Abstract
The solubility of orthophosphate (PO43−) in iron-rich sediments can be exceedingly low, limiting the bioavailability of this essential nutrient to microbial populations that catalyze critical biogeochemical reactions. Here we demonstrate that dissolved extracellular DNA can serve as a sole source of phosphorus, as well as carbon and energy, for metal-reducing bacteria of the genus Shewanella. Shewanella oneidensis MR-1, Shewanella putrefaciens CN32, and Shewanella sp. strain W3-18-1 all grew with DNA but displayed different growth rates. W3-18-1 exhibited the highest growth rate with DNA. While strain W3-18-1 displayed Ca2+-independent DNA utilization, both CN32 and MR-1 required millimolar concentrations of Ca2+ for growth with DNA. For S. oneidensis MR-1, the utilization of DNA as a sole source of phosphorus is linked to the activities of extracellular phosphatase(s) and a Ca2+-dependent nuclease(s), which are regulated by phosphorus availability. Mass spectrometry analysis of the extracellular proteome of MR-1 identified one putative endonuclease (SO1844), a predicted UshA (bifunctional UDP-sugar hydrolase/5′ nucleotidase), a predicted PhoX (calcium-activated alkaline phosphatase), and a predicted CpdB (bifunctional 2′,3′ cyclic nucleotide 2′ phosphodiesterase/3′ nucleotidase), all of which could play important roles in the extracellular degradation of DNA under phosphorus-limiting conditions. Overall, the results of this study suggest that the ability to use exogenous DNA as the sole source of phosphorus is widespread among the shewanellae, and perhaps among all prokaryotes, and may be especially important for nutrient cycling in metal-reducing environments.
Phosphorus (P) is a key element that often limits bacterial growth in various freshwater and marine habitats (13, 40, 44, 51, 53). Inorganic phosphate (Pi), or orthophosphate (PO43−), can serve as a direct source of P for essentially all physiological groups of microorganisms in both natural environments and laboratory media. Measurements of soluble phosphate in different aquatic environments, however, suggest that concentrations of bioavailable Pi are very low. Recent studies have revealed that Pi represents only a small fraction of soluble reactive P in natural waters and that even in eutrophic systems, its concentration may be as low as 27 pM (2, 20, 25). This is not surprising, since in a variety of aquatic systems, soils, and sediments, Pi bioavailability can be controlled by adsorption to metal oxides (3, 16, 59) and through chemical reactions with hydrous oxides, amorphous and crystalline complexes of Fe, Al, and Ca, and organic matter (2, 41, 49). In particular, the reaction of Pi with Fe(III) oxides such as goethite can result in the precipitation of tinticite [Fe6(PO4)4(OH)6 · 7H2O] (22) or griphite [Fe3Mn2(PO4)2 · 5(OH)2] (32) depending on the solution and surface conditions. The latter findings entail important physiological implications for dissimilatory metal-reducing bacteria residing in zones with high concentrations of Fe(III) and Mn(III, IV) oxides, where the levels of bioavailable Pi may significantly limit growth. In that respect, utilization of other bioavailable forms of P may be the key to the ecological success of these bacteria.
Alternative sources of P in freshwater and marine environments are organic phosphorus compounds that include phospholipids, phosphoproteins, and nucleic acids. It has been demonstrated that dissolved extracellular DNA constitutes a significant portion of the organic P pool, where DNA concentrations can range from 0.2 to 88 μg/liter (0.65 to 280 nM P) (7, 24, 36, 43). The main sources of dissolved nucleic acids in aquatic environments include phage-induced cell lysis and excretion by zooplankton feeding on bacteria (48, 52, 55). Earlier studies showed that the addition of DNA to natural water samples resulted in its degradation and incorporation into cells and sometimes in an increase in bacterial cell numbers (17, 23, 34, 42, 55).
To gain a better understanding of physiological and mechanistic aspects of microbial nucleic acid utilization by dissimilatory metal-reducing organisms, we investigated the abilities of Shewanella spp. to utilize DNA for growth and respiration. Physiological and biochemical measurements were carried out using Shewanella oneidensis MR-1, Shewanella putrefaciens CN32, and Shewanella sp. strain W-3-18-1, isolated from freshwater, terrestrial, and marine environments, respectively, to allow a broader representation of the various ecophysiotypes within the genus. Overall, the results indicate that DNA could be used as a sole source of carbon, nitrogen, and P by all three strains under aerobic and anaerobic conditions and could serve as an electron donor for metal reduction. Application of continuous-cultivation, proteomic, and biochemical methods suggested that the ability of Shewanella to utilize dissolved nucleic acids as the sole source of P depends, in part or in whole, on the extracellular phosphatase activities and, under certain conditions, on the extracellular nuclease activities.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
S. oneidensis MR-1 (54), S. putrefaciens CN-32 (16), and Shewanella sp. strain W3-18-1 (45) were routinely cultured aerobically at 30°C with shaking at 150 rpm using tryptic soy broth (TSB) or M1 minimal medium (pH 7.0) containing (per liter) 30 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 28 mM NH4Cl, 30 mM NaCl, 3 mM MgCl2 · 6H2O, 1.34 mM KCl, 6.8 μM CaCl2, 1 μM Na2SeO4, and 10 ml each of 10× Wolfe's vitamin solution and 10× mineral solution (26). When necessary, supplemental CaCl2 was added to the M1 medium to a final concentration of 1 mM. Either NaH2PO4 · H2O or DNA was used as a source of P. All growth experiments were performed using low-molecular-weight (average, 200 bp, as estimated by agarose gel electrophoresis) herring sperm DNA. For batch cultivation, the M1 medium was supplemented with 30 mM d,l-sodium lactate or 1 g/liter DNA as the sole source of carbon and energy. The modified M1 medium used for continuous cultivation contained 3 mM PIPES, 18 mM d,l-sodium lactate, 1 mM CaCl2, 10 μM ferric nitrilotriacetic acid [Fe(III)-NTA], and different concentrations of NaH2PO4 · H2O or DNA as a source of P. For anaerobic growth, the M1 medium was supplemented with 10 mM Fe(III)-NTA as an electron acceptor. The medium was purged with O2-free N2, anaerobically dispensed into N2-flushed Balch tubes or serum bottles, sealed with butyl rubber stoppers, and sterilized by autoclaving.
For DNA-dependent growth with lactate as the source of carbon and energy, DNA was added to the phosphate-free medium to final concentrations ranging from 20 to 300 mg/liter. The DNA was sterilized by filtering through a 0.22-μm-pore-size membrane or was added to the medium prior to autoclaving. To obtain phosphorus-starved Shewanella cultures, phosphorus-free M1 medium containing 30 mM lactate was inoculated with a 0.4% (vol/vol) overnight-grown TSB culture and was incubated for 48 h. To ensure that no residual P remained in the cultures, a second transfer into the same medium was performed. The cultures were considered phosphorus starved if no growth occurred after the second transfer. These phosphorus-starved cultures were used as inocula for growth experiments unless stated otherwise. All growth experiments were independently repeated at least twice and performed in triplicate.
Chemostat cultivation.
All chemostat experiments were performed using 6-liter New Brunswick Bioflow 3000 reactors (New Brunswick Scientific, Edison, NJ) operated at a 3-liter working volume at 30°C. Reactors were inoculated with 120-ml phosphorus-starved S. oneidensis MR-1 cultures and were maintained in a batch mode until late-logarithmic stage. The continuous mode was initiated and maintained at a dilution rate of 0.088 h−1. The gas flow rate and agitation were kept at 4 liters/min and 450 rpm, respectively, and the dissolved oxygen concentration was maintained at 5% of air saturation by changing the ratio of N2 and air in the gas mix. The pH was maintained at 7.0 by automatic addition of 2 M HCl. Periodically, 5- to 10-ml samples were taken from the reactors for further analyses.
DNA preparations.
Lyophilized herring sperm DNA was purchased from Sigma (St. Louis, MO; catalog no. D3159). Stock solutions were prepared by dissolving the DNA in M1 minimal medium (pH 7.0) immediately before use. For nuclease activity assays, we used a 1,526-bp PCR product that had been amplified from the pDS1779 plasmid using the M13 forward primer (5′-GTA AAA CGA CGG CCA G-3′) and reverse primer (5′-CAG GAA ACA GCT ATG AC-3′) and Taq polymerase. The pDS1779 plasmid consisted of two fragments of the S. oneidensis MR-1 genome (bases 1861034 to 1861707 and bases 1863885 to 1864489 from GenBank accession number NC_004347) that had been linked via a 20-bp linker (AGA GAC GAC CTA AGC CAG TA) and cloned into XcmI-digested pDS3.1 (54a). Following PCR amplification, the product was purified using a QIAquick PCR purification kit (Qiagen Inc., Valencia, CA) and was quantified spectrophotometrically by absorbance at 260 nm.
Preparation of extracellular proteins and proteome analysis.
For extracellular proteome analysis, 1-liter samples of chemostat grown MR-1 cultures were centrifuged at 9,200 × g (10 min, 4°C) and the resulting supernatant was passed through a 0.22-μm-pore-size membrane filter (Nalgene, Rochester, NY). The supernatant was further concentrated 100-fold by ultrafiltration through 5-kDa-cutoff Ultracel cellulose filters (Millipore, Billerica, MA) using Amicon Stirred Cell 8400. Concentrated protein samples were digested with 2 μg of modified porcine trypsin (Promega, Madison, WI). The resulting peptide samples were subjected to liquid chromatography-tandem mass spectrometry (LC-MS-MS) using an LCQ Deca XPPLUS ion-trap mass spectrometer (Thermo-Electron, San Jose, CA). Each sample was centrifuged at 16,000 × g for 15 min prior to dilution to 0.2 μg/μl with 1% acetic acid in distilled H2O, and 2 μg (10 μl) of each peptide sample was loaded by a Famos autosampler (Dionex, Sunnyvale, CA) onto a hand-packed reverse-phase C18 high-performance LC (HPLC) column for each LC-MS-MS experiment. The samples were subjected to 120-min HPLC (HP-1100; Agilent, Palo Alto, CA) gradients (5% plus 80% acetic acid in distilled H2O against 100% acetonitrile); the LC-MS setup has been described elsewhere (28). The mass spectrometer was operated in the data-dependent acquisition mode using Xcalibur (version 1.4) software. The experiment consisted of a single full-scan mass spectrum (400 to 2,000 m/z) followed by three data-dependent MS-MS scans. The database search program TurboSEQUEST (version 27, revision 12) (12) was used to identify peptides in the MS-MS spectra. Searches of spectra at charge states +1, +2, and +3 were conducted using the following search parameters: no enzyme specificity; peptide mass tolerance, 2.5; average molecular weight for both peptide and fragment ion masses; a b/y ion search including neutral losses from b/y ions; and variable modification at M = +15.995. The SEQUEST output was processed by the LIPS (logistic identification of peptide sequences) model (19) to generate peptide identification probabilities. Three or more peptides with probabilities above 0.95 were used for positive identification. Orthologs in other Shewanella spp. were identified using the BLAST programs provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and Gnare (Genome Analysis Research Environment; http://compbio.mcs.anl.gov/gnare/gnare_home.cgi). Genome neighborhood analysis was conducted using the Integrated Microbial Genomes environment (IMG; http://img.jgi.doe.gov/cgi-bin/pub/main.cgi).
Analytical methods.
Nuclease activity was assayed by filtering the culture supernatant through a 0.22-μm-pore-size Millex-GP membrane filter (Millipore) and then mixing a 230-μl aliquot of this filtrate with 20 μl of 1,526-bp PCR-amplified DNA (final concentration, 2 μg/ml). The reaction mixtures were typically incubated for 60 min at 30°C, and 25-μl aliquots were removed timewise from the reaction mixture. The DNA was visualized by agarose gel electrophoresis. If no DNA was visible on a gel, it was assumed that all DNA was digested. The nuclease activity was expressed as the amount of DNA digested per milliliter of filtered culture liquid per min.
Phosphatase activity was assayed by mixing 0.9 ml of 0.22-μm-pore-size-membrane-filtered culture supernatant with 0.1 ml of 100 mM p-nitrophenylphosphate (final concentration, 10 mM) and monitoring 4-nitrophenol accumulation by the change in the optical density at 418 nm (OD418) at 30°C with a Shimadzu UV-2101PC scanning spectrophotometer (Shimadzu Scientific Instruments, Columbia, MD). The slope of the OD418 change for the first 5 min (linear range) was taken to determine the phosphatase activity by using a calibration curve prepared with 4-nitrophenol.
Inorganic phosphate concentrations were measured using the previously established molybdate technique (35). The iron reduction rates were measured colorimetrically using the capture reagent ferrozine, which forms a chromophoric complex with Fe(II) that is detectable at 562 nm (47). To measure the total Fe, samples were treated with hydroxylamine hydrochloride to reduce Fe(III) to Fe(II) prior to analysis by the ferrozine assay (15). Cell numbers in experiments with Fe(III)-NTA as the electron acceptor were determined by acridine orange direct epifluorescent microscopic counting. Organic acids were quantified using an HP 1090 HPLC with a diode array detector at 210 nm. The mobile phase was 0.005 N H2SO4 at 0.5 ml/min, and the column was a 300- by 78-mm Rezex organic acid column (Phenomenex, Torrance, CA) at 60°C.
RESULTS
Shewanella grows under aerobic and anaerobic conditions with DNA as a sole source of P.
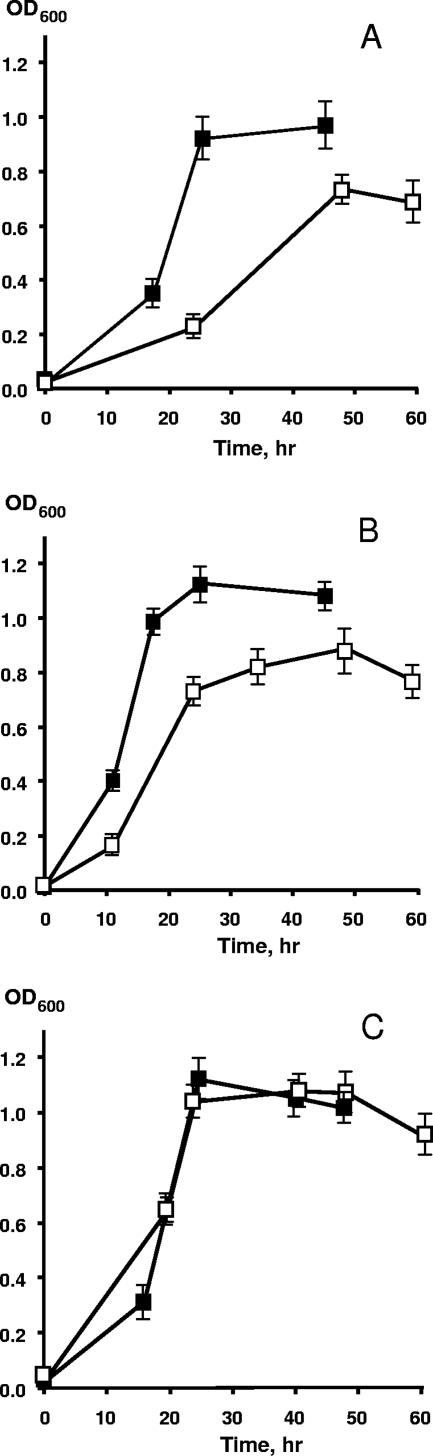
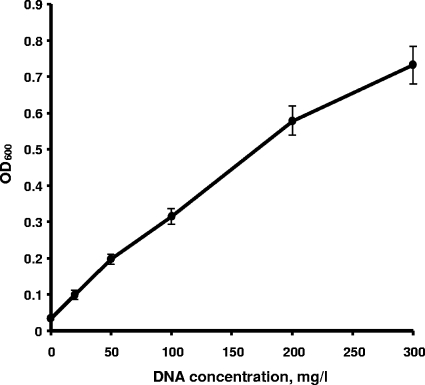
To probe the mechanisms of P assimilation by dissimilatory metal-reducing bacteria, we examined the aerobic growth of three Shewanella isolates, S. oneidensis MR-1, S. putrefaciens CN32, and Shewanella sp. strain W3-18-1, with lactate as the source of carbon and energy and DNA as the sole source of P. A series of experiments indicated that DNA supported vigorous aerobic growth of all three strains (Fig. 1). The growth rate and culture turbidity (OD600) differed for each strain, with W3-18-1 showing the fastest growth on both phosphate and DNA and having no preference for one P source over the other. By contrast, CN32 and MR-1 achieved consistently higher growth rates and final cell densities when NaH2PO4 · H2O was provided as the sole P source. As shown for MR-1, the increase in cell density during aerobic growth was nearly linear with increasing DNA concentrations (Fig. 2). Similar results were obtained for CN32 and W3-18-1 (data not shown). When DNA was autoclaved with the medium, the aerobic cultures of S. oneidensis MR-1 displayed higher growth yields, exceeding the cell densities observed with filter-sterilized DNA by 30 to 80%. This observation is consistent with the notion that autoclaving may cause partial DNA hydrolysis and is supported by our results showing that some DNA degradation to smaller fragments occurred as a result of autoclaving (data not shown), although free phosphate levels were still below detection (<0.25 μM).
FIG. 1.
Aerobic growth of S. oneidensis MR-1 (A), S. putrefaciens CN32 (B), and Shewanella sp. strain W3-18-1 (C) with NaH2PO4 (▪) or DNA (□) as the sole source of phosphorus. Approximately 5% of phosphorus-starved cultures were used as inocula for all strains. The DNA and NaH2PO4 · H2O concentrations in M1 medium were 300 mg/liter and 133.6 mg/liter, respectively, to achieve a final P concentration of 30 mg/liter. DNA was sterilized by autoclaving with the medium. The time course for growth was inferred from the OD600. The data points and error bars represent the means and standard deviations of triplicate cultures, respectively.
FIG. 2.
Relationship between S. oneidensis MR-1 aerobic growth for 48 h and initial DNA concentrations. DNA was added to the medium before autoclaving and served as the sole P source. Five percent of phosphorus-starved cultures were used as inocula.
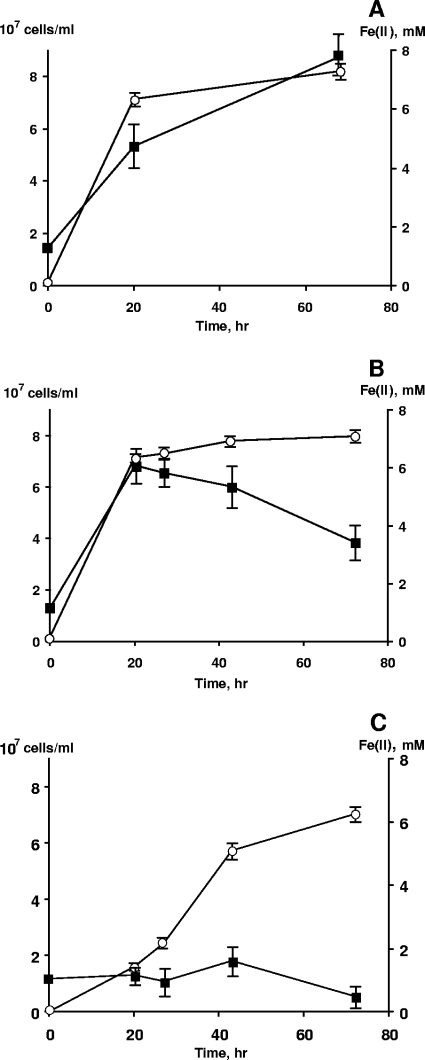
Next, we examined the abilities of three Shewanella strains to grow by reducing Fe(III)-NTA under anaerobic conditions using filter-sterilized DNA as a sole source of P and lactate as the electron donor. The ability to use DNA was assessed by five consecutive serial transfers (a 20-fold dilution factor was used) to fresh medium to minimize nutrient carryover from the inoculum. In each transfer, 90 to 98% of the total Fe(III) was reduced in 30 to 50 h. No formation of a secondary Fe(II) phase was observed during reduction by any of the three strains. Quantitative measurements indicate that within 20 h, in the presence of DNA as the sole source of P, S. oneidensis MR-1 reached the maximum cell count and achieved nearly complete reduction of Fe(III), while there was no growth and relatively slow reduction without any P source (Fig. 3). No significant differences in Fe(III) reduction rates were observed between cultures using DNA or NaH2PO4. The time to reach maximum cell density was longer with NaH2PO4 for reasons that are unclear.
FIG. 3.
Growth of S. oneidensis MR-1 in phosphate-free M1 medium with 8 mM Fe(III)-NTA as the electron acceptor and 30 mM lactate as the electron donor and carbon source. (A and B) NaH2PO4 · H2O (A) and DNA (B) were used at concentrations of 60 mg/liter and 100 mg/liter, respectively. Filter-sterilized DNA was added to the medium after autoclaving. (C) Growth and Fe(III) reduction in the absence of a P source. ▪, cell numbers; ○, Fe2+ concentrations.
Extracellular phosphatase activity is detected in S. oneidensis MR-1 culture filtrates.
To gain a basic understanding of the possible mechanisms of DNA utilization, we assayed extracellular nuclease and phosphatase activities in 0.22-μm-pore-size-filtered samples of S. oneidensis MR-1 cultures grown in the presence of DNA. Initial assays failed to detect cell-free nuclease activity in any of the MR-1 cultures grown with 150 or 300 mg of DNA/liter. In addition, agarose gel electrophoresis of cell supernatants revealed that the added DNA remained intact (data not shown), despite apparent utilization of P.
In contrast, phosphatase activity measured with p-nitrophenylphosphate reached 0.6 μmol/ml/min in cultures grown with 150 mg/liter of DNA for 48 h. To determine whether Pi is released as a result of extracellular phosphatase activity, we added DNA at final concentrations of 300 mg/liter to DNA-grown MR-1 culture filtrates (pore size, 0.22 μm) and incubated the mixture for 30 min at 30°C using a heat-inactivated filtrate as a negative control. Addition of a molybdate-based reagent for a P (PO4) assay (35) led to time-dependent development of a characteristic blue color due to the formation of a P-molybdate complex in the experimental tubes, whereas the controls remained colorless. Unfortunately, quantitative measurements of P (PO4) formation were not possible due to the formation of a precipitate in both the control and the experimental samples. We interpret these results as evidence for the production by S. oneidensis MR-1 of an extracellular phosphatase that is able to cleave phosphate groups from the 5′ end of DNA.
Effect of Ca2+ on DNA utilization by different Shewanella strains.
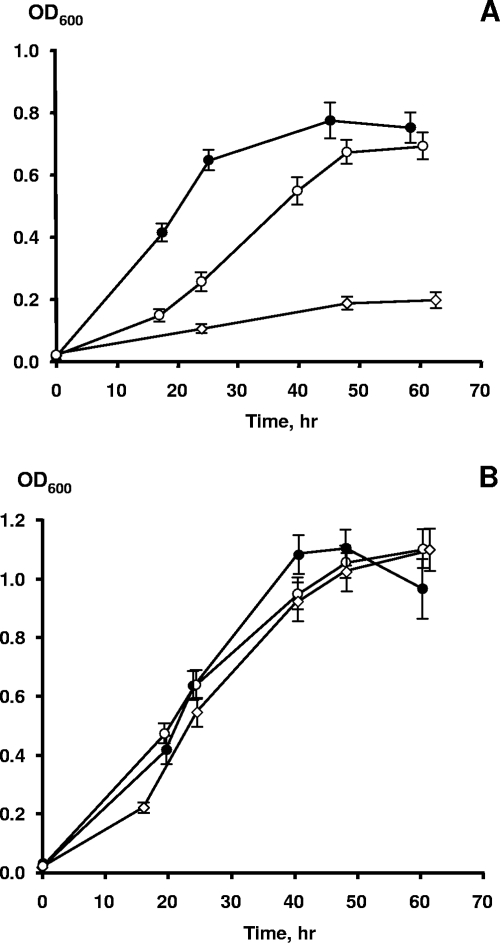
Although the growth of S. oneidensis MR-1 with DNA as a sole source of P was not accompanied by DNA hydrolysis and MR-1 displayed no extracellular nuclease activity, its genome encodes two proteins, SO1844 and SO1066, that belong to the COG2374 predicted extracellular-nuclease group (for complete genome sequence information, see http://www.ncbi.nlm.nih.gov) (18). Because some extracellular nucleases require divalent cations to be functional (4, 14, 31, 33, 38), we reasoned that the inability of MR-1 to cleave DNA molecules could be a result of the relatively low Ca2+ concentration (6.8 μM) in M1 growth medium. To address this question, we first examined the effect of CaCl2 on DNA assimilation by the three Shewanella strains. Addition of 1 mM CaCl2 greatly enhanced the ability of MR-1 (Fig. 4A) and CN32 (data not shown) to utilize DNA as the sole source of P for growth. In contrast, Shewanella sp. strain W3-18-1 grew equally well with either DNA or NaH2PO4, and additions of CaCl2 had no effect on growth kinetics (Fig. 4B), suggesting that DNA hydrolysis by this particular strain is not Ca2+ dependent.
FIG. 4.
Aerobic growth of S. oneidensis MR-1 (A) and Shewanella sp. strain W3-18-1 (B) with NaH2PO4 (•), DNA without supplemental CaCl2 (⋄), or DNA with 1 mM CaCl2 (○) as the sole source of phosphorus. Approximately 5% of phosphorus-starved cultures were used as inocula for all strains. The DNA and NaH2PO4 · H2O concentrations in M1 medium were 50 mg/liter and 22.3 mg/liter, respectively, to achieve a final P concentration of 5 mg/liter. The time course for growth was inferred from the OD600. The data points and error bars represent the means and standard deviations of triplicate cultures, respectively.
Analysis of MR-1 culture filtrates indicated that no residual extracellular DNA was detected after 48 h of incubation in the presence of 1 mM CaCl2. Additions of 1 mM Ca2+ to MR-1 cultures grown aerobically in the presence of 50 mg DNA/liter resulted in threefold biomass increases as indicated by optical density measurements (Fig. 4A). In contrast, when NaH2PO4 was provided as the sole P source, the addition of 1 mM CaCl2 did not stimulate growth (data not shown). Taken together, these results suggest that added exogenous DNA may be digested by a Ca2+-dependent nuclease(s) produced by growing cultures of MR-1 and CN32. Nevertheless, 0.22-μm-pore-size filtrates prepared from MR-1 cultures grown with DNA in the presence of 1 mM CaCl2 failed to exhibit nuclease activity (data not shown).
Growth studies and enzyme activities in continuous cultures of S. oneidensis MR-1 under phosphorus and carbon limitation.
Previous investigations have indicated that P availability can have a profound effect on the phosphorus assimilation pathways, since starvation or limitation may induce the expression of extracellular nuclease activities (21, 50). Although the lack of nuclease activity in batch culture supernatants could be the result of cell-bound enzymatic action, we hypothesized that the development of extracellular nuclease activity may also have been suppressed by the excess of DNA or products of its degradation in the medium. To investigate this, S. oneidensis MR-1 was grown in continuous culture with variable concentrations of P (0.1 mM, 0.5 mM, and 1.5 mM), which included PO43− or DNA in the presence of 1 mM CaCl2. Our results indicated that steady-state cultures grown with 0.1 mM P (PO4) or DNA were in fact P limited, as shown by the presence of acetate, the product of incomplete lactate oxidation, in the supernatant. Moreover, the cell densities in these cultures were about one-half of those obtained under carbon limitation with 1.5 mM P, and the concentrations of free P (PO4) in filtered supernatants were below 0.25 μM (Table 1). Nuclease and phosphatase assays using culture filtrates from both PO4 and DNA continuous cultures grown in the presence of 0.1 mM P indicated apparent time-dependent increases in the specific activities of both enzymes (Fig. 5A and C). In particular, nuclease activity increased 4- to 12-fold in DNA and PO4 reactors, respectively, after ∼7 reactor volume changes.
TABLE 1.
Biochemical and physiological parameters of S. oneidensis MR-1 growth in chemostat culturesa
| Concn of phosphorus in medium (mM) | OD600
|
Residual acetate concnb (mM)
|
P (PO4) concn in fermentor (μM)
|
Extracellular nuclease activity (μg/min/ml)
|
Extracellular phosphatase activity (nmol/min/ml)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| PO4 | DNA | PO4 | DNA | PO4 | DNA | PO4 | DNA | PO4 | DNA | |
| 0.1 | 0.355 | 0.355 | 4.67 | 4.76 | BD | BD | 0.8 | 0.8 | 5.5 | 5.3 |
| 0.5 | 0.735 | 0.759 | BD | BD | 2.27 | 1.12 | BD | 0.1 | 2.4 | 2.1 |
| 1.5 | 0.780 | 0.755 | BD | BD | 800 | ND | BD | BD | 0.34 | 2.2 |
Reactors were supplemented with NaH2PO4 or DNA as a P source (designated “PO4” and “DNA,” respectively). BD, below detection; ND, not detected.
Measured in the reactors after steady state was achieved. The concentrations of lactate and pyruvate were always below detection levels.
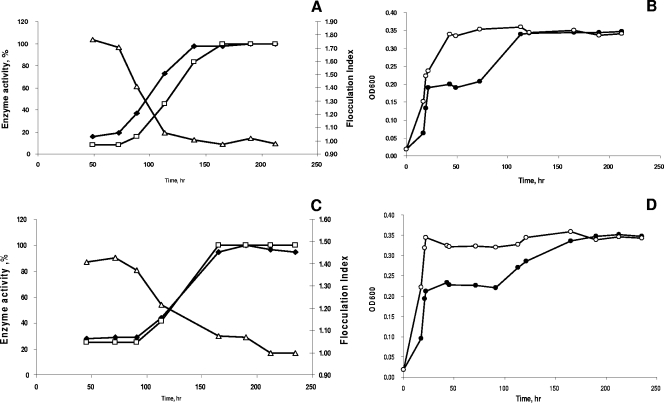
FIG. 5.
Growth and extracellular phosphatase and nuclease activities in chemostat cultures of MR-1 in the presence of NaH2PO4 (A and B) and DNA (C and D). The flocculation index (▵) was calculated based on the optical densities of cultures measured without (•) and with (○) 10 mM EDTA. Extracellular phosphatase (⧫) and nuclease (□) activities in chemostat cultures of S. oneidensis MR-1 were measured as described in Materials and Methods.
Elevating the P concentration to 0.5 mM resulted in a twofold increase in biomass, while the acetate concentrations declined below detection levels in culture filtrates. The P limitation was relieved in these cultures; the residual P (PO4) concentrations in both DNA- and PO4-supplied reactors increased to 1.12 μM and 2.27 μM, respectively. At 0.5 mM P, no extracellular nuclease activity was detected in the PO4 reactor, while the extracellular nuclease activity in the DNA-fed reactor decreased eightfold from that under P-limited conditions (Table 1). Increasing the P concentration further to 1.5 mM did not result in significant increases in cell density in either reactor. The extracellular nuclease activity in the DNA reactor decreased below detection levels, while free DNA was present in the culture filtrates (data not shown). Similarly, the concentrations of P (PO4) increased to 0.8 mM in filtrates from the PO4 continuous culture. Cell-free phosphatase activities in both the PO4 and DNA reactors decreased twofold with an increase in P concentration from 0.1 to 0.5 mM. Further increasing the P concentration to 1.5 mM did not affect phosphatase activity in the DNA culture, whereas in the PO4 reactor, the extracellular phosphatase activity decreased nearly 10-fold (Table 1).
It should be noted that P availability had a profound effect on the behavior of aerobic chemostat cultures of S. oneidensis MR-1, resulting in an intriguing physiological manifestation. In earlier studies, we have shown that aerobically grown MR-1 forms tight multicellular aggregates in the presence of millimolar concentrations of Ca2+ (J. S. McLean et al., unpublished data). The aggregates dissipate upon addition of EDTA, presumably due to the complexation of Ca2+, which serves as a bridging cation, resulting in an increase in optical density. Therefore, EDTA was used to estimate the biomass concentration of aggregated cultures. The ratio of OD600 values with and without 10 mM EDTA was designated the flocculation index (FI) and reflected the extent of aggregation. In this study, FI stability was one of the parameters used to define steady-state conditions in aerobic P-limited cultures. Our observations indicate that the aggregation of aerobic S. oneidensis MR-1 cultures was usually stable for at least 7 reactor volume changes when S. oneidensis was cultured aerobically in a carbon-limited chemostat with 1.5 mM NaH2PO4 in the presence of 1 mM CaCl2. Under these conditions, no extracellular nuclease activity was detected (Table 1). Under P limitation, however, the aerobic chemostat cultures of MR-1 gradually disaggregated, as reflected by the decrease in the FI. Our results show that the dissipation of cell aggregates coincided with a significant increase in extracellular nuclease and phosphatase specific activities (Fig. 5A to D).
Extracellular proteome analysis of P-limited chemostat cultures.
To investigate the identity of proteins that may contribute to the extracellular nuclease and phosphatase activities in MR-1, we isolated total extracellular proteins from a P (PO4)-limited chemostat culture and subjected them to LC-MS-MS analysis. Measurements of total extracellular protein concentrations showed 2.5 to 3.0 mg/liter under steady-state conditions when the P concentration was 0.1 mM. Table 2 contains a list of proteins that were detected with high confidence in filtered chemostat samples of S. oneidensis MR-1 grown under these conditions. Out of 22 proteins, all but 2, purine nucleoside phosphorylase (SO1221) and cysteine synthase A (SO2903), were predicted to be localized to the outside of the cytoplasmic membrane, indicating the absence of any significant cell lysis. Notably, the predicted functions of seven proteins (SO0990, SO1221, SO1560, SO1844, SO2001, SO2385, and SO3565) were related to P or DNA/nucleotide metabolism. Among these, SO1844 is predicted to encode an endonuclease and could account for the DNase activity observed in the supernatants of cells grown on Na2HPO4. The protein encoded by SO2385, originally annotated as a hypothetical protein, shares 42% identity with the recently discovered PhoX monomeric alkaline phosphatase of Pasteurella multocida (57). Both SO2385 and PhoX possess an N-terminal leader peptide that is characteristic of lipoproteins secreted across the cytoplasmic membrane by the twin arginine translocation system. PhoX is activated by calcium, suggesting that the increased growth of MR-1 on DNA observed in calcium-supplemented medium is due in part to activation of the phosphatase activity encoded by SO2385.
TABLE 2.
List of extracellular proteins identified in filtered supernatants of MR-1 chemostat cultures grown with 0.1 mM P (PO4)
| Locus taga | Functional annotation | Predicted localization | Peptide countb |
|---|---|---|---|
| SO0189 | Outer membrane fibronectin type III/cadherin domain protein | Outer membrane; β-barrel protein | 3 |
| SO0990 | Periplasmic amidohydrolase 3 family protein | Periplasm | 12 |
| SO1221 | Purine nucleoside phosphorylase (DeoD) | Cytoplasm | 4 |
| SO1557 | Outer membrane porin | Outer membrane; β-barrel protein | 20 |
| SO1560 | Phosphate-binding protein | Periplasm | 23 |
| SO1779 | Outer membrane decaheme cytochrome c (OmcA) | Outer membrane; lipoprotein | 5 |
| SO1822 | TonB-dependent receptor | Outer membrane; β-barrel protein | 11 |
| SO1844 | Outer membrane endonuclease/exonuclease/phosphatase | Outer membrane; β-barrel protein | 82 |
| SO2001 | Bifunctional UDP-sugar hydrolase/5′ nucleotidase (UshA) | Outer membrane; lipoprotein | 30 |
| SO2385 | Monomeric alkaline phosphatase (PhoX) | Extracellular | 35 |
| SO2903 | Cysteine synthase A (CysK) | Cytoplasm | 8 |
| SO2907 | TonB-dependent receptor | Outer membrane; β-barrel protein | 14 |
| SO3237 | Flagellin (FliC1) | Extracellular; flagella | 22 |
| SO3238 | Flagellin (FliC2) | Extracellular; flagella | 5 |
| SO3247 | Flagellar hook protein (FlgE) | Extracellular; flagella | 11 |
| SO3545 | Outer membrane porin | Outer membrane; β-barrel protein | 4 |
| SO3565 | Bifunctional 2′,3′ cyclic nucleotide 2′ phosphodiesterase/3′ nucleotidase (CpdB) | Outer membrane; lipoprotein | 9 |
| SO3896 | Outer membrane porin (Omp35) | Outer membrane; β-barrel protein | 8 |
| SO4317 | Cell surface calcium-binding protein with bacterial neuraminidase repeats | Extracellular | 11 |
| SO4652 | ABC sulfate/thiosulfate transporter, periplasmic ligand-binding subunit (Sbp) | Periplasm | 16 |
| SO4719 | ABC tungstate transporter, periplasmic ligand-binding subunit (TupA) | Periplasm | 8 |
| SOA0110 | Expressed lipoprotein | Outer membrane; lipoprotein | 5 |
Locus tag numbers correspond to the NCBI S. oneidensis MR-1 genome annotation (http://www.ncbi.nlm.nih.gov).
Number of peptides detected for a particular protein. Only nonoverlapping, fully tryptic peptides are included.
We have also identified two putative proteins, SO2001 and SO3565, that can catalyze the hydrolysis of 2′,3′ cyclic phosphates of adenosine, guanosine, cytosine, and uridine with the formation of a free phosphate group. SO3565 encodes a bifunctional 2′,3′ cyclic nucleotide 2′ phosphodiesterase/3′ nucleotidase similar to the CpdB protein in Escherichia coli. However, unlike the corresponding proteins in E. coli and other bacteria, which are predicted to be periplasmic, the putative CpdB from MR-1 is predicted to be localized to the outer membrane. Similarly, SO2001, which encodes a putative UDP-sugar hydrolase (UshA) protein, contains the N- and C-terminal 5′ nucleotidase domains. Furthermore, like CpdB, the Shewanella UshA proteins are predicted to be lipoproteins, while their counterparts in enteric bacteria are predicted to be periplasmic. Recent studies with Corynebacterium glutamicum showed that UshA is synthesized and secreted into the medium during phosphate starvation, suggesting involvement in the utilization of nucleotides and related compounds as sources of phosphorus (39). Collectively, the proteomic data indicate that S. oneidensis MR-1 excretes a number of extracellular proteins with putative functions in DNA degradation as a result of P limitation. Moreover, the extracellular nuclease and phosphatase activities and possibly nuclease localization are regulated by the P source and, most likely, P (PO4), availability.
Shewanella can use DNA as a sole source of carbon, energy, phosphorus, and nitrogen.
Shewanella strains are commonly isolated from freshwater or marine sediments and the water column, where DNA may be abundant (6, 7, 24, 36, 43). Given the results of our preliminary experiments demonstrating that some members of a bacterial population isolated from the upper layer of the Columbia River (Washington State) were able to grow with DNA as the sole nutrient, and the fact that MR-1 is able to use adenosine as a sole source of carbon and energy (M. Driscoll, unpublished data), we hypothesized that Shewanella can use DNA as a sole source of carbon and energy.
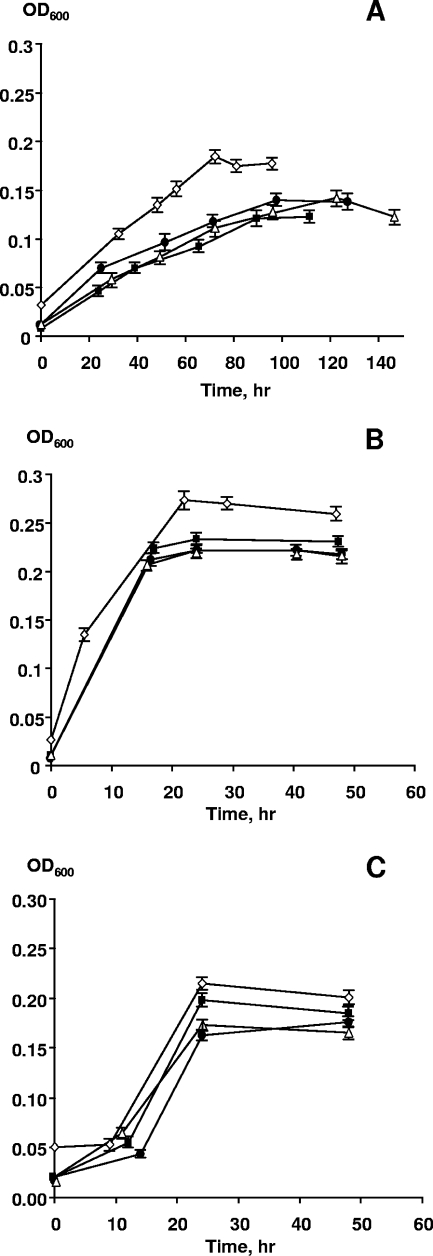
The results of our experiments presented in Fig. 6 show that all three strains were able to grow aerobically with DNA as the only C, N, P, and energy source for at least four transfers. Strain W3-18-1 showed the highest growth rates and final OD600, whereas MR-1 grew significantly more slowly (Fig. 6A and B). These results strongly suggest that these Shewanella strains are able to obtain nitrogen by catabolizing one or more of the nucleotides present in DNA. Anaerobic growth with filter-sterilized DNA as the sole nutrient was also examined in the presence of Fe(III)-NTA as the electron acceptor. Altogether, six consecutive serial transfers (using a 10-fold dilution factor) were performed; strains CN32 and W3-18-1 reduced 90% to 99% of Fe(III) in a period from 100 to 114 h upon each transfer. MR-1 also grew and reduced Fe(III) with DNA but required two to three times longer to achieve the same extent of reduction (data not shown). The Fe(III) reduction rates are consistent with the slower aerobic growth of MR-1 using DNA as the sole nutrient (Fig. 6). Hence, growth with DNA as the sole source of carbon, energy, nitrogen, and phosphorus under both aerobic and anaerobic conditions appears to be a common trait of metal-reducing Shewanella spp.
FIG. 6.
Aerobic growth of S. oneidensis MR-1 (A), Shewanella sp. strain W3-18-1 (B), and S. putrefaciens CN32 (C) with 1 g/liter of DNA as the sole source of carbon, energy, N, and P in phosphate- and nitrogen-free M1 medium. The inoculum consisted of a P-starved culture that was diluted 20-fold into M1 medium. The time course for growth was inferred from the OD600. The data points and error bars represent the means and standard deviations of triplicate cultures, respectively. ⋄, first transfer; ▪, second transfer; ▵, third transfer; •, fourth transfer.
DISCUSSION
The presence of extracellular DNA in water columns and sediments has been well documented, with concentrations often exceeding those of inorganic phosphate (2, 7, 20, 24, 25, 36, 43). DNA, as well as RNA and protein, associated with dead or moribund cells and insoluble organic matter settles through the water column in freshwater and marine environments and eventually reaches sediments, where it can potentially be assimilated by bacteria, including metal-reducing species. Analyses of marine sediments have shown that polymeric carbohydrates, proteins, and DNA may constitute most of the organic matter, because soluble and labile organic compounds are consumed by bacteria before particles reach the sea floor (5, 8, 30). Moreover, extracellular DNase activity was found in both sediments and seawater (8, 9, 30), suggesting ongoing DNA degradation. The ability to use DNA as a sole source of P may be especially beneficial for metal-reducing bacteria such as Shewanella, since phosphate complexation with Fe(II) or sorption to Fe(III) oxides (16, 22, 59) may limit Pi bioavailability.
In this work, we tested the abilities of three strains of Shewanella, S. oneidensis MR-1, S. putrefaciens CN32, and Shewanella sp. strain W3-18-1, isolated from freshwater lake sediment, the subsurface, and marine sediment, respectively, to use DNA as a sole source of P, and we probed for mechanisms involved in the utilization of this organic polymer. While their growth rates and requirements for Ca2+ differed, all three strains were capable of utilizing DNA not only as a P source but also as a sole source of nitrogen, carbon, and energy to maintain growth under both aerobic and anaerobic conditions. Of the three strains tested, S. oneidensis MR-1 showed the lowest rate of growth with DNA as a sole nutrient both aerobically and anaerobically. The reason for this is unclear, although the ability to slowly use DNA as an energy source may be beneficial for long-term survival when other carbon and electron sources in sediments are exhausted.
Using MR-1 as an example, we showed that the ability of Shewanella to utilize DNA as a P source was linked to extracellular nuclease and phosphatase activities. Interestingly, extracellular nuclease activity was absent in filtrates from batch cultures of S. oneidensis grown with DNA. On the one hand, it is likely that DNA utilization in batch cultures is a result of cell-bound nuclease activity. On the other hand, the development of extracellular nuclease activity could be repressed by high initial DNA concentrations and the accumulation of degradation products. To alleviate the limitations of batch cultivation, we grew MR-1 in chemostats to impose specific nutrient limitations. The results from these experiments indicated that P limitation promoted extracellular nuclease and phosphatase activities, either by increasing the expression levels or by releasing preexisting proteins to the extracellular milieu. Higher extracellular phosphatase and nuclease activities were measured when either Pi or DNA was the growth-limiting substrate in the chemostat cultures, suggesting that the specific activities of these enzymes were independent of the P source.
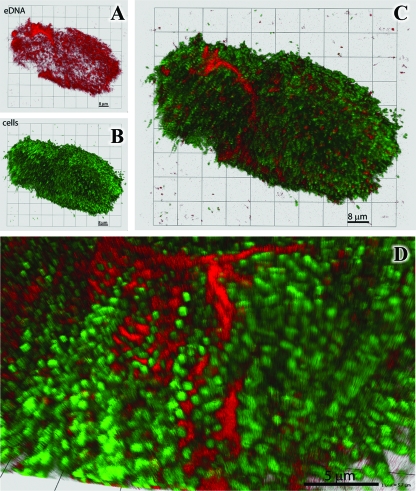
Chemostat cultivation of MR-1 under P limitation resulted in another interesting observation: dissipation of oxygen- and Ca2+-induced cell aggregation, which coincided with an increase in extracellular nuclease activity. Extracellular DNA has been identified in a number of biofilm systems (37, 46, 56) and appears to play a role as a structural component. Addition of DNase I to Pseudomonas aeruginosa PAO1 biofilms was reported to disaggregate the colonies (56). By analogy, we reasoned that extracellular DNA could be present in the MR-1 aggregate matrix and that its depolymerization by extracellular nuclease resulted in culture disaggregation. To evaluate this hypothesis, the MR-1 aggregates were investigated using the cell membrane-impermeant DNA stain 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one) (DDAO) as described in references 1 and 58 and the membrane-permeant DNA stain Syto 59. The confocal images indicate that extracellular DNA is in fact present throughout the structure and can be clearly seen in association with the intracellular matrix (Fig. 7).
FIG. 7.
Representative 3-dimensional confocal images of the MR-1 multicellular aggregates. (A to C) Differential staining using fluorescent dyes for extracellular DNA (DDAO; red) (A) and intercellular DNA (Syto 59; green) (B) and a merged image (C) reveal the spatial distribution and relative volumetric percentages of cells and matrix material. (D) Cells (green) and DDAO stain (red) at high resolution show extracellular DNA staining between cells.
Interestingly, W3-18-1, which is the only marine isolate among the three Shewanella strains used in this study, displayed Ca2+-independent utilization of DNA, whereas both CN32 and MR-1, which were isolated from the subsurface and freshwater sediments, respectively, depended on Ca2+ to fully digest DNA. From an ecological standpoint, this observation is somewhat counterintuitive, given that concentrations of Ca2+ in seawater are typically significantly higher than those in freshwater (27). Nonetheless, it indicates fundamental differences in the enzyme repertoire or DNA utilization pathways displayed by different strains of Shewanella. To probe this question, we investigated whether the genome sequences of strains CN32 and W3-18-1 contain any homologs of MR-1 genes SO1844 and SO1066, both of which belong to the COG2374 family of extracellular nucleases. Comparative analysis of the CN32 and W3-18-1 genomes indicated that both strains encode only two members of the COG2347 family orthologous to SO1066 and SO1844, of which only the latter was detected in supernatants of P-limited MR-1 cultures. While no unique known nucleases are found in the genome of W3-18-1, it is feasible to suggest that this bacterium encodes unknown novel Ca2+-independent extracellular nucleases.
It is also possible that the nuclease genes in W3-18-1 are regulated differently from those in MR-1 and that their expression under specific growth conditions contributes to Ca+2-independent utilization of DNA. In working with Shewanella strain W3-18-1, we have observed that this strain does not form biofilms on glass surfaces, nor does it form aggregates in the presence of Ca2+ (J. S. McLean, unpublished data). Previously, it was shown that the locus encoding a type I secretion system and the putative SO4317 adhesin detected in our P-limited supernatants are associated with biofilm and aggregate formation in MR-1 (11). In Shewanella sp. strain W3-18-1, however, the ATPase/C39 family cysteine peptidase component (Sputw3181_3731) of the analogous type I secretion system is interrupted by an insertion element. Consequently, we hypothesize that this strain is unable to secrete the SO4317 ortholog (Sputw3181_3730), leading to an inability to form biofilms or aggregate in the presence of Ca2+. Further evidence of biofilm formation deficiency in W3-18-1 is provided by the observation that three of the O-antigen biosynthesis genes are pseudogenes (Sputw3181_1473, -1475, and -1478) and that the putative pilN gene, encoding a type IV pilin biogenesis protein, is interrupted by an insertion element. Given this evidence, it is reasonable to propose that the loss of many functions associated with biofilm behavior in W3-18-1 may be balanced by changes in regulatory networks, leading to expression of different genes (e.g., the SO1066-type nuclease) in the absence of Ca2+ during growth as cell suspensions.
In light of earlier studies by Dell'Anno and Danovaro (10), which suggested that almost 50% of the P demand of deep-sea prokaryotic populations worldwide is supplied by DNA, the implications of the current work may be expanded to different environments. Indeed, independent experiments confirmed that the metal-reducing bacterium Geobacter sulfurreducens and the gram-positive bacterium Bacillus subtilis were able to use DNA as the sole source of P, while the prokaryotic population residing in the upper layer of the Columbia River grew rapidly in river water supplemented with 1 g of DNA/liter (unpublished data). Furthermore, the recently published results on DNA utilization by different groups of marine bacteria (29) support the notion that the ability to use exogenous DNA as a source of phosphorus, carbon, energy, and nitrogen is widespread among some groups of prokaryotes and may be especially important for nutrient cycling in PO4-limited environments in which metal-reducing bacteria reside.
Acknowledgments
We appreciate and gratefully acknowledge Christina Bilskis, Laine Riley, and Oleg Geydebrekht for help with growth experiments. We thank Eugene Kolker, Rodger Higdon, and Jason Hogan for the proteomic analysis of chemostat samples and Alexander Tsapin and Yuri Gorby for helpful discussions and advice regarding experimental procedures.
This work was supported by the Genomics:GTL Program via the Shewanella Federation consortium, Office of Biological and Environmental Research, U.S. Department of Energy. The Pacific Northwest National Laboratory is operated by the Battelle Memorial Institute for the DOE under contract DE-AC05-76RLO1830.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Allesen-Holm, M., K. B. Barken, L. Yang, M. Klausen, J. S. Webb, S. Kjelleberg, S. Molin, M. Givskov, and T. Tolker-Nielsen. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114-1128. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, D. S. 1998. Reactive “organic” phosphorus revisited. Water Res. 32:2265-2270. [Google Scholar]
- 3.Bjerrum, C. J., and D. E. Canfield. 2002. Ocean productivity before about 1.9 Gyr ago limited by phosphorus adsorption onto iron oxides. Nature 417:159-162. [DOI] [PubMed] [Google Scholar]
- 4.Burke, W. F., Jr., and B. S. Slinker. 1982. Extracellular exonuclease as a stage 0 biochemical marker in Bacillus subtilis sporulation. J. Bacteriol. 149:768-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danovaro, R., M. Armeni, A. Dell'Anno, M. Fabiano, E. Manini, D. Marrale, A. Pusceddu, and S. Vanucci. 2001. Small-scale distribution of bacteria, enzymatic activities, and organic matter in coastal sediments. Microb. Ecol. 42:177-185. [DOI] [PubMed] [Google Scholar]
- 6.Danovaro, R., A. Dell'Anno, A. Pusceddu, and M. Fabiano. 1999. Nucleic acid concentrations (DNA, RNA) in the continental and deep-sea sediments of the eastern Mediterranean: relationships with seasonally varying organic inputs and bacterial dynamics. Deep-Sea Res. Part I 46:1077-1094. [Google Scholar]
- 7.DeFlaun, M. F., J. H. Paul, and D. Davis. 1986. Simplified method for dissolved DNA determination in aquatic environments. Appl. Environ. Microbiol. 52:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dell'Anno, A., S. Bompadre, and R. Danovaro. 2002. Quantification, base composition, and fate of extracellular DNA in marine sediments. Limnol. Oceanogr. 47:899-905. [Google Scholar]
- 9.Dell'Anno, A., and C. Corinaldesi. 2004. Degradation and turnover of extracellular DNA in marine sediments: ecological and methodological considerations. Appl. Environ. Microbiol. 70:4384-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dell'Anno, A., and R. Danovaro. 2005. Extracellular DNA plays a key role in deep-sea ecosystem functioning. Science 309:2179. [DOI] [PubMed] [Google Scholar]
- 11.De Windt, W., H. Gao, W. Krömer, P. Van Damme, J. Dick, J. Mast, N. Boon, J. Zhou, and W. Verstraete. 2006. AggA is required for aggregation and increased biofilm formation of a hyper-aggregating mutant of Shewanella oneidensis MR-1. Microbiology 152:721-729. [DOI] [PubMed] [Google Scholar]
- 12.Eng, J. K., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 13.Farjalla, V. F., F. A. Esteves, R. L. Bozelli, and F. Roland. 2002. Nutrient limitation of bacterial production in clear water Amazonian ecosystems. Hydrobiologia 489:197-205. [Google Scholar]
- 14.Ferrieri, P., E. Gray, and L. Wannamaker. 1980. Biochemical and immunological characterization of the extracellular nucleases of group B streptococci. J. Exp. Med. 151:56-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredrickson, J. K., S. Kota, R. K. Kukkadapu, C. Liu, and J. M. Zachara. 2003. Influence of electron donor/acceptor concentrations on hydrous ferric oxide (HFO) bioreduction. Biodegradation 14:91-103. [DOI] [PubMed] [Google Scholar]
- 16.Fredrickson, J. K., J. M. Zachara, D. W. Kennedy, H. Dong, T. C. Onstott, N. W. Hinman, and S.-M. Li. 1998. Biogenic iron mineralization accompanying the dissimilatory reduction of hydrous ferric oxide by a groundwater bacterium. Geochim. Cosmochim. Acta 62:3239-3257. [Google Scholar]
- 17.Fujioka, R. S., P. C. Loh, and L. S. Lau. 1980. Survival of human enteroviruses in the Hawaiian ocean environment: evidence for virus-inactivating microorganisms. Appl. Environ. Microbiol. 39:1105-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidelberg, J., I. Paulsen, K. Nelson, E. Gaidos, W. Nelson, T. Read, J. Eisen, R. Seshadri, N. Ward, B. Methe, R. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. DeBoy, R. Dodson, A. Durkin, D. Haft, J. Kolonay, R. Madupu, J. Peterson, L. Umayam, O. White, A. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. Utterback, L. McDonald, T. Feldblyum, H. Smith, J. Venter, K. Nealson, and C. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 19.Higdon, R., N. Kolker, A. Picone, G. V. Belle, and E. Kolker. 2004. LIP index for peptide classification using MS/MS and SEQUEST search via logistic regression. Omics 8:357-369. [DOI] [PubMed] [Google Scholar]
- 20.Hudson, J. J., W. D. Taylor, and D. W. Schindler. 2000. Phosphate concentrations in lakes. Nature 406:54-56. [DOI] [PubMed] [Google Scholar]
- 21.Ishige, T., M. Krause, M. Bott, V. F. Wendisch, and H. Sahm. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 185:4519-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonasson, R. G., R. R. Martin, M. E. Giuliacci, and K. Tazaki. 1988. Surface reactions of goethite with phosphate. J. Chem. Soc. Faraday Trans. 84:2311-2315. [Google Scholar]
- 23.Jorgensen, N. O. G., and C. S. Jacobsen. 1996. Bacterial uptake and utilization of dissolved DNA. Aquat. Microb. Ecol. 11:263-270. [Google Scholar]
- 24.Karl, D. M., and M. D. Bailiff. 1989. The measurement and distribution of dissolved nucleic acids in aquatic environments. Limnol. Oceanogr. 34:543-558. [Google Scholar]
- 25.Karl, D. M., and K. Yanagi. 1997. Partial characterization of the dissolved organic phosphorus pool in the oligotrophic North Pacific Ocean. Limnol. Oceanogr. 42:1398-1405. [Google Scholar]
- 26.Kieft, T. L., J. K. Fredrickson, T. C. Onstott, Y. A. Gorby, H. M. Kostandarithes, T. J. Bailey, D. W. Kennedy, S. W. Li, A. E. Plymale, C. M. Spadoni, and M. S. Gray. 1999. Dissimilatory reduction of Fe(III) and other electron acceptors by a Thermus isolate. Appl. Environ. Microbiol. 65:1214-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kierek, K., and P. I. Watnick. 2003. The Vibrio cholerae O139 O-antigen polysaccharide is essential for Ca2+-dependent biofilm development in sea water. Proc. Natl. Acad. Sci. USA 100:14357-14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klammer, A. A., and M. J. MacCoss. 2006. Effects of modified digestion schemes on the identification of proteins from complex mixtures. J. Proteome Res. 5:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lennon, J. T. 2007. Diversity and metabolism of marine bacteria cultivated on dissolved DNA. Appl. Environ. Microbiol. 73:2799-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lochte, K., A. Boetius, and C. Petry. 2000. Microbial food webs under severe nutrient limitations: life in the deep sea, p. 95-102. In C. R. Bell, M. Brylinsky, and P. Johnson-Green (ed.), Microbial biosystems: new frontiers. Proceedings of the 8th International Symposium on Microbial Ecology. Atlantic Canada Society for Microbial Ecology, Halifax, Canada.
- 31.Maeda, M., and N. Taga. 1976. Extracellular nuclease produced by a marine bacterium. II. Purification and properties of extracellular nuclease from a marine Vibrio sp. Can. J. Microbiol. 22:1443-1452. [DOI] [PubMed] [Google Scholar]
- 32.Martin, R. R., R. S. C. Smart, and K. Tazaki. 1988. Direct observation of phosphate precipitation in the goethite phosphate system. Soil Sci. Soc. Am. J. 52:1492-1500. [Google Scholar]
- 33.Muro-Pastor, A. M., E. Flores, A. Herrero, and C. P. Wolk. 1992. Identification, genetic analysis and characterization of a sugar-non-specific nuclease from the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 6:3021-3030. [DOI] [PubMed] [Google Scholar]
- 34.Noble, R. T., and J. A. Fuhrman. 1999. Breakdown and microbial uptake of marine viruses and other lysis products. Aquat. Microb. Ecol. 20:1-11. [Google Scholar]
- 35.Parsons, T. R., Y. Maita, and C. M. Lalli. 1984. A manual of chemical and biological methods for seawater analysis. Pergamon Press, Oxford, United Kingdom.
- 36.Paul, J. H., S. C. Jiang, and J. B. Rose. 1991. Concentration of viruses and dissolved DNA from aquatic environments by vortex flow filtration. Appl. Environ. Microbiol. 57:2197-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petersen, F. C., L. Tao, and A. A. Scheie. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J. Bacteriol. 187:4392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rangarajan, S., and V. Shankar. 1999. Extracellular nuclease from Rhizopus stolonifer: purification and characteristics of - single strand preferential - deoxyribonuclease activity. Biochim. Biophys. Acta 1473:293-304. [DOI] [PubMed] [Google Scholar]
- 39.Rittmann, D., U. Sorger-Herrmann, and V. F. Wendisch. 2005. Phosphate starvation-inducible gene ushA encodes a 5′ nucleotidase required for growth of Corynebacterium glutamicum on media with nucleotides as the phosphorus source. Appl. Environ. Microbiol. 71:4339-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sala, M. M., F. Peters, J. M. Gasol, C. Pedros-Alio, C. Marrase, and D. Vaque. 2002. Seasonal and spatial variations in the nutrient limitation of bacterioplankton growth in the northwestern Mediterranean. Aquat. Microb. Ecol. 27:47-56. [Google Scholar]
- 41.Sharpley, A. N. 1995. Soil phosphorus dynamics: agronomic and environmental impacts. Ecol. Eng. 5:261-279. [Google Scholar]
- 42.Siuda, W., and R. J. Chrost. 2001. Utilization of selected dissolved organic phosphorus compounds by bacteria in lake water under non-limiting orthophosphate conditions. Pol. J. Environ. Stud. 10:475-483. [Google Scholar]
- 43.Siuda, W., and H. Gude. 1996. Determination of dissolved deoxyribonucleic acid concentration in lake water. Aquat. Microb. Ecol. 11:193-202. [Google Scholar]
- 44.Smith, E. M., and Y. T. Prairie. 2004. Bacterial metabolism and growth efficiency in lakes: the importance of phosphorus availability. Limnol. Oceanogr. 49:137-147. [Google Scholar]
- 45.Stapleton, R. D., Jr., Z. L. Sabree, A. V. Palumbo, C. L. Moyer, A. H. Devol, Y. Roh, and J. Zhou. 2005. Metal reduction at cold temperatures by Shewanella isolates from various marine environments. Aquat. Microb. Ecol. 38:81-91. [Google Scholar]
- 46.Steinberger, R. E., and P. A. Holden. 2005. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 71:5404-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stookey, L. L. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal. Chem. 42:779-781. [Google Scholar]
- 48.Stopar, D., A. Cerne, M. Zigman, M. Poljsak-Prijatelj, and V. Turk. 2004. Viral abundance and a high proportion of lysogens suggest that viruses are important members of the microbial community in the Gulf of Trieste. Microb. Ecol. 47:1-8. [DOI] [PubMed] [Google Scholar]
- 49.Stumm, W., and J. J. Morgan. 1996. Aquatic chemistry, 3rd ed. Wiley Interscience, New York, NY.
- 50.Suzuki, S., A. Ferjani, I. Suzuki, and N. Murata. 2004. The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in Synechocystis. J. Biol. Chem. 279:13234-13240. [DOI] [PubMed] [Google Scholar]
- 51.Thingstad, T. F., U. L. Zweifel, and F. Rassoulzadegan. 1998. P limitation of heterotrophic bacteria and phytoplankton in the northwest Mediterranean. Limnol. Oceanogr. 43:88-94. [Google Scholar]
- 52.Turk, V., A. Rehnstam, E. Lundberg, and A. Hagstrom. 1992. Release of bacterial DNA by marine nanoflagellates, an intermediate step in phosphorus regeneration. Appl. Environ. Microbiol. 58:3744-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vadstein, O., L. M. Olsen, A. Busch, T. Andersen, and H. R. Reinertsen. 2003. Is phosphorus limitation of planktonic heterotrophic bacteria and accumulation of degradable DOC a normal phenomenon in phosphorus-limited systems? A microcosm study. FEMS Microbiol. Ecol. 46:307-316. [DOI] [PubMed] [Google Scholar]
- 54.Venkateswaran, K., D. Moser, M. Dollhopf, D. Lies, D. Saffarini, B. MacGregor, D. Ringelberg, D. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed] [Google Scholar]
- 54a.Wan, X.-F., N. C. VanBerkmoes, L. A. McCue, D. Stanek, H. Connelly, L. J. Hauser, L. Wu, X. Liu, T. Yan, A. Leaphart, R. L. Hettich, J. Zhou, and D. K. Thompson. 2004. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J. Bacteriol. 186:8385-8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinbauer, M. G., D. Fuks, and P. Peduzzi. 1993. Distribution of viruses and dissolved DNA along a coastal trophic gradient in the Northern Adriatic sea. Appl. Environ. Microbiol. 59:4074-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 57.Wu, J.-R., J.-H. Shien, H. K. Shieh, C.-C. Hu, S.-R. Gong, L.-Y. Chen, and P.-C. Chang. 2007. Cloning of the gene and characterization of the enzymatic properties of the monomeric alkaline phosphatase (PhoX) from Pasteurella multocida strain X-73. FEMS Microbiol. Lett. 267:113-120. [DOI] [PubMed] [Google Scholar]
- 58.Yang, L., K. B. Barken, M. E. Skindersoe, A. B. Christensen, M. Givskov, and T. Tolker-Nielsen. 2007. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153:1318-1328. [DOI] [PubMed] [Google Scholar]
- 59.Zachara, J. M., J. K. Fredrickson, S.-M. Li, D. W. Kennedy, S. C. Smith, and P. L. Gassman. 1998. Bacterial reduction of crystalline Fe3+ oxides in single phase suspensions and subsurface materials. Am. Mineral. 83:1426-1443. [Google Scholar]