Abstract
Fibrobacter is a highly cellulolytic genus commonly found in the rumen of ruminant animals and cecum of monogastric animals. In this study, suppression subtractive hybridization was used to identify the genes present in Fibrobacter succinogenes S85 but absent from F. intestinalis DR7. A total of 1,082 subtractive clones were picked, plasmids were purified, and inserts were sequenced, and the clones lacking homology to F. intestinalis were confirmed by Southern hybridization. By comparison of the sequences of the clones to one another and to those of the F. succinogenes genome, 802 sequences or 955 putative genes, comprising approximately 409 kb of F. succinogenes genomic DNA, were identified that lack similarity to those of F. intestinalis chromosomal DNA. The functional groups of genes, including those involved in cell envelope structure and function, energy metabolism, and transport and binding, had the largest number of genes specific to F. succinogenes. Low-stringency Southern hybridization showed that at least 37 glycoside hydrolases are shared by both species. A cluster of genes responsible for heme, porphyrin, and cobalamin biosynthesis in F. succinogenes S85 was either missing from or not functional in F. intestinalis DR7, which explains the requirement of vitamin B12 for the growth of the F. intestinalis species. Two gene clusters encoding NADH-ubiquinone oxidoreductase subunits probably shared by Fibrobacter genera appear to have an important role in energy metabolism.
Fibrobacteres is recognized as a main division (phylum) within the group Bacteria that is closely related to the phyla of Bacteroides and Chlorobi (8, 19). Fibrobacter is the sole genus in this phylum. Bacteria of this genus are important anaerobic cellulose degraders and produce succinic acid as a major fermentation product (27). Fibrobacter strains were found in the rumen and ceca in cattle, sheep, horses, rats, pigs, and other fiber-consuming animals. Two species, Fibrobacter succinogenes and F. intestinalis, were identified by phylogenetic analysis of 16S rRNA as well as phenotypic characterization. Considerable genetic diversity between the two species is apparent, since they have 92% 16S rRNA similarity and less than 20% DNA-DNA similarity (4).
F. succinogenes S85 had been studied extensively because of its higher cellulolytic activity and important position in plant cell wall digestion in the rumen (9). More than 100 carbohydrate-active enzymes, including cellulases, xylanases, polysaccharide lyase, and esterases, have been identified in the recently sequenced genome of F. succinogenes S85 (29). Recently, the gene coding for a major endoglucanase (cel9B) was identified, and three novel cellulases and two acetylxylan esterases were characterized (17, 32). Synergistic interactions were detected among the cellulases (32) and between a xylanase and the two acetylxylan esterases (17). In addition, genes coding for 13 cellulose-binding proteins, which may be important for cellulose degradation, were identified in a proteomics study (16).
Previous studies of F. intestinalis identified two endoglucanases, one cellodextrinase and two xylanases, with highest similarity to those in F. succinogenes S85 (7, 18). In addition, as seen with F. succinogenes S85 (11), a series of cellulose-binding proteins were also identified in F. intestinalis (26). Furthermore, an in vivo 13C nuclear magnetic resonance study of glucose and cellobiose metabolism in F. intestinalis and F. succinogenes has revealed marked homogeneity in their carbon metabolism (23). However, there are also some key differences between the species, notably, the sites of colonization within the gastrointestinal tract that are favored by the two species, with F. intestinalis principally found or recovered from the ceca or hindgut of nonruminant animals, including mice (3).
Suppression subtractive hybridization (SSH) (2) had been used to identify genes present in F. intestinalis DR7 that are absent from F. succinogenes S85 (33). Fifty-five unique sequences were identified in F. intestinalis that do not exhibit detectable similarity to proteins in either F. succinogenes or GenBank. That study also showed that F. intestinalis encodes at least 30 related plant cell wall-degrading proteins, including 18 cellulases or xylanases, which have the highest similarity to those from F. succinogenes. Ninety of the sequences (including those of at least 30 transposases and six genes encoding restriction modification systems) exhibit low or no homology to sequences of the F. succinogenes S85 genome. Furthermore, extensive genome reorganization was detected in F. intestinalis compared to F. succinogenes.
However, this study identified only genes that exist in F. intestinalis but are absent from F. succinogenes. To acquire a more in-depth appreciation of the genetic relatedness between the two species and unique features of the Fibrobacter genus, a further set of SSH experiments were conducted to identify genes in F. succinogenes that either do not exist in or have low similarity to those in F. intestinalis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
F. succinogenes S85 (ATCC 19169) and F. intestinalis DR7 (ATCC 43855) were grown under anaerobic conditions in a chemically defined medium (CDM) with 0.3% glucose as a carbon source and carbon dioxide as a gas phase at 37°C as previously described (33).
To test the requirement for vitamin B12 by F. intestinalis DR7, 0.45 ml of CDM culture (optical density at 675 nm [OD675], ∼1.0) was inoculated into 9 ml of CDM without vitamin B12 and was subcultured in the latter medium n times (n ranged from 4 to 5; see Results) until the cells stopped growing. Cells from the n = 1 subculture were also inoculated into chemically defined media in which vitamin B12 was replaced by either of the two precursors 5-aminolaevulinic acid (Sigma) or porphobilinogen (Sigma) in the same molar concentration as the vitamin B12. The biotin requirement of F. succinogenes S85 was tested in CDM without biotin. In a separate trial both F. intestinalis and F. succinogenes were inoculated into the medium without either vitamin B12 or biotin and were subcultured in the same medium 10 times. Growth of the cells was monitored, in triplicate, by measuring the OD675 in a Lambda 2 spectrophotometer with a 1 cm cuvette.
DNA preparation.
Genomic DNA was isolated from F. succinogenes S85 and F. intestinalis DR7 by the cetyltrimethylammonium bromide procedure as described by Wilson (38). Standard recombinant DNA techniques were performed as described by Sambrook and Russell (36).
Suppressive subtractive hybridization.
SSH (2) was performed by using a Clontech PCR-Select bacterial genome subtraction kit as recommended in the manufacturer's instructions (Clontech, Palo Alto, CA). The resultant PCR amplicons from SSH that were enriched in F. succinogenes S85-specific sequences were purified by phenol-chloroform extraction and ethyl alcohol precipitation followed by TA cloning into pGEM-T Easy vector (Promega) and transformation into Escherichia coli JM109. The E. coli strains were then grown in LB medium supplemented with 100 μg/ml (wt/vol) ampicillin and screened for clones by use of plates containing 1.6% agar 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). White colonies with inserts were restreaked for purity, and the plasmid inserts were sequenced at The Institute of Genome Research (TIGR [now the J. Craig Venter Institute]) (Rockville, MD) as described by Qi et al. (33).
DNA dot blot analysis.
DNA inserts from all the colonies selected were subjected to DNA dot blot analysis to identify false positives. Each of the clones was inoculated into 2 ml of LB medium containing ampicillin at 100 μg/ml and incubated at 37°C for 16 h at 150 rpm. Inserts from all of the SSH clones were amplified by PCR using nested primers 1 and 2R (Clontech protocol) and Platinum Taq DNA high-fidelity polymerase (Invitrogen) and 1 μl quantities of 16 h E. coli clones as the templates. The reactions were performed in 30 μl volumes for 25 cycles with an annealing temperature of 52°C. The PCR amplicons were purified by phenol-chloroform extraction as well as ethyl alcohol precipitation, and the concentration was determined using PicoGreen reagent (Molecular Probes) (1, 33). A 120 ng quantity of PCR amplicons from each clone was mixed with 400 μl 0.5 M NaOH and heated at 100°C and then spotted on duplicate Hybond N+ nylon membranes (Amersham) by use of a Bio-Dot SF microfiltration apparatus (Bio-Rad, Hercules, CA). Genomic DNA from F. intestinalis DR7 was digested with restriction enzyme RsaI (Roche) and labeled with digoxigenin (DIG) by use of a digoxigenin High-Prime labeling mixture (Roche). The hybridization and detection procedures that followed were carried out using 400 ng of the DIG-labeled genomic DNA. DNA dot blots were prehybridized and hybridized at 60°C for high stringency or 37°C for low stringency as described by Sambrook and Russell (36). The DIG detection was conducted following the manufacturer's instructions (Roche). Hybridization of PCR amplicons with the F. intestinalis genomic DNA probes at high stringency indicated their high level of similarity to F. intestinalis genes. These sequences were treated as false positives and were discarded. Results revealing amplicons that did not hybridize to F. intestinalis genomic DNA under high-stringency conditions but hybridized under low-stringency conditions indicated that they had low sequence similarity to genes in F. intestinalis. Amplicons that did not hybridize to F. intestinalis genomic DNA at low stringency were treated as absent from F. intestinalis.
Sequence analysis.
Insert sequences were used to query the F. succinogenes S85 genome by using the TIGR BLAST search program (http://www.tigr.org/tdb/ruminomics/) and were subsequently mapped onto the genome. The translated amino acid sequences from the carbohydrate-active enzyme genes in F. succinogenes were used for searches of the GenBank nonredundant amino acid database (http://www.ncbi.nlm.nih.gov). The BLAST results were parsed and analyzed using Microsoft Excel 2000 and Visual Basic for Application, version 6.3. Theoretical library sizes were estimated as described by Nesbø et al. (30).
RESULTS
Summary of the SSH library.
A total of 1,152 clones were sequenced from both the 5′ and 3′ ends (Table 1). Sequences were assembled, and redundant sequences were removed. By dot blot analysis, 70 clones that hybridized with the F. intestinalis genomic DNA probe at high stringency (see Materials and Methods) were treated as false positives and were removed. This resulted in 802 nonredundant sequences with an average length of 509.8 bp (409 kb in total). All of the sequences had 100% identity to those in the F. succinogenes genome. The G+C content of the unique clones was 47.3% (Table 1), which is slightly lower than the overall G+C content of the F. succinogenes S85 (48.1%). Based on the incidence of duplicate clones, the theoretical library was estimated as 4,100 clones, with about 3,100 unique clones. Therefore, assuming that the average length of the clones was 510 bp, the DNA sequence present in F. succinogenes that was different from F. intestinalis was 1,581 kb, or 41%, taking the size of the genome (3,843 kb) of F. succinogenes S85 into account.
TABLE 1.
Summary of F. succinogenes cloned DNA fragments enriched by SSH that did not hybridize to F. intestinalis DNA under the conditions used
| No. of sequences | No. of unique sequences | Average sequence length (nta) | Total length of unique sequences (nt) | Estimated library size (no. of clones) | No. of unique sequences in estimated library | Total length of unique sequences in estimated library (k nt) | GC % |
|---|---|---|---|---|---|---|---|
| 1,082 | 802 | 509.8 | 408,823 | 4,100 | 3,100 | 1,581 | 47.3 |
nt, nucleotides.
Each of the SSH clones spanned up to three open reading frames in F. succinogenes except for 10 that resided in nonencoding regions. Altogether, 955 open reading frames were identified for the SSH clones (see Table S1 in the supplemental material). Among the genes identified, 51.5% were not assigned any function. Of the remaining genes, those responsible for cell envelope metabolism and energy metabolism as well as transport and binding were the most numerous (Table 2; see also Table S1 in the supplemental material).
TABLE 2.
Roles of the unique F. succinogenes genes identified by SSH that exhibit low similarity to those in F. intestinalis
| Functional role(s) or description | No. (%) of genesa |
|---|---|
| Amino acid biosynthesis | 17 (19.1) |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 23 (26.7) |
| Cell envelope | 137 (33.8) |
| Cellular processes | 21 (33.3) |
| Central intermediary metabolism | 8 (29.6) |
| DNA metabolism | 22 (23.4) |
| Energy metabolism | 84 (40.2) |
| Fatty acid and phospholipid metabolism | 10 (30.3) |
| Mobile and extrachromosomal element functions | 5 (62.5) |
| Protein fate | 19 (25.0) |
| Protein synthesis | 26 (19.8) |
| Purines, pyrimidines, nucleosides, and nucleotides | 12 (24.0) |
| Regulatory functions | 22 (30.6) |
| Signal transduction | 16 (59.3) |
| Transcription | 13 (33.3) |
| Transport and binding proteins | 55 (32.5) |
| Unknown function | 497 (28.4) |
| Total no. of genes identifiedb | 955 (29.4) |
This table lists the functional roles of (each of) the genes identified, which shows the comprehensive random coverage of the genome by SSH. See Table S1 in the supplemental material for genes in each category. Numbers in parentheses are the percentages of the genes identified versus total genes of each class in the sequenced genome of F. succinogenes.
The total number of genes identified was less than the sum of the genes included in each function role (988), because some proteins had more than one role.
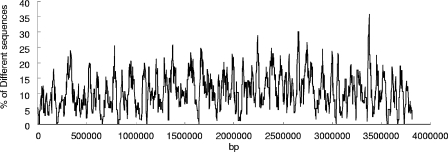
The SSH sequences were mapped on the F. succinogenes genome (Fig. 1) to determine whether there were any large contiguous regions of the genome that were absent from F. intestinalis. Most of the SSH sequences seemed to be randomly distributed along the chromosome, while some regions contained higher percentages of SSH sequences, indicating possible mutation or recombination hotspots.
FIG. 1.
The distribution of the F. succinogenes-specific genes on the chromosome of F. succinogenes. The x axis represents positions on the chromosome. The y axis represents the percentages of the SSH sequences in each of several continuous 20-kb regions on the F. succinogenes chromosome.
Genes involved in energy metabolism.
Eighty-four genes in the functional category of energy metabolism showed low DNA similarity to the F. intestinalis genome. There were 69 SSH sequences that coded for 61 enzymes involved in plant cell wall degradation, including 19 cellulases and 16 hemicellulases (Table 3; see also Table S1 in the supplemental material). To determine whether the cellulases and hemicellulases identified by reverse SSH also exist in F. intestinalis despite the observation that they diverged greatly in DNA sequence, a Southern dot blot analysis of the glycoside hydrolase genes at a low hybridization temperature was performed. Southern hybridization at low stringency showed that 32 of the 35 sequences identified by SSH hybridized to the F. intestinalis genomic DNA, indicating their existence in the latter genome. Only three glycoside hydrolase genes, which overlapped in sequences RSGB_542 (F. succinogenes open reading frame no. 2294 [FSU2294], which belongs to glycoside hydrolase family 10 [GH10]), RSGB_424 (FSU1894, GH45), and RSGB_704 (FSU2912, GH9), did not hybridize with the F. intestinalis genomic DNA at low stringency. Previously we had identified 18 cellulases or xylanases in F. intestinalis (33). Thirteen of them were also identified in the present study. As shown by assembling the results of the two SSH experiments, a minimum of 37 glycoside hydrolase genes may be present in the F. intestinalis genome (Table 3). The nonredundant GenBank database was searched by the BLASTP program using the translated SSH cellulose gene sequences in the query. The names of organisms that contained glycoside hydrolases with the highest similarity to the respective F. succinogenes proteins are listed (Table 3).
TABLE 3.
Cellulase and xylanase genes in F. succinogenes and F. intestinalis identified by reverse and forward SSH using genomic DNA from F. intestinalis and F. succinogenes, respectively, as the drivers
| FSU gene identification no. | Protein encoded by genea | Prior protein nameb (reference[s]) | F. succinogenes gene(s) showing similarity to an F. intestinalis genec | Gene unique to F. succinogenesd | F. intestinalis gene showing similarity to F. succinogenes genee | Non-Fibrobacter organism showing highest BLAST match scoref |
|---|---|---|---|---|---|---|
| 2622 | Cel16A | RSGB_633 | Bacillus circulans | |||
| 1893 | Cel45A | RSGB_423 | Cellvibrio japonicus | |||
| 1894 | Cel45B | RSGB_424 | Cellvibrio japonicus | |||
| 1947 | Cel45C | + | Cellvibrio japonicus | |||
| 0382 | Cel51A | CelF (22, 24) | RSGB_074 | + | Alicyclobacillus acidocaldarius | |
| 1685 | Cel5A | RSGB_380 | Cytophaga hutchinsonii | |||
| 2070 | Cel5C | CedA (13) | RSGB_476, RSGB_477 | Unidentified bacterium | ||
| 2290 | Cel5E | Cel5Kg | Saccharophagus degradans | |||
| 2534 | Cel5F | RSGB_612 | + | Orpinomyces joyonii | ||
| 2772 | Cel5G | Cel-3 (25) | RSGB_666, RSGB_667 | + | Cytophaga hutchinsonii | |
| 2914 | Cel5H | + | Orpinomyces joyonii | |||
| 1346 | Cel5K | CelG (14) | RSGB_289 | Saccharophagus degradans | ||
| 2866 | Cel74A | RSGB_694 | + | Cytophaga hutchinsonii | ||
| 1680 | Cel8A | + | Cytophaga hutchinsonii | |||
| 2303 | Cel8B | RSGB_549 | Cytophaga hutchinsonii | |||
| 3149 | Cel8C | RSGB_771 | + | Cytophaga hutchinsonii | ||
| 2013 | Cel9A | RSGB_459 | Xanthomonas axonopodis | |||
| 2361 | Cel9B | CelE (21, 32) | RSGB_564 | + | Pseudomonas sp. | |
| 2362 | Cel9C | CelDg (21) | Cytophaga hutchinsonii | |||
| 2558 | Cel9D | RSGB_618 | Vibrio parahaemolyticus | |||
| 2912 | Cel9E | RSGB_704,705 | Cytophaga hutchinsonii | |||
| 0134 | Cel9F | RSGB_018 | + | Bacillus pumilus | ||
| 0451 | Cel9G | EGBg (6) | Cytophaga hutchinsonii | |||
| 0809 | Cel9H | RSGB_149 | + | Clostridium cellulovorans | ||
| 0810 | Cel9I | RSGB_150, RSGB_151 | + | Acetivibrio cellulolyticus | ||
| 0257 | Cel10A | ClCBase (12) | RSGB_042 | Cytophaga hutchinsonii | ||
| 0226 | Lic16C | LicA (37) | + | Cytophaga hutchinsonii | ||
| 2181 | Man26B | RSGB_513 | + | Clostridium thermocellum | ||
| 1165 | Man26C | RSGB_231 | Piromyces sp. | |||
| 1166 | Man5B | RSGB_231 | Saccharophagus degradans | |||
| 2292 | Xyn10A | XynD (15) | RSGB_541 | Saccharophagus degradans | ||
| 2293 | Xyn10B | XynEg (15) | Cytophaga hutchinsonii | |||
| 2294 | Xyn10C | XynB (15) | RSGB_542 | Saccharophagus degradans | ||
| 2851 | Xyn10D | RSGB_686, RSGB_687 | Cytophaga hutchinsonii | |||
| 1195 | Xyn10F | RSGB_237 | + | Thermotoga neapolitana | ||
| 0777 | Xyn11C | XynCg (31) | Piromyces sp. | |||
| 2265 | Xyn30A | RSGB_532, RSGB_533 | Cytophaga hutchinsonii | |||
| 0206 | Xyn30B | RSGB_032 | Clostridium thermocellum | |||
| 1373 | Xyn39A | RSGB_298 | Clostridium acetobutylicum | |||
| 2263 | Xyn43B | RSGB_530 | Clostridium thermocellum | |||
| 2264 | Xyn43C | RSGB_531 | + | Clostridium thermocellum | ||
| 2520 | Xyn43G | RSGB_606 | Bacteroides thetaiotaomicron | |||
| 2622 | Xyn43J | + | Clostridium thermocellum | |||
| 0651 | Xyn8B | RSGB_123 | Cytophaga hutchinsonii | |||
| 0889 | Xyn8C | RSGB_172 | Cytophaga hutchinsonii |
The names of the genes as per the genome of F. succinogenes S85.
Previous names of the genes and references are provided where applicable.
Cellulase and xylanase genes identified in this study with F. succinogenes S85 as tester and F. intestinalis DR7 as driver that hybridized to DIG-labeled F. intestinalis genomic DNA under low-stringency conditions (hybridization at 37°C) but not under high-stringency conditions (hybridization at 60°C) in dot-Southern hybridization experiments. These genes are likely to be present in F. intestinalis but show low DNA sequence similarity to their counterparts in F. succinogenes. The clone names of the genes identified are shown.
Data are the same as described for column 4 in footnote c except that these genes did not hybridize to an F. intestinalis genomic DNA probe in either high- or low-stringency Southern blotting analysis, indicating they were absent from the genome of F. intestinalis DR7.
Cellulase and xylanase genes identified in F. intestinalis in the forward SSH experiment with F. intestinalis DR7 as tester and F. succinogenes S85 as driver (33). Although they had low similarity at the DNA sequence level, the amino acid sequences were quite similar to their counterparts in F. succinogenes.
The amino acid sequences encoded were used for BLAST searches of the NCBI nonredundant amino acid database; the non-Fibrobacter-genus organisms containing a glycoside hydrolase with the highest match score are listed.
These genes were not identified in either of the SSH experiments. However, they are included to give a complete list of the cellulase and xylanase genes reported to date.
Besides the glycoside hydrolase sequences, genes identified in the role of energy metabolism coded for proteins that are responsible for electron transport and fermentation. Among these, 8 of 13 NADH dehydrogenase I subunits, which were in a cluster (FSU2661 to FSU2673 [cluster A]), seemed either to be absent or to have diverged greatly in F. intestinalis (see Table S1 in the supplemental material). Genes encoding the subunits of succinate dehydrogenase (FSU3061, FSU3062, and FSU3070) were also identified.
Genes involved in transport and binding.
Fifty-five genes encoded proteins that are involved in transport and binding. These included 4 proteins that are responsible for amino acid, peptide, and amine transport, 6 involved in anion transport, 2 involved in carbohydrate transport, 20 responsible for cation and iron transport, and 21 other transporters of unknown substrate specificity.
Genes involved in DNA metabolism.
The genes involved in DNA metabolism included genes responsible for DNA replication, recombination, and repair. Furthermore, there was a hemolytic unit family DNA-binding protein that is known to be associated with the chromosome. However, no proteins responsible for DNA degradation were identified despite the previous characterization of an endonuclease in F. succinogenes S85 (20).
Genes involved in cell envelope metabolism.
The largest category of genes that differed from those in F. intestinalis encoded proteins for cell envelope metabolism. This included 6 genes involved in peptidoglycan biosynthesis and 35 involved in biosynthesis and degradation of surface polysaccharides and lipopolysaccharides (including at least 11 glycosyl transferases); remarkably, there were 68 genes annotated as lipoproteins with unknown functions.
Genes involved in the biosynthesis of cofactors, prosthetic groups, and carriers.
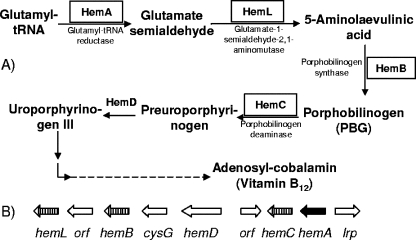
Twenty-three genes were identified that encoded proteins involved in the synthesis of cofactors such as biotin, folic acid, cobalamin (vitamin B12), ubiquinone, pantothenate, pyridoxine, and thiamine. Among these, a cluster of four genes that encoded enzymes responsible for cobalamin biosynthesis in F. succinogenes were identified (Fig. 2). Three of them, hemB (FSU0299), hemC (FSU0303), and hemL (FSU0297), were shown to also exist in F. intestinalis DR7, as documented by low-stringency Southern hybridization. However, the gene that encoded glutamyl-tRNA reductase (hemA) was not detected even at low stringency. Glutamyl-tRNA reductase catalyzes the conversion of glutamyl-tRNA to glutamate semialdehyde, which is known as the first step for cobalamin biosynthesis (34, 35) (Fig. 2). In addition to the cobalamin biosynthesis cluster, genes encoding biotin synthase (bioB; FSU1052) and para-aminobenzoate synthetase (pabB; FSU2014) were also identified.
FIG. 2.
(A) Vitamin B12 biosynthetic pathway and genes involved in the first five steps of biosynthesis. (B) Gene cluster for vitamin B12 biosynthesis in F. succinogenes. The genes highlighted by boxes (A) or cross-hatched arrows (B) are those with low homology to their respective counterparts in F. intestinalis. The hemA gene (filled arrow) was not detected under low-stringency Southern blotting conditions.
Growth of F. intestinalis DR7 and F. succinogenes in the absence of vitamin B12 and/or biotin.
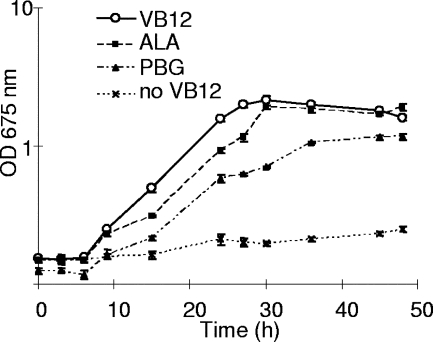
A supplementation experiment was carried out to determine whether the absence of a functioning hemA gene is the basis for the inability of F. intestinalis DR7 to synthesize vitamin B12 (Fig. 3). F. intestinalis DR7 was transferred from chemically defined medium to vitamin B12-free medium. After four subcultures in the latter medium (1:20 inoculum), F. intestinalis DR7 stopped growing. However, when the downstream product of glutamyl-tRNA reductase, 5-aminolaevulinic acid, or porphobilinogen was added to the vitamin B12-free medium, growth of F. intestinalis DR7 was restored. A similar experiment was done with F. succinogenes S85 except using biotin-free medium. After five subcultures were grown, growth under these conditions ceased as well. However, when F. intestinalis DR7 and F. succinogenes S85 cells were mixed (1:1, as determined by the OD675 value) and inoculated into the same medium without either vitamin B12 or biotin, the culture maintained growth even after 10 subcultures. The cells were harvested and subjected to PCR using both F. succinogenes and F. intestinalis DR7-specific primers and cells from the culture as the template. Both primer sets amplified DNA fragments with the corresponding sizes, indicating that the two strains coexisted in the culture and provided for cross-feeding of the missing intermediates.
FIG. 3.
Growth curve of F. intestinalis DR7 in vitamin B12-free medium containing 5-aminolaevulinic acid (ALA; -▪-) and porphobilinogen (PBG; -▴-); results from experiments using a negative-control vitamin B12-free medium (-×-) and a positive-control complete medium with vitamin B12 (VB12; -O-) are also shown. Cells were subcultured in vitamin B12-free medium three times before being inoculated into the respective media. The growth rates for media with VB12, ALA, and PBG were 0.171 h−1, 0.137 h−1, and 0.117 h−1, respectively. The means ± standard deviations are illustrated by error bars.
DISCUSSION
The strains F. intestinalis DR7 and F. succinogenes S85 were shown to have 92% sequence similarity in 16S rRNA (27). The present study suggested that approximately 41% of genes in F. succinogenes were either absent from or exhibited low similarity to those in F. intestinalis. Our previous study indicated that 33% of genes were specific to F. intestinalis DR7 (33). These results indicated that there are large differences between the genomes of the two species.
Despite the major differences between the DNA sequences of the two species, the cellulases and xylanases within F. succinogenes and F. intestinalis are well conserved. In our previous study (33), we reported that most of the genes identified in F. intestinalis exhibited greater homology to those in F. succinogenes than to those in other organisms, which also indicates the close relationship between the two species. This conclusion is further supported by this study in that most of the F. succinogenes cellulases and xylanases identified in this study hybridized to F. intestinalis total genomic DNA at low stringency. Recently, we characterized several glycoside hydrolases that demonstrated synergistic interactions with cellulose degradation (32). Homologues of these F. succinogenes enzymes, including the two major endoglucanases Cel9B (endoglucanase 1) and Cel51A (endoglucanase 2), a family 5 endoglucanase (Cel5H), a family 8 endoglucanase (Cel8B), and the chloride-stimulated cellobiosidase Cel10A were all identified in our SSH studies (Table 3 and reference 33). Synergistic interaction may occur with these F. intestinalis cellulases as well. The conservation of the cellulases in the two species emphasizes the unique nature of the glycoside hydrolase system of the Fibrobacter genus.
Adhesion of F. succinogenes cells to cellulose appears to be a prerequisite for cellulose hydrolysis. Recently, 13 cellulose-binding proteins, which are thought to be important for cellulose adhesion, were identified in this organism (16). Seven of these cellulose-binding proteins were found to be present in F. intestinalis (data not shown). These findings indicate that the cellulose adhesion mechanism of the two species may be conserved as well.
Many of the glycoside hydrolases identified in F. succinogenes have highest similarity to genes of Cytophaga hutchinsonii, Clostridium thermocellum, and Saccharophagus degradans, which belong to three distinct phyla, Bacteroides, Firmicutes, and Proteobacteria, respectively.
Besides the 16S rRNA sequence and genomic content differences, there are several known phenotypic differences between the two species (27, 28). These differences include the vitamin requirements of different strains. The SSH experiment identified four genes in a cobalamin biosynthesis gene cluster that either are missing from or have diverged greatly in F. intestinalis DR7. Figure 2 shows the cobalamin biosynthetic pathway that has been found in many bacteria. Cobalamin is involved as a cofactor in a variety of enzymatic reactions and is synthesized by some bacteria and archaea (35). In the genome of F. succinogenes, the gene cluster responsible for uroporphyrinogen III biosynthesis and several genes in heme biosynthesis pathway were identified (Fig. 2), addressing the capability of F. succinogenes to synthesize vitamin B12 via this pathway. The genes encoding glutamyl tRNA reductase (hemA), glutamate-1-semialdehyde-2,1-aminomutase (hemL), porphobilinogen synthase (hemB), and porphobilinogen deaminase (hemC) were found to be missing from or to have greatly diverged in F. intestinalis DR7. At low stringency, hemA did not hybridize with the F. intestinalis genomic DNA, which indicates that it might be absent from the genome. The missing genes would cause a block in synthesis of uroporphyrinogen III in F. intestinalis, which is the precursor of cobalamin (35). This finding was supported by restoration of growth of F. intestinalis by inclusion of either 5-aminolaevulinic acid or porphobilinogen in the medium. Interestingly, addition of porphobilinogen did not fully restore F. intestinalis growth, which may be due to the lack of a specific transporter(s) or permease.
A large number of genes that differed from those in F. intestinalis encoded proteins for cell envelope metabolism. In addition, many genes involved in biosynthesis and biodegradation of cell surface polysaccharides and lipopolysaccharides were also identified in this and previous studies (33). These suggested substantial differences in the surface structure of the two species.
NADH:ubiquinone oxidoreductase complex I is the first complex of the respiratory chain which provides the proton motive force required for energy-consuming processes such as the synthesis of ATP (10). There are two sets of NADH dehydrogenase I subunits that form clusters (cluster A, FSU2661 to FSU2674; cluster B, FSU2886 to FSU2895) in the genome of F. succinogenes (29). The two clusters were on two different strands of the genome and were separated by approximately 200 kb. Interestingly, all eight genes that encode subunits B, C, D, E, G, H, I, and M as identified by SSH belonged to the same cluster (cluster A) whereas no genes in cluster B were identified. This may indicate that the genes in cluster B are highly conserved within the members of the Fibrobacter genus.
Our present and previous SSH experiments demonstrated that SSH is an effective approach for identification of species-specific genes in the Fibrobacter genus. The strain-specific sequences identified may account for the different niches occupied by the two bacterial strains. The high level of similarity of cellulases and hemicellulases identified in F. intestinalis to those in F. succinogenes indicates that bacteria in the Fibrobacter genus probably share similar mechanisms of plant cell wall degradation. Finally, studies of the proteins with unknown functions, especially those conserved in the Fibrobacter genus but with no homology to those of other organisms, will also help us to understand additional features of this unique phylum.
Supplementary Material
Acknowledgments
This research was supported by the Initiative for Future Agriculture and Food Systems grant 2000-52100-9618 from USDA-CSREES awarded to the North American Consortium for Genomics of Fibrolytic Ruminal Bacteria and by the Natural Science and Engineering Research Council of Canada. The Fibrobacter succinogenes genome sequencing project was conducted at The Institute for Genomic Research (now the J. Craig Venter Institute) and was also supported by the USDA-CSREES funds listed above.
Footnotes
Published ahead of print on 21 December 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahn, S. J., J. Costa, and J. R. Emanuel. 1996. PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Res. 24:2623-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., C. H. Lin, R. Key, L. Montgomery, and D. A. Stahl. 1992. Diversity among Fibrobacter isolates—towards a phylogenetic classification. Syst. Appl. Microbiol. 15:23-31. [Google Scholar]
- 5.Reference deleted.
- 6.Béra-Maillet, C., V. Broussolle, P. Pristas, J. P. Girardeau, G. Gaudet, and E. Forano. 2000. Characterisation of endoglucanases EGB and EGC from Fibrobacter succinogenes. Biochim. Biophys. Acta 1476:191-202. [DOI] [PubMed] [Google Scholar]
- 7.Béra-Maillet, C., Y. Ribot, and E. Forano. 2004. Fiber-degrading systems of different strains of the genus Fibrobacter. Appl. Environ. Microbiol. 70:2172-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsberg, C. W., E. Forano, and A. Chesson. 2000. Microbial adherence to plant cell wall and enzymatic hydrolysis, p. 79-88. In P. B. Cronje (ed.), Ruminant physiology digestion, metabolism, growth and reproduction. CABI Publishing, Wallingford, Oxfordshire, United Kingdom.
- 10.Friedrich, T., and D. Scheide. 2000. The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 479:1-5. [DOI] [PubMed] [Google Scholar]
- 11.Gong, J. H., E. E. Egbosimba, and C. W. Forsberg. 1996. Cellulose binding proteins of Fibrobacter succinogenes and the possible role of a 180-kDa cellulose-binding glycoprotein in adhesion to cellulose. Can. J. Microbiol. 42:453-460. [Google Scholar]
- 12.Huang, L., C. W. Forsberg, and D. Y. Thomas. 1988. Purification and characterization of a chloride-stimulated cellobiosidase from Bacteroides succinogenes S85. J. Bacteriol. 170:2923-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyo, A. H., and C. W. Forsberg. 1994. Features of the cellodextrinase gene from Fibrobacter succinogenes S85. Can. J. Microbiol. 40:592-596. [DOI] [PubMed] [Google Scholar]
- 14.Iyo, A. H., and C. W. Forsberg. 1996. Endoglucanase G from Fibrobacter succinogenes S85 belongs to a class of enzymes characterized by a basic C-terminal domain. Can. J. Microbiol. 42:934-943. [DOI] [PubMed] [Google Scholar]
- 15.Jun, H. S., J. K. Ha, L. M. Malburg, Jr., G. A. Verrinder, and C. W. Forsberg. 2003. Characteristics of a cluster of xylanase genes in Fibrobacter succinogenes S85. Can. J. Microbiol. 49:171-180. [DOI] [PubMed] [Google Scholar]
- 16.Jun, H. S., M. Qi, J. Gong, E. E. Egbosimba, and C. W. Forsberg. 2007. Outer membrane proteins of Fibrobacter succinogenes with potential roles in adhesion to cellulose and in cellulose digestion. J. Bacteriol. 189:6806-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kam, D. K., H. S. Jun, J. K. Ha, G. D. Inglis, and C. W. Forsberg. 2005. Characteristics of adjacent family 6 acetylxylan esterases from Fibrobacter succinogenes and the interaction with the Xyn10E xylanase in hydrolysis of acetylated xylan. Can. J. Microbiol. 51:821-832. [DOI] [PubMed] [Google Scholar]
- 18.Lin, C., and D. A. Stahl. 1995. Comparative analyses reveal a highly conserved endoglucanase in the cellulolytic genus Fibrobacter. J. Bacteriol. 177:2543-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludwig, W., and K. H. Schleifer. 2001. Overview: a phylogenetic backbone and taxonomic framework for prokaryotic systematics, p. 49-65. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology. Springer-Verlag, Berlin, Germany. (In German.)
- 20.MacLellan, S. R., and C. W. Forsberg. 2001. Properties of the major non-specific endonuclease from the strict anaerobe Fibrobacter succinogenes and evidence for disulfide bond formation in vivo. Microbiology 147:315-323. [DOI] [PubMed] [Google Scholar]
- 21.Malburg, L. M., Jr., A. H. Iyo, and C. W. Forsberg. 1996. A novel family 9 endoglucanase gene (celD), whose product cleaves substrates mainly to glucose, and its adjacent upstream homolog (celE) from Fibrobacter succinogenes S85. Appl. Environ. Microbiol. 62:898-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malburg, S. R., L. M. Malburg, Jr., T. Liu, A. H. Iyo, and C. W. Forsberg. 1997. Catalytic properties of the cellulose-binding endoglucanase F from Fibrobacter succinogenes S85. Appl. Environ. Microbiol. 63:2449-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matheron, C., A. M. Delort, G. Gaudet, and E. Forano. 1998. In vivo 13C NMR study of glucose and cellobiose metabolism by four cellulolytic strains of the genus Fibrobacter. Biodegradation 9:451-461. [DOI] [PubMed] [Google Scholar]
- 24.McGavin, M., and C. W. Forsberg. 1988. Isolation and characterization of endoglucanases 1 and 2 from Bacteroides succinogenes S85. J. Bacteriol. 170:2914-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGavin, M. J., C. W. Forsberg, B. Crosby, A. W. Bell, D. Dignard, and D. Y. Thomas. 1989. Structure of the cel-3 gene from Fibrobacter succinogenes S85 and characteristics of the encoded gene product, endoglucanase 3. J. Bacteriol. 171:5587-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miron, J., and C. W. Forsberg. 1999. Characterisation of cellulose-binding proteins that are involved in the adhesion mechanism of Fibrobacter intestinalis DR7. Appl. Microbiol. Biotechnol. 51:491-497. [DOI] [PubMed] [Google Scholar]
- 27.Montgomery, L., B. Flesher, and D. Stahl. 1988. Transfer of Bacteroides succinogenes (Hungate) to Fibrobacter gen. nov. as Fibrobacter succinogenes comb. nov. and description of Fibrobacter intestinalis sp. nov. Int. J. Syst. Bacteriol. 38:430-435. [Google Scholar]
- 28.Montgomery, L., and J. M. Macy. 1982. Characterization of rat cecum cellulolytic bacteria. Appl. Environ. Microbiol. 44:1435-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison, M., K. E. Nelson, I. K. O. Cann, C. W. Forsberg, R. I. Mackie, J. B. Russell, B. A. White, K. Amava, B. Cheng, M. Qi, H. Jun, S. Mulligan, K. Tran, H. A. Carty, H. Khouri, W. Nelson, S. Daugherty, and C. M. Fraser. 2003. The Fibrobacter succinogenes strain S85 genome sequencing project. Abstr. 3rd ASM-TIGR Conf. Microb. Genomes, p. 33.
- 30.Nesbø, C. L., K. E. Nelson, and W. F. Doolittle. 2002. Suppressive subtractive hybridization detects extensive genomic diversity in Thermotoga maritima. J. Bacteriol. 184:4475-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paradis, F. W., H. Zhu, P. J. Krell, J. P. Phillips, and C. W. Forsberg. 1993. The xynC gene from Fibrobacter succinogenes S85 codes for a xylanase with two similar catalytic domains. J. Bacteriol. 175:7666-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi, M., H. S. Jun, and C. W. Forsberg. 2007. Characterization and synergistic interactions of Fibrobacter succinogenes glycoside hydrolases. Appl. Environ. Microbiol. 73:6098-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi, M., K. E. Nelson, S. C. Daugherty, W. C. Nelson, I. R. Hance, M. Morrison, and C. W. Forsberg. 2005. Novel molecular features of the fibrolytic intestinal bacterium Fibrobacter intestinalis not shared with Fibrobacter succinogenes as determined by suppressive subtractive hybridization. J. Bacteriol. 187:3739-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raux, E., H. L. Schubert, and M. J. Warren. 2000. Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell. Mol. Life Sci. 57:1880-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth, J. R., J. G. Lawrence, and T. A. Bobik. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50:137-181. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY.
- 37.Teather, R. M., and J. D. Erfle. 1990. DNA sequence of a Fibrobacter succinogenes mixed-linkage β-glucanase (1,3-1,4-β-d-glucan 4-glucanohydrolase) gene. J. Bacteriol. 172:3837-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson, K. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl, Current protocols in molecular biology. Greene Publishing Associates and John Wiley & Sons, Inc., New York, NY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.