Abstract
Bioinformatics analysis and transcriptional response information for Pyrococcus furiosus grown on α-glucans led to the identification of a novel isomaltase (PF0132) representing a new glycoside hydrolase (GH) family, a novel GH57 β-amylase (PF0870), and an extracellular starch-binding protein (1,141 amino acids; PF1109-PF1110), in addition to several other putative α-glucan-processing enzymes.
Pyrococcus furiosus is a heterotrophic, hyperthermophilic archaeon that uses a range of glucans as carbon/energy sources (5). α-Glucan utilization in P. furiosus has been examined using transcriptional response analysis, providing the basis for proposed mechanisms of starch utilization (8). Bioinformatic analysis based on domain homology (9) and domain families (12, 14) in the P. furiosus genome identified additional open reading frames (ORFs) that had putative functions related to α-glucan utilization. P. furiosus was grown anaerobically at 80°C in a sea salt-based medium supplemented with 3.3 g/liter of glycogen, maltose, pullulan, or starch (13). RNA was harvested and subjected to cDNA microarray analysis, as described previously (8, 16). Based on the leads identified from these complementary approaches, genes encoding candidate enzymes/proteins were cloned and expressed in Escherichia coli (see Table S1 in the supplemental material). While no activity or function could be assigned to several of the ORFs identified, biochemical analysis of PF0132, PF0870, and PF1109-PF1110 provided insights into the roles that these proteins play in α-glucan processing (Table 1).
TABLE 1.
ORFs encoding putative proteins associated with α-glucan processing
| ORF | Molecular mass (kDa) | Operona | Microarray fold changeb
|
Annotation | Biochemical activities testedc | ||
|---|---|---|---|---|---|---|---|
| Gly-Mal | Pul-Mal | Sta-Mal | |||||
| PF0132 | 55.0 | PF0132-PF0133 | NC | 3.2 | NC | Hypothetical protein | Pan, IM, IP, Mal3, T, Mal, pαGlc, pαGal |
| PF0133 | 21.7 | PF0132-PF0133 | NC | 4.3 | NC | Hypothetical protein | Sta, Gly, Pul, Gly |
| PF0870 | 69.6 | PF0870-PF0872 | NCe | −3.3e | NCe | GHF57 | pMalt5, pαGlc, pMalt, pMan, pβGlc, pXyl, pGal, Mal, Sta, Gly, Pul, Mal3, Mal4, Mal5, Mal6 |
| PF1108 | 46.9 | None | NC | NC | NC | Esterase/hydrolase | pMalt5, pαGlc, pMalt, pMan, pβGlc, pXyl, pGal, Stad, Puld, Amyd, Glyd, CD, pA, pPrp, pC, pPal, pB |
| PF1109 | 106.5 | None | NC | NC | NC | Cell adhesion domain | Sta, Cel |
| PF1110 | 19.3 | None | 2.0 | 2.5 | 2.0 | Hypothetical protein | Sta, Cel |
| PF1393 | 74.5 | None | NC | NC | NC | GHF57, α-amylase | pMalt5, pαGlc, pMalt, pMan, pβGlc, pXyl, pGal, Stad, Puld, Amyd, Glyd |
| PF1746 | 72.7 | Mal-I ABC transporter (PF1739-PF1747) | −10.0 | −2.0 | −5.0 | Glycogen debranching | pMalt5, pαGlc, pMalt, pMan, pβGlc, pXyl, pGal, Stad, Puld, Amyd, Glyd |
| PF1747 | 38.1 | Mal-I ABC transporter (PF1739-PF1747) | −10.0 | NC | −5.0 | β-Fructosidase | pMalt5, pαGlc, pMalt, pMan, pβGlc, pXyl, pGal, Stad, Puld, Amyd, Glyd, Suc |
Operons determined by the technique of Ermolaeva et al. (4).
NC, no change.
Amy, amylose; Cel, cellulose; CD, β-cyclodextrin; Gly, glycogen; IM, isomaltose; IP, isopanose; Mal, maltose; Mal3, maltotriose; Mal4, maltotetraose; Mal5, maltopentaose; Mal6, maltohexaose; pA, pNP-acetate; pGal, pNP-α-galactopyranoside; pαGlc, pNP-α-glucopyranoside; pB, pNP-butyrate; pβGlc, pNP-β-glucopyranoside; pC, pNP-caprate; pMalt, pNP-α-maltopyranoside; pMalt5, pNP-α-maltopentaoside; pMan, pNP-α-mannopyranoside; pPal, pNP-palmitate; pPrp, pNP-propionate; pXyl, pNP-β-xylopyranoside; Pan, panose; Pul, pullulan; Sta, starch; Suc, sucrose; T, turanose. Assays were performed at 90°C in sodium phosphate buffer, pH 5.6 and 7.2. Bold indicates a substrate hydrolyzed by the gene product.
Tested with and without 1-kDa filtered cellular extract.
Determined by quantitative PCR.
Recombinant PF0870 was specific for short α-glucans (Table 1). Indeed, bioinformatic analysis identified a domain in PF0870 related to glycoside hydrolase family 57 (17). Transcriptional analysis of PF0870 by quantitative PCR (10) showed similar responses to maltose, glycogen, and starch but not to pullulan. PF0870 was most active on pNP-α-maltopyranoside (kcat and Km were 962 s−1 and 82.7 μM, respectively, at 90°C, pH 7.0), less active on para-nitrophenyl (pNP)-α-galactoside, and not active on other substrates. The presence or absence of various divalent cations or EDTA had a minimal effect on enzyme activity. No transglycosylation activity was noted, and PF0870 did not hydrolyze starch, glycogen, pullulan, or large maltooligosaccharides. The biochemical properties of PF0870 most closely resemble those of β-amylases found in plants, fungi, and some bacteria; these progressively cleave maltose from the nonreducing ends of starch, amylase, and maltodextrins. PF0870 is the most thermostable β-amylase known (optimum temperature, 110°C). PF0870 primarily hydrolyzes maltotriose into glucose and maltose (data not shown), likely functioning in vivo as the final processing step prior to intracellular maltodextrin hydrolysis before the transfer of a glucose unit from maltose to maltodextrin by PF0272 (8).
The transcriptional response of PF0132 indicated that it is likely involved in processing α-1,6-glucans. Since a recombinant version could not be produced, the native form of PF0132 was purified 92-fold, essentially as described previously (1), from P. furiosus cells grown on pullulan; its identity was confirmed by mass spectroscopy (40.2% coverage). PF0132, identical to an α-glucosidase previously studied in our laboratory (1), has broad substrate specificity for α-glucan carbohydrates (Table 1). The native form of PF0132 was determined to have comparable catalytic efficiencies for panose and maltose (Table 2). PF0132 and its homologs in other hyperthermophiles (e.g., Pyrococcus horikoshii, Pyrococcus abyssi, Thermococcus kodakarensis, and Aeropyrum pernix) represent a new glycosyl hydrolase family (7). PF0132, functioning as an isomaltase with broad substrate specificity, is ideal for the final processing step in generating glucose from α-glucans for high-fructose corn syrup production and ethanol fermentation processes (2). Previously, Lee et al. (8) proposed a starch utilization pathway for P. furiosus. An issue not considered in that earlier analysis was how pullulan (an α-1,6-linked glucan) could be used by P. furiosus. Based on the new information here, pullulan is likely degraded extracellularly by an amylopullulanase (PF1934-PF1935) (3, 8) and further hydrolyzed by PF0132 after intracellular transport; indeed, the number of transcripts for PF0132 was 3.2-fold higher on pullulan than on maltose or starch.
TABLE 2.
Kinetic parameters of PF0132
| Substrate | kcat (s−1) | Km (mM) | kcat/Km (s−1·mM−1) |
|---|---|---|---|
| pNP-α-glucopyranoside | 21.9 | 0.051 | 429 |
| Maltose | 1,600 | 7.1 | 225 |
| Panose | 278 | 3.2 | 85.6 |
| Isomaltose | 58.8 | 5.0 | 11.8 |
PF1109 and PF1110 were annotated as separate hypothetical proteins in the P. furiosus genome. However, the PF1109-PF1110 locus is very similar to that for a single hypothetical protein (PAB1790) encoded in the P. abyssi genome. In fact, the PF1109-PF1110 locus was found to PCR amplify as a single ORF corresponding to 1,141 amino acids, including a signal peptide. This gene, when cloned and expressed in E. coli (without the signal peptide), resulted in a single protein with a molecular mass of approximately 120 kDa. No enzymatic activity could be detected for PF1109-PF1110. A glucan binding assay (11), however, showed that the protein binds starch (2% raw corn starch) but not cellulose (Avicel) (data not shown). The role of PF1109-PF1110 in extracellular α-glycoside utilization is not clear, but it likely plays a role in sequestering extracellular α-glucans for subsequent hydrolysis by amylolytic enzymes.
Several genes identified by bioinformatics analysis as “α-glucan-related” (the PF1108, PF1393, PF1460, and PF1746-PF1747 genes) were not responsive to the conditions tested. These genes are either constitutively transcribed or stimulated under growth conditions not considered here. Note that a mixture of PF1108, PF1393, PF1746, and PF1747 hydrolyzed starch and glycogen, although none of these proteins individually were active against these substrates (D. A. Comfort and R. M. Kelly, unpublished data).
Starch, glycogen, and pullulan induced a transcriptional “polysaccharide” effect: 118 ORFs were upregulated twofold or more, relative to growth on maltose (Fig. 1; see Table S2 in the supplemental material). Of these, 49 were induced on all three α-glucan polysaccharides; in particular, 38 of the 49 ORFs were within the PF0682-PF0783 locus, which encodes a number of hypothetical proteins as well as dehydrogenases and reductases. Many putative proteins encoded by the PF0682 to PF0736 loci are unique to P. furiosus and are not found in other Thermococcales, such as P. horikoshii, P. abyssi, or T. kodakarensis (see Table S2 in the supplemental material). Of the 49 α-glucan polysaccharide-induced genes, T. kodakarensis contained the most homologs, with 32. The PF0776 gene (upregulated ca. fourfold on all polysaccharides) encodes a VapC toxin (and putative RNase) and is paired in an operon with the PF0775 gene, encoding its cognate VapB antitoxin (PF0775); toxin-antitoxin loci are thought to be involved in RNA management functions during stress (6, 15). The fact that the PF0776 locus was differentially transcribed on α-glucan polysaccharides implies that toxins-antitoxins play a role in RNA management during nonstress conditions. The PF0777 and PF0782-PF0783 genes (upregulated two- to fivefold on all polysaccharides) encode putative heteropolysaccharide biosynthesis-related proteins, possibly involved in the production of capsular polysaccharide or exopolysaccharide.
FIG. 1.
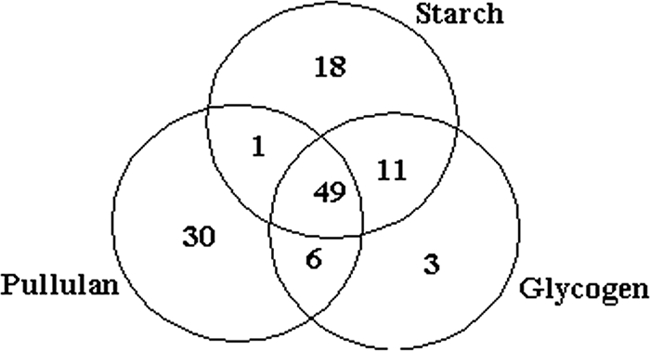
Venn diagram of P. furiosus transcriptional response during growth on polysaccharides relative to maltose. Those genes upregulated twofold or more relative to maltose are represented.
The results reported here show that P. furiosus (and perhaps other fermentative anaerobes) use an array of novel enzymes and proteins in α-glucoside utilization, a potentially important consideration in efforts related to biofuel production.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the DOE Energy Biosciences Program (DE-FG02-96ER20219) and the NSF Biotechnology Program (CBET-0317886 and CBET-0617272). D.A.C. and A.L.V. acknowledge support from a Department of Education GAANN Fellowship.
Clones for several enzymes were generously provided by Michael Adams, University of Georgia.
Footnotes
Published ahead of print on 21 December 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Costantino, H. R., S. H. Brown, and R. M. Kelly. 1990. Purification and characterization of an α-glucosidase from a hyperthermophilic archaebacterium, Pyrococcus furiosus, exhibiting a temperature optimum of 105 to 115°C. J. Bacteriol. 172:3654-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crabb, W. D., and J. K. Shetty. 1999. Commodity scale production of sugars from starches. Curr. Opin. Microbiol. 2:252-256. [DOI] [PubMed] [Google Scholar]
- 3.Dong, G., C. Vieille, and J. G. Zeikus. 1997. Cloning, sequencing, and expression of the gene encoding amylopullulanase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl. Environ. Microbiol. 63:3577-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ermolaeva, M. D., O. White, and S. L. Salzberg. 2001. Prediction of operons in microbial genomes. Nucleic Acids Res. 29:1216-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiala, G., and K. O. Stetter. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100 degrees C. Arch. Microbiol. 145:56-61. [Google Scholar]
- 6.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 7.Henrissat, B., and G. Davies. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7:637-644. [DOI] [PubMed] [Google Scholar]
- 8.Lee, H.-S., K. R. Shockley, G. J. Schut, S. B. Conners, C. I. Montero, M. R. Johnson, C.-J. Chou, S. L. Bridger, N. Wigner, S. D. Brehm, F. E. Jenney, Jr., D. A. Comfort, R. M. Kelly, and M. W. W. Adams. 2006. Transcriptional and biochemical analysis of starch metabolism in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 188:2115-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. Q. He, D. I. Hurwitz, J. D. Jackson, Z. X. Ke, C. J. Lanczycki, C. A. Liebert, C. L. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. C. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rashid, N., J. Cornista, S. Ezaki, T. Fukui, H. Atomi, and T. Imanaka. 2002. Characterization of an archaeal cyclodextrin glucanotransferase with a novel C-terminal domain. J. Bacteriol. 184:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shockley, K. R., D. E. Ward, S. R. Chhabra, S. B. Conners, C. I. Montero, and R. M. Kelly. 2003. Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 69:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonnhammer, E. L. L., S. R. Eddy, and R. Durbin. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405-420. [DOI] [PubMed] [Google Scholar]
- 15.Tachdjian, S., and R. M. Kelly. 2006. Dynamic metabolic adjustments and genome plasticity are implicated in the heat shock response of the extremely thermoacidophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 188:4553-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh, P. Bushel, C. Afshari, and R. S. Paules. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8:625-637. [DOI] [PubMed] [Google Scholar]
- 17.Zona, R., F. Chang-Pi-Hin, M. J. O'Donohue, and S. Janecek. 2004. Bioinformatics of the glycoside hydrolase family 57 and identification of catalytic residues in amylopullulanase from Thermococcus hydrothermalis. Eur. J. Biochem. 271:2863-2872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


