Abstract
In the vast number of random mutagenesis experiments that have targeted protein thermostability, single amino acid substitutions that increase the apparent melting temperature (Tm) of the enzyme more than 1 to 2°C are rare and often require the creation of a large library of mutated genes. Here we present a case where a single beneficial mutation (R236F) of a hemp fiber-processing pectate lyase of Xanthomonas campestris origin (PLXc) produced a 6°C increase in Tm and a 23-fold increase in the half-life at 45°C without compromising the enzyme's catalytic efficiency. This success was based on a variation of sequence alignment strategy where a mesophilic amino acid sequence is matched with the sequences of its thermophilic counterparts that have established Tm values. Altogether, two-thirds of the nine targeted single amino acid substitutions were found to have effects either on the thermostability or on the catalytic activity of the enzyme, evidence of a high success rate of mutation without the creation of a large gene library and subsequent screening of clones. Combination of R236F with another beneficial mutation (A31G) resulted in at least a twofold increase in specific activity while preserving the improved Tm value. To understand the structural basis for the increased thermal stability or activity, the variant R236F and A31G R236F proteins and wild-type PLXc were purified and crystallized. By structure analysis and computational methods, hydrophobic desolvation was found to be the driving force for the increased stability with R236F.
Improving the thermostability of a protein or enzyme is desirable for commercially viable bioprocesses that prolong product shelf life, increase energy efficiency, and save costs (30). Unfortunately, there are no simple rules for improving thermostability, the mechanisms of which include the formation of disulfide bridges, hydrophobic or aromatic interactions, contact order, hydrogen bonding, ion pairing, dimer-dimer interaction, and a preponderance of glutamine usage (32, 36, 41). A critical and challenging task in thermostability engineering is, therefore, choosing a method whereby promising results can be obtained quickly and efficiently. Both rational (structure-based, including molecular modeling) and randomized or irrational (directed evolution or random and combinatorial engineering) approaches to protein thermostability improvement have been applied with varying degrees of success (11, 19, 30). Notable single amino acid substitutions that have led to dramatic increases in melting temperatures (Tm; the temperature at which 50% of the protein is unfolded), by 20°C and 24°C, respectively, were identified in the Drosophila protein drk SH3 domain (24), and in a bacterial malate dehydrogenase (4). However, these successes were made possible only through a good understanding of the thermodynamics and kinetics of folding, or through knowledge of the electrostatic interactions in the dimer-dimer interface of the tetrameric protein. Otherwise, single amino acid substitutions that increase the apparent Tm of a given enzyme more than 1 to 2°C are rare (19).
In this study, we selected a bacterial-genome-mined pectate lyase sequence as a template in order to evolve added thermostability and/or catalytic activity using a newly improvised strategy that does not require the construction of a large library of clones, as in directed evolution experiments, or specialized computational skills, as in rational protein engineering approaches. We further characterized the beneficial variants at the biochemical and structural levels. Pectate lyases (EC 4.2.2.2) are secreted enzymes produced by a variety of plant-pathogenic bacteria and therefore are best studied as virulence factors (for a review, see reference 15). Biotechnologically, these pectinolytic enzymes, as well as various endopolygalacturonases, are useful retting or bioscouring reagents for the processing of natural bast fibers (e.g., hemp and flax), and cotton fabric (1, 2, 28, 35).
MATERIALS AND METHODS
Molecular cloning and site-directed mutagenesis.
Standard methods were used for isolation of plasmid DNA, cloning, and transformation (33). The Xanthomonas campestris pectate lyase II (PLXc) gene (NP_638163) was amplified from its genomic DNA (ATCC 33913) and cloned into pSD80, an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression vector, at the EcoRI and HindIII restriction sites (34). This cloning is part of a larger genome-mining effort for pectinase-encoding sequences (Z. Xiao, J. Boyd, S. Grosse, E. Coupe, and P. C. K. Lau, unpublished data). The recombinant plasmid (pSD80PLII) and resultant derivatives were transformed into Escherichia coli strain Rosetta 2 (Novagen). Site-directed mutagenesis was performed using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA) according to its instruction manual. The mutagenic oligonucleotide primers are listed in Table S1 in the supplemental material. The expected mutations were confirmed by gene sequencing using a BigDye DNA sequencing kit (Applied Biosystems) with an automated DNA sequencer (model 377; ABI Prism).
Multiple sequence alignment.
The ClustalW program (http://www.ebi.ac.uk/clustalw/) was used to align the X. campestris pectate lyase II (NP_638163) sequence (9) with the four available thermostable pectate lyase sequences, selected by screening of more than a thousand candidates in the NCBI database for their established thermostability in reported literature. The thermostable candidate sequences are accession numbers CAD56882 from Bacillus licheniformis (3), BAA96478 from Bacillus sp. strain P-4-N (14), BAB40336 from Bacillus sp. strain TS-47 (37), and AAD35518 from Thermotoga maritima MSB8 (18), with optimal temperatures of 65 to 69, 70, 70, and 90°C, respectively.
Calculation of the success rate of the mutation.
The success rate of the mutation, expressed as a percentage, is calculated as (number of improved variants × 100)/(total number of screened clones) (see Table S2 in the supplemental material).
Protein expression and fermentation.
Single colonies of E. coli Rosetta 2 harboring the X. campestris pectate lyase II gene and the desired mutations (V26A, A31G, L64I, Y66V, K69T, F70I, Q123R, V187I, R236F, and the A31G R236F double mutation) were taken from plates (LB medium containing 50 μg/ml ampicillin and 30 μg/ml chloramphenicol) and grown overnight at 30°C in the same medium (starting with 5 ml; afterwards the colonies were transferred to 500 ml of medium). Fermentations to produce appropriate amounts of pectate lyase (up to 1 g of protein for each variant) were carried out in a New Brunswick Scientific BioFlo 110 Benchtop fermentor system using defined medium (31). Fermentors with a working volume of 6 liters were inoculated with 300 ml of each preculture (giving a starting optical density at 600 nm of about 0.3), which had been grown overnight at 30°C on LB medium. Glucose (0.7%) was used as the sole carbon source. The pH was maintained automatically using NH4OH (29%) and H3PO4 (5 M). One milliliter of antifoam was added after sterilization. After a batch phase of about 9 h, a feed with 0.22 ml/min of glucose (50% solution) and MgSO4 (20 g/liter) was started. Culture was induced a few minutes later by addition of 1 mM IPTG, and the temperature was reduced to 25°C. After 16 h (optical density at 600 nm, about 25), the fermentors were brought to 10°C and cells were harvested subsequently. Cells were centrifuged (at 6,000 × g for 20 min at 4°C), resuspended in phosphate buffer (20 mM; pH 7.0), and broken via two passages in a microfluidizer operating at 12,000 lb/in2. Unbroken cells and debris were removed by filtration using a hollow fiber membrane (pore size, 0.22 μm; on an Amicon DC 10 ultrafiltration system), and the filtrate was kept at 4°C for further processing.
Protein purification.
The cleared cell extract (about 1 liter) was loaded onto an SP-Sepharose FF column (XK50/20) (GE Healthcare) previously equilibrated with 20 mM sodium phosphate buffer (pH 7.0). The column was washed with the same buffer, and bound protein was eluted using a linear NaCl gradient from 0 to 200 mM. Active fractions were pooled and concentrated using a YM 3 membrane in an Amicon stirring cell. Protein levels during chromatography were monitored at 280 nm, and concentrations were determined using the Bradford assay (5).
Pectate lyase assay and kinetics.
The pectate lyase activity was determined by measuring the increase in the absorbance at 232 nm of polygalacturonic acid (PGA) (8). Fifty microliters of enzyme and 50 μl of 0.5% PGA in PL buffer (50 mM Tris-HCl buffer [pH 8.5] containing 0.5 mM CaCl2) were incubated at 50°C for 30 min. The reaction was stopped by addition of 20 μl of 0.35 M HCl. One hundred microliters of reaction mixture was used to determine the absorbance at 232 nm. One unit of pectate lyase was defined as the amount of enzyme producing 1 μmol of unsaturated product in 1 min under the assay conditions. The amount of degraded products of pectin was determined by the increase in absorbance at 232 nm (8). The molar extinction coefficient for the unsaturated product at 232 nm is 4,600 M−1 cm−1. All data were averages of triplicate measurements. The kinetic parameters Km, Vmax, and kcat were determined at the optimal temperatures of the purified enzymes.
Gel electrophoresis and zymogram staining.
Two hundred nanograms of protein samples was heated for 10 min at 45°C in sample loading buffer before being applied to a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel containing 0.1% PGA. After electrophoresis, protein bands were stained with Coomassie blue. For zymogram staining, the gel was soaked in 2.5% Triton X-100 for 30 min and washed with PL buffer for 30 min. After 20 h of incubation at 40°C in PL buffer containing 0.1% PGA, the gel was stained with 0.05% (wt/vol) ruthenium red for 20 min and washed with water for 60 min.
Stability measurement.
To determine the half-life of inactivation, the pectate lyases were incubated in PL buffer at 45°C. Samples were taken at various time points, followed by the pectate lyase assay. Purified proteins were adjusted to about 0.5 mg/ml in 10 mM phosphate buffer, pH 7.0, for determining the circular dichroism (CD) spectrum and Tm. Thermal denaturation experiments were performed at a temperature increase of 40°C per h. The data were collected with a JASCO J-710 CD spectrometer at a wavelength of 222 nm, where maximal signal difference was observed.
Protein crystallization and structure determination.
Initial crystallization conditions were identified using the PEGs (Nextal, Montreal, Canada), Classic Suite I and II (Qiagen), screens, with the sitting drop vapor diffusion set up at 20°C. Numerous chemical conditions yielded triclinic crystals overnight. The conditions were reproduced and optimized using the hanging drop vapor diffusion method in 24-well Linbro plates (Hampton Research). The best conditions found were 20 to 30% (wt/vol) polyethylene glycol 3350 and various buffers with pHs ranging from 5.5 to 8.5. The crystals belong to the primitive triclinic space group P1, with unit cell dimensions of 47.2 Å for a, 53.2 Å for b, 73.0 Å for c, 71.7° for α, 80.0° for β, 69.0° for γ, and two protein molecules in the asymmetric unit. The Matthews coefficient (23) Vm is 2.20 Å3 Da−1, corresponding to a solvent content of 44%. The cryoprotectant solution used consisted of mother liquor supplemented with 10 to 12% (wt/vol) glycerol, and data were collected at 100 K. Diffraction data extended to 2.0, 2.1, and 1.9 Å resolution for wild-type, R236F, and A31G R236G proteins, respectively. The images were processed with the HKL2000 program package (27). The structure of wild-type PLXc was solved by molecular replacement using the MolRep program (38) and the structure of Erwinia chrysanthemi pectate lyase (22) (PDB entry 1AIR) as a search model. Free atom density modification was implemented using ARP/wARP (29) to improve the electron density maps, and the protein model was built manually using COOT (12). The model was refined using REFMAC5 (26) with the TLS option, with each of the monomers treated separately. The structures of the R236F and A31G R236F variants were solved and refined in the same manner as for wild-type PLXc, whose structure was used as a search model for molecular replacement. Refinement statistics are summarized in Table S4 in the supplemental material for the three structures solved. The models were validated with PROCHECK (20).
Computational method.
The stabilities (i.e., the free energies of folding) of pectate lyase variants relative to that of the wild-type protein were calculated with the FOLD-X program (13), using the crystal structure of the wild-type protein determined in this study (chain A). The FOLD-X program employs a first-principle-based energy function that was shown to be able to predict folding free energies with a squared-correlation coefficient of 0.69 and a standard deviation of 0.81 kcal/mol, within a data set of more than 1,000 mutations from various proteins. The effect of the R236F mutation on stability was further verified by performing a FOLD-X calculation using the crystal structure of the R236F variant (chain A) and simulating its mutation to the wild-type protein. All crystallographic water molecules and observed phosphate ions were removed. Possible water bridges between protein atoms were implicitly taken into account during FOLD-X calculations. Point mutations were introduced by the FOLD-X program with the mutate function of the WHAT IF program (42), which optimizes the mutated side chains with respect to rotamer distributions (7) and hydrogen bond networking (16). The analyses of relative stabilities in terms of contributions from van der Waals, hydrogen bonding, electrostatic, and water bridge interaction energies, hydrophobic and polar desolvation, and main-chain and side-chain entropies, were based on the FOLD-X calculations. Clash energies were not included in the calculation of folding free energies.
Protein structure accession numbers.
The coordinates of the structures of wild-type, R236F, and A31G R236F PLXc have been deposited in the Protein Data Bank with the codes 2OX3, 2OXZ, and 2OY1, respectively.
RESULTS AND DISCUSSION
Sequence characteristics of a new pectate lyase in comparison to its thermophilic counterparts.
The predicted pectate lyase NP_638163 entry of Xanthomonas campestris pv. campestris ATCC 33913 in the CAZy database (PLXc) is a biochemically uncharacterized entity except for its annotated sequence classification as a member of the polysaccharide lyase family 1 of proteins (http://www.cazy.org/fam/PL1.html). We established the Tm of PLXc to be 48°C, certifying it as a mesophilic protein (see below). Figure 1 shows a multiple alignment of the predicted amino acid sequence of PLXc with four available sequences of thermostable PLs, whose optimal temperatures have been established as ranging from 65 to 90°C. Interestingly, only nine amino acid positions were invariant in all four thermostable PLs but not in PLXc. Pairwise comparison of the subject and database PLXc sequences ranging from 212 to 301 amino acids gave only 25 to 36% identity, indicating low sequence homology among this group of proteins.
FIG. 1.
Multiple alignment of protein sequences of thermostable pectate lyases and thermolabile PLXc in family PL1. The signal peptide sequences of the respective proteins are not shown. The numbering of each sequence starts from the initiation codon. Conserved catalytic sites (K199, R230, R235), conserved calcium binding sites (D137, D175, D179), the core structure of the parallel β-helix (vWiDH region), and sites conserved in all thermostable PL1 pectate lyases (CAD56882 from Bacillus licheniformis, BAA96478 from Bacillus sp. strain P-4-N, BAB40336 from Bacillus sp. strain TS-47, AAD35518 from Thermotoga maritima MSB8) but variant in PLXc (NP_638163) are shaded.
Characterization of PLXc and its variants.
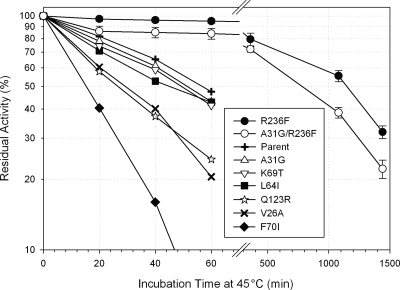
We set out to test whether any of the nine variable positions could contribute to added thermostability and/or catalytic activity in PLXc when they were replaced by the corresponding residue of a thermophilic counterpart (Table 1). A PGA assay was used to determine the activity and thermal inactivation of PLXc and its variants in crude cell extracts. One mutation, R236F, increased the Tm by 6°C and extended the half-life at 45°C 23-fold. Three other mutations, F70I, Q123R, and V187I, reduced thermostabilities by 18 to 53% (Fig. 2). Thus, four out of the nine positions appeared to be crucial for the thermostability of PLXc. Of the other mutations, the Y66V substitution resulted in a loss of activity, while the A31G variant was fivefold more active than its parent enzyme, although its thermostability was not substantially changed. Combining the best attributes of A31G and R236F in a double variant, the A31G R236F variant, resulted in a 5-fold improvement in activity and a 12-fold increase in thermostability over those of the wild-type enzyme (Table 1). All in all, two-thirds of the amino acid substitutions (A31G, Y66V, F70I, Q123R, V187I, and R236F) substantially affected either the thermostability or the catalytic activity of the PLXc enzyme, indicating a high success rate of identifying susceptible residues without having to generate a large library.
TABLE 1.
Thermostabilities and activities of PLXc and its variants
| Enzyme variant | DNA mutation | Half-life of inactivation at 45°C (min)a | Activity in crude cell extract (U/mg protein)a |
|---|---|---|---|
| Parent | 54.2 ± 5.9 | 0.64 ± 0.01 | |
| V26A | GTC→GCC | 38.8 ± 8.5 | 0.53 ± 0.04 |
| A31G | GCC→GGC | 51.9 ± 8.8 | 3.37 ± 0.09 (5×) |
| L64I | CTG→ATC | 44.6 ± 2.8 | 0.54 ± 0.01 |
| Y66V | TAC→GTC | NA | No activity |
| K69T | AAG→ACG | 48.2 ± 7.4 | 0.87 ± 0.03 |
| F70I | TTC→ATC | 14.0 ± 1.5 | 0.81 ± 0.03 |
| Q123R | CAG→CGG | 29.0 ± 1.3 | 0.62 ± 0.08 |
| V187I | GTC→ATC | <10 | 0.55 ± 0.08 |
| R236F | CGT→TTT | 1,292 ± 139 (23×) | 1.42 ± 0.17 (2×) |
| A31G R236F | GCC→GGC, CGT→TTT | 659 ± 41 (12×) | 3.48 ± 0.20 (5×) |
Fold improvement is given in parentheses. NA, not applicable.
FIG. 2.
Thermostability of PLXc and its variants.
Enzyme purification and kinetics.
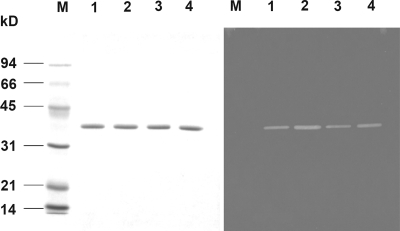
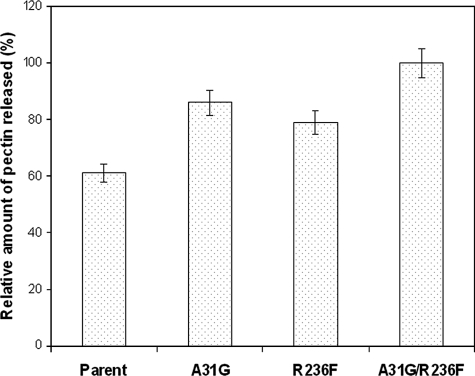
The wild-type enzyme and the A31G, R236F, and A31G R236F variants were characterized further by purification to apparent homogeneity with a single cation-exchange SP-Sepharose column yielding 40% recovery of the respective proteins. These proteins appeared as a single band on an SDS-PAGE gel with an apparent molecular mass of 35 kDa, in good agreement with the theoretical mass of 35,239 Da. Zymogram analyses of the purified enzymes indicated that they are active pectate lyases (Fig. 3). The intensities of the clear zones of the respective enzymes were consistent with the measured specific activities. The Km values of the purified A31G, R236F, and A31G R236F proteins with PGA as a substrate were determined and found not to differ substantially from each other and from that of the parent enzyme (Table 2). However, the catalytic efficiencies for both the A31G and A31G R236F variants were about twofold greater than that for the parent enzyme. The activity of the most thermostable variant, the R236F variant, was the same as that of the parent. Overall, the specific activities of the purified enzymes are consistent with their kcat/Km values. Similar results were observed when citrus pectin was used as a substrate (Table 3). Additionally, these variants were found to be 20 to 40% more effective than the parent enzyme in releasing pectin from natural hemp fiber (Fig. 4).
FIG. 3.
Purified X. campestris pectate lyase II and its variants on an SDS-PAGE gel (left) and a ruthenium red-stained zymogram gel containing 0.1% PGA (right). Lanes: M, molecular standard; 1, parent enzyme; 2, A31G variant; 3, R236F variant; 4, A31G R236F variant.
TABLE 2.
Kinetic parameters of the parent PLXc and variants with PGA as the substrate
| Enzyme | Km (g·liter−1) | kcat (s−1) | kcat/Km (liter·g−1·s−1) | Sp act (U/mg) |
|---|---|---|---|---|
| Parent | 0.98 ± 0.20 | 114 ± 11 | 116 ± 17 | 196 ± 20 |
| A31G | 0.73 ± 0.11 | 194 ± 29 | 266 ± 40 | 333 ± 53 |
| R236F | 0.83 ± 0.15 | 121 ± 7 | 146 ± 16 | 208 ± 27 |
| A31G R236F | 0.93 ± 0.19 | 216 ± 12 | 232 ± 19 | 370 ± 22 |
TABLE 3.
Kinetic parameters of the variant pectate lyases with citrus pectin as the substrate
| Enzyme | Km (g·liter−1) | kcat (s−1) | kcat/Km (liter·g−1·s−1) | Sp act (IU/mg) |
|---|---|---|---|---|
| Parent | 0.80 ± 0.12 | 61 ± 7 | 76 ± 9 | 105 ± 12 |
| A31G | 0.68 ± 0.10 | 117 ± 10 | 172 ± 21 | 200 ± 25 |
| R236F | 0.59 ± 0.08 | 67 ± 7 | 114 ± 12 | 115 ± 15 |
| A31G R236F | 0.55 ± 0.07 | 139 ± 11 | 253 ± 30 | 238 ± 18 |
FIG. 4.
Amounts of pectin released from natural hemp fiber by PLXc and its variants.
CD study.
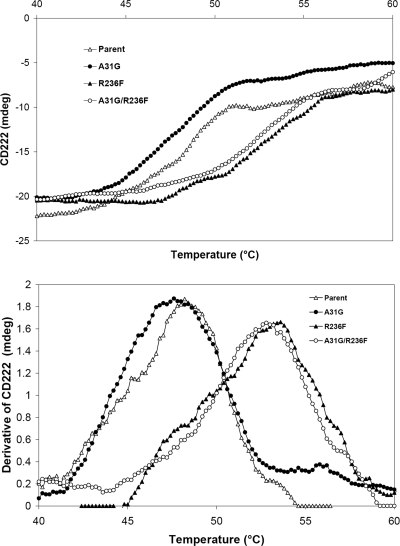
The structures of the wild-type enzyme and the A31G, R236F, and A31G R236F variant enzymes showed no discernible differences in their α-helical structure contents by CD spectroscopy (see Fig. S1 in the supplemental material). However, the Tms of the four proteins were established to be 48, 47.5, 54, and 53°C, respectively (Fig. 5). The 5 to 6°C increase in the Tms of the R236F variant and the double variant are consistent with the increases in the half-lives of the proteins.
FIG. 5.
Tms of the wild-type and variant enzymes determined by CD. (A) Original CD data; (B) derivative of CD signal.
Structures of wild-type PLXc and variant proteins.
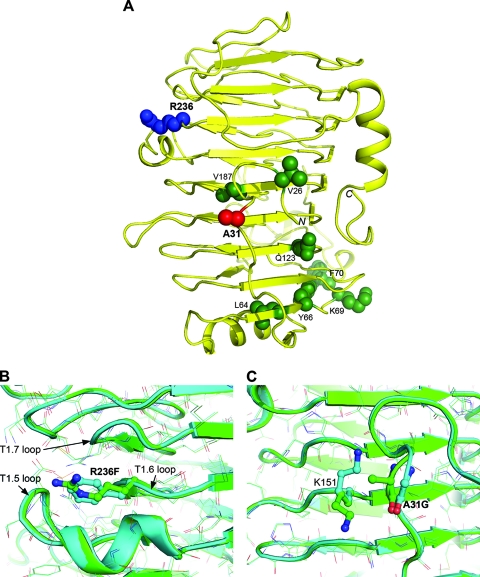
To understand the structural basis for the thermal stability of the variant proteins, R236F, A31G R236F, and wild-type PLXc were crystallized and their structures solved by molecular replacements. These structures have the same right-handed parallel β-helix architecture as that described for Erwinia chrysanthemi (presently classified as Dickeya dadantii) pectate lyase PelC (43) (Fig. 6A; see also Fig. S2, the supplementary note, and Table S3 in the supplemental material). Residue R236 is partially solvent exposed and is located in the T1.6 loop (the 6th loop of turn T1), which stacks against the T1.5 loop, containing a helical structure previously shown to enhance the catalytic activity of related pectate lyases (10). Its guanidinium group is 10 Å, 17 Å, and 22 Å away from the putative active sites R235, R230, and K199, respectively. This may explain the similar activities of the wild-type enzyme and the R236 variant. The R236F substitution caused no structural change in the enzyme. Although bulkier, the F236 side chain occupies the same region of space as the wild-type R236; no neighboring residues are affected and no major electrostatic interactions interrupted (Fig. 6B). However, the aromatic side chain in this position allows hydrophobic interactions with several adjacent side chains, particularly M258 from the T1.7 loop, the aliphatic portions of T210 and N212 from the T1.5 loop, and the neighboring R235. Hydrophobic stacking between the T1.5 and T1.7 loops appears to confer higher stability on the T1.6 loop. A structure-based first-principle energetic analysis (13) (see Fig. S3 and Table S4 in the supplemental material) predicts a favorable change in folding free energy of −1.7 kcal/mol for the stabilizing R236F mutation, underscoring the gain afforded by hydrophobic desolvation (−2.8 kcal/mol) as driving the increase in stability. This more than offsets the major destabilizing contribution, i.e., the loss of hydrogen bonding energy upon mutation (1.4 kcal/mol), between the carbonyl group of D207 and the guanidinium group of R236.
FIG. 6.
Crystal structures of wild-type PLXc and its A31G R236F variant. (A) Mapping of the nine mutation sites of wild-type PLXc on its crystal structure. The protein is rendered as a yellow ribbon. Side chains at mutation positions are shown as CPK (Corey Pauling Koltun) models and color coded by mutation outcomes: blue, stabilizing; red, activating; green, others. (B and C) Close-up views of structural changes introduced upon mutation at positions 236 and 31, respectively. Superimposed wild-type PLXc and its A31G R236F variant are colored green and cyan, respectively. Mutated residues and local structural changes are represented as sticks.
The double mutation A31G R236F also does not introduce major variations relative to PLXc wild-type and R236F variant structures (Fig. 6B). A noticeable change is a peptide flip at the A31G mutation site, bringing the backbone carbonyl of A30 closer to the side chain of K151, which undergoes a concerted conformational change (Fig. 6C). The A31G mutation resides in the N-terminal region of the enzyme on the opposite side of the β-helix relative to the catalytic site. Thus, the twofold increase in catalytic efficiency upon the A31G mutation cannot be accounted for by the present structural data. Mutations distant from catalytic sites are documented to be as effective as close mutations in improving enzymatic activity (25), but the underlying factors are often subtle and elude structural interpretations. Computational predictions yielded a marginal destabilizing effect (0.2 kcal/mol) for the A31G mutation, in agreement with the experimental data (Table 1; see also Fig. S3 in the supplemental material).
The remaining single point mutations had either marginal or destabilizing effects on thermal stability. A retrospective structural and computational analysis can rationalize why these single amino acid substitutions did not materialize into improved thermostability of the target mesophilic enzyme (see the supplementary note, Fig. S3, and Table S4 in the supplemental material).
Concluding remarks.
This study characterized a new bacterial pectate lyase that otherwise remains a hypothetical sequence in the vast microbial genome database. From this basic sequence, more thermostable and active variants were produced that have potential applications in the retting of natural fibers. A new approach to protein/enzyme thermostabilization was conceived that made selective use of those thermophilic sequences that have established Tm values to identify invariant residues (besides those of catalytic residues or binding sites) as possible sites for mutagenesis on a mesophilic sequence. Although based on sequence alignment, this approach differs greatly from the “consensus” approach for thermostability engineering of proteins whereby a synthetic gene is used to replace as many as 38 amino acids to arrive at improved thermostability (21). This “consensus” concept for thermostability engineering of proteins was also based on highly homologous sequences (>80% identity), as opposed to the relatively low degree of sequence identity (25% to 36.5%) among the candidate sequences used in this study. The present strategy also differs from the work of van den Burg et al. (39, 40), where the respective roles of three amino acid residues, as a result of sequence alignment of two highly homologous (99% identity) neutral protease sequences, were analyzed by site-directed mutagenesis for their possible contributions to thermostability. In the present study, a success rate of at least 11% for identifying “thermosusceptible” residues was evident. In some directed evolution experiments, for example, a success rate of 0.05 to 0.06% was observed (see Table S2 in the supplemental material). Our model study is encouraging enough to be applied to other protein families to provide the first possible best candidate for thermostabilization, which may then require the application of additional strategies, including saturation mutagenesis (6, 44). Finally, it is acknowledged that proteins, even among members of the same families, are individuals (17). Until universal rules governing protein thermostability emerge, proteins need to be studied individually, and new approaches or strategies such as that described in this study can help in gaining new understanding and facilitate potential industrial applications.
Supplementary Material
Acknowledgments
This research was largely supported by the Climate Change Technology and Innovation Biotechnology Program of the Canadian Biomass Innovation Network of Natural Resources Canada.
We thank A. Matte for critical reading of the manuscript.
This article is issued as NRCC publication 49095.
Footnotes
Published ahead of print on 21 December 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akin, D. E., G. Henriksson, J. D. Evans, A. P. S. Adamsen, J. A. Foulk, and R. B. Dodd. 2004. Progress in enzyme-retting of flax. J. Natural Fibers 1:21-47. [Google Scholar]
- 2.Antonov, V., J. Msarek, M. Bjelkova, P. Smirous, and H. Fischer. 2007. Easily available enzymes as natural retting agents. Biotechnol. J. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 3.Berensmeier, S., S. A. Singh, J. Meens, and K. Buchholz. 2004. Cloning of the pelA gene from Bacillus licheniformis 14A and biochemical characterization of recombinant, thermostable, high-alkaline pectate lyase. Appl. Microbiol. Biotechnol. 64:560-567. [DOI] [PubMed] [Google Scholar]
- 4.Bjørk, A., B. Dalhus, D. Mantzilas, R. Sirevag, and V. G. H. Eijsink. 2004. Large improvement in the thermal stability of a tetrameric malate dehydrogenase by single point mutations at the dimer-dimer interface. J. Mol. Biol. 341:1215-1226. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Chaparro-Riggers, J. F., K. M. Polizzi, and A. S. Bommarius. 2007. Better library design: data-driven protein engineering. Biotechnol. J. 2:180-191. [DOI] [PubMed] [Google Scholar]
- 7.Chinea, G., G. Padron, R. W. Hooft, C. Sander, and G. Vriend. 1995. The use of position-specific rotamers in model building by homology. Proteins 23:415-421. [DOI] [PubMed] [Google Scholar]
- 8.Collmer, A., J. L. Ried, and M. S. Mount. 1988. Assay methods for pectic enzymes. Methods Enzymol. 161:329-335. [Google Scholar]
- 9.da Silva, A. C. R., et al. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 10.Dehdashti, S. J., C. N. Doan, K. L. Chao, and M. D. Yoder. 2003. Effect of mutations in the T1.5 loop of pectate lyase A from Erwinia chrysanthemi EC16. Acta Crystallogr. D 59:1339-1342. [DOI] [PubMed] [Google Scholar]
- 11.Eijsink, V. G. H., S. Gaseidnes, T. V. Borchert, and T. van den Burg. 2005. Directed evolution of enzyme stability. Biomol. Eng. 22:21-30. [DOI] [PubMed] [Google Scholar]
- 12.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 13.Guerois, R., J. E. Nielsen, and L. Serrano. 2002. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 320:369-387. [DOI] [PubMed] [Google Scholar]
- 14.Hatada, Y., T. Kobayashi, and S. Ito. 2001. Enzymatic properties of the highly thermophilic and alkaline pectate lyase Pel-4B from alkaliphilic Bacillus sp. strain P-4-N and the entire nucleotide and amino acid sequences. Extremophiles 5:127-133. [DOI] [PubMed] [Google Scholar]
- 15.Herron, S. R., J. A. E. Benen, R. D. Scavetta, J. Visser, and F. Jurnak. 2000. Structure and function of pectic enzymes: virulence factors of plant pathogens. Proc. Natl. Acad. Sci. USA 97:8762-8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooft, R. W., C. Sander, and G. Vriend. 1996. Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins 26:363-376. [DOI] [PubMed] [Google Scholar]
- 17.Jaenicke, R. 2000. Stability and stabilization of globular proteins in solution. J. Biotechnol. 79:193-203. [DOI] [PubMed] [Google Scholar]
- 18.Kluskens, L. D., G. van Alebeek, A. Voragen, W. M. de Vos, and J. van der Oost. 2003. Molecular and biochemical characterization of the thermoactive family 1 pectate lyase from the hyperthermophilic bacterium Thermotoga maritima. Biochem. J. 370:651-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuchner, O., and F. H. Arnold. 1997. Directed evolution of enzyme catalysts. Trends Biotechnol. 15:523-530. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 21.Lehmann, M., et al. 2002. The consensus concept for thermostability engineering of proteins: further proof of concept. Protein Eng. 15:403-411. [DOI] [PubMed] [Google Scholar]
- 22.Lietzke, S. E., R. D. Scavetta, M. D. Yoder, and F. Jurnak. 1996. The refined three-dimensional structure of pectate lyase E from Erwinia chrysanthemi at 2.2 Å resolution. Plant Physiol. 111:73-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews, B. W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33:491-497. [DOI] [PubMed] [Google Scholar]
- 24.Mok, Y., E. L. Elisseeva, A. R. Davidson, and J. D. Forman-Kay. 2001. Dramatic stabilization of an SH3 domain by a single substitution: roles of the folded and unfolded states. J. Mol. Biol. 307:913-928. [DOI] [PubMed] [Google Scholar]
- 25.Morley, K., and R. Kazlauskas. 2005. Improving enzyme properties: when are closer mutations better? Trends Biotechnol. 23:231-237. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 28.Ouajai, S., and R. A. Shanks. 2005. Morphology and structure of hemp fibre after bioscouring. Macromol. Biosci. 5:124-134. [DOI] [PubMed] [Google Scholar]
- 29.Perrakis, A., M. Harkiolaki, K. S. Wilson, and V. S. Lamzin. 2001. ARP/wARP and molecular replacement. Acta Crystallogr. D 57:1445-1450. [DOI] [PubMed] [Google Scholar]
- 30.Polizzi, K. M., A. S. Bommarius, J. M. Broering, and J. F. Chaparro-Riggers. 2007. Stability of biocatalysts. Curr. Opin. Chem. Biol. 11:220-225. [DOI] [PubMed] [Google Scholar]
- 31.Riesenberg, D. 1991. High-cell-density cultivation of Escherichia coli. Curr. Opin. Biotechnol. 2:380-384. [DOI] [PubMed] [Google Scholar]
- 32.Robinson-Rechavi, M., and A. Godzik. 2005. Structural genomics of Thermotoga maritima proteins shows that contact order is a major determinant of protein thermostability. Structure 13:857-860. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Smith, S. P., K. R. Barber, S. D. Dunn, and G. S. Shaw. 1996. Structural influence of cation binding to recombinant human brain S100b: evidence for calcium-induced exposure of a hydrophobic surface. Biochemistry 35:8805-8814. [DOI] [PubMed] [Google Scholar]
- 35.Solbak, A., et al. 2005. Discovery of pectin-degrading enzymes and directed evolution of a novel pectate lyase for processing cotton fabric. J. Biol. Chem. 280:9431-9438. [DOI] [PubMed] [Google Scholar]
- 36.Sterner, R., and W. Liebl. 2001. Thermophilic adaptation of proteins. Crit. Rev. Biochem. Mol. Biol. 36:39-106. [DOI] [PubMed] [Google Scholar]
- 37.Takao, M., T. Nakaniwa, K. Yoshikawa, T. Terashita, and T. Sakai. 2001. Molecular cloning, DNA sequence, and expression of the gene encoding for thermostable pectate lyase of thermophilic Bacillus sp. TS 47. Biosci. Biotechnol. Biochem. 65:322-329. [DOI] [PubMed] [Google Scholar]
- 38.Vagin, A., and A. Teplyakov. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022-1025. [Google Scholar]
- 39.van den Burg, B., H. G. Enequist, M. E. van der Haar, V. G. Eijsink, B. K. Stulp, and G. Venema. 1991. A highly thermostable neutral protease from Bacillus caldolyticus: cloning and expression of the gene in Bacillus subtilis and characterization of the gene product. J. Bacteriol. 173:4107-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van den Burg, B., B. W. Dijkstra, G. Vriend, B. Van Dar Vinne, G. Venema, and V. G. Eijsink. 1994. Protein stabilization by hydrophobic interactions at the surface. Eur. J. Biochem. 220:981-985. [DOI] [PubMed] [Google Scholar]
- 41.Vieille, B. R., and G. J. Zeikus. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65:1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52-56. [DOI] [PubMed] [Google Scholar]
- 43.Yoder, M. D., N. T. Keen, and F. Jurnak. 1993. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science 260:1503-1507. [DOI] [PubMed] [Google Scholar]
- 44.Zhao, H. 2007. Directed evolution of novel protein functions. Biotechnol. Bioeng. 98:313-317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.