Abstract
Bacterial contamination of touch surfaces poses a serious threat for public health. The use of bactericidal surface materials, such as copper and its alloys, might constitute a way to aid the use of antibiotics and disinfectants, thus minimizing the risk of emergence and spread of multiresistant germs. The survival of Escherichia coli on metallic copper surfaces has been studied previously; however, the mechanisms underlying bacterial inactivation on copper surfaces have not been elucidated. Data presented in this study suggest that bacteria are killed rapidly on dry copper surfaces. Several factors, such as copper ion toxicity, copper chelators, cold, osmotic stress, and reactive oxygen species, but not anaerobiosis, influenced killing rates. Strains deleted in copper detoxification systems were slightly more sensitive than was the wild type. Preadaptation to copper enhanced survival rates upon copper surface exposure. This study constitutes a first step toward understanding the reasons for metallic copper surface-mediated killing of bacteria.
Copper, both in its metallic and ionic forms, has been exploited empirically since ancient times for medical uses in countless cultures around the globe. Later, the therapeutic powers of this metal were accredited to its antimicrobial properties for the curing of various wound and skin diseases. In recent times, copper has been introduced into clothing, bedding, and other articles, providing them with biocidal properties (4).
The toxicity of copper is largely due to its tendency to alternate between its cuprous, Cu(I), and cupric, Cu(II), oxidation states, differentiating copper from other trace metals, such as zinc or nickel. Under aerobic conditions, this redox cycling leads to the generation of highly reactive hydroxyl radicals that readily and efficiently damage biomolecules, such as DNA, proteins, and lipids. The underlying Fenton-like reactions involving reactive oxygen species (ROS) can be described as copper-catalyzed Haber-Weiss reactions (reviewed in reference 17). While the reaction of dihydrogen peroxide with superoxide primarily has a negligible rate constant (equation 1), this rate is greatly accelerated in the presence of copper (or iron). Copper ions are believed to catalyze this reaction (equations 2 and 3): Cu(II) is initially reduced by superoxide (equation 2), followed by reoxidation by dihydrogen peroxide (equation 3), resulting in a net production of the hydroxyl radical.
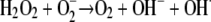 |
(1) |
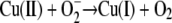 |
(2) |
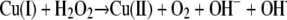 |
(3) |
Bacteria such as Escherichia coli possess several specific cellular resistance systems for defense against copper toxicity. The P-type ATPase CopA transports Cu(I) from the cytoplasm into the periplasm using ATP hydrolysis (23). Periplasmic Cu(I) is transported out of the cell by the proton-driven Cus system, a transport system that also involves a periplasmic copper chaperon, CusF (11). A second periplasmic defense system is the multicopper oxidase CueO that, among other reactions, oxidizes Cu(I) to Cu(II) (26). In this enzyme, molecular oxygen is the acceptor for the electrons originating from Cu(I). Thus, CueO is a detoxification system under aerobic conditions. Enzymatic activity of CueO is also dependent on the presence of copper. All four multicopper oxidase Cu centers have to be occupied, and CueO additionally possesses a fifth copper-binding site that is important for catalysis (24).
Regulation of the copper response in E. coli is governed by two independent circuits: cus is induced by a two-component system, CusRS, which likely senses periplasmic copper ions, while copA and cueO are under the control of CueR, a cytoplasmic MerR-like repressor that is converted into an activator upon Cu(I) binding (22, 27).
In contrast to Cus, CueO, and CopA, which are encoded on the chromosome, some E. coli strains have acquired additional plasmid-encoded copper resistance determinants that provide for higher resistance levels of the host. The best studied is the Pco system of plasmid pRJ1004 isolated from a piggery in the United Kingdom (5). Pco comprises PcoA, a multicopper oxidase distantly related to CueO; two periplasmic copper chaperones, PcoC and PcoE; and two proteins of uncertain function, PcoB and PcoD. Copper-dependent expression is achieved by the two-component system PcoRS and also by its paralog CusRS (5).
Previously, a series of studies were conducted that investigated the survival of different bacteria, such as E. coli O157 (19, 30), Listeria monocytogenes (31), Staphylococcus aureus (28), Enterococcus faecalis (25), Salmonella enterica, Campylobacter jejuni (9), and Klebsiella pneumoniae (32), on metallic copper surfaces. It was found that exposure to metallic copper rapidly killed the bacteria. Killing on various copper alloys was monitored at low (4°C) and room (20°C) temperatures to mimic conditions of food handling and production industries or hospital environments, respectively. For testing, bacteria were diluted in physiological buffers and applied to copper surfaces. The interaction of the organisms with the metallic copper in this setup was rather indirect, since the sample required up to 65 min before all the liquid evaporated and the cells made direct contact to the surface (19). Furthermore, all prior studies were of a descriptive nature. The killing efficiencies of alloys with various copper contents were tested and compared, yet no experiments were conducted that addressed the mechanism of killing of bacteria by metallic copper and its alloys.
In this report, we investigated why E. coli dies on metallic copper surfaces and the contribution of copper resistance mechanisms for survival. An improved test method was developed that minimized the time of aqueous suspension of the cells and better mimicked bacterial contamination of copper touch surfaces.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and alloys.
The strains used in this work are listed in Table 1. E. coli cells on Luria-Bertani (LB) agar plates were incubated overnight at 37°C. For each experiment, a single colony of E. coli from a fresh LB agar plate was inoculated in LB broth for 16 h at 37°C until it reached stationary growth phase. Trimethoprim (25 μg/ml) was added to maintain pco plasmid pPA173 in strain W3110. Copper alloy coupons (2.5 × 2.5 cm) used in this study were “copper” (C11000, 99.9% Cu), the cupronickel “nickel silver” (C75200, 62% maximum Cu, 18% maximum Ni, and 21% maximum Zn), the brass Muntz metal, which is used in shipbuilding because of its antifouling properties (C28000, 59 to 62% Cu and 38 to 41% Zn), and for comparison, austenitic stainless steel (AISI 304, approximately 67 to 72% Fe, 17 to 19.5% Cr, and 8 to 10.5% Ni). All copper alloy coupons were treated prior to each experiment to standardize the surface properties. Coupons were incubated for 30 s in 3% NaOH solution at 70°C and rinsed in distilled water. After transfer to 10% sulfuric acid solution for 5 s at room temperature, coupons were immediately washed with distilled water. All coupons were sterilized and cleaned by immersion in ethanol and kept in a sterile container. To prevent surface reoxidation, cleaned coupons were not flamed after immersion in ethanol.
TABLE 1.
E. coli strains used in this study
| Strain | Relevant genotype | Relevant phenotype | Reference |
|---|---|---|---|
| W3110 | Wild type (K-12 derivative) | Intermediate copper resistant | 1 |
| W3110(pPA173) | pcoABCDRSE from plasmid pRJ1004 | Copper resistant | 5 |
| GR5 | W3110 ΔcusCFBAa | Intermediate copper resistant | 12 |
| GR1 | W3110 ΔcueOa | Intermediate copper sensitive | 12 |
| EC949 | W3110 ΔcopA | Intermediate copper sensitive | This study |
| GR15 | W3110 ΔcusCFBA ΔcueOa | Intermediate copper sensitive | 12 |
| GR17 | W3110 ΔcusCFBA ΔcopA ΔcueOa | Copper sensitive | 12 |
Resistance cassette of the original strain was removed using plasmid pCP20 (7).
Survival on metal surface assay.
For determination of the survival of E. coli on dry metal surfaces, overnight cultures were concentrated 10-fold by centrifugation and cells were resuspended in phosphate-buffered saline (PBS). A 40-μl sample (equivalent to approximately 2.8 × 1010 cells) was applied to a sterile cotton swab and spread evenly across a 2.5- by 2.5-cm metal coupon. The overall amount of liquid applied on the surface was 2.83 ± 0.17 μl (7% of the total volume), containing 1.4 × 109 cells (4.3% of the initial cell number). This discrepancy in percentages is probably due to the binding of cells to cotton fibers; however, the cell numbers were highly reproducible. All samples dried completely within 5 s after contact with the surfaces. To avoid contamination from the laboratory environment, coupons were incubated in glass petri dishes at 23°C or 5.5°C for different times. Coupons were removed in 10 ml PBS with approximately 20 glass beads (2 mm; Roth, St. Leon-Rot, Germany) and vortexed vigorously for 1 min. Samples were diluted in PBS and plated on LB agar. Surviving bacteria were counted as CFU by using an automatic counter (aCOLyte; Synbiosis, Cambridge, United Kingdom) and its associated software (version 2.0.8).
For an evaluation of different potentially protecting substances, cells (2.8 × 1010) were resuspended in supplemented PBS prior to being spread on metal coupons. The compounds and enzymes tested comprised 10% sucrose, 20 mM d-mannitol, 10 to 500 mM EDTA, 20 mM bathocuproine disulfonate (BCS), 20 mM 4,5-dihydroxy-1,3-benzene disulfonic acid (Tiron), catalase (500 U/ml), or superoxide dismutase (SOD) (500 U/ml).
To investigate survival under anaerobic conditions, the standard procedure was followed with the exception that the application of cells onto metal surfaces and the transfer into PBS were performed in an anaerobic Sekuroka glove chamber (I2R, Cheltenham, PA), which was then purged and filled with N2.
For liquid, acute copper toxicity survival experiments (copper shock), 1.2 × 109 cells of overnight cultures of E. coli were diluted into different CuCl2 concentrations. This is about the same number of cells that was applied on metallic copper coupons. After incubation for 1 min, samples were withdrawn in 10 ml PBS with approximately 20 glass beads and then serially diluted into PBS and plated.
RESULTS
E. coli cells exposed to dry copper surfaces are killed rapidly.
Recent studies demonstrated that 2 × 107 E. coli cells are inactivated on copper surfaces in a time period of 75 to 90 min (19, 30). When the killing of bacteria on metallic copper surfaces was investigated, the cells were usually diluted in physiological buffer, the solutions were applied onto the surfaces, and the samples were dried by passive evaporation. This process takes about 65 min (19), during which there is ample time for Cu(0) to be oxidized and dissolved as Cu(I) or Cu(II). Thus, under such corrosive conditions, the bacteria are finally most probably challenged by dissolved copper cations. However, this wet exposure does not quite correspond to contamination of copper touch surfaces with bacteria, which are usually dry. To examine the toxic effect of metallic copper on living cells, an assay that mimics bacterial exposure to touch surfaces was developed. Using this method, drying of the applied sample was achieved in about 5 s, a 780-fold decrease in the time in which copper can go into solution compared to the time with the previous method (19).
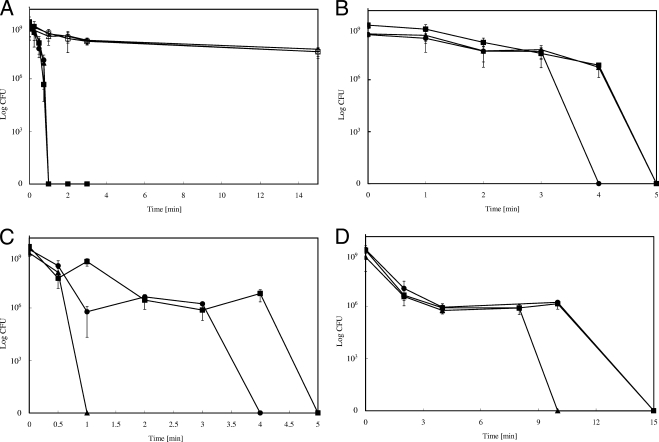
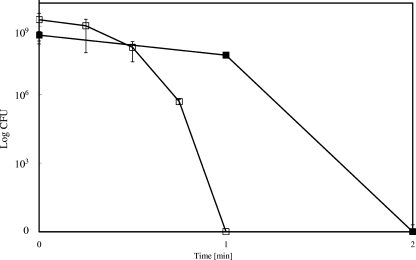
Unexpectedly, this reduced time period in aqueous solution led to a decrease in survival of E. coli. Figure 1A demonstrates that E. coli wild-type strain W3110 exposed to 99% pure copper (copper alloy C11000) resulted in complete killing after only 1 min at 23°C. In contrast, cells applied on stainless steel did not suffer a dramatic decrease in viability under the same conditions.
FIG. 1.
Survival of copper-resistant or -sensitive E. coli strains on copper surfaces and those of its alloys and stainless steel. Cells of E. coli wild-type strain W3110 (▪), its copper-sensitive derivative GR17 (ΔcopA Δcus ΔcueO) (▴) or W3110(pPA173) harboring the high-level copper resistance system Pco (•) were streaked on copper alloy surfaces (filled symbols) or stainless steel (open sympols). After the indicated time periods at ambient conditions (23°C [A, C, and D] or 5.5°C [B]) cells were removed from metal surfaces, diluted, and plated on LB agar. Surviving cells were counted as CFU after 16 h at 37°C. The alloys were pure copper (C11000, 99.9%) (A and B), “nickel-silver” (C75200, maximum of 62% Cu) (C), Muntz metal (C28000, maximum of 62% Cu) (D), and stainless steel (AISI 304) (A). Shown are averages with standard deviations (error bars) from three independent experiments.
Likewise, when this experiment was repeated at 5.5°C, a similar result was obtained. As expected, the rate of killing was slightly decreased in comparison to the rate when the experiment was conducted at 23°C. However, bacteria were completely killed on copper after 5 min (Fig. 1B). This result indicated that exposure to metallic copper, even without extended aqueous incubation, rapidly inactivates E. coli. Furthermore, survival on stainless steel rules out desiccation or osmotic stress as underlying mechanisms.
The observed inactivation of the bacteria occurred very rapidly. Therefore, killing efficacies of other alloys containing lower percentages of copper, such as “nickel silver” (C75200) and Muntz metal (C28000), were tested as well. We expected similar rates of killing by these alloys because they contain approximately the same percentage of copper (about 60%). However, this was not the case; complete inactivation of E. coli W3110 was observed after 5 or 15 min for the copper-nickel-zinc alloy or for the brass Muntz metal, respectively (Fig. 1C and D). It is interesting to note that there is a quick 2- to 3-log decrease in viability within the first 2 min of exposure. After that, a plateau is reached where no further significant killing was observed. Finally, all remaining live cells were inactivated within 2 min, a decrease of 106. A similar biphasic effect, yet in a compressed manner, can also be seen with pure copper (Fig. 1A). Stainless steel also caused a small 10-fold decrease in cell number within the first 2 min. Therefore, this initial decrease can most probably be attributed to plating stress. The sharp decline in viability on copper surfaces within the last 2 min might be caused by the effects of a stressor that has to reach a lethal concentration before efficient killing of the cell can commence. It is possible that this stressor is the copper ion. This initial inactivation effect, however, was not observed on pure copper at 5.5°C (Fig. 1B).
E. coli is resistant against short-term exposure to copper cations.
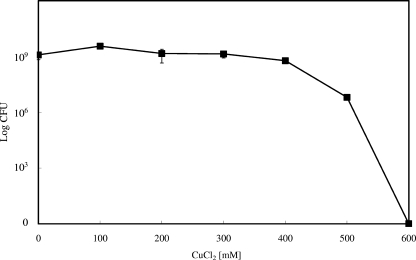
If dissolved copper ions were the underlying reason for copper surface-mediated killing, this fact could be used to calibrate the concentration of copper necessary for complete inactivation of bacteria in a given time period. Therefore, a series of solutions of known CuCl2 concentrations were prepared and cells diluted therein. Incubation for 1 min on copper surfaces completely inactivated E. coli (Fig. 1A). Thus, cells were also incubated for 1 min in each CuCl2 solution before they were diluted and plated. Figure 2 shows that exposure to concentrations of up to 400 mM CuCl2 did not significantly affect the survival of E. coli. However, bacteria were completely killed at 600 mM. This extended range of tolerance against copper between 100 and 400 mM CuCl2 before total inactivation is accomplished resembles the biphasic survival curves on metallic copper (Fig. 1), indicating that copper ions are the lethal stressor. However, it is hardly conceivable that during the short time period of liquid application (5 s), oxidation of the copper coupons resulted in the release of 600 mM of copper from the surface. It should also be mentioned that during metallic copper oxidation, the relatively insoluble and aerobically unstable cuprous ion is generated. Because of these unfavorable properties of the cuprous ion, no liquid Cu(I) survival assay was performed. Thus, the observed killing might be caused by the effects not only of cupric copper but also of cuprous copper or of a combination of both.
FIG. 2.
Killing of E. coli by aqueous CuCl2. Cells of E. coli wild-type strain W3110 were diluted in different CuCl2 solutions and incubated at 23°C for 1 min. Samples were diluted and plated on LB agar. Surviving cells were counted as CFU after 16 h at 37°C. Shown are averages with standard deviations (error bars) from three independent experiments.
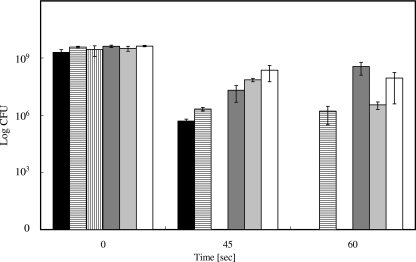
Metal chelators, quenchers of ROS, or osmolytes protect E. coli from metallic copper surface-mediated toxicity.
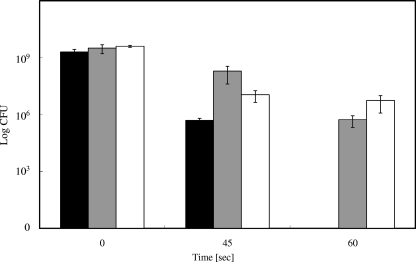
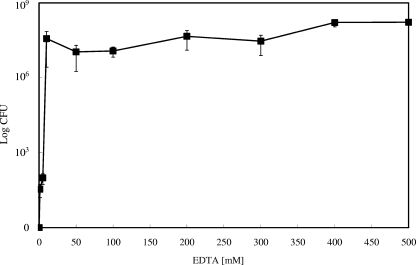
Even though it is an elusive species, under aerobic conditions, the concentration of free cuprous ions can be diminished by suitable chelators. BCS is frequently used as a specific chelator of the soft Lewis acid Cu(I), whereas EDTA efficiently complexes the borderline metal Cu(II). If the different copper species have different partitions on the killing of E. coli, the addition of specific chelators to the copper surface assay should indicate which species, Cu(I) or Cu(II), is responsible. Figure 3 shows that the addition of chelators did not significantly alter the decrease in viable cell numbers during the first 45 s of exposure. Thus, this effect can probably be attributed not to copper but to general plating stress. In contrast, 60 s represented a time after which cells without chelator are completely inactivated (Fig. 1A), but both BCS and EDTA efficiently protected E. coli, with BCS being slightly more effective (Fig. 3). Therefore, if the cells were killed by copper ions released from the metallic copper surface, most probably a mixture of cuprous and cupric ions was responsible. To explore the relationship between copper and copper chelators in more detail, an EDTA dose-response survival assay on copper surfaces was conducted. Increasing concentrations of EDTA were mixed with E. coli challenged on copper surfaces. Counterintuitively, already comparatively low concentrations (10 mM) of EDTA were sufficient to fully protect the cells from copper toxicity (Fig. 4). Conversely, Cu(II) diminished the survival of E. coli only above a concentration of 400 mM and killed at 600 mM (Fig. 2). EDTA and Cu(II) form 1:1 complexes with a stability constant (logK) of 18.8. It is presently unclear how copper chelation by 10 mM EDTA can have such a strong influence on survival.
FIG. 3.
Protective effects of metal chelators for survival of E. coli on copper surfaces. Cells were treated as described in the legend for Fig. 1 except samples were withdrawn only after 0, 45, and 60 s. Cells were mixed with Cu(II) chelator EDTA (gray bars), Cu(I) chelator BCS (white bars), or no chelator (black bars) prior to application onto copper surfaces. Shown are averages with standard deviations (error bars) from three independent experiments.
FIG. 4.
EDTA concentration-dependent protection of E. coli on copper surfaces. Cells of E. coli W3110 were treated as described in the legend for Fig. 1 except cells were mixed with Cu(II) chelator EDTA at the indicated concentrations and all samples were withdrawn after 60 s. Shown are averages with standard deviations (error bars) from three independent experiments.
Copper oxidation may lead not only to the release of copper ions but also to the generation of ROS. To explore the possibility that ROS take part in copper-mediated killing, the protective properties of hydroxyl radical and superoxide quenchers were studied. Mannitol, a hydroxyl radical quencher, or Tiron, a quencher of superoxide, was added in copper surface assays, and its effect on survival was measured after 45 and 60 s. Tiron did not protect cells; rather, it accelerated killing (Fig. 5). After 45 s, all cells were inactivated. At the concentration used, Tiron was not toxic to E. coli when tested in the absence of copper surfaces (data not shown). In contrast, mannitol efficiently prevented the killing of cells exposed to copper surfaces. This result suggested that hydroxyl radicals were the responsible lethal agent. To corroborate this finding, copper surface assays were also conducted in the presence of catalase or SOD. Both catalase and SOD effectively protected E. coli from ROS-mediated killing. After 60 s of exposure to copper surfaces, catalase was the most potent protectant, with approximately 100-fold more survivors than the number with SOD or mannitol (Fig. 5).
FIG. 5.
Effects of ROS quenchers or sucrose on copper surface-mediated killing of E. coli. Cells of E. coli W3110 were treated as described in the legend for Fig. 1 except cells were mixed with ROS quenchers mannitol (horizontally striped bars), Tiron (vertically striped bars), catalase (dark gray bars), or SOD (light gray bars) or sucrose (white bars) or no additive (black bars). All samples were withdrawn after 0, 45, or 60 s. Shown are averages with standard deviations (error bars) from three independent experiments.
During the initial time period after application of cultures on copper surfaces at 23°C, we observed a significant decrease in viability, which was probably caused by plating stress (Fig. 1 and 5). Previously, the osmolyte sucrose was shown to protect E. coli from oxidative stress by raising the osmolarity of the medium (2). Therefore, the addition of sucrose to the copper surface assay probably results in the diminution of this initial stress condition. Unexpected, however, was not only that sucrose prevented initial inactivation of E. coli but also that cells were still viable after 1 min of copper surface exposure. Under these conditions, cells were completely killed without sucrose (Fig. 1A and 5). Since sucrose is known neither as a copper chelator nor as an oxygen radical quencher, this result suggests that sucrose indirectly diminished copper surface toxicity through the elimination of osmotic stress.
Anaerobiosis does not significantly protect E. coli from copper surface toxicity.
To gain more insight into the toxic properties of metallic copper surfaces and the contribution of copper speciation herein, E. coli cells were challenged by metallic copper surfaces under anaerobiosis. Anaerobic conditions should abolish stress by copper-mediated generation of ROS. Thus, improved survival of a facultative anaerobic organism such as E. coli was expected when cells were copper surface exposed in an N2 atmosphere. Unexpectedly, there was only a small increase in survival under these conditions compared to the level of survival under an oxic atmosphere (Fig. 6). Possibly, contact with cells might directly oxidize the copper surface and release Cu(I). Cu(I) is not very soluble and not stable under aerobic conditions, but Cu(I) is considered more toxic than Cu(II). As a consequence, anaerobic conditions do not significantly improve survival on metallic copper.
FIG. 6.
Survival of E. coli strains on copper surfaces under anaerobiosis. Cells of E. coli W3110 were treated as described in the legend for Fig. 1 except that exposure to copper surfaces was conducted under an N2 atmosphere (filled symbols) or air (open symbols). Error bars indicate standard deviations.
E. coli cells lacking copper ion resistance systems are more susceptible to copper surface-mediated killing.
E. coli possesses three major systems for protection against copper cation-mediated toxicity. CopA and the Cus system actively efflux Cu(I), and CueO oxidizes periplasmic Cu(I) to Cu(II) (21, 26). Hitherto, whether these resistance systems play a role in the survival of E. coli on metallic copper surfaces was not known. To investigate whether cells deleted in copA, cusCFBA, and cueO suffer increased susceptibility, the survival rate of the mutant on copper surfaces was determined and compared with that of its parental wild-type strain, E. coli W3110. Additionally, E. coli strain W3110(pPA173), carrying the high-level copper resistance system Pco, was tested. On 99% pure copper, virtually no difference in survival rates among the three strains was observed. Complete killing was accomplished after 1 min, regardless of the presence or absence of resistance systems (Fig. 1A). In contrast, at 5.5°C, cells lacking CopA, Cus, and CueO were inactivated slightly faster than were wild-type cells with or without Pco (Fig. 1B). However, this difference was apparent only after 3 min of exposure to the copper surface. Prior to that time, mutant and wild-type survival rates were indistinguishable. After that time, a sharp 108-fold decrease in cell number was observed and, within 1 min, mutant cells were killed completely.
Because 99% pure copper has such superior killing properties compared to those of other alloys, the experiment was repeated with “nickel silver” and with Muntz metal, both sharing similar copper contents of about 60%. Using these less efficient alloys, we observed a clear difference in survival between the wild type and the copA cusCFBA cueO deletion mutant strain (Fig. 1C and D). Mutant cells were killed completely after 1 min on “nickel silver,” only one-fifth of the time needed for the wild type. The difference between the mutant and the wild type was less pronounced on Muntz metal, on which the mutant was fully inactivated after 10 min and the wild type after 15 min, a difference of one-third. The presence of the pco system did not confer higher resistance than that of strain W3110 without pco; rather, pco-bearing cells were inactivated slightly faster on “nickel silver.” Thus, under the conditions tested, the chromosomally encoded CopA, Cus, and CueO systems, but not the plasmid-encoded pco operon, contributed to survival on metallic copper plates. Because CopA, Cus, and CueO are related to copper ion detoxification, this result suggests that ionic copper is the primary lethal agent.
Induction of cellular copper resistance mechanisms only slightly improves survival of E. coli on metallic copper.
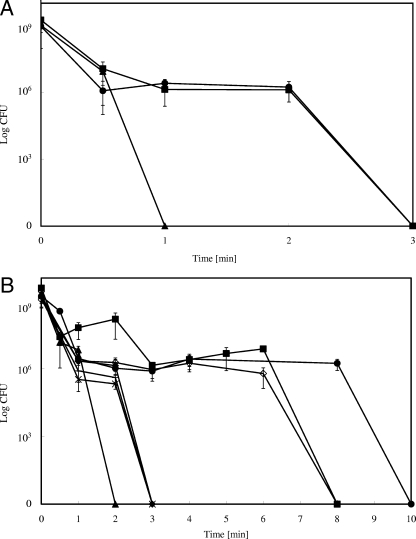
When bacterial cells were exposed to metallic copper, the fraction of survivors rapidly declined. However, in all previous reports, bacterial cells were shocked, but not previously adapted to copper (19, 30, 31). We reasoned that the induction of copper resistance systems of E. coli might enhance tolerance against toxicity mediated by copper surfaces. Cells were thus challenged with subtoxic concentrations of copper ions (100 μM CuCl2) to induce the expression of copA, cus, cueO, and other unknown copper ion resistance systems prior to contact with metallic copper surfaces. The survival rate of the preadapted copA cus cueO mutant on 99% copper (Fig. 7A) was not different from the survival rate of nonadapted cells (Fig. 1A). In contrast, strain E. coli W3110 and its Pco-bearing derivative survived three times longer on copper and were inactivated only after 3 min.
FIG. 7.
Survival of preadapted E. coli strains on copper alloy surfaces. E. coli cultures were grown overnight in the presence of nontoxic concentrations of CuCl2 to induce copper detoxification systems. Washed cells of E. coli wild-type strain W3110 (▪) or its copper-sensitive derivatives GR17 (ΔcopA Δcus ΔcueO) (▴), GR15 (Δcus ΔcueO) (⧫), EC949 (ΔcopA) (+), GR1 (ΔcueO) (X), GR5 (Δcus) (⋄), or W3110 (pPA173) harboring the high-level copper resistance system Pco (•) were streaked on 99.9% copper (A) or “nickel silver” alloy (C75200, maximum of 62% Cu) (B) surfaces and treated as described in the legend for Fig. 1. Shown are averages with standard deviations (error bars) from three independent experiments.
On “nickel silver,” the preadapted ΔcopA Δcus ΔcueO triple mutant experienced an extended time of survival compared to survival times of nonadapted cells (Fig. 7B). The time needed for full inactivation approximately doubled compared to the times needed for nonadapted cells of all strains (Fig. 1C). Under these conditions, the strain expressing pco was the most resistant, the only instance where the presence of Pco was advantageous. Therefore, the preadaptation of E. coli allowed cells to withstand copper toxicity to a higher degree on copper and its alloys, such as “nickel silver” with diminished copper content. In addition, the factors of time needed for full inactivation (adapted/nonadapted) are similar for pure copper and for the “nickel silver” alloy of lower copper concentration.
To evaluate which of the three copper resistance systems, Cus, CueO, and CopA, contributed the most to survival on copper surfaces, single gene mutants were also tested. Additionally, mutants lacking periplasmic (Δcus ΔcueO) or cytoplasmic (ΔcopA) copper resistance systems were included to investigate which compartment is more susceptible to copper stress. “Nickel silver” was used because the time span until complete inactivation between the wild type and the triple mutant was at maximum using this alloy. Furthermore, cells were preincubated with subtoxic copper concentrations to amplify these differences in survival rates. Figure 7B shows that neither the single mutants nor the double mutant was as sensitive to exposure to copper surfaces as was the Δcus ΔcueO ΔcopA triple mutant. The Δcus mutant exhibited a survival rate similar to that of the wild-type strain. This result suggested that both cytoplasmic and periplasmic copper resistance systems are necessary to diminish copper stress. The Cus system that detoxifies periplasmic copper ions was not necessary under the conditions tested because the periplasmic copper resistance protein CueO can compensate for the lack of the Cus system.
In summary, these results suggested that preadaptation could be a significant factor for survival on metallic copper surfaces. This might not be relevant for the use of copper touch surfaces, however, since upon contact, cells are copper shocked, with their resistance systems not well expressed.
DISCUSSION
Doorknobs and other touch surfaces, such as push plates on doors, were found to be notorious sources of nosocomial infections. These infections can easily lead to fatalities in intensive care units or nurseries (29). Replacing stainless steel surfaces with surfaces made of copper or its alloys might provide a way to avoid this health hazard. Using the biocidal properties of copper surfaces would be an easy way to counter the spread of pathogens via human carriers in medical facilities. In fact, an ongoing hospital study is exploring this possibility (www.cda.org.uk/antimicrobial/).
The present study, however, was conducted to gain a deeper understanding of the mechanism of direct copper surface contact-mediated bacterial killing. It is striking that in contrast to the various studies describing the antimicrobial properties of metallic copper surfaces (9, 19, 25, 28, 30-32), to our knowledge, no investigation was conducted on the contribution of bacterial copper resistance mechanisms herein. To what extent cellular copper resistance mechanisms take part in survival is not known for either copper water-pipe systems or short-term exposure of buffer-diluted cells on copper and its alloys. Frequently, however, copper-resistant microorganisms can be isolated from copper piping (6) and other water-exposed copper surfaces (15) where stable biofilms develop. Inadvertently, heterotrophic bacteria isolated from copper plumbing biofilms, such as Acidovorax delafieldii, Flavobacterium sp., Corynebacterium sp., Pseudomonas sp., and Stenotrophomonas maltophilia, were also found to be capable of cuprosolvency (6). Thus, copper ions were efficiently released from copper surfaces by these bacteria, while at the same time, they were resistant against copper ions.
In contrast, on dry touch surfaces, bacteria do not have time to develop biofilms and the stress and survival conditions are different from those of aqueous systems. Stress is directly related to copper under these dry conditions, as we (Fig. 1) and others (9, 19, 31, 32) have demonstrated that cells exposed to other metallic surfaces, such as stainless steel, were not inactivated. Therefore, desiccation stress is not the major underlying mechanism.
Bacteria suffering from diverse stressful conditions frequently go through a physiological differentiation known as the viable but nonculturable (VBNC) state (20). In the VBNC state, bacteria are still viable and show metabolic activity and respiration, but cannot be detected as CFU by conventional plate counts. Possibly, bacteria evade metallic copper surface stress in this way. Bacteria exposed to metallic copper surfaces, however, do not enter the VBNC state; instead, they are completely inactivated (31).
E. coli cells exposed to copper surfaces are thought to be killed by dissolved copper ions. The amount of ionic copper released from a copper surface has previously been determined to be 2.01 × 10−2 μg/cm3 Cu(II) during 24 h at 25°C (32). Consequently, a concentration of about 0.3 μM Cu(II) can be calculated. This is far too little, even for a time period of 24 h, to be toxic for E. coli. Since our coupons dried within 5 s, a copper release of about 17 pM can be approximated. Previously, we and others grew E. coli strain W3110 in liquid medium up to concentrations of >1 mM before significant growth inhibition was observed (12, 21). In short-term exposure (copper shock), E. coli showed a remarkable resistance to ionic copper (Fig. 2), also contradicting ion release from copper surface as the lethal agent.
Nevertheless, cells lacking the major copper detoxification systems CopA, Cus, and CueO showed some decrease in their survival rates on copper surfaces (Fig. 1 and 7). The proactive effect of these systems was especially evident for copper alloys with lower copper contents, for which a prolonged residence time was achieved. Remarkably, all three systems, CopA, Cus, and CueO, in one way or another, detoxify cuprous ions. Periplasmic CueO is an oxygen-dependent multicopper oxidase (26), whereas the Cus system is responsible for periplasmic Cu(I) detoxification, preferably under anaerobic conditions (21). These facts might also explain why a cus deletion strain exhibited a killing rate on copper alloy coupons in air that was similar to the rate of the wild type. CopA detoxifies the cytoplasm under any condition (21). Interestingly, all the copper-demanding enzymatic reactions in E. coli occur in the periplasm or at the cytoplasmic membrane. There is no cytoplasmic enzyme known that uses copper as a cofactor. Accordingly, the broad activity of CopA for cytoplasmic Cu(I) detoxification makes sense physiologically.
In accordance with the activity of the copper detoxification systems, the cellular compartment most affected by copper toxicity is not the cytoplasm but the periplasm (18). Nevertheless, in the present study, mutants of periplasmic or cytoplasmic copper defense systems were almost equally susceptible to killing by copper surfaces. Figure 7B indicates that the lack of either periplasmic (CueO) or cytoplasmic (CopA) protection from copper toxicity had a severe impact on survival on metallic copper surfaces. The Cus system, which was shown to be important for survival under anaerobic copper stress (21), did not significantly contribute to survival on copper alloy surfaces (Fig. 7B).
The mechanisms by which copper ions inhibit or kill overloaded cells are not known (18), but a redox cycling system of cupric ions, dihydrogen peroxide, and ascorbate severely impaired the respiration of E. coli cells (8). The general biocidal properties of copper can be described. Copper toxicity originates not only from the generation of oxidative stress but also from the high affinity of the soft [Cu(I)] or the borderline [Cu(II)] Lewis acid for amino acid side chains, such as that of cysteine or histidine. This likely results in misfolded proteins or the displacement of other transition metal cations from native active sites (16). Furthermore, recent advances in the understanding of copper toxicity have revealed that in vivo, copper does not catalyze significant DNA damage. Instead, the current model suggests that copper decreased the rate at which hydrogen peroxide damages DNA by a mechanism that is not yet understood (18). The rapid inactivation of cells by pure copper observed in the present study (Fig. 1A) also favors the hypothesis that the lethality of copper does not primarily originate from the targeting of DNA.
The combination of cupric ions plus reducing agents markedly increases dihydrogen peroxide-mediated oxidative degradation of various biological compounds (13). In the case of iron, the actual toxic oxidants might be the hydroxyl radical, formed by a one-electron reduction of dihydrogen peroxide, the instable higher valent ferryl ion (cupryl in case of copper), or peroxy complexes as well as secondary radicals generated by reactions with medium components (8).
Dihydrogen peroxide has been shown to be generated from electroplated copper coatings. This ROS is produced by the emission of electrons from the surface of the metal in contact with aqueous electrolytes in the presence of atmospheric oxygen. However, the dihydrogen peroxide measured has been produced by a metal surface incubated in an aqueous buffer system (32). Because we used a quasi dry experimental setup, conditions cannot be easily compared. It is possible that the time the surface is electrolyte exposed is too short for dihydrogen peroxide production through corrosion. Coupon surfaces were dried after 5 s, whereas cells were only fully inactivated after 60 s on 99% copper. Nevertheless, ROS do contribute to killing on dry copper surfaces, as indicated by the protective properties of the ROS quencher, catalase or SOD (Fig. 5).
Recent new data from Macomber et al. suggest that at least in E. coli, the toxicity of copper ions does not originate from DNA-damaging Fenton-like reactions (18). The authors found that both catalase and SOD were strongly induced in cells exposed to copper, indicative of dihydrogen peroxide and superoxide stress. Targets of these ROS probably constitute specific metalloenzymes that can be damaged directly (10, 14); however, the crucial target of the ROS generated is unknown (18). Future studies will address the contribution of the cellular ROS detoxification mechanism for survival on copper surfaces.
Considering Haber-Weiss-mediated generation of ROS by copper, it was surprising that E. coli cells did not exhibit a markedly prolonged survival time on copper surfaces under anaerobiosis (Fig. 6). Cu(I) is more toxic than Cu(II) to cells when under anaerobiosis, and both the reduction of Cu(II) to Cu(I) and the oxidation of Cu(I) to Cu(II) occur under anaerobic conditions (3). The accumulation of Cu(I) might be the underlying reason why we failed to observe much improved survival rates of E. coli in an N2 atmosphere (Fig. 6) in which we expected Cu oxidation and Cu redox cyling to be prevented. Probably, overall metallic copper oxidation was diminished even so, while at the same time, the Cu(II)/Cu(I) ratio was shifted in favor of the more toxic cuprous ion. Nevertheless, both cuprous-ion- and cupric-ion-mediated bactericidal mechanisms involve the inhibition of cellular energy-transducing capabilities and accumulated damage at multiple, yet unknown cellular sites (8).
Acknowledgments
This work was supported by the International Copper Association (ICA) to D.H.N. and G.G. C.E.S. was supported by the Academic Mobility of Higher Education Program (ERASMUS) of the European Union.
We thank Ayla Nies for help with experiments and Christopher Rensing for critically reading the manuscript.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Bachmann, B. J. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36:525-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benov, L., N. M. Kredich, and I. Fridovich. 1996. The mechanism of the auxotrophy for sulfur-containing amino acids imposed upon Escherichia coli by superoxide. J. Biol. Chem. 271:21037-21040. [DOI] [PubMed] [Google Scholar]
- 3.Beswick, P. H., G. H. Hall, A. J. Hook, K. Little, D. C. McBrien, and K. A. Lott. 1976. Copper toxicity: evidence for the conversion of cupric to cuprous copper in vivo under anaerobic conditions. Chem. Biol. Interact. 14:347-356. [DOI] [PubMed] [Google Scholar]
- 4.Borkow, G., and J. Gabbay. 2004. Putting copper into action: copper-impregnated products with potent biocidal activities. FASEB J. 18:1728-1730. [DOI] [PubMed] [Google Scholar]
- 5.Brown, N. L., S. R. Barrett, J. Camakaris, B. T. Lee, and D. A. Rouch. 1995. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 17:1153-1166. [DOI] [PubMed] [Google Scholar]
- 6.Critchley, M. M., N. J. Cromar, N. C. McClure, and H. J. Fallowfield. 2003. The influence of the chemical composition of drinking water on cuprosolvency by biofilm bacteria. J. Appl. Microbiol. 94:501-507. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elzanowska, H., R. G. Wolcott, D. M. Hannum, and J. K. Hurst. 1995. Bactericidal properties of hydrogen peroxide and copper or iron-containing complex ions in relation to leukocyte function. Free Radic. Biol. Med. 18:437-449. [DOI] [PubMed] [Google Scholar]
- 9.Faundez, G., M. Troncoso, P. Navarrete, and G. Figueroa. 2004. Antimicrobial activity of copper surfaces against suspensions of Salmonella enterica and Campylobacter jejuni. BMC Microbiol. 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flint, D. H., J. F. Tuminello, and M. H. Emptage. 1993. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 268:22369-22376. [PubMed] [Google Scholar]
- 11.Franke, S., G. Grass, C. Rensing, and D. H. Nies. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grass, G., and C. Rensing. 2001. Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 183:2145-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halliwell, B., and J. M. Gutteridge. 1990. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 186:1-85. [DOI] [PubMed] [Google Scholar]
- 14.Jang, S., and J. A. Imlay. 2007. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 282:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kielemoes, J., and W. Verstraete. 2001. Influence of copper-alloying of austenitic stainless steel on multi-species biofilm development. Lett. Appl. Microbiol. 33:148-152. [DOI] [PubMed] [Google Scholar]
- 16.Koch, K. A., M. M. Pena, and D. J. Thiele. 1997. Copper-binding motifs in catalysis, transport, detoxification and signaling. Chem. Biol. 4:549-560. [DOI] [PubMed] [Google Scholar]
- 17.Liochev, S. I., and I. Fridovich. 2002. The Haber-Weiss cycle—70 years later: an alternative view. Redox Rep. 7:55-57, 59-60. [DOI] [PubMed] [Google Scholar]
- 18.Macomber, L., C. Rensing, and J. A. Imlay. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 189:1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noyce, J. O., H. Michels, and C. W. Keevil. 2006. Use of copper cast alloys to control Escherichia coli O157 cross-contamination during food processing. Appl. Environ. Microbiol. 72:4239-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliver, J. D. 2005. The viable but nonculturable state in bacteria. J. Microbiol. 43:93-100. [PubMed] [Google Scholar]
- 21.Outten, F. W., D. L. Huffman, J. A. Hale, and T. V. O'Halloran. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670-30677. [DOI] [PubMed] [Google Scholar]
- 22.Outten, F. W., C. E. Outten, J. Hale, and T. V. O'Halloran. 2000. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J. Biol. Chem. 275:31024-31029. [DOI] [PubMed] [Google Scholar]
- 23.Rensing, C., B. Fan, R. Sharma, B. Mitra, and B. P. Rosen. 2000. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. USA 97:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts, S. A., G. F. Wildner, G. Grass, A. Weichsel, A. Ambrus, C. Rensing, and W. R. Montfort. 2003. A labile regulatory copper ion lies near the T1 copper site in the multicopper oxidase CueO. J. Biol. Chem. 278:31958-31963. [DOI] [PubMed] [Google Scholar]
- 25.Robine, E., L. Boulange-Petermann, and D. Derangere. 2002. Assessing bactericidal properties of materials: the case of metallic surfaces in contact with air. J. Microbiol. Methods 49:225-234. [DOI] [PubMed] [Google Scholar]
- 26.Singh, S. K., G. Grass, C. Rensing, and W. R. Montfort. 2004. Cuprous oxidase activity of CueO from Escherichia coli. J. Bacteriol. 186:7815-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoyanov, J. V., J. L. Hobman, and N. L. Brown. 2001. CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol. Microbiol. 39:502-511. [DOI] [PubMed] [Google Scholar]
- 28.Tolba, O., A. Loughrey, C. E. Goldsmith, B. C. Millar, P. J. Rooney, and J. E. Moore. 2007. Survival of epidemic strains of nosocomial- and community-acquired methicillin-resistant Staphylococcus aureus on coins. Am. J. Infect. Control 35:342-346. [DOI] [PubMed] [Google Scholar]
- 29.Wenger, P. N., J. I. Tokars, P. Brennan, C. Samel, L. Bland, M. Miller, L. Carson, M. Arduino, P. Edelstein, S. Aguero, C. Riddle, C. O'Hara, and W. Jarvis. 1997. An outbreak of Enterobacter hormaechei infection and colonization in an intensive care nursery. Clin. Infect. Dis. 24:1243-1244. [DOI] [PubMed] [Google Scholar]
- 30.Wilks, S. A., H. Michels, and C. W. Keevil. 2005. The survival of Escherichia coli O157 on a range of metal surfaces. Int. J. Food Microbiol. 105:445-454. [DOI] [PubMed] [Google Scholar]
- 31.Wilks, S. A., H. T. Michels, and C. W. Keevil. 2006. Survival of Listeria monocytogenes Scott A on metal surfaces: implications for cross-contamination. Int. J. Food Microbiol. 111:93-98. [DOI] [PubMed] [Google Scholar]
- 32.Zhao, Z. H., Y. Sakagami, and T. Osaka. 1998. Toxicity of hydrogen peroxide produced by electroplated coatings to pathogenic bacteria. Can. J. Microbiol. 44:441-447. [PubMed] [Google Scholar]