Abstract
The changes in bacterial communities associated with the marine sponge Mycale laxissima on transfer to aquaculture were studied using culture-based and molecular techniques. M. laxissima was maintained alive in flowthrough and closed recirculating aquaculture systems for 2 years and 1 year, respectively. The bacterial communities associated with wild and aquacultured sponges, as well as the surrounding water, were assessed using 16S rRNA gene clone library analysis and denaturing gradient gel electrophoresis (DGGE). Bacterial richness and diversity were measured using DOTUR computer software, and clone libraries were compared using S-LIBSHUFF. DGGE analysis revealed that the diversity of the bacterial community of M. laxissima increased when sponges were maintained in aquaculture and that bacterial communities associated with wild and aquacultured M. laxissima were markedly different than those of the corresponding surrounding water. Clone libraries of bacterial 16S rRNA from sponges confirmed that the bacterial communities changed during aquaculture. These communities were significantly different than those of seawater and aquarium water. The diversity of bacterial communities associated with M. laxissima increased significantly in aquaculture. Our work shows that it is important to monitor changes in bacterial communities when examining the feasibility of growing sponges in aquaculture systems because these communities may change. This could have implications for the health of sponges or for the production of bioactive compounds by sponges in cases where these compounds are produced by symbiotic bacteria rather than by the sponges themselves.
Marine sponges have been recognized as hosts for many microorganisms. Sponges are filter feeders; numerous tiny pores on the surface allow water to enter and circulate through a series of aquiferous channels where microorganisms and organic matter are filtered out (27). In the presence of appropriate growth conditions, microorganisms that can resist the sponge digestive process and immune response may successfully colonize the sponge. Microorganisms can constitute up to 60% of the sponge biomass (21, 63, 69). In addition to those bacteria serving as a food source, the sponge microbial community is comprised of a transient seawater population that is coincidentally present in the sponge, microbes that grow in the mesohyl, and symbionts that live inside the sponge cells. Sponge-microbe associations involve a diverse range of heterotrophic bacteria, cyanobacteria, facultative anaerobes, unicellular algae, and archaea (19-21, 33, 57, 60, 65). Sponge-bacteria interactions are widely distributed and sometimes host specific (57). The large numbers of bacteria within sponges and the specific nature of some of these relationships strongly suggest that symbiotic interactions exist between sponges and microorganisms. Several studies have shown the presence of common microbial communities between different sponge species from different geographic regions (16, 19, 33, 70). Recently, Enticknap and coworkers reported the presence of a group of closely related alphaproteobacteria affiliated with Pseudovibrio denitrificans in seven genera of marine sponges from several geographic locations (11).
Marine sponges are sessile invertebrates that have developed effective strategies to protect themselves against viruses, bacteria, and eukaryotic predators. One of these defense mechanisms is the production of secondary metabolites (48, 52). Sponges are known as prolific sources of bioactive compounds that can potentially be used to treat various human diseases (13, 15). In some cases, the primary producers of bioactive compounds are symbiotic microorganisms hosted by marine sponges (14, 25, 46). The low yield of metabolites originating from marine sponges is one of the major obstacles for the completion of clinical studies and the development of promising compounds and has been termed the “supply problem” (36, 41). Careful harvesting of sponges from the marine environment without damaging the wild population is a possible option for rapidly growing and abundant species, but not for rare species. Chemical synthesis is not an option for many marine natural compounds due to their structural complexity. When compounds of interest are produced by bacterial symbionts rather than by the sponges, it may be possible in some cases to isolate producer microbes and ensure an economic, sustainable supply of compounds by growing the microbes in fermentation systems. If this is not achievable, culturing of the entire sponge and its microbial consortium in aquaculture systems is another option. In situ cultivation of marine sponges (mariculture) and cell culture approaches have been explored as possibilities for large-scale production of sponge-derived compounds (A. Duckworth, presented at World Aquaculture, Raleigh, NC, 14 to 18 June 2007; 34, 36, 40, 55, 64, 71). One promising strategy is the ex situ culture of sponges in closed or semiclosed systems. Aquaculture in tanks might be preferable to in situ mariculture because it is reliable and provides the possibility of switching from seasonal growth to continuous growth during the year (8, 55). Also, maintenance of marine sponges in aquaculture provides a potential model to study sponge-microbe interactions. However, very little is known about the optimal environmental conditions and ecological needs of sponges, and this makes the optimization of sponge growth and compound production a difficult process (9, 41, 64).
In order to examine the potential of ex situ cultivation of marine sponges as a solution for the supply problem, it is crucial to determine whether the microbial communities change upon culturing. Mycale laxissima was chosen as our model sponge due to its high capability of adapting to aquaculture conditions compared to this ability in other sponges from the same reef environment examined in preliminary trials. Also, M. laxissima is a representative of the genus Mycale that is of considerable interest as a source of metabolites with a wide range of bioactivities, including significant cytotoxic, antiviral, antitumor, and antimitotic activities (18, 24, 30, 31, 38, 39, 43-45, 49, 61, 62, 66, 67). A number of these cytotoxins have been isolated from Mycale hentscheli sponges (3) from New Zealand waters. Perloruside A, the most recent cytotoxic metabolite, was isolated from M. hentscheli sponges collected from the North coast of the South Island. It is a microtubule stabilizer with a potency and mode of action similar to those of the major anticancer drug paclitaxel (Taxol) and the epothilones (42).
The first objective of this study was to characterize the microbial communities associated with wild and aquacultured M. laxissima sponges using culture-dependent and molecular techniques. The second objective was to compare the bacterial communities of wild sponges to those in aquaculture systems. The following questions were addressed. (i) Is there a sponge-specific community in M. laxissima sponges that is different from the bacterioplankton community in the water column? (ii) Does the transfer of the sponges into aquaculture cause a change in the bacterial community?
MATERIALS AND METHODS
Sponge collection and taxonomic identification.
Individual M. laxissima sponges were collected by scuba diving at Conch Reef, Key Largo, FL, in July 2001 and June 2004 in water depths of ca. 15 m (latitude 24° 57.11′ N, longitude 80° 27.57′ W). The water salinity was 36 ppt, and the temperature was 26.7°C. Chemically characterized voucher specimens were registered with the Natural History Museum (formerly the British Museum of Natural History) (11). Water samples were collected near the sponges at a depth of ca. 15 m in sterile 20-liter containers, and 15 to 20 liters were filtered through 0.22-μm-pore-size Sterivex filters (Millipore) for each water sample. The Sterivex filters were frozen immediately and stored at −20°C for isolation of nucleic acids. The sponge samples and Sterivex filters were transferred to Baltimore, MD, on dry ice and stored at −80°C. The sponges collected for maintenance in aquaculture were transported in containers filled with aerated seawater. The water (50% volume) was changed every 4 to 6 h and was kept aerated by battery-operated air pumps and airstones.
Sponge aquaculture.
Two different aquaculture systems were designed.
(i) Flowthrough system.
A closed flowthough system was constructed using a 360-liter head tank and three 80-liter fish tanks fitted with small external filters and siphons (see Fig. S1 in the supplemental material). The flow rate was controlled by using a valve set to 4 liters per h, giving a turnover rate of 2.4 times every 24 h.
The constant influx of new sterile artificial seawater and removal of old salt water helps maintain water quality. Sponges were fed the microalga Nanochloropsis sp. with the addition of 40 ml of a 4 × 106 cell ml−1 culture every 2 to 3 days. Four sponge individuals were collected in 2001. One was processed as a wild sponge, one was kept in this aquaculture system for 6 months, and two were kept in the aquaculture system for 2 years. The health of the aquacultured sponges was monitored visually during this period, and digital images were taken. After 6 months, one individual sponge was sacrificed and processed immediately for microbiology. Specimens were stored at −80°C for the isolation of nucleic acids. The procedure was repeated for the two remaining sponges after 2 years in the aquaculture system. At 2 years, 4 liters of the water were filtered through Sterivex filters for the isolation of nucleic acids (designated AW03).
(ii) Recirculating system.
A large-scale recirculation aquaculture system was designed and constructed to house the 2004 sponge collection (see Fig. S1 in the supplemental material). The 800-liter system was constructed using four 80-liter tanks and two 160-liter tanks. These tanks were drilled with a 1.5-cm hole about 9.5 cm from the bottom, and bulkheads were attached. Bulkheads were connected with 1.5-cm polyvinyl chloride pipe to allow water to drain from the tanks. These pipes emptied into a sump where the waste passed through a 100-μm-mesh bag filter. A protein skimmer was installed in the sump to remove organic material from the water. From the sump, the water was pumped into a biofilter and an algal turf scrubber. The system had actinic and 10,000K lights set on a normal daily light regime. Five sponges were collected in 2004. Three were processed as wild sponges. Two were maintained in the recirculating aquaculture system for 3 months, and their health condition was inspected visually during this period.
Sponge processing for isolation of culturable bacteria.
Immediately after the sponges were collected, samples were rinsed thoroughly three times with sterile artificial seawater to remove any transient bacteria, algae, or mucus attached to the surface of the sponge. Sponge tissue (1 cm3) was ground in artificial seawater, and 10-fold serial dilutions were plated on Difco marine agar 2216 (BD Biosciences, Franklin Lakes, NJ). The plates were incubated at 30°C for a week. Serial dilutions of water samples were processed similarly for bacterial isolation.
Identification of isolates by 16S rRNA gene sequence analysis.
One representative of each bacterial morphotype was selected from each sample for further purification and sequencing. Single pure colonies of each isolate were transferred to 20 ml of marine broth 2216 (BD BioSciences) and incubated overnight at 30°C in a shaking incubator at 100 rpm. DNA was extracted from these isolates by using an Ultra-Clean microbial kit (MoBio Laboratories, Carlsbad, CA). Isolates were stored at −80°C in marine broth 2216 supplemented with 30% glycerol. The 16S rRNA gene was PCR amplified using universal primers 27F and 1492R (26) as described by Enticknap et al. (11).
DNA extraction from sponges and surrounding water samples.
Freeze-dried sponge tissue (1 cm3) was ground using a sterile mortar and pestle. Total genomic DNA was extracted using the method described by Pitcher et al. (47). The protocol was modified for sponge tissues (12). DNA was extracted from the filters obtained from seawater and aquarium water samples using the protocol described by Somerville et al. (56).
Denaturing gradient gel electrophoresis (DGGE) of bacterial communities.
A 195-bp region corresponding to positions 341 and 534 in the 16S rRNA gene of Escherichia coli was PCR amplified from genomic DNA extracted from sponges and water samples using P2 and P3 primers (37). DGGE was performed by using a DCode system (Bio-Rad, Hercules, CA) on a 6% (wt/vol) polyacrylamide gel with a denaturing gradient of 40 to 70% in 1× Tris-acetate-EDTA. Electrophoresis was performed for 17 h at 60 V at 60°C. Gels were stained in a staining bath of Sybr green in 1× Tris-acetate-EDTA and visualized with a Typhoon 9410 image system (Amersham Biosciences, Piscataway, NJ).
PCR amplification of genomic DNA, cloning, and sequencing.
16S rRNA gene fragments were PCR amplified from the total genomic DNA isolated from sponge and water samples using the same protocol described for culturable isolates. Cycling conditions were as described previously, but the PCR was terminated after 15, 20, 25, or 30 cycles, with 30 cycles for the negative control sample (11). Amplification products were visualized by agarose gel electrophoresis. Visible bands of approximately 1,500 bp from the reactions with the least number of cycles were cut and gel purified. Corresponding areas from the negative control samples were excised and taken through the cloning procedure to provide strict negative controls. Purified PCR products were ligated into the pCR-XL-TOPO vector and transformed into OneShot TOP 10 chemically competent Escherichia coli cells using a TOPO XL PCR cloning kit (Invitrogen Life Technologies, Carlsbad, CA). Plasmid DNA was isolated from individual clones and purified using a SprintPrep 384 HC kit (Agencourt Bioscience, Beverly, MA). Sequencing was done using an ABI PRISM 3130xl genetic analyzer (Applied Biosystems, Foster City, CA) and M13 forward and reverse sequencing primers.
Phylogenetic analysis.
16S rRNA gene sequences from isolates were analyzed by using the BLASTn tool at the National Center for Biotechnology Information website. Isolates were presumptively identified according to the identity of the closest cultured relative in the top BLAST hits. 16S rRNA gene sequences from clone libraries were edited using PreGap4 and Gap4 from the Staden package and analyzed initially by using the BLAST tool to aid in the selection of the closest reference sequences. Chimeric sequences were identified by using the CHECK_CHIMERA program of the Ribosomal Database Project (29). Phylogenetic analyses of clone libraries were performed by using the ARB software package (28), and sequences were aligned by using the positional tree server with a data set containing the nearest relative matches. Trees were constructed using the neighbor-joining (Jukes-Cantor correction) (50) algorithms implemented in ARB. The robustness of the inferred tree topologies was evaluated after 1,000 bootstrap replicates of the neighbor-joining data. Bootstrap values were generated using Phylip version 3.6 (J. Felsenstein, Department of Genetics, University of Washington, Seattle, WA).
Statistical analyses of clone libraries and estimation of microbial diversity.
S-LIBSHUFF version 1.22 was used to compare libraries statistically (54). It compares more than two libraries at once with the same distance matrix to determine whether two libraries were drawn from the same population. DOTUR (distance-based operational taxonomic unit [OTU] and richness) version 1.53 (53) was used to assign sequences to OTUs and to calculate collector's curves for observed unique OTUs, Chao1, and ACE (abundance-based coverage estimator) richness estimators. The Shannon and Simpson's diversity indices were also calculated (22). Rarefaction analysis was done to determine the number of observed OTUs as a function of the distance between sequences and the number of sequences sampled. The rarefaction curve data were obtained by using DOTUR.
Nucleotide sequence accession numbers.
16S rRNA gene sequences from isolates were submitted to GenBank under accession no. EF629829 to EF629882. 16S rRNA gene sequences from clone libraries were submitted to GenBank under accession no. EF629883 to EF630353 and EU340240.
RESULTS
Maintenance of M. laxissima sponges in two aquaculture systems.
Individual M. laxissima sponges were successfully maintained in two aquaculture systems, a flowthrough system and a closed recirculating system (see Fig. S1 in the supplemental material). Sponge health was assessed visually by observing size, color, and the appearance of necrotic spots. No significant growth of sponges was observed in either aquaculture system. Sponges maintained integrity and showed no necrosis or fouling, although there was some color change from black to gray, possibly indicating a loss of dark-pigmented cyanobacteria. Sponges remained viable throughout the study period, as shown by a sponge cell aggregation assay (1, 4, 35). Manually dispersed sponge tissue reaggregated spontaneously when the sponge cells were alive. Digital images were taken routinely.
DGGE.
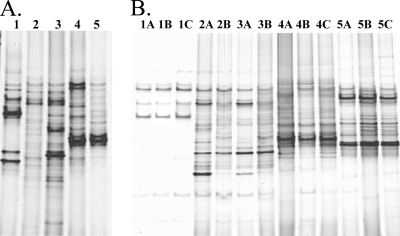
DGGE analysis revealed that the bacterial communities varied substantially between wild sponges and surrounding seawater and between aquacultured sponges and aquarium water under both aquaculture conditions (Fig. 1). DGGE banding patterns were generally consistent between sponges sampled at the same time point. The marked differences in overall DGGE patterns indicate that the bacterial communities associated with M. laxissima were clearly different from those in the surrounding water. A few DGGE bands were shared between both sponge and water samples, suggesting the commonality of some bacteria. The diversity of the microbial community associated with M. laxissima, assessed by the number of bands present in DGGE, increased in both aquaculture systems.
FIG. 1.
DGGE fingerprints of the bacterial communities associated with M. laxissima individual sponges from the following sources. (A) Flowthrough aquaculture system and a wild sponge. Lanes: wild sponge (1) collected at the same time as the 6-month-aquacultured (2) and 2-year-aquacultured (3) sponges and 2001 seawater (4), and 2004 seawater (5) samples. (B) Recirculating system and wild sponges. Lanes: wild individuals (1A to C) collected at the same time as the 1-month- (2A and B) and 3-month-aquacultured (3A and B) sponges, seawater samples from the surrounding vicinity of freshly collected M. laxissima sponges (4A to C), and aquarium water samples from the recirculating system (5A to C). The denaturing gradient was from 40% to 70%.
Phylogenetic analysis of 16S rRNA gene clone libraries.
In order to determine the stability of the microbial communities upon transfer of the M. laxissima into aquaculture, seven 16S rRNA gene clone libraries were generated. One clone library from a representative sponge was constructed at each selected time point, described in detail below. This was based on the general consistency of DGGE banding patterns between individuals sampled at the same time point (Fig. 1). Chimeric clones were excluded from the analysis. DOTUR was used to assign sequences to OTUs based on the genetic distance between sequences.
(i) Flowthrough system.
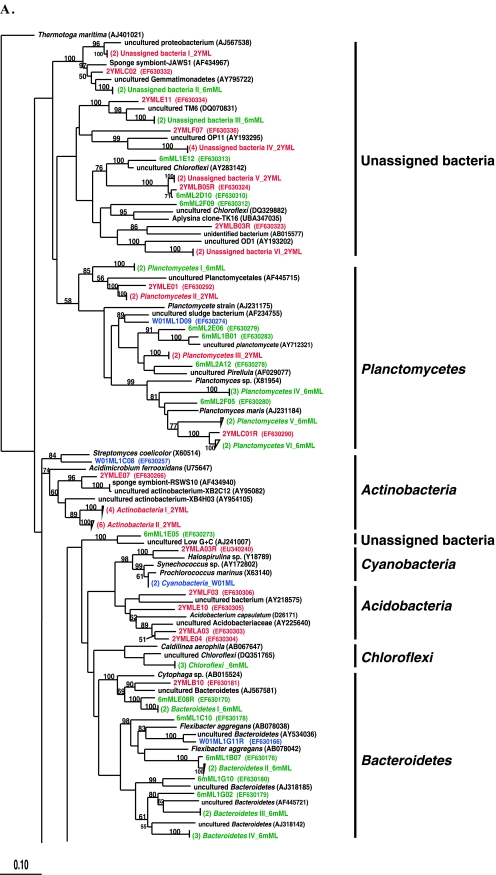
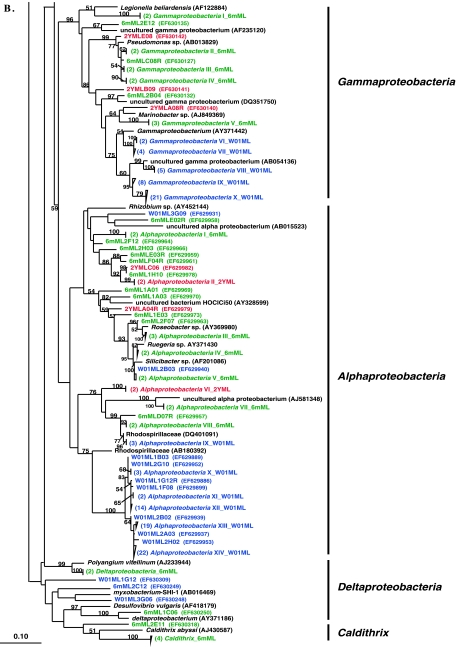
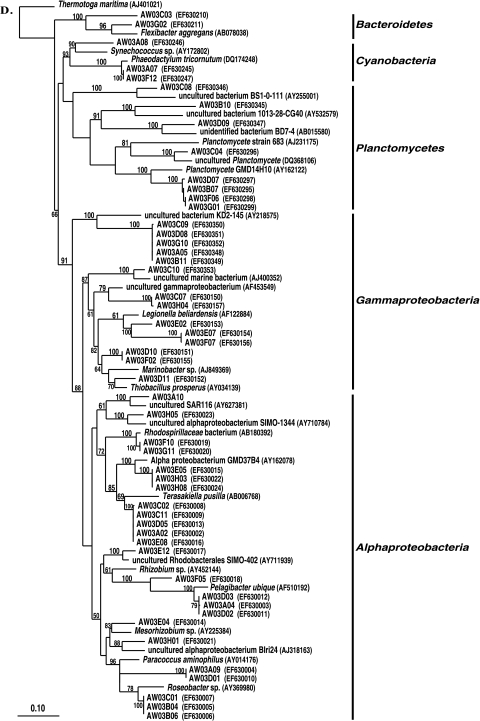
In this system, 6-month- and 2-year-aquacultured M. laxissima sponges were compared to M. laxissima sponges collected from the wild in 2001. Clone libraries, designated W01ML and 6mML, were constructed from one wild sponge and one sponge maintained in aquaculture for 6 months, respectively. Two sponge samples were processed at the 2-year time point for DGGE analysis and culturing of isolates. However, a clone library (designated 2YML) was constructed from the bacterial community in only one sponge, judged to be representative of communities in both sponges based on the similarity of the DGGE banding patterns (data not shown). A total of 119 16S rRNA gene clones were analyzed from the wild sponge. This generated 67 unique OTUs which fell within seven bacterial phyla (Alpha-, Gamma-, and Deltaproteobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, and Planctomycetes). A total of 85 16S rRNA gene clones were analyzed from the 6-month-aquacultured sponge. This generated 75 unique OTUs, which fell into eight phyla (Alpha-, Gamma-, and Deltaproteobacteria, Bacteroidetes, Caldithrix, Chloroflexi, Planctomycetes, and unassigned bacteria). A total of 47 16S rRNA gene clones were analyzed from the 2-year-aquacultured sponge. This generated 35 unique OTUs, which fell into eight phyla (Alpha- and Gammaproteobacteria, Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Planctomycetes, and unassigned bacteria). A phylogenetic tree showing the relationships between sequences from these three libraries is shown in Fig. 2A and B.
FIG. 2.
Rooted neighbor-joining phylogenetic tree of partial 16S rRNA gene sequences of clones that were recovered from the flowthrough system and a wild sponge, including the wild sponge (prefixed W01ML, presented in blue) collected at the same time as the 6-month-aquacultured (prefixed 6mML, presented in green) and 2-year-aquacultured (2YML, presented in red) sponges (A and B), seawater sample collected in the vicinity of wild sponges at Key Largo (prefixed WW01) (C), and water from the flowthrough aquaculture system (prefixed AW03) (D). Bootstrap confidence values of >50% are shown at the nodes. The tree was constructed using ARB. The tetragons represent the clones that are >99.5% similar (see Table S1 in the supplemental material); the numbers listed in bold before the group names indicate the numbers of clones. Thermatoga maritima (NCBI accession no. AJ401021) was used as the outgroup in the analysis. Scale bar indicates 0.10 substitutions per nucleotide position. Reference sequences are shown with GenBank accession numbers listed after each sequence name. Major bacterial groups found in the three libraries are indicated in bold on the right-hand side of the tree.
A total of 38 16S rRNA gene clones were analyzed from the seawater, generating 31 unique OTUs which fell into eight phyla (Alpha- and Gammaproteobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Planctomycetes, uncultured TM7, and Verrucomicrobia). A phylogenetic tree showing sequences from the seawater library is shown in Fig. 2C. A total of 51 16S rRNA gene clones were analyzed from the aquarium water, generating 40 unique OTUs which fell within five phyla (Alpha- and Gammaproteobacteria, Bacteroidetes, Cyanobacteria, and Planctomycetes). A phylogenetic tree showing sequences from the aquarium water library is shown in Fig. 2D.
(ii) Recirculating system.
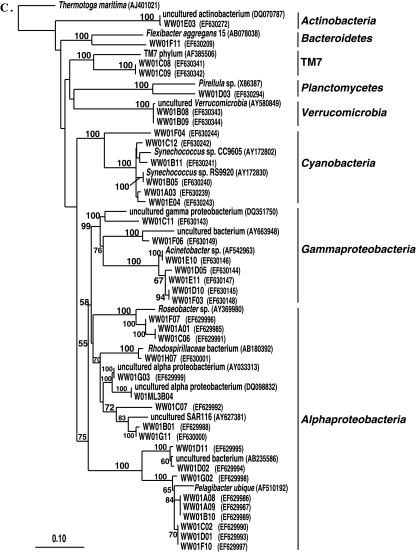
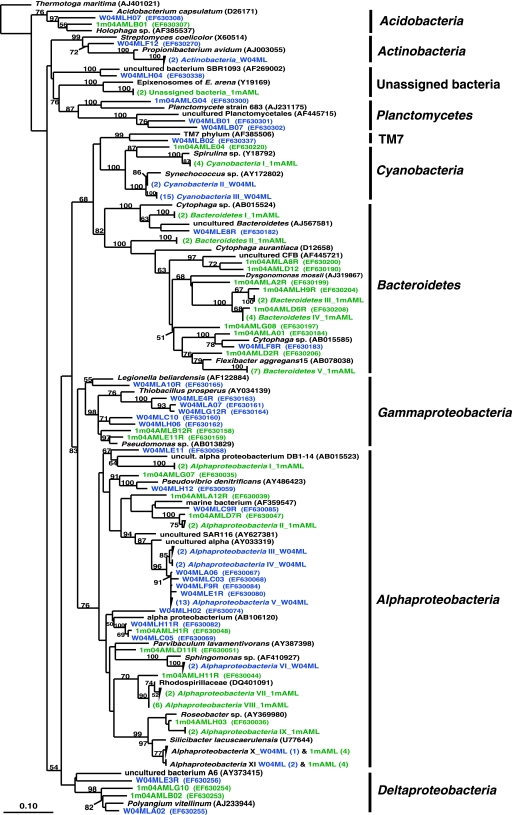
Two sponges were compared in this analysis, an M. laxissima sponge collected from the wild in 2004 and an M. laxissima sponge maintained in the recirculating aquaculture system for 1 month. A clone library designated 1m04AML was constructed from a sample of one representative sponge maintained for 1 month in aquaculture. This sample was regarded as a representative sample based on the similarity of the DGGE banding patterns of the two 1-month-aquacultured sponges (Fig. 1B). A total of 67 16S rRNA gene clones were analyzed from the wild sponge. This generated 59 unique OTUs which fell into 10 bacterial phyla (Alpha-, Gamma-, and Deltaproteobacteria, Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Planctomycetes, uncultured TM7, and unassigned bacteria). An equal number of 16S rRNA gene clones from the 1-month-aquacultured sponge were analyzed. This generated 52 unique OTUs which fell into eight phyla (Alpha-, Gamma-, and Deltaproteobacteria, Acidobacteria, Bacteroidetes, Cyanobacteria, Planctomycetes, and unassigned bacteria). A phylogenetic tree showing comparisons of sequences from these two libraries is shown in Fig. 3. The distribution of OTUs within the major phylogenetic groups detected in sponges from both aquaculture systems is shown in Fig. 4.
FIG. 3.
Rooted neighbor-joining phylogenetic tree of partial 16S rRNA gene sequences of clones that were recovered from wild sponges (prefixed W04ML, shown in blue) collected at the same time as 1-month-aquacultured sponges (prefixed 1m04AML, shown in green) from the recirculating aquaculture system. Bootstrap confidence values of >50% are shown at the nodes. The tree was constructed using ARB. The tetragons represent clones that are >99.5% similar (see Table S2 in the supplemental material); the numbers listed in bold before the group names indicate the numbers of clones. Thermatoga maritima was used as the outgroup in the analysis. Scale bar indicates 0.10 substitutions per nucleotide position. Reference sequences are shown with GenBank accession numbers listed after each sequence name. Major bacterial groups found in both libraries are indicated in bold on the right-hand side of the tree.
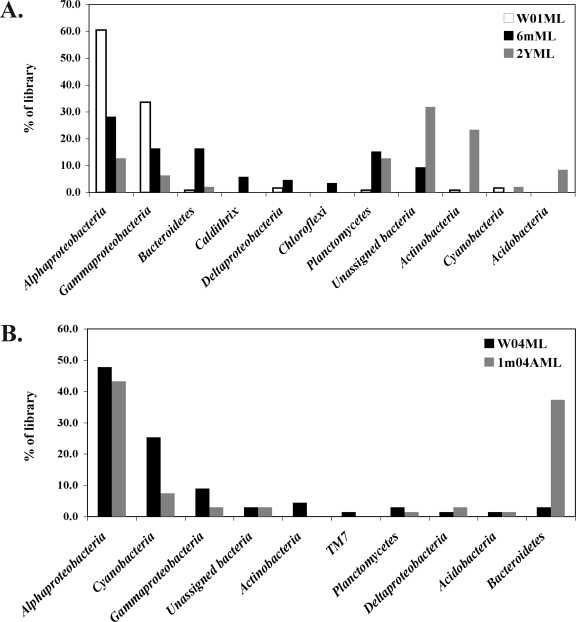
FIG. 4.
Distribution of bacterial 16S rRNA gene sequences within the phylogenetic groups detected in the clone libraries from individual M. laxissima sponges from both aquaculture systems. (A) Flowthrough system. Wild M. laxissima sponges (W01ML) were collected at the same time as 6-month- (6mML) and 2-year- (2YML) aquacultured sponges. (B) Recirculation system. Wild M. laxissima sponges (W04ML) were collected at the same time as 1-month-aquacultured (1m04AML) sponges.
Phylogenetic analysis of isolates.
Culturing techniques were used to isolate heterotrophic bacteria from sponge samples. Isolates were identified based on the top BLAST hits of ca. 700 bp of the 16S rRNA gene sequence. Alpha- and gammaproteobacterial isolates dominated the culturable bacterial assemblages in wild sponges and seawater samples (Table 1). Isolates affiliated with Acidobacteria were isolated only from sponges maintained under flowthrough aquaculture conditions. In the recirculating system, a diverse assemblage of isolates affiliated with Alphaproteobacteria and the Bacteroidetes group was obtained from all sponges, with an increase in the number of culturable Gammaproteobacteria from sponges maintained for 3 months.
TABLE 1.
16S rRNA gene sequence identities of isolates from sponges and surrounding water samples
| Source (yr) | Isolate; GenBank accession no. | Phylum | Closest cultured organism; GenBank accession no. | % Identity |
|---|---|---|---|---|
| Flowthrough aquaculture system | ||||
| Wild M. laxissima (2001) | KLH10; EF629829 | Alphaproteobacteria | Stappia sp. strain M8; AY307927 | 99 |
| KLH11; EF629830 | Alphaproteobacteria | Ruegeria sp. strain AS-36; AJ391197 | 98 | |
| 2-Yr-aquacultured M. laxissima | N2yML1; EF629831 | Bacteroidetes | Flexibacteraceae bacterium UST030701-097; DQ080995 | 95 |
| N2yML2; EF629832 | Gammaproteobacteria | Xanthomonas sp. strain ML-122; AF139997 | 87 | |
| N2yML3; EF629833 | Alphaproteobacteria | Mesorhizobium sp. strain BNC1; CP000390 | 96 | |
| N2yML4; EF629834 | Acidobacteria | Holophaga foetida strain TMBS4-T; X77215 | 80 | |
| N2yML5; EF629835 | Alphaproteobacteria | Rhodospirillaceae bacterium CL-UU02; DQ401091 | 89 | |
| N2yML6; EF629836 | Alphaproteobacteria | Mesorhizobium sp. strain NH-14; AB196496 | 95 | |
| Seawater (2001) | SWKLH6; EF629837 | Alphaproteobacteria | Sulfitobacter sp. strain KMM 3457; AY682197 | 99 |
| SWKLH7; EF629838 | Alphaproteobacteria | Erythrobacter sp. strain JL-378; DQ285076 | 100 | |
| SWKLH8; EF629839 | Alphaproteobacteria | Erythrobacter sp. strain JL1020; DQ985038 | 100 | |
| SWKLH14; EF629840 | Alphaproteobacteria | Roseobacter sp. strain RED68; AY136132 | 97 | |
| SWKLH15; EF629841 | Gammaproteobacteria | Pseudoalteromonas sp. strain S511-1; AB029824 | 99 | |
| SWKLH16; EF629842 | Gammaproteobacteria | Shewanella putrefaciens; U91549 | 95 | |
| Aquarium water (2003) | N03AW1; EF629843 | Bacteroidetes | Cytophaga sp. strain J18-M01; AB017046 | 97 |
| N03AW2; EF629844 | Alphaproteobacteria | Roseobacter sp. strain DSS-8; AF098493 | 98 | |
| N03AW3; EF629845 | Acidobacteria | Holophaga foetida strain TMBS4-T; X77215 | 67 | |
| N03AW4; EF629846 | Alphaproteobacteria | Roseobacter sp. strain JL-126; AY745859 | 99 | |
| Recirculating aquaculture system | ||||
| Wild M. laxissima (2004) | JE022; DQ097257 | Alphaproteobacteria | Pseudovibrio denitrificans; AY486423 | 99 |
| DQ097258 | Alphaproteobacteria | Pseudovibrio denitrificans; AY486423 | 100 | |
| JE025; DQ097259 | Alphaproteobacteria | Pseudovibrio denitrificans; AY486423 | 99 | |
| N04ML1; EF629847 | Bacilli | Bacillus cereus; AY689066 | 99 | |
| N04ML2; EF629848 | Alphaproteobacteria | Silicibacter sp. strain JC1077; AF201086 | 99 | |
| N04ML4; EF629849 | Alphaproteobacteria | Ruegeria sp. strain AS-36; AJ391197 | 98 | |
| N04ML5; EF629850 | Alphaproteobacteria | Ruegeria atlantica; AB255399 | 98 | |
| N04ML6; EF629851 | Alphaproteobacteria | Ruegeria sp. strain AS-36; AJ391197 | 98 | |
| N04ML7; EF629852 | Bacteroidetes | Flavobacteriaceae bacterium LA8; AF513435 | 95 | |
| N04ML8; EF629853 | Bacteroidetes | Flavobacteriaceae bacterium LA8; AF513435 | 95 | |
| N04ML9; EF629854 | Alphaproteobacteria | Ruegeria sp. strain AS-36; AJ391197 | 98 | |
| N04ML10; EF629855 | Bacteroidetes | Flexibacteraceae bacterium UST030701-097; DQ080995 | 95 | |
| N04ML11; EF629856 | Alphaproteobacteria | Silicibacter sp. strain JC1077; AF201086 | 99 | |
| 1-Mo-aquacultured M. laxissima | N1mML3; EF629857 | Bacteroidetes | Flavobacteriaceae bacterium KE2-02; AJ784113 | 93 |
| N1mML4; EF629858 | Bacteroidetes | Flexibacteraceae bacterium UST030701-097; DQ080995 | 93 | |
| N1mML5; EF629859 | Alphaproteobacteria | Silicibacter sp. strain E932; AY369990 | 98 | |
| N1mML6; EF629860 | Alphaproteobacteria | Stappia sp. strain M8; AY307927 | 98 | |
| N1mML7; EF629861 | Alphaproteobacteria | Silicibacter sp. strain E923; AY369990 | 99 | |
| N1mML8; EF629862 | Alphaproteobacteria | Silicibacter sp. strain E923; AY369990 | 100 | |
| N1mML9; EF629863 | Alphaproteobacteria | Ruegeria atlantica; DQ888840 | 98 | |
| N1mML10c; EF629864 | Bacteroidetes | Flexibacteraceae bacterium UST030701-097; DQ080995 | 95 | |
| N1mML11; EF629865 | Alphaproteobacteria | Ruegeria atlantica; DQ888840 | 98 | |
| N1mML12; EF629866 | Bacteroidetes | Flavobacteriaceae bacterium UST030701-097; AF513435 | 94 | |
| N1mML13; EF629867 | Gammaproteobacteria | Oceanospirillum beijerinckii; AB006760 | 89 | |
| N1mML14; EF629868 | Alphaproteobacteria | Ruegeria atlantica; DQ888840 | 99 | |
| 3-Mo-aquacultured M. laxissima | N3mML1; EF62986 | Bacteroidetes | Flexibacteraceae bacterium UST030701-097; DQ080995 | 94 |
| N3mML2; EF629870 | Alphaproteobacteria | Ruegeria sp. strain N286; AY369984 | 98 | |
| N3mML3; EF629871 | Alphaproteobacteria | Silicibacter sp. strain E923; AY369990 | 99 | |
| N3mML4; EF629872 | Alphaproteobacteria | Silicibacter sp. strain E923; AY369990 | 99 | |
| N3mML5; EF629873 | Alphaproteobacteria | Ruegeria sp. strain N354; AY371430 | 98 | |
| N3mML6; EF629874 | Bacteroidetes | Flexibacteraceae bacterium UST030701-097; DQ080995 | 94 | |
| N3mML7; EF629875 | Alphaproteobacteria | Silicibacter sp. strain E923; AY369990 | 99 | |
| N3mML8; EF629876 | Alphaproteobacteria | Roseivivax sp. strain K376; AY368571 | 100 | |
| N3mML9; EF629877 | Gammaproteobacteria | Vibrio neptunius; AY620979 | 99 | |
| N3mML11; EF629878 | Gammaproteobacteria | Vibrio sp. strain R-14968; AJ316168 | 99 | |
| N3mML12; EF629879 | Alphaproteobacteria | Pseudovibrio denitrificans; AY486423 | 100 | |
| N3mML13; EF629880 | Gammaproteobacteria | Vibrio sp. strain R-14968; AJ316168 | 99 | |
| N3mML14; EF629881 | Gammaproteobacteria | Pseudoalteromonas ruthenica; AY723742 | 100 | |
| N3mML15; EF629882 | Gammaproteobacteria | Ferrimonas futtsuensis; AB245515 | 98 |
Rarefaction analysis.
Rarefaction curves at the estimated phylum level (distance = 0.20) reached saturation for all of the seven libraries, suggesting that the sampling effort was sufficient to reveal all phyla present in the samples. Only the clone library of the 2001 wild sponge reached saturation at the estimated species level (distance = 0.03). Further sampling from the other six libraries may have revealed more diversity at the species level. The bacterial species richness in sponges maintained in both aquaculture systems was greater than that in sponges collected from the wild, indicated by steeper inclines in rarefaction curves (see Fig. S2 and S3 in the supplemental material).
Statistical analysis of bacterial diversity.
The computer program LIBSHUFF was used to compare the libraries of wild and aquacultured sponges. This program is designed to compare undersampled 16S rRNA gene libraries. Evolutionary distances were calculated using the neighbor-joining algorithm in ARB. In the flowthrough system, the libraries of the wild, 6-month-, and 2-year-aquacultured sponges were significantly different at the 99% confidence level (P ≤ 0.01). Similarly, the library from the sponge maintained for 1 month in the recirculating system was significantly different from the library of the wild sponge collected in 2004 (99% confidence level). Additional measures of diversity and richness were obtained (Table 2). These indices were calculated using DOTUR. The input files were in the form of distance matrices generated by using ARB. DOTUR uses the furthest-neighbor method to collapse similar sequences into groups at arbitrary levels of taxonomic similarity and then computes the Shannon, Chao, and ACE statistics for that taxonomic level (53). Maintenance of M. laxissima in the flowthrough system increased the bacterial richness at both phylum and species levels. This was consistent with the higher number of OTUs observed using rarefaction analyses. The values of the Shannon and Simpson indices were higher for aquacultured sponges than for sponges collected from the wild. The richness and diversity estimates changed slightly after M. laxissima was kept for 1 month in the recirculating system.
TABLE 2.
Richness and diversity estimates for bacterial 16S rRNA gene clone libraries from wild and aquacultured M. laxissima sponges from both aquaculture systems and from surrounding water samples
| Source (na) | Distanceb | Richnessc | ACEd | Chao1e | Shannonf | 1/Simpsong |
|---|---|---|---|---|---|---|
| Flowthrough aquaculture system | ||||||
| Wild M. laxissima (2001) (n = 119) | 0.2 | 8 | 14 | 10 | 0.97 | 2.2 |
| 0.03 | 18 | 37 | 36 | 2.2 | 6.3 | |
| 6-Mo-aquacultured sponge (n = 85) | 0.2 | 22 | 28 | 26 | 2.6 | 9.3 |
| 0.03 | 52 | 89 | 85 | 3.8 | 71.4 | |
| 2-Yr-aquacultured sponge (n = 47) | 0.2 | 16 | 22 | 23 | 2.4 | 9.7 |
| 0.03 | 28 | 60 | 50 | 3.1 | 30 | |
| Recirculating aquaculture system | ||||||
| Wild M. laxissima (2004) (n = 67) | 0.2 | 13 | 29 | 24 | 1.9 | 5.2 |
| 0.03 | 28 | 71 | 66 | 2.6 | 8.3 | |
| 1-Mo-aquacultured sponge (n = 67) | 0.2 | 11 | 13 | 11 | 1.8 | 4.2 |
| 0.03 | 32 | 77 | 59 | 3.1 | 21.7 | |
| Water samples | ||||||
| Seawater (n = 37) | 0.2 | 11 | 13 | 12 | 2.2 | 8.6 |
| 0.03 | 21 | 46 | 34 | 2.8 | 20 | |
| Aquarium (n = 51) | 0.2 | 14 | 17 | 16 | 2.4 | 10.8 |
| 0.03 | 28 | 48 | 45 | 3.1 | 33.3 |
n, number of gene sequences analyzed.
80% identity was estimated as the phylum-level distance (D = 0.20), and 97% identity was estimated as the species-level distance (D = 0.03).
Richness is based on observed unique OTUs.
Nonparametric statistical prediction of total richness of different OTUs based on distribution of abundant (>10) and rare (≤10) OTUs.
Nonparametric statistical predictions of total richness of OTUs based on distribution of singletons and doubletons.
Shannon diversity index. A higher number represents more diversity.
Reciprocal of Simpson's diversity index. A higher number represents more diversity.
DISCUSSION
We used two aquaculture systems to examine the feasibility of maintaining the marine sponge M. laxissima in ex situ closed systems under controlled environmental conditions with ecological parameters similar to those in the sponges' natural habitat. The aquaculture of sponges in closed or semiclosed systems is a promising strategy to overcome the supply problem for sponge-derived compounds. This strategy offers good control of environmental conditions, such as light levels and periods, temperature, food supply, and possible precursors of important secondary metabolites (55). However, aquaculture of marine sponges in completely closed systems is still challenging (2, 6, 10, 32, 40). Sponges generally do not have fast growth rates, and growth has rarely been obtained in aquaculture systems (55). It was therefore not surprising that the sponges did not grow under aquaculture conditions. Further optimization of the aquaculture system is required for it to be useful in terms of the production of sponge biomass for harvesting natural products. M. laxissima showed high capability of adapting to aquaculture conditions in comparison to this ability in other sponges from the same reef environment examined in preliminary trials. M. laxissima was successfully maintained in the flowthrough system for 2 years. The recirculating system was designed to give a well-controlled steady-state system with no reliance on a continual input of fresh seawater. Our study is one of a few reports of monitoring the microbial communities associated with marine sponges in aquaculture (17, 23; N. M. Mohamed, V. Rao, M. T. Hamann, M. Kelly, and R. T. Hill, submitted for publication; L. T. Isaacs, J. Kan, L. Nguyen, P. Videau, M. A. Anderson, T. L. Wright, and R. T. Hill, submitted for publication). Hoffmann and coworkers (23) used fluorescent in situ hybridization to study the stability and specificity of microbes associated with the marine cold-water sponge Geodia barretti during cultivation for 8 months in an open recirculation system. They suggested that the explants which survived aquaculture conditions have developed effective buffer systems to prevent infection by foreign sulfate-reducing bacteria during the critical phase of cultivation. In agreement with the results of our study, members of the Alpha- and Gammaproteobacteria were maintained during the period of cultivation. Friedrich and coworkers (17) found that a large fraction of the microbial community of the Mediterranean sponge Aplysina aerophoba remained stable during starvation of sponges or antibiotic exposure over the 11 days in recirculating seawater aquariums.
Changes in the microbial community associated with M. laxissima sponges on maintenance in the flowthrough system included the presence of clones from the Actinobacteria that were highly enriched after 2 years in the aquaculture system (Fig. 2A and 4A). Actinobacteria were not detected after 6 months in aquaculture. This may be due to their absence or to a decrease in abundance to numbers below our detection limit. Acidobacteria were detected only from a sponge maintained for 2 years in this system. This was consistent with the culture-based approach where an isolate (N2yML4) affiliated with the Acidobacteria was recovered from the 2-year-aquacultured sponge. Interestingly, sequences affiliated with Acidobacteria were absent in the library of the aquarium water. This indicates that these strains might be sponge specific and have increased in numbers from levels undetectable in the wild sponges to detectable numbers after long-term maintenance in aquaculture. After acclimation to aquaculture environmental conditions, the growth of some bacteria may be favored, resulting in these groups becoming major components of the microbial communities of aquacultured sponges. Changes in the microbial community associated with the M. laxissima sponges on maintenance in the recirculating system included a decrease in the dominance of Alpha- and Gammaproteobacteria, as was seen in sponges maintained in the flowthrough system (Fig. 3 and 4). Actinobacteria were not detected in aquaculture, and the Bacteroidetes group was significantly enriched.
The library representing bacterial communities found in wild sponges was significantly different from the library of the bacterial community from the surrounding seawater, based on LIBSHUFF results. In addition, there were marked differences in overall DGGE patterns of bacterial communities associated with M. laxissima sponges and those in the surrounding water. This suggests that the bacterial community associated with wild M. laxissima sponges is sponge specific rather than simply comprising a transient population from the water column. After maintenance in aquaculture for 2 years, the bacterial community in an aquacultured sponge was different than the library of the bacterial community in the aquaculture system water. This suggests that M. laxissima maintains a distinct bacterial community different from that in the surrounding water filtered by the sponges, both in the wild and in aquaculture. This is consistent with other reports (19, 51, 57-59, 68) showing that sponges harbor different bacteria than those in the surrounding water, but this is the first time that this has been shown for sponges maintained in aquaculture systems.
The total number of OTUs in bacterial communities was calculated using nonparametric estimators. Chao1 richness estimates were based on singletons and doubletons as described by Chao (5), while ACE was based on the distribution of abundant (>10) and rare (≤10) species. The Shannon index and the reciprocal of Simpson's index were used as diversity indices. Higher numbers indicate greater diversity. Small sample size may affect the performance of diversity estimators. We predict, based on the rarefaction analyses that indicate that further sampling from almost all libraries would reveal more diversity at the species level (see Fig. S2 and S3 in the supplemental material), that the diversity that was observed is an underestimate and additional sampling would lead to an increased estimate of total diversity.
The richness and the diversity of the bacterial communities increased in the flowthrough aquaculture system. This was somewhat unexpected, as we had anticipated a decrease in the bacterial diversity on long-term maintenance of sponges in aquaculture systems. A loss in bacterial diversity might have had adverse consequences for sponge health and for the production of bioactive compounds, such as antibiotic, antifungal, and antifouling compounds by bacterial symbionts. An increase in the bacterial diversity of the sponge-associated communities raises the interesting possibility that additional novel bacteria could be cultured from aquacultured sponges compared with the bacteria in wild sponges. The maintenance of sponges in aquaculture may provide a means for assessing new culturable bacterial diversity from sponges. This possibility is supported by our successful isolation of an Acidobacterium strain (N2yML4) from M. laxissima after maintenance of this sponge in aquaculture, although we have not yet shown that these novel cultured bacteria are sources of new bioactive compounds. To our knowledge, Acidobacteria has not previously been cultivated from marine sponges. This strain is a potential candidate for a genomics approach that may reveal aspects of its metabolic capabilities and importance for the sponge host. We were successful in culturing several additional novel strains that were distantly (≤95% 16S rRNA gene identity) related to previously cultured strains with sequences deposited in GenBank. These included four Flavobacteriaceae, six Flexibacteriaceae, two Gammaproteobacteria, and two Alphaproteobacteria strains (Table 1). All of these strains warrant description as new species or genera.
We were able to maintain M. laxissima alive in closed aquaculture systems. The bacterial community of M. laxissima changed substantially on transfer into aquaculture. Based on the results from both flowthrough and recirculating systems, there was a permanent component of the bacterial community that was present in wild sponges and was maintained in sponges in aquaculture (Fig. 2, 3, and 4). This fraction included members of the Alpha-, Gamma-, and Deltaproteobacteria, Bacteroidetes, and Planctomycetes. This suggests that specific strains in this stable component may be essential for the health of the sponge and possibly play a role in essential symbiotic roles, such as the production of antifouling agents and antimicrobial agents that prevented the growth of pathogenic bacteria in aquacultured sponges.
Supplementary Material
Acknowledgments
We are grateful to Michelle Kelly of the National Institute of Water and Atmospheric Research in New Zealand for confirming the taxonomic identification of the M. laxissima sponge samples and for review of the manuscript. We thank Matthew Anderson, Naomi Montalvo, and Olivier Peraud for their kind assistance in sponge collection and maintenance in aquaculture. Matthew Anderson kindly provided the schematic diagrams of the aquaculture systems. We thank Marcelino Suzuki for his invaluable assistance with ARB. Two anonymous reviewers are thanked for helpful and constructive comments. We acknowledge the National Undersea Research Center (NURC), University of North Carolina at Wilmington, for providing sampling opportunities in Key Largo, Florida.
This study was supported by the Microbial Observatory Program, National Science Foundation (MCB-0238515).
Footnotes
Published ahead of print on 21 December 2007.
Contribution no. 06148 from the Center of Marine Biotechnology.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bagby, R. M. 1972. Formation and differentiation of the upper pinacoderm in reaggregation masses of the sponge Microciona prolifera (Ellis and Solander). J. Exp. Zool. 180:217-225. [DOI] [PubMed] [Google Scholar]
- 2.Belarbi, E. H., M. R. Domînguez, M. C. C. Garcîa, A. C. Gômez, F. G. Camacho, and E. M. Grima. 2003. Cultivation of explants of the marine sponge Crambe crambe in closed systems. Biomol. Eng. 20:333-337. [DOI] [PubMed] [Google Scholar]
- 3.Bergquist, P. R., and P. J. Fromont. 1988. The marine fauna of New Zealand: Porifera, Demospongiae. Part 4 (Poecilosclerida), New Zealand Oceanographic Institute Memoir 96. New Zealand Oceanographic Institute, Wellington, New Zealand.
- 4.Blumbach, B., Z. Pancer, B. Diehl-Seifert, R. Steffen, J. Munkner, I. Muller, and W. E. Muller. 1998. The putative sponge aggregation receptor. Isolation and characterization of a molecule composed of scavenger receptor cysteine-rich domains and short consensus repeats. J. Cell Sci. 111:2635-2644. [DOI] [PubMed] [Google Scholar]
- 5.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:783-791. [Google Scholar]
- 6.de Caralt, S., G. Agell, and M. J. Uriz. 2003. Long-term culture of sponge explants: conditions enhancing survival and growth, and assessment of bioactivity. Biomol. Eng. 20:339-347. [DOI] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.Duckworth, A. R., and C. N. Battershill. 2003. Developing farming structures for production of biologically active sponge metabolites. Aquaculture 217:139-156. [Google Scholar]
- 9.Duckworth, A. R., and C. N. Battershill. 2003. Sponge aquaculture for the production of biologically active metabolites: the influence of farming protocols and environment. Aquaculture 221:311-329. [Google Scholar]
- 10.Duckworth, A. R., G. A. Samples, A. E. Wright, and S. A. Pomponi. 2003. In vitro culture of the tropical sponge Axinella corrugata (Demospongia): effect of food cell concentration on growth, clearance rate and biosynthesis of stevensine. Mar. Biotechnol. 5:519-527. [DOI] [PubMed] [Google Scholar]
- 11.Enticknap, J. J., M. Kelly Shanks, O. Peraud, and R. T. Hill. 2006. Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl. Environ. Microbiol. 72:4105-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enticknap, J. J., R. Thompson, O. Peraud, J. E. Lohr, M. T. Hamann, and R. T. Hill. 2004. Molecular analysis of a Florida Keys sponge: implications for natural products discovery. Mar. Biotechnol. 6:S288-S293. [Google Scholar]
- 13.Faulkner, D. J. 2002. Marine natural products. Nat. Prod. Rep. 19:1-48. [DOI] [PubMed] [Google Scholar]
- 14.Faulkner, D. J., M. K. Harper, M. G. Haygood, C. E. Salomon, and E. W. Schmidt. 2000. Symbiotic bacteria in sponges: sources of bioactive substances, p. 107-119. In N. Fusetani (ed.), Drugs from the sea. Karger, Basel, Switzerland.
- 15.Faulkner, D. J., M. K. Harper, C. E. Salomon, and E. W. Schmidt. 1999. Localization of bioactive metabolites in marine sponges. Mem. Queensl. Mus. 44:167-173. [Google Scholar]
- 16.Fieseler, L., M. Horn, M. Wagner, and U. Hentschel. 2004. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 70:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich, A. B., I. Fischer, P. Proksch, J. Hacker, and U. Hentschel. 2001. Temporal variation of the microbial community associated with the Mediterranean sponge Aplysina aerophoba. FEMS Microbiol. Ecol. 38:105-113. [Google Scholar]
- 18.Fusetani, N., T. Sugawara, and S. Matsunaga. 1991. Cytotoxic metabolites of the marine sponge Mycale adhaerens Lambe. J. Org. Chem. 56:4971-4974. [Google Scholar]
- 19.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hentschel, U., K. M. Usher, and M. W. Taylor. 2006. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55:167-177. [DOI] [PubMed] [Google Scholar]
- 21.Hill, M., A. Hill, N. Lopez, and O. Harriott. 2006. Sponge-specific bacterial symbionts in the Caribbean sponge, Chondrilla nucula (Demospongiae, Chondrosida). Mar. Biol. 148:1221-1230. [Google Scholar]
- 22.Hill, M. O. 1973. Diversity and evenness: a unifying notation and its consequences. Ecology 54:427-432. [Google Scholar]
- 23.Hoffmann, F., H. Rapp, and J. Reitner. 2006. Monitoring microbial community composition by fluorescence in situ hybridization during cultivation of the marine cold-water sponge Geodia barretti. Mar. Biotechnol. 8:373-379. [DOI] [PubMed] [Google Scholar]
- 24.Hood, K. A., L. M. West, P. T. Northcote, M. V. Berridge, and J. H. Miller. 2001. Induction of apoptosis by the marine sponge (Mycale) metabolites mycalamide A and pateamine. Apoptosis 6:155-166. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, M. 2000. Search for biologically active substances from marine sponges, p. 46-58. In N. Fusetani (ed.), Drugs from the sea. Karger, Basel, Switzerland.
- 26.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 27.Lee, Y. K., J.-H. Lee, and H. K. Lee. 2001. Microbial symbiosis in marine sponges. J. Microbiol. 39:254-264. [Google Scholar]
- 28.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsunaga, S., Y. Nogata, and N. Fusetani. 1998. Thiomycalolides: new cytotoxic trisoxazole-containing macrolides isolated from a marine sponge Mycale sp. J. Nat. Prod. 61:663-666. [DOI] [PubMed] [Google Scholar]
- 31.Matsunaga, S., T. Sugawara, and N. Fusetani. 1998. New mycalolides from the marine sponge Mycale magellanica and their interconversion. J. Nat. Prod. 61:1164-1167. [DOI] [PubMed] [Google Scholar]
- 32.Mendola, D. 2003. Aquaculture of three phyla of marine invertebrates to yield bioactive metabolites: process developments and economics. Biomol. Eng. 20:441-458. [DOI] [PubMed] [Google Scholar]
- 33.Montalvo, N. F., N. M. Mohamed, J. J. Enticknap, and R. T. Hill. 2005. Novel actinobacteria from marine sponges. Antonie van Leeuwenhoek 87:29-36. [DOI] [PubMed] [Google Scholar]
- 34.Müller, W. E., V. A. Grebenjuk, G. Le Pennec, H. C. Schroder, F. Brummer, U. Hentschel, I. M. Muller, and H. J. Breter. 2004. Sustainable production of bioactive compounds by sponges-cell culture and gene cluster approach: a review. Mar. Biotechnol. 6:105-117. [DOI] [PubMed] [Google Scholar]
- 35.Müller, W. E., I. Muller, and R. K. Zahn. 1974. Two different aggregation principles in reaggregation process of dissociated sponge cells (Geodia cydonium). Experientia 30:899-902. [DOI] [PubMed] [Google Scholar]
- 36.Munro, M. H., J. W. Blunt, E. J. Dumdei, S. J. Hickford, R. E. Lill, S. Li, C. N. Battershill, and A. R. Duckworth. 1999. The discovery and development of marine compounds with pharmaceutical potential. J. Biotechnol. 70:15-25. [DOI] [PubMed] [Google Scholar]
- 37.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimuraa, S., S. Matsunagaa, S. Yoshida, Y. Nakaoa, H. Hirotab, and N. Fusetania. 2005. Structure-activity relationship study on 13-deoxytedanolide, a highly antitumor macrolide from the marine sponge Mycale adhaerens. Bioorg. Med. Chem. 13:455-462. [DOI] [PubMed] [Google Scholar]
- 39.Northcote, P. T., J. W. Blunt, and M. H. G. Munro. 1991. Pateamine: a potent cytotoxin from the New Zealand marine sponge. Tetrahedron Lett. 32:6411-6414. [Google Scholar]
- 40.Osinga, R., E. H. Belarbi, E. M. Grima, J. Tramper, and R. H. Wijffels. 2003. Progress towards a controlled culture of the marine sponge Pseudosuberites andrewsi in a bioreactor. J. Biotechnol. 100:141-146. [DOI] [PubMed] [Google Scholar]
- 41.Osinga, R., J. Tramper, and R. H. Wijffels. 1999. Cultivation of marine sponges. Mar. Biotechnol. 1:509-532. [DOI] [PubMed] [Google Scholar]
- 42.Page, M. J., L. M. West, P. T. Northcote, C. N. Battershill, and M. Kelly. 2005. Spatial and temporal variability of cytotoxic metabolites in populations of the New Zealand sponge Mycale hentscheli. J. Chem. Ecol. 31:1161-1174. [DOI] [PubMed] [Google Scholar]
- 43.Pattenden, G., D. J. Critcher, and M. Remuiñán. 2004. Total synthesis of (-)-pateamine A, a novel immunosuppressive agent from Mycale sp. Can. J. Chem. 82:353-365. [Google Scholar]
- 44.Perry, N. B., J. W. Blunt, M. H. G. Munro, and L. K. Pannell. 1988. Mycalamide A, an antiviral compound from a New Zealand sponge of the genus Mycale. J. Am. Chem. Soc. 110:4850-4851. [Google Scholar]
- 45.Phuwapraisirisan, P., S. Matsunaga, and N. Fusetani. 2005. Mycapolyols A-F, new cytotoxic metabolites of mixed biogenesis from the marine sponge Mycale zuensis. Org. Lett. 7:2233-2236. [DOI] [PubMed] [Google Scholar]
- 46.Piel, J., D. Hui, G. Wen, D. Butzke, M. Platzer, N. Fusetani, and S. Matsunaga. 2004. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA 101:16222-16227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 48.Proksch, P. 1994. Defensive roles for secondary metabolites from marine sponges and sponge-feeding nudibranchs. Toxicon 32:639-655. [DOI] [PubMed] [Google Scholar]
- 49.Rzasa, R. M., H. A. Shea, and D. Romo. 1998. Total synthesis of the novel, immunosuppressive agent (-)-pateamine A from Mycale sp. employing a β-lactam-based macrocyclization. J. Am. Chem. Soc. 120:591-592. [Google Scholar]
- 50.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 51.Santavy, D. L., P. Willenz, and R. R. Colwell. 1990. Phenotypic study of bacteria associated with the Caribbean sclerosponge, Ceratoporella nicholsoni. Appl. Environ. Microbiol. 56:1750-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarma, A. S., T. Daum, and W. E. G. Muller. 1993. Secondary metabolites from marine sponges. Akademie gemeinnütziger Wissenschaften zu Erfurt, Berlin, Germany.
- 53.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sipkema, D., R. Osinga, W. Schatton, D. Mendola, J. Tramper, and R. H. Wijffels. 2005. Large-scale production of pharmaceuticals by marine sponges: sea, cell, or synthesis? Biotechnol. Bioeng. 90:201-222. [DOI] [PubMed] [Google Scholar]
- 56.Somerville, C. C., I. T. Knight, W. L. Straube, and R. R. Colwell. 1989. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl. Environ. Microbiol. 55:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor, M. W., R. Radax, D. Steger, and M. Wagner. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor, M. W., P. J. Schupp, I. Dahllof, S. Kjelleberg, and P. D. Steinberg. 2004. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6:121-130. [DOI] [PubMed] [Google Scholar]
- 59.Taylor, M. W., P. J. Schupp, R. de Nys, S. Kjelleberg, and P. D. Steinberg. 2005. Biogeography of bacteria associated with the marine sponge Cymbastela concentrica. Environ. Microbiol. 7:419-433. [DOI] [PubMed] [Google Scholar]
- 60.Thacker, R. W. 2005. Impacts of shading on sponge-cyanobacteria symbioses: a comparison between host-specific and generalist associations. Integr. Comp. Biol. 45:369-376. [DOI] [PubMed] [Google Scholar]
- 61.Thompson, A. M., J. W. Blunt, M. H. G. Munro, and B. M. Clark. 1994. Chemistry of the mycalamides, antiviral and antitumor compounds from a marine sponge. Part 4. Reactions of mycalamide A and alkyl derivatives with basic nucleophiles. J. Chem. Soc. Perkin Trans. 1:1025-1031. [Google Scholar]
- 62.Tsukamoto, S., K. Koimaru, and T. Ohta. 2005. Secomycalolide A: a new proteasome inhibitor isolated from a marine sponge of the genus Mycale. Mar. Drugs 3:29-35. [Google Scholar]
- 63.Vacelet, J., and C. Donadey. 1977. Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol. 30:301-314. [Google Scholar]
- 64.van Treeck, P., M. Eisinger, J. Müller, M. Paster, and H. Schuhmacher. 2003. Mariculture trials with Mediterranean sponge species. The exploitation of an old natural resource with sustainable and novel methods. Aquaculture 218:439-455. [Google Scholar]
- 65.Webster, N. S., and R. T. Hill. 2001. The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an alpha-Proteobacterium. Mar. Biol. 138:843-851. [Google Scholar]
- 66.West, L. M., P. T. Northcote, and C. N. Battershill. 2000. Peloruside A: a potent cytotoxic macrolide isolated from the New Zealand marine sponge Mycale sp. J. Org. Chem. 65:445-449. [DOI] [PubMed] [Google Scholar]
- 67.West, L. M., P. T. Northcote, K. A. Hood, J. H. Miller, and M. J. Page. 2000. Mycalamide D, a new cytotoxic amide from the New Zealand marine sponge Mycale species. J. Nat. Prod. 63:707-709. [DOI] [PubMed] [Google Scholar]
- 68.Wilkinson, C. R. 1978. Microbial associations in sponges. I. Ecology, physiology and microbial populations of coral reef sponges. Mar. Biol. 49:161-167. [Google Scholar]
- 69.Wilkinson, C. R. 1978. Microbial associations in sponges. II. Numerical analysis of sponge and water bacterial populations. Mar. Biol. 49:169-176. [Google Scholar]
- 70.Wilkinson, C. R., M. Nowak, B. Austin, and R. R. Colwell. 1981. Specificity of bacterial symbionts in Mediterranean and Great Barrier Reef sponges. Microb. Ecol. 7:13-21. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, X., X. Cao, W. Zhang, X. Yu, and M. Jin. 2003. Primmorphs from archaeocytes-dominant cell population of the sponge hymeniacidon perleve: improved cell proliferation and spiculogenesis. Biotechnol. Bioeng. 84:583-590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.