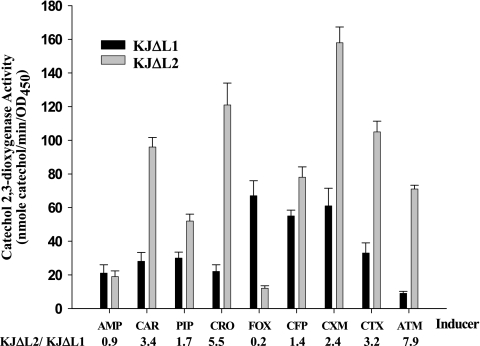
Figure 3 shows the induction of the L1 and L2 genes by various β-lactams. At a concentration of 50 μg/ml, most of the inducers hardly affected the growth of KJΔL1 or KJΔL2, with the exception of cefoperazone against KJΔL1 and aztreonam against KJΔL2. Consequently, induction experiments using cefoperazone and aztreonam as the inducers were also performed using lower concentrations, i.e., 16 μg/ml cefoperazone and 8 μg/ml aztreonam. In general, the induced C23O activity of KJΔL2 was higher than that of KJΔL1 against a specific inducer, except ampicillin and cefoxitin. The ratio of induction for KJΔL2 to that for KJΔL1 ranged from 0.2 to 7.9. Consequently, the induction potencies toward the L1 and L2 genes differed significantly for different inducers.
FIG. 3.
Induction of C23O activity by various β-lactam antibiotics in S. maltophilia KJΔL1 and KJΔL2. Inducers (shown on the x axis): AMP, ampicillin; CAR, carbenicillin; PIP, piperacillin; CRO, ceftriaxone; FOX, cefoxitin; CFP, cefoperazone; CXM, cefuroxime; CTX, cefotaxime; ATM, aztreonam. The induction ratio of KJΔL2 to KJΔL1 for each inducer is also included. The concentration of each inducer is 50 μg/ml, except that cefoperazone's is 16 μg/ml and aztreonam's is 8 μg/ml. The error bars indicate standard deviations (n = 3). OD450, optical density at 450 nm.
In conclusion, based on the results of induction experiments with KJΔL1 and KJΔL2 for the aspects of the induction course, the inducer type, and its concentration, this study suggests that the L1 and L2 genes are differentially regulated during induction.
Nucleotide sequence accession numbers.
The nucleotide sequences of the L1 and L2 PCR amplicons have been deposited in the GenBank database under accession numbers EF601224 and EF601225, respectively.
Acknowledgments
This study was supported in part by grant NSC 95-2320-B-039-022 from the National Science Council and by grant CMU-95-155 from the China Medical University.
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Avison, M. B., C. S. Higgins, C. J. von Heldreich, P. M. Bennett, and T. R. Walsh. 2001. Plasmid location and molecular heterogeneity of the L1 and L2 β-lactamase genes of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avison, M. B., C. S. Higgins, P. J. Ford, C. J. von Heldreich, T. R. Walsh, and P. M. Bennett. 2002. Differential regulation of L1 and L2 β-lactamase expression in Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 49:387-389. [DOI] [PubMed] [Google Scholar]
- 3.Bebrone, C., C. Moali, F. Mahy, S. Rival, J. D. Docquier, G. M. Rossolini, J. Fastrez, R. Pratt, J.-M. Frere, and M. Galleni. 2001. CENTA as a chromogenic substrate for studying β-lactamases. Antimicrob. Agents Chemother. 45:1868-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowder, M. W., T. R. Walsh, L. Banovic, M. Pettit, and J. Spencer. 1998. Overexpression, purification, and characterization of the cloned metallo-β-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 42:921-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felici, A., G. Amicosante, A. Oratore, R. Strom, P. Ledent, B. Joris, L. Fanuel, and J.-M. Frere. 1993. An overview of the kinetic parameters of class B β-lactamases. Biochem. J. 291:151-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goethals, K., M. Van Montagu, and M. Holsters. 1992. Conserved motifs in a divergent nod box of Azothizobium caulinodans ORS571 reveals a common structure in promoters regulated by LysR-type proteins. Proc. Natl. Acad. Sci. USA 89:1646-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gootz, T. D., and C. C. Sanders. 1983. Characterization of β-lactamase induction in Enterobacter cloacae. Antimicrob. Agents Chemother. 23:91-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karkhoff-Schweizer, R. R., and H. P. Schweizer. 1994. Utilization of a mini-Dlac transposable element to create an alpha-complementation and regulated expression system for cloning in Pseudomonas aeruginosa. Gene 140:7-15. [DOI] [PubMed] [Google Scholar]
- 9.Lindstrom, E. B., H. G. Boman, and B. B. Steele. 1970. Resistance of Escherichia coli to penicillins. VI. Purification and characterization of the chromosomally mediated penicillinase present in ampA-containing strains. J. Bacteriol. 101:218-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 11.Mett, H., S. Rosta, B. Schacher, and R. Frei. 1988. Outer membrane permeability and β-lactamase content in Pseudomonas maltophilia clinical isolates and laboratory mutants. Rev. Infect. Dis. 10:765-769. [DOI] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 2006. Performance standards for antimicrobial susceptibility testing of bacteria; 14th informational supplement M07-A7. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 13.Nordstrom, K., and R. B. Sykes. 1974. Induction kinetics of β-lactamase biosynthesis in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 6:734-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paton, R., R. S. Miles, and S. G. B. Amyes. 1994. Biochemical properties of inducible β-lactamases from Xanthomonas maltophilia. Antimicrob. Agents Chemother. 38:2143-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosta, S., and H. Mett. 1989. Physiological studies of the regulation of β-lactamase expression in Pseudomonas maltophilia. J. Bacteriol. 171:483-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saino, Y., F. Kobayashi, M. Inoue, and S. Mitsuhashi. 1982. Purification and properties of inducible penicillin β-lactamase isolated form Pseudomonas maltophilia. Antimicrob. Agents Chemother. 22:564-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saino, Y., M. Inoue, and S. Mitsuhashi. 1984. Purification and properties of an inducible cephalosporinase from Pseudomonas maltophilia GN12873. Antimicrob. Agents Chemother. 25:362-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanschagrin, F., J. Dufresne, and R. C. Levesque. 1998. Molecular heterogeneity of L1 metallo-β-lactamase family from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 42:1245-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 20.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 21.Trepanier, S., A. Prince, and A. Huletsky. 1997. Characterization of the penA and penR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysR-type transcriptional regulator. Antimicrob. Agents Chemother. 41:2399-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh, T. R., A. P. MacGowen, and P. M. Bennett. 1997. Sequence analysis and enzyme kinetics of the L2 β-lactamase from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41:1460-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh, T. R., L. Hall, S. J. Assinder, W. W. Nichols, S. J. Cartwright, A. P. MacGowan, and P. M. Bennett. 1994. Sequence analysis of the L1 metallo-β-lactamase from Xanthomonas maltophilia. Biochim. Biophys. Acta 1218:199-201. [DOI] [PubMed] [Google Scholar]
- 24.Weng, S. F., J. W. Lin, C. H. Chen, Y. Y. Chen, Y. H. Tseng, and Y. H. Tseng. 2004. Constitutive expression of a chromosomal class A (BJM group 2) β-lactamase in Xanthomonas campestris. Antimicrob. Agents Chemother. 48:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, L., X.-Z. Li, and K. Poole. 2000. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob. Agents Chemother. 44:287-293. [DOI] [PMC free article] [PubMed] [Google Scholar]