Abstract
We characterized the selective advantage profiles of a panel of hepatitis C virus (HCV) NS3 protease mutants with three HCV protease inhibitors (PIs), BILN-2061, ITMN-191, and VX-950, using a genotype 1b HCV replicon system. Selective advantage curves were generated by a novel mathematical method that factors in the degree of drug susceptibility provided by the mutation, the base-level replication capacity of the mutant in the absence of drugs, and the overall viral replication levels as a function of drug concentration. Most of the mutants showed significantly increased selective advantages over the wild-type species upon drug treatment. Each drug is associated with unique selective advantage profiles that reflect its antiviral activity and mutant susceptibility. Five mutants (R155K/Q, A156T, and D168A/V) showed significant levels of selective advantage after treatment with >10 nM (∼7 times the wild-type 50% effective concentration [EC50]) of BILN-2061. R155K displayed dominant levels of selective advantage over the other mutants upon treatment with ITMN-191 over a broad range of concentrations. Upon VX-950 treatment, various mutants (A156T, A156S, R155K, T54A, V170A, V36M/R155K, and R155Q) exhibited high levels of selective advantage in different drug concentration ranges, with A156T and A156S being the dominant mutants at >3 μM (∼10 times the wild-type EC50) of VX-950. This method provides more accurate estimates of the behavior of various mutants under drug pressure than replication capacity analysis. We noted that the R155K mutant shows reduced susceptibility to all three PIs and significant selective advantage, raising concern over the potential emergence of R155K as a multidrug-resistant, highly fit mutant in HCV patients treated with PIs.
Hepatitis C virus (HCV) is the leading cause of chronic liver disease in the United States and many other countries and has become a major burden on public health (19). It is estimated to infect ∼170 million people globally, including almost 4 million Americans. Long-term HCV infection frequently leads to cirrhosis and hepatocellular carcinoma in chronic patients. Current therapies for hepatitis C patients are based on a combination of pegylated recombinant interferons and ribavirin and result in viral clearance in less than 60% of treated patients, while being limited by significant side effects and high costs (10, 16). Novel, more specific and potent antiviral drugs are needed to meet the medical challenges of HCV infection.
The HCV viral enzymes, especially the NS3/4A serine protease and the NS5B RNA-dependent RNA polymerase, offer attractive targets for designing novel viral inhibitors (16, 24). Numerous classes of drug molecules targeting these enzymes are currently in various phases of research and development pipelines (15). The first HCV protease inhibitor (PI) molecule to enter human clinical trials and provide proof-of-concept antiviral activity was BILN-2061, a peptidomimetic inhibitor of NS3/4A serine protease (4). Other HCV PIs that have entered clinical trials include VX-950 (18, 20), SCH503034 (21), and ITMN-191 (23), and also peptidomimetic molecules. These HCV PIs caused significant inhibition of HCV replication both in cell culture systems and in HCV-infected patients. However, drug-resistant mutants may preexist in the HCV quasispecies (12), and studies have shown that mutant viral species resistant to these molecules could emerge and be quickly selected both in vitro and in patients (Table 1 and references therein). This highlights the importance of understanding the selection, evolution, and survival of these drug-resistant mutant viruses during drug treatment. Previous studies of drug-resistant mutants have benefited from the use of cell culture-based viral infection/replication model systems (11, 17).
TABLE 1.
List of drug-resistant NS3 protease mutant replicons used in this study
| Residue | Mutation | PI compound(s) reported | Reference(s) |
|---|---|---|---|
| 36 | V36M | VX950 | 25, 26 |
| 54 | T54A | VX950 | 25, 26 |
| SCH6 | 29 | ||
| 155 | R155Q | BILN-2061 | 8 |
| R155K | VX950 | 25, 26 | |
| 156 | A156T | BILN-2061 | 6, 7, 8 |
| A156S | VX950 | 6, 7, 25, 26 | |
| SCH6 | 29 | ||
| A156V | BILN-2061 | 6, 7, 16 | |
| VX950 | 6, 7, 25, 26 | ||
| 168 | D168V | BILN-2061 | 6, 7, 8, 16 |
| ITMN191 | 27 | ||
| D168A | BILN-2061 | 6, 7 | |
| ITMN191 | 27 |
Important determinants of the outgrowth of drug-resistant mutant viruses include the extent of drug susceptibility and replication capacity (RC) relative to the predominant wild-type viral population (i.e., the relative fitness in the presence of a given concentration of drug). Previous studies have shown that mutant viral species that are drug resistant often suffer from reduced fitness in the absence of drugs relative to the wild-type species (7, 13, 22). However, there is evidence that drug-resistant mutant viral species are not always unfit relative to the wild-type population, and multiple fitness peaks could exist within the fitness landscape of a quasispecies virus such as HCV (25, 27). These species may display various degrees of susceptibility to a viral inhibitor while being able to replicate equally well (27). In addition, it has been noted that the relative fitness levels of drug-resistant mutant viral species may be significantly influenced by drug treatment (9). Under drug pressure, the drug-resistant mutants gain RC relative to the drug-sensitive wild type and display a selective advantage by outgrowing the otherwise more robust wild-type species.
In this study, a panel of drug-resistant HCV NS3 protease mutants (Table 1 and Fig. 1) were created in a genotype 1b N strain replicon. All of these mutants have been reported to be resistant to HCV NS3 PIs in previous studies (Table 1 and references therein). Five of these mutants (R155Q, A156T, A156V, D168V, and D168A) were reported to be resistant to BILN-2061 (5-7, 13). Seven of the mutants (V36M, T54A, R155K, A156T, A156S, A156V, and the V36M/R155K double mutant) were claimed to confer resistance against VX-950 (5, 6, 20, 22). Two mutants at residue 168 (D168V and D168A), among others, have been identified as resistant mutants against ITMN-191 (23). In this study we systematically analyzed the PI susceptibility and relative RC of this panel of NS3 protease mutants using a transient-replication assay. In particular, we characterized the relative RC and selective advantage profiles of these mutants with various drug concentrations of the three HCV PIs, using both existing methods and a novel mathematical model that adjusts the mutants' selective advantage curves in accordance with the decrease in overall viral replication with increasing drug pressure. Our findings suggest that the selective advantage conferred by drug-resistant mutants depends on their relative drug susceptibility, relative RC compared to the wild-type virus, and the concentration of drug. Different PIs caused unique selective advantage profiles that may correlate with their resistance profiles.
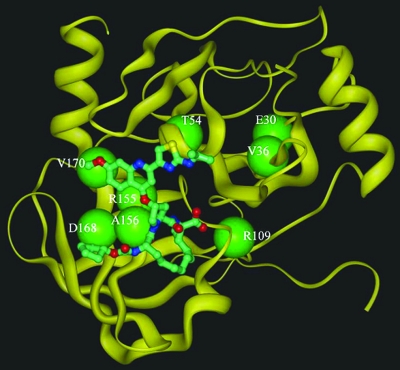
FIG. 1.
Structural modeling of HCV 1b NS3 protease (yellow ribbon) in complex with BILN-2061 (ball and stick). Locations of residues associated with drug resistance mutations reported in the literature are indicated (large green balls).
MATERIALS AND METHODS
Construction of mutant replicon clones.
The genotype 1b N strain HCV replicon construct carrying the NS3-NS5B region of the HCV genome and a firefly luciferase reporter gene has been described in previous reports (7, 13). Mutant replicons carrying the R155Q, A156T, and D168V substitutions were created in a previous study (7). The other NS3 mutant replicons (V36M, T54A, R155K, A156S, A156V, D168A, V170A, and V36M+R155K) were created by site-directed mutagenesis (Stratagene) using oligonucleotide primers (IDT) listed in Table 2. All of the NS3 mutant replicons were confirmed by direct sequencing (ABI) of the plasmid DNA.
TABLE 2.
Oligonucleotide primers used in this study to create NS3 protease mutant replicons by site-directed mutagenesis
| Mutation | Original sequence (5′-3′)a | Primer (5′-3′)b | Strandc |
|---|---|---|---|
| V36M | GG GAG GTT CAA GTG GTC TCC ACC GC | GGGAGGTTCAAATGGTCTCCACCGC | S |
| GCGGTGGAGACCATTTGAACCTCCC | AS | ||
| T54A | GC GTA TGT TGG ACT GTC TAC CAT GG | GCGTATGTTGGGCTGTCTACCATGG | S |
| CCATGGTAGACAGCCCAACATACGC | AS | ||
| R155K | G GGC GTC TTC CGG GCC GCT GTA TGC | GGGCGTCTTCAAGGCCGCTGTATGC | S |
| GCATACAGCGGCCTTGAAGACGCCC | AS | ||
| A156S | GC GTC TTC CGG GCC GCT GTA TGC AC | GCGTCTTCCGGTCCGCTGTATGCAC | S |
| GTGCATACAGCGGACCGGAAGACGC | AS | ||
| A156V | GC GTC TTC CGG GCC GCT GTA TGC AC | GCGTCTTCCGGGTCGCTGTATGCAC | S |
| GTGCATACAGCGACCCGGAAGACGC | AS | ||
| D168A | GCA AAG GCG GTG GAT TTT GTC CCC G | GCAAAGGCGGTGGCTTTTGTCCCCG | S |
| CGGGGACAAAAGCCACCGCCTTTGC | AS | ||
| V170A | CG GTG GAT TTT GTC CCC GTT GAG TC | CGGTGGATTTTGCCCCCGTTGAGTC | S |
| GACTCAACGGGGGCAAAATCCACCG | AS |
Bold indicates the codons to be mutated.
The mutagenesis sites are underlined.
S, sense; AS, antisense.
Transient-replication assays.
The transient-replication assay method has been described previously (7, 13). Briefly, HCV replicon RNA transcripts were prepared (Ambion) from plasmid DNA carrying either wild-type or mutant replicon constructs and transfected into Huh7 cells by electroporation (Bio-Rad). The transfected cells were diluted with Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (Atlanta) and 1× penicillin-streptomycin-glutamine (Invitrogen) and plated into 96-well tissue culture plates (Costar). Firefly luciferase assays (Promega) were performed on one plate set of transfected cells using a Victor plate reader (Perkin-Elmer) at 4 hours after transfection to monitor transfection and translation levels. Various concentrations of drugs were diluted into culture medium and added to a second plate set of transfected cells that were incubated in a CO2 incubator at 37°C for 4 days before being harvested for firefly luciferase assays. The 4-day firefly luciferase assays were performed on triplicate samples in each experiment, and the assay signals had standard deviations less than 20% of the mean values for all RNA samples based on results from the signal control wells on each assay plate.
Calculation of EC50 values, drug susceptibility levels, and relative replication capacity values.
The 50% effective concentration (EC50) values of drugs were calculated with the GraphPad Prism 4.0 software as previously described (7, 13), using means of assay signals from triplicate samples in each experiment. Drug susceptibility levels of the mutant replicons were calculated by dividing a mutant's EC50 by the wild-type replicon's EC50 value against the same drug and were expressed as the change of resistance. Relative RC values of the mutant replicons were generated according to the following formula, as previously described (7, 13): (Luc4Dm/Luc4Dwt)/(Luc4Hm/Luc4Hwt), where Luc4Dm is the mean luciferase value of the mutant after 4 days of incubation, Luc4Dwt is the mean luciferase value of the wild type after 4 days of incubation, Luc4Hm is the mean luciferase value of the mutant after 4 h of incubation, and Luc4Dwt is the mean luciferase value of the wild type after 4 h of incubation.
The relative RC value of each mutant in the absence of drugs is defined as its base-level RC. The relative RC values of the mutants with drug treatment were graphed as a function of drug concentration to generate relative RC curves. The drug susceptibility and base-level RC results shown in this study are average values from at least three independent experiments. For RC profiles and selective advantage curves, the experiments were done in triplicate and the mean assay values were used for analysis.
Generation of selective advantage curves.
We modified a previously published method (9) to construct selective advantage curves as follows. Two-parameter logistic regression models were used to fit inhibition-concentration curves based on the relative fitness at each drug concentration. Negative relative fitness values were set to zero before fitting the logistic regression model for the wild type and for each mutant replicon. The ratio of percent replication (defined as the complement of the fitted percent inhibition) between mutant and wild type was calculated after adjusting the mutant replicon for its RC. High values of this ratio are not relevant at very high drug concentrations. Therefore, the resulting relative fitness curve was multiplied by an adjustment factor that accounts for decrease of overall replication levels with increasing drug concentrations, specifically, the point-wise maximum of the residual wild-type replication rate or the RC-adjusted residual mutant replication rate. Such an adjustment factor should leave the ratio unadjusted in the absence of drug, and it should shrink the ratio toward 1 (i.e., shrink the log of the ratio toward 0) as the drug concentration increases. We chose the maximum of the residual wild-type replication rate or the residual mutant replication rate after adjustment for its RC as a factor meeting both of these criteria. The value of the selective advantage curve at each concentration is thus given by the expression exp{log[(Rmut × RC)/(Rwt)] × max(Rwt, Rmut × Rc)}, in which Rwt and Rmut are the residual replication levels of the wild-type and the mutant, respectively, at a given drug concentration.
HCV protease inhibitors.
The HCV protease inhibitors used in this study (BILN-2061, ITMN-191, and VX-950) were synthesized in-house.
RESULTS
Basal-level replication capacity and PI susceptibility of HCV NS3 protease mutant replicons.
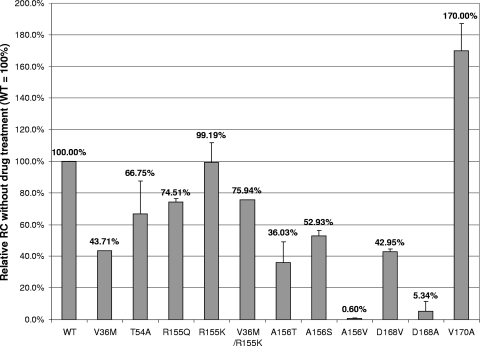
The relative RC values of the panel of NS3 protease mutant replicons were determined from 4-day transient assays without drug treatment (Fig. 2). Our results show that most NS3 protease mutant replicons displayed reduced RC relative to the wild-type control. In particular, A156V and D168A showed severely reduced RC levels (less than 1% and 10% of the wild-type control, respectively) under the assay conditions. It is noteworthy that all the mutants with substitutions at residues 156 and 168 (A156T/S/V and D168V/A) showed reduced RC levels, suggesting that drug resistance mutations at these two residues are associated with fitness loss. However, the R155K mutant, which displayed reduced susceptibility to all three HCV PIs (Fig. 3), exhibited RC equivalent to the wild type. Another residue 155 mutant replicon, R155Q, also showed an RC (75%) similar to the wild type. Interestingly, a V170A mutant selected with SCH503034 showed even higher RC levels than the wild-type replicon in this study, indicating that multiple fitness peaks may exist in an HCV quasispecies population. Our results suggest that residues 155 and 170 might be more tolerant to mutations than the other positions, at least in the 1b N strain.
FIG. 2.
Relative RC levels of NS3 protease mutant replicons in 4-day transient-replication assays without drug treatment. Mean values of results from at least three experiments are shown, and the errors bars represent standard deviations.
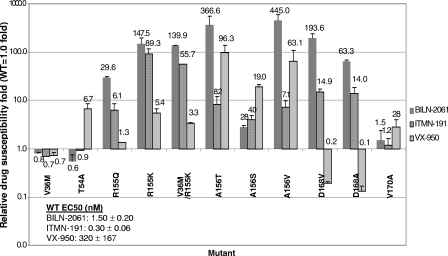
FIG. 3.
Drug susceptibility levels of NS3 protease mutant replicons to BILN-2061, ITMN-191, and VX-950. Mean values of results from at least three experiments are shown, and the errors bars represent standard deviations. The inset shows the PI mean EC50 values against the wild-type replicon ± the standard deviation from at least three experiments.
The PI susceptibility levels of this panel of NS3 protease mutant replicons were also determined from 4-day transient assays. BILN-2061 and ITMN-191 showed EC50 values around 1.5 nM and 0.3 nM, respectively, while VX-950 showed much higher EC50 values (mean, 320 nM) against the wild-type replicon (Fig. 3, inset). The drug titration curves and EC50 values were reproducible between experiments, with standard deviations no more than 50% of the mean values (Fig. 3, inset). Due to the higher EC50 values of VX-950 relative to the other two PIs, this compound was tested at a higher concentration range than the others, as reflected by the difference in concentration scales of relative RC and selective advantage graphs later in this paper. The mutants' EC50 values were compared to that of the wild-type control to determine the change in drug resistance conferred by each mutation. Four single mutants (R155K, A156T, A156V, and D168V) showed a more-than-100-fold decrease in susceptibility to BILN-2061, relative to the wild-type control (Fig. 3). In particular, A156T and A156V showed around 400-fold-reduced susceptibility, indicating that A156 is a key residue determining resistance to BILN-2061. Two more mutants, R155Q and D168A, showed 30- and 60-fold changes in susceptibility to BILN-2061. It is noteworthy that residues 155, 156, and 168 are either in direct contact with or close to BILN-2061 in the enzyme-inhibitor complex structure, and changes in these residues are likely to confer drug resistance (2) (Fig. 1). Indeed, our previous replicon selection study with BILN-2061 revealed mutations at these three residues (7). On the other hand, V36M, T54A, and V170A, three mutants selected with VX-950 and/or SCH503034 (20, 22, 25), showed no resistance to BILN-2061, and these three residues are more distant from BILN-2061 in the enzyme-inhibitor complex structure (2) (Fig. 1).
R155K gave the greatest reduction in susceptibility (89-fold) against ITMN-191. On the other hand, R155Q showed only a sixfold loss of susceptibility. These results suggest the nature of residue 155 is a key determinant of resistance toward ITMN-191. All the mutants with substitutions at residues 156 and 168 also showed loss in susceptibility against ITMN-191, ranging from 4- to 14-fold. The three mutants with substitutions at residue 156 (A156T, A156V, and A156S) showed the greatest reductions in susceptibility against VX-950 (96-, 63-, and 19-fold, respectively) among this panel of NS3 mutants, suggesting that residue 156 is the key determinant of high-level resistance toward VX-950, consistent with results from other studies (5, 6, 20, 22). T54A and R155K also showed a severalfold loss in susceptibility against VX-950. It is noteworthy that R155K, a resistant mutant selected from patients treated with VX-950 (20, 22), is also highly resistant to both BILN-2061 and ITMN-191, two macrocyclic PIs with structures different from VX-950. On the other hand, T54A is sensitive to both macrocyclic PIs. Interestingly, the two mutants with substitutions at D168, while being resistant to both macrocyclic molecules, became significantly more sensitive to VX-950 than the wild-type control, consistent with observations from earlier studies (5, 6). These results indicate that different PI classes may partially complement each other by cross-inhibiting different mutants.
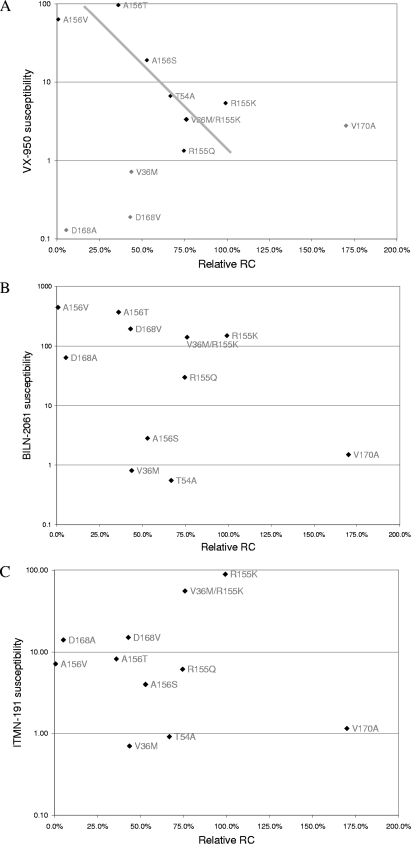
A previous study (22) reported an inverse correlation between the relative fitness of NS3 protease mutant viruses in patients and the corresponding mutant protease enzyme's resistance levels against VX-950, suggesting that higher drug resistance is associated with reduced viral fitness in patients. Does such a correlation exist in the HCV replicon system, and could it be used to predict RC of mutant replicons? The relative RC and drug susceptibility levels of the NS3 mutant replicons in this study are graphed in a similar manner, in an effort to identify any correlation pattern. An inverse correlation was observed between VX-950 susceptibility levels and relative RC scores of several mutant replicons, after excluding mutants that showed no loss of susceptibility toward VX-950 (V36M and D168A/V) and V170A (Fig. 4A). However, no correlation seems to exist between relative RCs of these mutants and their loss of susceptibility to the other two PIs, BILN-2061 and ITMN-191 (Fig. 4B and C). These results suggest that there is no universal correlation between the relative RC and drug susceptibility levels of HCV mutants in the replicon system. Since relative RC is also affected by drug treatment (9), which in turn changes the overall viral replication level, the relative RC value is not able to reflect these changes in viral replication upon drug treatment. So, a better method is required to characterize the relative RC of drug-resistant HCV mutants under various drug pressures.
FIG. 4.
Correlation between relative RC and drug susceptibility levels of NS3 protease mutant replicons. Replicons were treated with BILN-2061 (A), ITMN-191 (B), or VX-950 (C).
Replication capacity profiles and selective advantage curves of HCV NS3 protease mutant replicons with PI treatment.
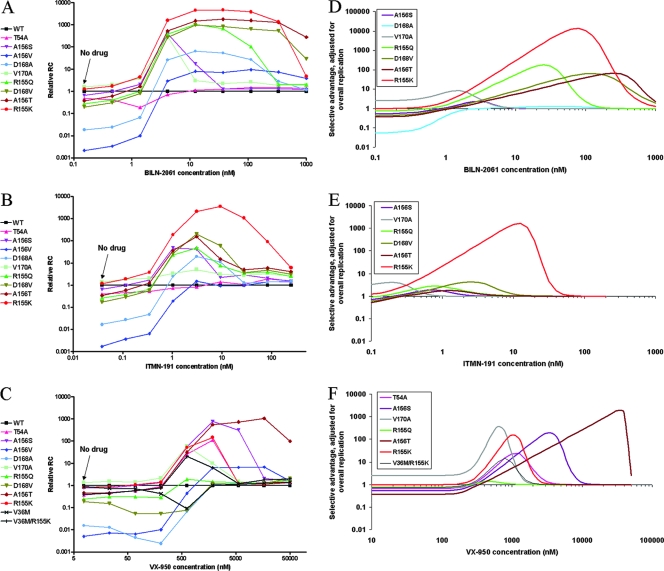
It has been demonstrated that relative fitness values of mutant viral species are regulated by the presence of viral inhibitors and dependent on drug concentrations (9). Drug-resistant mutant viruses gain relative fitness in the presence of antiviral drugs relative to the wild type; in other words, they gain a selective advantage. As described in Materials and Methods and shown in Fig. 5, the NS3 protease mutant replicons were treated with various concentrations of each of the three HCV PIs during transient-replication assays, and the RC of each mutant replicon relative to the wild type was determined at each drug concentration. As discussed above, most of the NS3 protease mutants replicate less well than the wild-type control when there is no drug pressure. Only V170A and R155K showed RCs similar to the wild-type control without drug treatment. However, the relative RC of most mutant replicons increased significantly in various concentration ranges of the three HCV PIs (Fig. 5A to C). As suggested in a previous study (9), this assay method may be used to model drug-resistant mutant profiles and to predict the emergence and evolution of mutant viruses upon treatment with drugs. However, in addition to changes in relative ratios of mutant and wild-type species upon drug treatment, there are also changes in overall replication levels of the entire viral population. The relative RC calculation methods used in the previous study (9) and above fail to take into account changes in overall replication levels with drug treatment. As a result, the relative RC curves may not clearly differentiate between mutants and may overestimate the selective advantage of less fit mutants, especially at high drug concentrations, where overall replication levels are low. In order to provide a more accurate predictor of the mutants' selective advantage under drug pressure, the relative RC curves were modified with an adjustment factor to account for decreases in overall replication levels with addition of drugs. The results are shown in Fig. 5D to F. As an adjustment factor for overall replication levels, we chose the larger of the replication rates for the wild type or for the mutant after adjusting by its basal-level RC. This combination of factors reflects the residual relative replication level of the more-fit replicon at a given drug concentration. Only mutants that show any selective advantage after adjustment are shown in the graphs for clarity. These selective advantage curves reflect the combinatorial effects of the mutants' drug susceptibility levels, RC, and overall viral replication levels as a function of drug concentration.
FIG. 5.
Relative RC (A to C) and selective advantage (D to F) profiles of NS3 protease mutant replicons with PI treatment. Replicons were treated with BILN-2061 (A and D), ITMN-191 (B and E), or VX-950 (C and F). Selective advantage profiles (D to F) show only those curves that exceed zero meaningfully. The relative RC curves are based on mean values of triplicate samples.
With BILN-2061 treatment, several mutants (R155K, R155Q, A156T, D168V, and V170A) showed at least a 1-log peak selective advantage. The first four mutants, at residues 155, 156, and 168, showed at least a 2-log peak selective advantage at higher drug concentrations (between 10 nM and 1,000 nM, or 7- to 660-fold of the wild-type EC50), highlighting the importance of these mutants in causing resistance against BILN-2061. On the other hand, T54A and A156V, two mutants that showed very low relative RC levels during BILN-2061 treatment (Fig. 5A), did not show significant selective advantage and are omitted from the graph (Fig. 5D). Of the mutants that showed significant selective advantage, the concentration range over which the peak of this advantage occurs varies by over 100-fold. Thus, this representation of the response to an inhibitor demonstrates both the extent to which a mutation provides a replication advantage and the concentration range over which this advantage occurs. The selective advantage curves (Fig. 5D) differentiate the various mutants from each other and define the optimal drug concentrations for their respective selective advantage curves. On the other hand, the corresponding RC curves that do not take into account overall replication levels (Fig. 5A) tend to overlap or parallel each other, especially for those mutants with high RC values (R155K, A156T, D168V, and R155Q), and thus do not provide as much insight as the selective advantage curves.
With ITMN-191 treatment, six mutants (R155K/Q, A156S/T, D168V, and V170A) showed observable levels of selective advantage (Fig. 5E). In particular, R155K showed much higher selective advantage values than all the other mutants, especially at higher drug concentrations (up to 3 logs at >10 nM, or 33-fold above the wild-type EC50), highlighting its potential to emerge as a dominant resistant mutant species during ITMN-191 treatment. On the other hand, three mutants (T54A, A156V, and D168A) that showed low relative RC values (Fig. 5B) did not show significant selective advantage and are omitted in the final graph (Fig. 5E). Under VX-950 drug pressure (Fig. 5F), several mutants (T54A, R155K/Q, A156S/T, V170A, and V36M/R155K) showed selective advantage levels greater than 1 log. In particular, the two mutants with substitutions at residue 156 (A156S/T) appear as the dominant mutant species at higher drug concentrations (>3 μM, or ∼10-fold of the wild-type EC50), reaching 2 to 3 logs of selective advantage in a drug concentration range where the other mutants show no advantage. This is consistent with previous observations that these mutants are the dominant mutants at the population sequence level in replicon selection studies on VX-950 (5, 6). On the other hand, the three mutants showing low relative RCs (A156V and D168A/V) failed to show significant selective advantage values and are ignored in the graph.
Overall, the three PIs are associated with distinctive selective advantage profiles, each preferentially favoring different mutants and resulting in different levels of selective advantage of the mutants. Both BILN-2061 and ITMN-191 favor selective advantage of mutants with substitutions at residues 155, 156, and 168, while VX-950 favors a selective advantage of mutants with substitutions at residues 54, 155, 156, and 170, but not those with substitutions at residue 168, which are more sensitive to VX-950 than the wild type. In particular, the R155K mutant shows much higher selective advantage over the other mutants for both BILN-2061 and ITMN-191 over a broad range of drug concentrations, suggesting that these two macrocyclic inhibitors might be more susceptible to the R155K mutant than VX-950. On the other hand, VX-950 treatment is associated with multiple mutants that show similar levels of selective advantage but at different drug concentrations, suggesting that the dominant mutant against VX-950 varies as a function of drug pressure. Interestingly, ITMN-191 is associated with fewer mutants with high selective advantage values (>1.0 log) than the other two compounds (only one mutant, R155K, for ITMN-191, versus five mutants for both BILN-2061 and VX-950), and the advantage occurred at lower drug concentrations, indicating that ITMN-191 might experience less resistance than the other two PIs at similar drug concentrations and thus might be more potent than the earlier drug candidates.
DISCUSSION
Antiviral drug treatment favors selection and growth of drug-resistant mutant viruses. Both drug resistance and relative fitness levels contribute to the replication behavior of mutants, and these two parameters are usually determined in studies on drug-resistant mutant viruses or replicons (7, 13). Since relative fitness of mutants is also affected by drug treatment, Mammano et al. (9) developed a method by which they determined relative fitness values of human immunodeficiency virus (HIV) mutants at various drug concentrations. The resultant HIV mutant fitness profiles were found to be consistent with observations in drug-treated HIV patients, supporting the usefulness of this analytical approach. However, as this method does not take into account the decrease in overall viral replication levels with drug treatment, the relative fitness curves may not be predictive at high drug concentrations where overall replication levels are low. In this study, in addition to determining relative RC curves of the HCV protease mutants, a novel analysis method was developed to generate the selective advantage curves of these mutants as well. These selective advantage curves demonstrate that the behavior of a mutant is affected by its basal RC value in the absence of drug treatment, the mutant's susceptibility to drug, the drug concentration, and the overall viral replication level at that drug concentration. The three PIs resulted in very different selective advantage profiles of these mutants, which were presumably determined by the different interactions of these drugs with the mutants. Overall, the selective advantage curves tend to highlight the mutant species with the highest RC at a certain drug concentration range, while ignoring the less fit ones, especially in the case of ITMN-191 treatment (Fig. 5E). This method might provide a more informative system for predicting the dominant mutants at a given drug pressure than the relative RC curves. For example, most of the mutants showing significant levels of selective advantage (T54A, R155K, A156S/T, and V36M/R155K) upon VX-950 treatment have been reported in previous studies to be resistant mutants against VX-950 in hepatitis C patients treated with the drug (20, 22), indicating a potential for this analysis method to predict the emergence of resistant mutants in drug-treated patients. In patients treated with VX-950, the mutations at residue 156 confer more drug resistance than other single mutations (20, 22), consistent with their selective advantage profiles (Fig. 5F). In another case, three mutants (155Q, 156T, and 168V) showing high levels of selective advantage upon BILN-2061 treatment were selected in our previous replicon selection study (7), again suggesting the value for these selective advantage curves in predicting the selection of drug-resistant mutants. Interestingly, R155K, the mutant that showed the highest selective advantage value (up to ∼4 logs) in this system, has not been observed in genotype 1b replicon selection studies using BILN-2061. This may be a reflection of codon usage in genotype 1b. Based on a consensus NS3 RNA sequence developed from 134 HCV genotype 1b isolates (data not shown), the change from Arg to Lys at position 155 would require two base changes, whereas the change to Gln would only require one change. A more complete evaluation of this analysis method awaits additional results from clinical evaluations of anti-HCV drugs.
Since there is no effective infection system for most HCV genotypes and strains except for a genotype 2a strain (3), we chose to study HCV protease mutants in the context of an HCV genotype 1b replicon system. The usefulness of the replicon system for HCV mutant study has been demonstrated by previous work, and overall there is a good correlation between in vitro selection of drug-resistant variants in the replicon and mutants selected in drug-treated patients (1, 5, 6, 13, 20, 22). Still, the results of this study are based on a genotype 1b N strain replicon system which does not represent the genetic backgrounds of all other strains and genotypes, and so the RC profiles of these mutants might change in different genetic backgrounds. Ideally, the fitness profiles of viral mutants should be determined in the same genetic backgrounds from which they are isolated. Due to the lack of robust infection/replication systems for most HCV genotypes and strains, fitness assays using the mutants' natural genetic backgrounds cannot be done in most cases. However, with the development of replicon-based “shuttle vector” systems (8, 12, 26), it is possible to study individual genes from patient samples and select drug-resistant mutant species from these samples. It will be interesting to clone the NS3 protease genes from HCV-infected patient samples and perform selective advantage analysis with resistant mutants selected with various HCV PIs. Since the HCV replicon system only reproduces a part of the HCV life cycle and does not involve infectious virus particles, caution should be exercised in the interpretation of replicon RC results and their correlation with fitness results from infectious and in vivo systems.
Some previous studies suggested that competition-based assay systems might provide a more robust measurement of fitness. Using this approach, fitness is measured by mixing the mutant and the wild type in the same infection or replication system and monitoring ratios of their replication levels after a certain assay period (11, 17). We also analyzed the RC profiles of these NS3 protease mutants in competition experiments, by mixing and cotransfecting a mutant replicon RNA encoding firefly luciferase with a wild-type replicon RNA encoding Renilla luciferase into the same pool of Huh7 cells, followed by dual-luciferase assays (Promega) after 4 days of incubation, to monitor the replication of both replicon species within the same cell population. Relative RC values of the mutant were calculated for each drug concentration as described in Materials and Methods. However, the competition-based assays generated RC profiles of these mutants very similar to those from assays without competition (data not shown), suggesting our current assay system is sufficient for RC profiling of these mutants.
In this study, the R155K mutant showed significant loss in susceptibility against all the three PIs and exhibited an RC equivalent to the wild-type 1b N replicon. The R155K mutant also gave a relative RC level (80%) comparable to the wild-type control in the Con1 strain replicon (29). In addition, R155K was estimated to be relatively fit (∼40% fitness of wild-type virus) in VX-950-treated patients (20, 22) and became the dominant, persistent mutant in a PI-treated chimpanzee (14), further supporting its high fitness level. These observations raise the concern that R155K may emerge as a multidrug-resistant, highly fit mutant in PI-treated patients. The V170A mutant also displayed high levels of RC in the replicon system. It is noteworthy that this mutant appeared at least as fit as the wild type in replicon cells and outgrew other mutant species in a long-term drug treatment study (25). These results suggest that drug-resistant HCV mutants are not necessarily less fit than wild-type species, and mutants that are both drug resistant and fit may pose problems for future HCV therapy.
The drug susceptibility and relative RC levels of the same mutants can vary between different HCV genotypes and strains, and so do their selective advantage values. Such variation may even exist between HCV strains belonging to the same subtype, such as 1b N and Con1, two popular genotype 1b lab strains. It is noteworthy that V36M, a low-level resistant mutant observed in VX-950-treated patients (20, 22), showed no resistance to any of the three PIs tested, including VX-950, in our 1b N strain replicon. Also, when combined with R155K, V36M did not significantly affect drug susceptibility levels of the latter mutant. However, in a recent study (28) utilizing the Con1 strain replicon, V36M showed a 7.0-fold loss in susceptibility to VX-950, and the V36M/R155K double mutant showed an increased resistance level to VX-950 compared to either of the single mutants. It appears that the drug susceptibility phenotype of V36M depends on the genetic background. The V36M mutant alone is less fit (44%) than the wild type in our assay system and caused a slight decrease in the relative RC level of the V36M/R155K double mutant relative to the R155K single mutant, which is similar to observations made in the Con1 strain replicon (28, 29). We suggest that it is important to assess the phenotype of drug-resistant mutants in different genetic backgrounds, especially those resembling the clinical samples.
Some limitations of this mathematical method for analyzing the selective advantage of mutant viruses should be noted. The fitted inhibition-concentration curves are based on data that are subject to measurement error, as are the computed replication capacity values. While this limitation is mitigated by the use of three or more independent experiments, it is difficult to represent these combined errors in the selective advantage curves. In addition, the value we used to adjust for decreasing replication with increasing drug concentration was arbitrarily chosen. While it meets the obvious requirements for such an adjustment factor, other possible adjustments could be employed. Still, this method may provide an important tool for resistance profiling. For example, this method could be used to model the emergence, selection, and competition of various mutant viruses at expected drug concentrations, to predict identities of potential dominant mutant viruses for a certain drug, and to estimate the drug concentration ranges in which various mutant viruses may survive and proliferate. Overall, the protease mutants' fitness profiles shown in this study largely agree with drug-resistant mutant selection results from previous studies with either HCV replicon cells or drug-treated HCV patients (5-7, 13, 20, 22), supporting the usefulness of this analysis method.
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Chen, C. M., Y. He, L. Lu, H. B. Lim, R. L. Tripathi, T. Middleton, L. E. Hernandez, D. W. Beno, M. A. Long, W. M. Kati, T. D. Bosse, D. P. Larson, R. Wagner, R. E. Lanford, W. E. Kohlbrenner, D. J. Kempf, T. J. Pilot-Matias, and A. Molla. 2007. Activity of a potent hepatitis C virus polymerase inhibitor in the chimpanzee model. Antimicrob. Agents Chemother. 51:4290-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Courcambeck, J., M. Bouzidi, R. Perbost, B. Jouirou, N. Amrani, P. Cacoub, G. Pepe, J. M. Sabatier, and P. Halfon. 2006. Resistance of hepatitis C virus to NS3-4A protease inhibitors: mechanisms of drug resistance induced by R155Q, A156T, D168A and D168V mutations. Antivir. Ther. 11:847-855. [PubMed] [Google Scholar]
- 3.Kato, T., T. Date, A. Murayama, K. Morikawa, D. Akazawa, and T. Wakita. 2006. Cell culture and infection system for hepatitis C virus. Nat. Protoc. 1:2334-2339. [DOI] [PubMed] [Google Scholar]
- 4.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 5.Lin, C., C. A. Gates, B. G. Rao, D. L. Brennan, J. R. Fulghum, Y. P. Luong, J. D. Frantz, K. Lin, S. Ma, Y. Y. Wei, R. B. Perni, and A. D. Kwong. 2005. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J. Biol. Chem. 280:36784-36791. [DOI] [PubMed] [Google Scholar]
- 6.Lin, C., K. Lin, Y. P. Luong, B. G. Rao, Y. Y. Wei, D. L. Brennan, J. R. Fulghum, H. M. Hsiao, S. Ma, J. P. Maxwell, K. M. Cottrell, R. B. Perni, C. A. Gates, and A. D. Kwong. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508-17514. [DOI] [PubMed] [Google Scholar]
- 7.Lu, L., T. J. Pilot-Matias, K. D. Stewart, J. T. Randolph, R. Pithawalla, W. He, P. P. Huang, L. L. Klein, H. Mo, and A. Molla. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 48:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludmerer, S. W., D. J. Graham, E. Boots, E. M. Murray, A. Simcoe, E. J. Markel, J. A. Grobler, O. A. Flores, D. B. Olsen, D. J. Hazuda, and R. L. LaFemina. 2005. Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay. Antimicrob. Agents Chemother. 49:2059-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mammano, F., V. Trouplin, V. Zennou, and F. Clavel. 2000. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J. Virol. 74:8524-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manns, M. P., H. Wedemeyer, and M. Cornberg. 2006. Treating viral hepatitis C: efficacy, side effects, and complications. Gut 55:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maree, A. F., W. Keulen, C. A. Boucher, and R. J. De Boer. 2000. Estimating relative fitness in viral competition experiments. J. Virol. 74:11067-11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Middleton, T., Y. He, T. Pilot-Matias, R. Tripathi, H. B. Lim, A. Roth, C. M. Chen, G. Koev, T. I. Ng, P. Krishnan, R. Pithawalla, R. Mondal, T. Dekhtyar, L. Lu, H. Mo, W. M. Kati, and A. Molla. 2007. A replicon-based shuttle vector system for assessing the phenotype of HCV NS5B polymerase genes isolated from patient populations. J. Virol. Methods 145:137-145. [DOI] [PubMed] [Google Scholar]
- 13.Mo, H., L. Lu, T. Pilot-Matias, R. Pithawalla, R. Mondal, S. Masse, T. Dekhtyar, T. Ng, G. Koev, V. Stoll, K. D. Stewart, J. Pratt, P. Donner, T. Rockway, C. Maring, and A. Molla. 2005. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 49:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsen, D. B., S. S. Carroll, L. Handt, S. W. Ludmerer, D. J. Graham, C. Fandozzi, J. DeLuca, N. Liverton, J. Vacca, and D. J. Hazuda. 2007. 42nd Annu. Meet. EASL, Barcelona, Spain, abstr. 791.
- 15.Pawlotsky, J. M., S. Chevaliez, and J. G. McHutchison. 2007. The hepatitis C virus life cycle as a target for new antiviral therapies. Gastroenterology 132:1979-1998. [DOI] [PubMed] [Google Scholar]
- 16.Pawlotsky, J. M., and R. G. Gish. 2006. Future therapies for hepatitis C. Antivir. Ther. 11:397-408. [PubMed] [Google Scholar]
- 17.Quinones-Mateu, M. E., and E. J. Arts. 2002. Fitness of drug resistant HIV-1: methodology and clinical implications. Drug Resist. Update 5:224-233. [DOI] [PubMed] [Google Scholar]
- 18.Reesink, H. W., S. Zeuzem, C. J. Weegink, N. Forestier, A. van Vliet, J. van de Wetering de Rooij, L. McNair, S. Purdy, R. Kauffman, J. Alam, and P. L. Jansen. 2006. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology 131:997-1002. [DOI] [PubMed] [Google Scholar]
- 19.Sarbah, S. A., and Z. M. Younossi. 2000. Hepatitis C: an update on the silent epidemic. J. Clin. Gastroenterol. 30:125-143. [DOI] [PubMed] [Google Scholar]
- 20.Sarrazin, C., T. L. Kieffer, D. Bartels, B. Hanzelka, U. Muh, M. Welker, D. Wincheringer, Y. Zhou, H. M. Chu, C. Lin, C. Weegink, H. Reesink, S. Zeuzem, and A. D. Kwong. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767-1777. [DOI] [PubMed] [Google Scholar]
- 21.Sarrazin, C., R. Rouzier, F. Wagner, N. Forestier, D. Larrey, S. K. Gupta, M. Hussain, A. Shah, D. Cutler, J. Zhang, and S. Zeuzem. 2007. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology 132:1270-1278. [DOI] [PubMed] [Google Scholar]
- 22.Sarrazin, C., T. Kieffer, D. Bartels, B. Hanzelka, U. Muh, G. Rao, M. Welker, D. Wincheringer, C. Lin, S. Purdy, C. Weegink, H. Reesink, S. Zeuzem, and A. D. Kwong. 2005. 56th Annu. Meet. AASLD, San Francisco, CA, abstr. LB06.
- 23.Seiwert, S., J. Hong, S. Lim, H. Tan, K. Kossen, and L. Blatt. 2006. 1st Int. Workshop Hepatitis C: Resistance and New Compounds, Boston, MA, abstr. 2.
- 24.Stauber, R. E., and V. Stadlbauer. 2006. Novel approaches for therapy of chronic hepatitis C. J. Clin. Virol. 36:87-94. [DOI] [PubMed] [Google Scholar]
- 25.Tong, X., R. Chase, A. Skelton, T. Chen, J. Wright-Minogue, and B. A. Malcolm. 2006. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antivir. Res. 70:28-38. [DOI] [PubMed] [Google Scholar]
- 26.Tripathi, R. L., P. Krishnan, Y. He, T. Middleton, T. Pilot-Matias, C. M. Chen, D. T. Lau, S. M. Lemon, H. Mo, W. Kati, and A. Molla. 2007. Replication efficiency of chimeric replicon containing NS5A-5B genes derived from HCV-infected patient sera. Antivir. Res. 73:40-49. [DOI] [PubMed] [Google Scholar]
- 27.van Maarseveen, N. M., D. de Jong, C. A. Boucher, and M. Nijhuis. 2006. An increase in viral replicative capacity drives the evolution of protease inhibitor-resistant human immunodeficiency virus type 1 in the absence of drugs. J. Acquir. Immune Defic. Syndr. 42:162-168. [DOI] [PubMed] [Google Scholar]
- 28.Zhou, Y., D. J. Bartels, B. L. Hanzelka, U. Muh, Y. Wei, H. M. Chu, A. M. Tigges, D. L. Brennan, B. G. Rao, L. Swenson, A. D. Kwong, and C. Lin. 2007. Phenotypic characterization of Val36 resistant variants of hepatitis C virus Ns3-4a serine protease. Antimicrob. Agents Chemother. 52:110-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou, Y., U. Muh, B. L. Hanzelka, D. J. Bartels, Y. Wei, B. G. Rao, D. L. Brennan, A. M. Tigges, L. Swenson, A. D. Kwong, and C. Lin. 2007. Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha. J. Biol. Chem. 282:22619-22628. [DOI] [PubMed] [Google Scholar]