Abstract
The recently finished genome sequence of Ralstonia eutropha H16 harbors nine genes that are thought to encode functions for intracellular depolymerization (mobilization) of storage poly(3-hydroxybutyrate) (PHB). Based on amino acid similarities, the gene products belong to four classes (PhaZa1 to PhaZa5, PhaZb, PhaZc, and PhaZd1/PhaZd2). However, convincing direct evidence for the in vivo roles of the gene products is poor. In this study, we selected four candidate genes (phaZa1, phaZb, phaZc, and phaZd1) representing the four classes and investigated the physiological function of the gene products (i) with recombinant Escherichia coli strains and (ii) with R. eutropha null mutants. Evidence for weak but significant PHB depolymerase activity was obtained only for PhaZa1. The physiological roles of the other potential PHB depolymerases remain uncertain.
Polyhydroxyalkanoates (PHA) are typical storage compounds of carbon and energy and are found widely in prokaryotes. The most common PHA is poly(3-hydroxybutyrate) (PHB), and this polymer can accumulate up to 90% of the cellular dry weight of some bacteria. PHA are thermoplasts, and despite their relatively high production costs, biologically produced PHA have entered the industrial market.
PHA can be degraded extracellularly by many types of bacteria that are able to secrete specific extracellular PHA depolymerases into the environment or by the intracellular mobilization of PHA in the accumulating strain itself. Considerable knowledge of the biochemical properties of the respective extracellular PHA depolymerases has accumulated (14, 17, 18). Intracellular mobilization of PHA differs from extracellular degradation because of the differences between the biophysical conformations of extracellular (denatured) PHA and those of intracellular (native) PHA. (For definitions of the terms “denatured” and “native PHA,” see references 15, 25, and 26.) Several groups have reported on the identification of potential intracellular PHB depolymerases (1, 5, 8, 19, 20, 33, 40, 43; for an overview see references 15, 17, and 34). These data, together with the genome sequence of Ralstonia eutropha, suggested that R. eutropha H16 might have as many as nine PHB depolymerases/oligomer hydrolases (29). Five putative PHB depolymerase isoenzymes (PhaZa1 to PhaZa5), two 3-hydroxybutyric acid (3HB) oligomer hydrolases (PhaZb and PhaZc [PhaY1 and PhaY2]), and two isoenzymes of a recently found new type of putative intracellular PHB depolymerase (PhaZd1 and PhaZd2) have been described (1). The last two have amino acid sequences that are significantly similar to those of the catalytic domain of extracellular PHB depolymerases (16, 36). We were surprised to find evidence for so many PHB depolymerases in one organism, and we wondered whether all of these proteins were physiologically important for intracellular PHB mobilization. Clarification of this point appeared necessary because independent evidence for the function of the above-mentioned proteins such as the physiological PHB depolymerases exists only for one of them, namely PhaZa1 (8, 32, 43). Unfortunately, an in vitro assay of intracellular PHB depolymerase activity is difficult to perform, and to our knowledge no publication shows high in vitro activity of an intracellular PHB depolymerase by using the natural substrate native PHB (nPHB) granules. Abe et al. reported on a new type of intracellular PHB (iPHB) depolymerase (PhaZd) with high activity toward artificial PHB but with low specific activity toward nPHB granules (0.43 U/mg) (1).
Interestingly, the production of large amounts of 3HB from glucose-grown cells of recombinant Escherichia coli by coexpression of the PHB biosynthetic operon phaCAB of R. eutropha together with the putative PHB depolymerase gene phaZa1 (all genes present on pSYL105red) was reported (23, 28), and a patent claiming the production of ∼14 g/liter 3HB from 20 g/liter glucose by the use of this method was granted (Fig. 4 and 5 of reference 21). Inspired by these successful in vivo data, we thought that the expression of other PHB depolymerases/oligomer hydrolase genes, such as phaZb, phaZc, and phaZd1, together with PHB biosynthetic genes from recombinant E. coli could be a valuable tool with which to check other genes for their potential roles in PHB mobilization, and we intended to use the same principle for the detection of PHB depolymerase activity in vivo by the determination of the amount of 3HB and/or 3HB oligomers released. In a first attempt to elucidate the roles of the respective depolymerase proteins, we chose (i) PhaZa1 as the representative of gene products PhaZa1 to PhaZa5, (ii) PhaZb, (iii) PhaZc, and (iv) PhaZd1 as the representative of PhaZd1/PhaZd2 for further investigation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are given in Table 1. Bacteria were grown on NB (R. eutropha strains) or LB medium (E. coli) at 30 or 37°C, respectively. For some experiments, a mineral salt-based medium (R medium [22]) supplemented with thiamine (20 μg/ml) and glucose (20 g/liter) was used according to the composition reported by Lee and Lee (23), as follows (per liter): KH2PO4, 13.5 g; (NH4)2HPO4, 4.0 g; MgSO4·7H2O, 1.4 g; citric acid, 1.7 g (pH 6.8 [NaOH]); trace metal solution, 10.0 ml trace metal solution per liter of 0.5 N HCl [FeSO4·7H2O, 10.0 g; CaCl2·2H2O, 2.0 g; ZnSO4·7H2O, 2.2 g; MnSO4·4H2O, 0.5 g; CuSO4·5H2O, 1.0 g; (NH4)6Mo7O24·4H2O, 0.1 g; Na2B4O7·10H2O, 0.02 g]. Antibiotics were added according to the presence of plasmid-borne resistance genes. Five milliliters of seed culture (LB) or 20 ml of R medium culture was used for inoculation. When strains harboring the pHGW640 plasmid or derivates of this plasmid were cultivated, 2% (wt/vol) rhamnose was added to the LB medium. Alternatively, a combination of 0.2% (wt/vol) rhamnose and 0.4% (wt/vol) acetate was used to induce the expression of PHB biosynthetic genes and to promote PHB formation. Plasmid-encoded PHB depolymerase expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG, 0.2 mM) during exponential growth phase. IPTG was added at about 1 h after the addition of rhamnose. Bacterial cells were used for nPHB granule isolation or for the determination of the release of 3HB. For the latter, bacteria were resuspended in 1 volume of 0.1 M potassium phosphate buffer (pH 7). Cells were incubated at 30°C (R. eutropha) or at 37°C (E. coli) without shaking.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli BL21(DE3) | Expression strain | Novagen |
| E. coli XL1-Blue | Host for pSYL105red | |
| R. eutropha H16 | Wild type | Accession no. DSMZ428 |
| R. eutropha D1 | PhaZa1 null mutant | 32 |
| R. eutropha OH1 | PhaZb null mutant | 19 |
| R. eutropha TK0130 | PhaZc null mutant | 20 |
| R. eutropha ReDZd1 | PhaZd null mutant | 1 |
| Plasmids | ||
| pSYL105red | Plasmid carrying phaCAB and phaZa1 | 23 |
| pSYL105 | pSYL105red derivative without phaZa1 | 23; this study |
| pSYL105redNEW | Plasmid carrying phaCAB and phaZa1 | This study |
| pBHR68 | Source of phaCAB operon | 39 |
| pAE171 | R. eutropha pUC18 carrying phaZa1 | 32 |
| pHWG640 | Rhamnose-inducible vector | J. Altenbuchner |
| pHWG640::phaCAB | pHWG640 harboring phaCAB under rhamnose promoter control | This study |
| pET23b | Expression vector for T7 promoter | Novagen |
| pET171H | pET23b harboring phaZa1 under T7 promoter | 32 |
| pETOH | pET23b harboring phaZb under T7 promoter | 19 |
| pE3ReZc | pET23b harboring phaZc under T7 promoter | 20 |
| pE3ReZd1 | pET23b harboring phaZd under T7 promoter | 1 |
Construction of recombinant E. coli strains.
A 1.7-kbp HindIII-SalI DNA fragment of pAE171 harboring phaZa1 was cloned into the major DNA fragment of the HindIII/XhoI-digested pSYL105red plasmid to give pSYL105redNEW. DNA sequencing confirmed the presence of one phaZa1 copy plus 129 bp of the 5′ upstream region and 302 bp of the downstream region of phaZa1. The plasmid pBHR68 (39) was used as a source for the R. eutropha H16 phaCAB operon: the 5′ end of phaC was cloned into pHWG640 as an NdeI-BglII DNA fragment by PCR. The 3′ end of phaC including phaAB was cloned into the former construct as a BglII-HindIII fragment that was obtained from pBHR68, resulting in pHWG640, with phaCAB under the control of the rhamnose promoter. E. coli strains harboring pHWG640::phaCAB produced PHB granules, as revealed by the appearance of fluorescent globular structures after cells were stained with Nile red. For details of fluorescence microscopy see reference 12.
In vivo hydrolysis of PHB and secretion of 3HB.
Based on reports that the in vivo hydrolysis of PHA and the secretion of 3-hydroxyalkanoic acids by different bacteria can be efficient at a pH level of 10 to 11 (30, 42) or at a pH level of 3 to 4 (24), cells were washed to limit the source of secreted 3HB to intracellularly accumulated PHB and resuspended in the same volume of 0.1 M of potassium phosphate buffer (pH 7), morpholineethanesulfonic acid-NaOH buffer (pH 4), or Tris-HCl buffer (pH 10). After cell-free samples were incubated at 30°C, they were analyzed for secreted 3HB. In some experiments, culture supernatants were treated with alkali to hydrolyze 3HB oligomers to monomeric 3HB, as described previously (23), as follows: 500 μl of supernatant was alkalized with 500 μl of 10 N NaOH and incubated at 95°C for 2 h. After the supernatant was cooled to room temperature, 500 μl of 10 N HCl was added, and the content of 3HB was determined enzymatically.
Other methods.
The concentration of 3HB was determined using a NAD+-dependent 3HB dehydrogenase assay at an E value at 340 nm (38). In brief, the reaction mixture contained 3.3 mM NAD+, 1 mM MgCl2 in 100 mM Tris-HCl (pH 8.0). The reaction was started by the addition of 2.5 μl of 3HB dehydrogenase (10 mg/ml). If the ΔE340 value was above 0.2, the assay was repeated with a diluted sample to ensure that complete conversion of the substrate had occurred. 3HB content was also determined by high-performance liquid chromatography (HPLC) after the samples underwent derivatization with bromophenacylbromide, as described previously (6). This method allowed the detection of 3HB oligomers. Glucose was determined enzymatically with hexokinase and glucose-6-phosphate dehydrogenase. Western blotting analysis of the expression of PhaZa1, PhaZb, PhaZc, and PhaZd1 was performed by the standard procedure using polyclonal antisera, as described previously (1, 20, 32, 33). nPHB granules were isolated from cells broken by French press and two subsequent steps of glycerol density gradient centrifugation (6, 15). The numbers and sizes of PHB granules in PHB-accumulating bacteria were determined by fluorescence microscopy after the samples were stained with Nile red (12). PHB content was also determined according to the method described in reference 2.
RESULTS
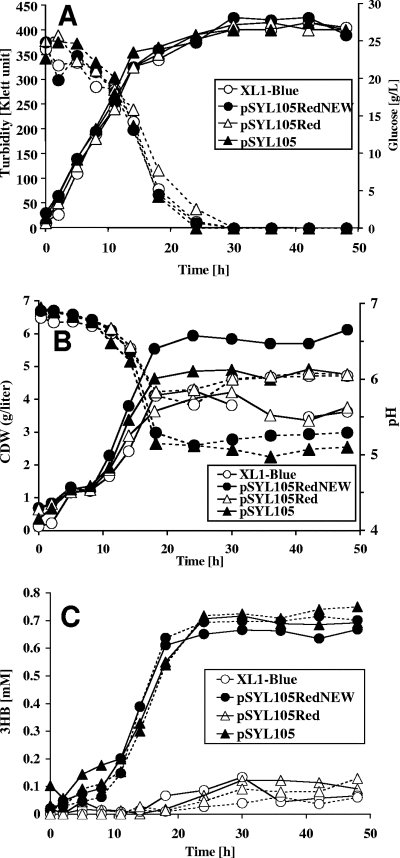
Inspired by the detection of large amounts of secreted 3HB (23) obtained after the coexpression of phaZa1 together with phaCAB in recombinant E. coli, we selected this in vivo system for the detection of potential PHB depolymerase/hydrolase activities of the candidate PHB depolymerase genes (phaZb, phaZc, and phaZd1). To test this concept, E. coli XL1-Blue (control), E. coli XL1-Blue(pSYL105) (carrying phaCAB only), E. coli XL1-Blue(pSYL105red) (carrying phaCAB/phaZa1), and E. coli XL1-Blue(pSYL105redNEW) (the new version of pSYL105red, carrying phaCAB/phaZa1) were grown in R medium with 2% glucose for 48 h at 37°C under the conditions described in reference 23. The pSLY105redNEW plasmid was constructed because preliminary experiments with pSYL105red failed to reproduce the data for 3HB production. Cell growth (Klett units), cellular dry weight, pH, glucose concentration, the concentration of 3HB by 3HB dehydrogenase, and the concentration levels of 3HB, 3HB oligomers, and acetate were determined by HPLC analysis (Fig. 1). The presence of PHB was determined by fluorescence microscopy after samples were stained with Nile red (12) and by gas chromatography after methanolysis. All four strains showed almost the same growth behavior in terms of Klett units (final Klett unit density, ∼400 Klett units) and complete glucose consumption within 30 h (Fig. 1A). The concentration levels of cellular dry weight ranged from less than 4 g/liter for the control strain and for the E. coli strain harboring pSYL105red to about 6 g/liter for the E. coli strain harboring pSYL105redNEW or E. coli with pSYL105 (Fig. 1B). The latter two strains produced the most PHB, as revealed by the appearance of one to a few cell pole-located PHB granules. E. coli harboring pSYL105red produced only a little PHB, resulting in a relatively low concentration of cellular dry matter, comparable to that of the XL1-Blue control strain. The E. coli XL1-Blue control strain produced no PHB, as expected. All strains secreted acids, as indicated by the decrease in pH from a neutral value to a pH range of 5.2 to 6.2 (Fig. 1B). Enzymatic determination of the 3HB concentration in a cell-free supernatant by using NAD+-dependent 3HB dehydrogenase showed that the XL1-Blue control strain and the E. coli strain harboring pSYL105red produced traces of 3HB (≤0.1 mM). Both E. coli XL1-Blue (pSYL105) (carrying only phaCAB) and E. coli XL1-Blue(pSYL105redNEW) (carrying phaCAB and phaZa1) produced little more 3HB (Fig. 1C, 0.6 to 0.7 mM 3HB), but a correlation between 3HB secretion and the presence of phaZa1 was not obvious. Microscopical inspection of Nile red-stained cells did not indicate a significant decrease in PHB content between 24 and 48 h in any of the strains investigated. Treating the samples with alkali before the 3HB dehydrogenase assay resulted in a marginal or no increase in 3HB concentration, suggesting that only small amounts of 3HB oligomers were produced (Fig. 1C). The concentration of 3HB produced (after alkali treatment) by phaZa1-containing strains varied between 0.5 and 1.5 mM (0.05 to 0.16 g/liter) in different experiments. This result was confirmed by HPLC analysis after the derivatization of the sample with bromophenacylbromide: only small amounts of 3HB and no significant amounts of 3HB oligomers were detected. Acetate concentration varied between 0.05 and 0.35% (not shown). Western blotting analysis using antibodies raised against PhaZa1 confirmed the fact that PhaZa1 was present in the cell extracts of phaZa1-harboring strains and excluded the possibility that the absence of PHB depolymerase expression could be the reason for low 3HB production (not shown). Apparently, PhaZa1 has only a low level of PHB depolymerase activity under the conditions used. In conclusion, we were not able to reproduce the production of 3HB to a level as high as ∼9.6 g/liter or even to 14 g/liter (21, 23), and our results do not support the finding that phaZa1 can be used to promote the secretion of large amounts of 3HB with recombinant E. coli under the conditions described.
FIG. 1.
Growth (turbidity) and glucose consumption (A); cellular dry weight (CDW)/liter and pH profile (B); and secretion of 3HB (C) with NaOH treatment for the determination of total 3HB concentration (dotted lines) or without NaOH treatment (solid lines) by E. coli XL1-Blue harboring different combinations of R. eutropha pha genes (Table 1). Growth was measured in R medium with 20 g/liter glucose without pH control as described in references 21 and 23. Three independent experiments have been performed. A typical result is shown.
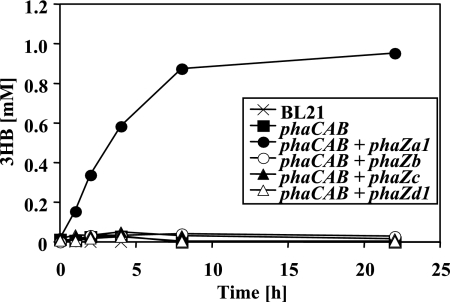
The “pSYL105red system,” in which the pha genes are transcribed from their own promoters, did not work well in our hands; therefore, another system was chosen to evaluate candidate depolymerases: a two-plasmid system with inducible promoters was constructed as described in Materials and Methods. The PHB synthesizing ability was supplied by one plasmid harboring the R. eutropha phaCAB operon under rhamnose promoter control, and the candidate PHB depolymerases to be evaluated were introduced by the other plasmid under lac promoter control. The growth of E. coli was performed in LB medium supplemented with 2% (wt/vol) rhamnose and IPTG (0.2 mM) for the induction of PHB accumulation and PHB depolymerase expression, respectively. Cells harboring phaCAB with or without the depolymerase (phaZa1, phaZb, phaZc, or phaZd1) accumulated roughly 15 to 30% of PHB after 18 h of growth. The expression of depolymerase protein was confirmed by Western blotting analysis using depolymerase-specific antibodies. Similar amounts of PHB content were observed when rhamnose concentration was reduced to 0.2%, and 0.4% acetate was added as a second carbon source to provide precursor molecules for PHB synthesis. No significant reduction in the numbers and sizes of PHB granules and PHB content was detectable with any of the strains during incubation prolonged for up to 48 h (not shown). The cells were centrifuged, washed with buffer, and incubated in buffers of different pH levels. None of the strains investigated secreted significant amounts of 3HB at low pH (pH 4 [3HB], ≤0.05 mM). At an alkaline pH level (pH 10), only E. coli strains containing phaZa1 and phaCAB secreted significant amounts of 3HB (0.4 mM). No 3HB was secreted by cells expressing phaZb, phaZc, or phaZd1 at pH 10 (≤0.05 mM; data not shown). Significant amounts of 3HB were detected in E. coli strains expressing phaCAB and phaZa1 incubated at pH 7: the secretion of 3HB increased almost continuously and reached 0.88 mM within 8 h, after which the concentration increased only marginally to 0.95 mM at 22 h (Fig. 2). Only a small amount of 3HB was detected (≤0.05 mM) in the E. coli cells incubated at pH 7 that contained only the PHB biosynthetic operon or in those in which phaZa1 was replaced by any of the other three putative PHB depolymerase/hydrolase genes (phaZb, phaZc, or phaZd) (Fig. 2). These data suggested that only phaZa1 can function as an intracellular PHB depolymerase, resulting in the release of 3HB to the culture fluid in recombinant E. coli; however, in our hands, the amount was about 2 orders of magnitudes lower than that reported in references 21 and 23.
FIG. 2.
Secretion of 3HB by washed cells of recombinant E. coli BL21 harboring different combination of pha genes (Table 1). Bacteria were grown in the presence of rhamnose and IPTG as described in the text. Washed cells were incubated in buffer (100 mM potassium phosphate [pH 7]) at 37°C. Cell-free samples taken at the time points shown were assayed for amounts of 3HB released, by 3HB dehydrogenase assay. Three independent experiments have been performed. A typical result is shown.
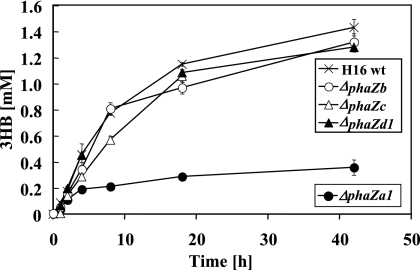
To find independent evidence for the function of PhaZa1 as an intracellular PHB depolymerase, the R. eutropha wild-type strain and the phaZa1, phaZb, phaZc, or phaZd1 null mutant were constructed and investigated for the production of 3HB. PHB-accumulated cells of the corresponding mutant strains were incubated in 0.1 M potassium phosphate buffer (pH 7) at 30°C, and the concentration of the 3HB released was determined by enzymatic assay and by HPLC after derivatization in cell-free samples. The presence of significant amounts of 3HB oligomers could be excluded for all strains by using HPLC analysis after strains underwent derivatization with bromophenacylbromide. The R. eutropha wild-type strain and the ΔphaZb, ΔphaZc, and ΔphaZd1 mutant strains all secreted significant amounts of monomeric 3HB (1.2 to 1.5 mM) into the culture medium. However, the ΔphaZa1 mutant secreted significantly less 3HB ( ≤0.3 mM; Fig. 3), confirming that of the four genes tested, only phaZa1 contributed significantly to 3HB secretion in vivo. When the R. eutropha H16 wild type and the four null mutants were cultivated in NB medium and inspected for the numbers and sizes of PHB granules by fluorescence microscopy, only the ΔphaZa1 strains showed significantly more and bigger PHB granules between 12 and 18 h of incubation (B. Gebauer and D. Jendrossek, unpublished results). However, after 30 h of incubation, all strains, including the ΔphaZa1 mutant, had completely reutilized the intermediately accumulated PHB. These results are in agreement with the depolymerase function of PhaZa1 and suggest the presence of at least one additional physiologically important PHB depolymerase.
FIG. 3.
Secretion of 3HB by washed cells of R. eutropha H16 and phaZ null mutants as indicated. Bacteria were grown in mineral salts medium (35) with gluconate. Washed cells were incubated in buffer (100 mM potassium phosphate [pH 7]) at 30°C. Cell-free samples taken at the time points shown were assayed for the amounts of 3HB released, by 3HB dehydrogenase assay.
DISCUSSION
The biochemical mechanism by which intracellularly accumulated PHB can be reutilized (mobilized) is poorly understood. The literature contains frequent reports of putative iPHB depolymerases (iPhaZs) (1, 5, 8, 32, 40, 43) or medium-chain-length iPHA (iPHAMCL) depolymerases (3, 4, 13). However, a convincing in vitro assay system for iPhaZs does not exist, and unfortunately, researchers have used differently prepared substrates for assays (6, 15). Artificial PHB granules most often have been used as substrates. Artificial PHB granules, unlike nPHB granules, do not contain phasin proteins or any other proteins but surfactants (e.g., sodium dodecyl sulfate, cholate, or others) at the polymer surface. Some confusion also exists about the term “native” for describing nPHB granules. We have suggested restricting the term “native” to those natural PHB granules that have been purified by density gradient centrifugation only (15, 26). It is difficult to judge the physiological importance of the in vitro activity observed with PhaZa1 (32) or PhaZd1 (1) with artificial PHB granules because both enzymes have only very little or no (0.43 U/mg) depolymerase activity with the natural substrate nPHB. A bacterial cell has many esterases/hydrolases, some of which might have the ability to hydrolyze ester bonds of artificial PHB in an in vitro system, but such an activity would not indicate whether this enzyme is also able to hydrolyze the densely protein-covered polymer of the native PHB granules in vivo. The importance of the proteinaceous PHB surface layer to the susceptibility of PHB granules for in vitro hydrolysis has been described for a PHB depolymerase purified from Rhodospirillum rubrum (9-11, 26, 27). The intracellular expression of the PHB depolymerase PhaZ7 of Paucimonas lemoignei, an extracellular PHB depolymerase with unusual specificity for proteinaceous and amorphous nPHB granules (7), in a PHB-accumulating background resulted in an increase of secreted 3HB (37). Therefore, the high depolymerase activity of cautiously isolated nPHB granules could be indicative of a physiological function of the respective protein. In this context, the in vivo system for PHB hydrolysis in recombinant E. coli strains harboring the PHB biosynthetic pathway (phaCAB) together with the iPhaZ (phaZa1) described by Lee and Lee (21, 23, 28) appeared very attractive, and we thought that this in vivo system might be a valuable tool for the detection of potential physiological PHB depolymerase activity of other putative iPhaZs. However, we could not reproduce the data. In our hands, only marginal amounts of 3HB, together with other acids such as acetate, were detected in the culture supernatant of PhaZa1- and PhaCAB-expressing strains of E. coli.
The production of 9.6 g/liter 3HB equivalents (92 mM) from 20 g/liter glucose (111 mM) according to reference 23 is a high value: the biochemical pathway allows conversion of one glucose molecule into one molecule of 3HB. The maximum theoretical value of 3HB produced from 111 mM of glucose is, therefore, 111 mM. However, a significant portion of glucose is used for the synthesis of cellular biomass, for the maintenance of metabolism, and for overflow metabolism (acetate production). Therefore, the production of 92 mM of 3HB (83% of the theoretical maximum) is a surprisingly high value. The production of 14 g/liter 3HB (135 mM) from 20 g/liter glucose (111 mM), as shown in Fig. 5 and 6 of the patent for 3HB production (21), is not possible based on theoretical considerations, even if the 9 mM of citrate of the R medium is additionally taken into consideration.
Although the expression of PhaZa1 did not enable recombinant E. coli strains to hydrolyze the accumulated PHB completely to 3HB, washed cells were able to secrete significantly more 3HB (range of 1 mM 3HB) than cells of control strains. The finding that strains expressing phaCAB also secreted more 3HB than the background (E. coli without plasmid) indicated that elevated levels of 3HB-coenzyme A can be used partially for the secretion of 3HB. Only very small amounts of 3HB were secreted when phaZa1 was replaced with other putative iPhaZs or oligomer hydrolases (phaZb, phaZc, or phaZd1) (Fig. 2), indicating that PhaZa1 is the most likely candidate for a physiological iPHB depolymerase. Recent in vitro data obtained from our laboratory suggest that intracellular PHB depolymerase PhaZa1 can cleave accumulated PHB by thiolysis, resulting in the formation of 3HB-coenzyme A instead of the free acid (41). The secretion of free 3HB would therefore require high thioesterase activity.
Recently, several reports were published showing that PHA-accumulating bacteria can rapidly hydrolyze accumulated PHA and secrete 3HB or 3HAMCL (3, 24, 30, 31) simply by incubating PHA-rich bacteria in buffers. Mutants of Pseudomonas putida defective in intracellular PHA depolymerase secreted considerably less 3HAMCL, suggesting that iPHA depolymerase activity is responsible for 3HAMCL secretion (42). In R. eutropha, the ΔphaZa1 mutant was still able to secrete ∼0.3 mM 3HB. Since no soluble exogenous carbon sources were available, R. eutropha apparently also has at least one other physiological iPHB depolymerase. This conclusion is in agreement with earlier reports (8, 32, 43). The results with R. eutropha cells are complementary to and in agreement with the results obtained with recombinant E. coli strains (Fig. 2). However, the concentration of secreted 3HB was rather low (1 to 2 mM) and significantly lower than the secretion of 3HAMCL by P. putida (3, 30, 31, 42). In conclusion, PhaZa1 is a physiologically important iPHB depolymerase in R. eutropha but apparently is only little suited for the efficient biotechnological production of 3HB.
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to D.J.
We thank Birgit Gebauer and Manuela Unsin for technical assistance with some experiments and Y. Lee and S.-Y. Lee for providing the plasmid pSYL105red and for discussion.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Abe, T., T. Kobayashi, and T. Saito. 2005. Properties of a novel intracellular poly(3-hydroxybutyrate) depolymerase with high specific activity (PhaZd) in Wautersia eutropha H16. J. Bacteriol. 187:6982-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandl, H., R. A. Gross, R. W. Lenz, and R. C. Fuller. 1988. Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl. Environ. Microbiol. 54:1977-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Eugenio, L. I., P. Garcia, J. M. Luengo, J. M. Sanz, J. S. Roman, J. L. Garcia, and M. A. Prieto. 2007. Biochemical evidence that phaZ gene encodes a specific intracellular medium chain length polyhydroxyalkanoate depolymerase in Pseudomonas putida KT2442: characterization of a paradigmatic enzyme. J. Biol. Chem. 282:4951-4962. [DOI] [PubMed] [Google Scholar]
- 4.Foster, L. J. R., R. W. Lenz, and R. C. Fuller. 1999. Intracellular depolymerase activity in isolated inclusion bodies containing polyhydroxyalkanoates with long alkyl and functional substituents in the side chain. Int. J. Biol. Macromol. 26:187-192. [DOI] [PubMed] [Google Scholar]
- 5.Gao, D., A. Maehara, T. Yamane, and S. Ueda. 2001. Identification of the intracellular polyhydroxyalkanoate depolymerase gene of Paracoccus denitrificans and some properties of the gene product. FEMS Microbiol. Lett. 196:159-164. [DOI] [PubMed] [Google Scholar]
- 6.Gebauer, B., and D. Jendrossek. 2006. Assay of poly(3-hydroxybutyrate) depolymerase activity and product determination. Appl. Environ. Microbiol. 72:6094-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handrick, R., S. Reinhardt, M. L. Focarete, M. Scandola, G. Adamus, M. Kowalczuk, and D. Jendrossek. 2001. A new type of thermoalkalophilic hydrolase of Paucimonas lemoignei with high specificity for amorphous polyesters of short chain-length hydroxyalkanoic acids. J. Biol. Chem. 276:36215-36224. [DOI] [PubMed] [Google Scholar]
- 8.Handrick, R., S. Reinhardt, and D. Jendrossek. 2000. Mobilization of poly(3-hydroxybutyrate) in Ralstonia eutropha. J. Bacteriol. 182:5916-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handrick, R., S. Reinhardt, P. Kimmig, and D. Jendrossek. 2004. The “intracellular” poly(3-hydroxybutyrate) (PHB) depolymerase of Rhodospirillum rubrum is a periplasm-located protein with specificity for native PHB and with structural similarity to extracellular PHB depolymerases. J. Bacteriol. 186:7243-7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Handrick, R., S. Reinhardt, D. Schultheiss, T. Reichart, D. Schuler, V. Jendrossek, and D. Jendrossek. 2004. Unraveling the function of the Rhodospirillum rubrum activator of polyhydroxybutyrate (PHB) degradation: the activator is a PHB-granule-bound protein (phasin). J. Bacteriol. 186:2466-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Handrick, R., U. Technow, T. Reichart, S. Reinhardt, T. Sander, and D. Jendrossek. 2004. The activator of the Rhodospirillum rubrum PHB depolymerase is a polypeptide that is extremely resistant to high temperature (121 degrees C) and other physical or chemical stresses. FEMS Microbiol. Lett. 230:265-274. [DOI] [PubMed] [Google Scholar]
- 12.Hermawan, S., and D. Jendrossek. 2007. Microscopical investigation of poly(3-hydroxybutyrate) granule formation in Azotobacter vinelandii. FEMS Microbiol. Lett. 266:60-64. [DOI] [PubMed] [Google Scholar]
- 13.Huisman, G. W., E. Wonink, R. Meima, B. Kazemier, P. Terpstra, and B. Witholt. 1991. Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. Identification and sequences of genes and function of the encoded proteins in the synthesis and degradation of PHA. J. Biol. Chem. 266:2191-2198. [PubMed] [Google Scholar]
- 14.Jendrossek, D. 2002. Extracellular polyhydroxyalkanoate depolymerases: the key enzymes of PHA degradation, p. 41-83. In Y. Doi and A. Steinbüchel (ed.), Biopolymers, vol. 3b. Polyesters II. Wiley-VCH, Hoboken, NJ. [Google Scholar]
- 15.Jendrossek, D. 2007. Peculiarities of PHA granules preparation and PHA depolymerase activity determination. Appl. Microbiol. Biotechnol. 74:1186-1196. [DOI] [PubMed] [Google Scholar]
- 16.Jendrossek, D., A. Frisse, A. Behrends, M. Andermann, H. D. Kratzin, T. Stanislawski, and H. G. Schlegel. 1995. Biochemical and molecular characterization of the Pseudomonas lemoignei polyhydroxyalkanoate depolymerase system. J. Bacteriol. 177:596-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jendrossek, D., and R. Handrick. 2002. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 56:403-432. [DOI] [PubMed] [Google Scholar]
- 18.Jendrossek, D., A. Schirmer, and H. G. Schlegel. 1996. Biodegradation of polyhydroxyalkanoic acids. Appl. Microbiol. Biotechnol. 46:451-463. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, T., M. Shiraki, T. Abe, A. Sugiyama, and T. Saito. 2003. Purification and properties of an intracellular 3-hydroxybutyrate-oligomer hydrolase (PhaZ2) in Ralstonia eutropha H16 and its identification as a novel intracellular poly(3-hydroxybutyrate) depolymerase. J. Bacteriol. 185:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi, T., K. Uchino, T. Abe, Y. Yamazaki, and T. Saito. 2005. Novel intracellular 3-hydroxybutyrate-oligomer hydrolase in Wautersia eutropha H16. J. Bacteriol. 187:5129-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, S.-Y., and Y. Lee. June 2003. (R)-Hydroxycarboxylic acid producing recombinant microorganisms and process for preparing (R)-hydroxycarboxylic acid using the same. WIPO patent WO03/046159.
- 22.Lee, S. Y., and N. H. Chang. 1993. High cell density cultivation of Escherichia coli using sucrose as a carbon source. Biotechnol. Lett. 15:971-974. [Google Scholar]
- 23.Lee, S. Y., and Y. Lee. 2003. Metabolic engineering of Escherichia coli for production of enantiomerically pure (R)-(−)-hydroxycarboxylic acids. Appl. Environ. Microbiol. 69:3421-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, S. Y., Y. Lee, and F. Wang. 1999. Chiral compounds from bacterial polyesters: sugars to plastics to fine chemicals. Biotechnol. Bioeng. 65:363-368. [DOI] [PubMed] [Google Scholar]
- 25.Merrick, J. M., and M. Doudoroff. 1964. Depolymerization of poly-β-hydroxybutyrate by intracellular enzyme system. J. Bacteriol. 88:60-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merrick, J. M., D. G. Lundgren, and R. M. Pfister. 1965. Morphological changes in poly-beta-hydroxybutyrate granules associated with decreased susceptibility to enzymatic hydrolysis. J. Bacteriol. 89:234-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merrick, J. M., R. Steger, and D. Dombroski. 1999. Hydrolysis of native poly(hydroxybutyrate) granules (PHB), crystalline PHB, and artificial amorphous PHB granules by intracellular and extracellular depolymerases. Int. J. Biol. Macromol. 25:129-134. [DOI] [PubMed] [Google Scholar]
- 28.Park, S. J., S. Y. Lee, and Y. Lee. 2004. Biosynthesis of R-3-hydroxyalkanoic acids by metabolically engineered Escherichia coli. Appl. Biochem. Biotechnol. 113-116:373-379. [PubMed]
- 29.Pohlmann, A., W. F. Fricke, F. Reinecke, B. Kusian, H. Liesegang, R. Cramm, T. Eitinger, C. Ewering, M. Potter, E. Schwartz, A. Strittmatter, I. Voss, G. Gottschalk, A. Steinbuchel, B. Friedrich, and B. Bowien. 2006. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 24:1257-1262. [DOI] [PubMed] [Google Scholar]
- 30.Ren, Q., A. Grubelnik, M. Hoerler, K. Ruth, R. Hartmann, H. Felber, and M. Zinn. 2005. Bacterial poly(hydroxyalkanoates) as a source of chiral hydroxyalkanoic acids. Biomacromolecules 6:2290-2298. [DOI] [PubMed] [Google Scholar]
- 31.Ruth, K., A. Grubelnik, R. Hartmann, T. Egli, M. Zinn, and Q. Ren. 2007. Efficient production of (R)-3-hydroxycarboxylic acids by biotechnological conversion of polyhydroxyalkanoates and their purification. Biomacromolecules 8:279-286. [DOI] [PubMed] [Google Scholar]
- 32.Saegusa, H., M. Shiraki, C. Kanai, and T. Saito. 2001. Cloning of an intracellular poly[d(−)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 183:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saegusa, H., M. Shiraki, and T. Saito. 2002. Cloning of an intracellular d(−)-3-hydroxybutyrate-oligomer hydrolase gene from Ralstonia eutropha H16 and identification of the active site serine residue by site-directed mutagenesis. J. Biosci. Bioeng. 94:106-112. [DOI] [PubMed] [Google Scholar]
- 34.Saito, T., and T. Kobayashi. 2002. Intracellular degradation of PHAs, p. 23-40. In Y. Doi and A. Steinbüchel (ed.), Biopolymers, vol. 3b. Polyesters II. Wiley-VCH, Hoboken, NJ. [Google Scholar]
- 35.Schlegel, H. G., G. Gottschalk, and R. Von Bartha. 1961. Formation and utilization of poly-beta-hydroxybutyric acid by Knallgas bacteria (Hydrogenomonas). Nature 191:463-465. [DOI] [PubMed] [Google Scholar]
- 36.Shinohe, T., M. Nojiri, T. Saito, T. Stanislawski, and D. Jendrossek. 1996. Determination of the active sites serine of the poly(3-hydroxybutyrate) depolymerases of Pseudomonas lemoignei (PhaZ5) and of Alcaligenes faecalis. FEMS Microbiol. Lett. 141:103-109. [DOI] [PubMed] [Google Scholar]
- 37.Shiraki, M., T. Endo, and T. Saito. 2006. Fermentative production of (R)-(−)-3-hydroxybutyrate using 3-hydroxybutyrate dehydrogenase null mutant of Ralstonia eutropha and recombinant Escherichia coli. J. Biosci. Bioeng 102:529-534. [DOI] [PubMed] [Google Scholar]
- 38.Shuster, C. W., and M. Doudoroff. 1962. A cold-sensitive d(−)beta-hydroxybutyric acid dehydrogenase from Rhodospirillum rubrum. J. Biol. Chem. 237:603-607. [PubMed] [Google Scholar]
- 39.Spiekermann, P., B. H. Rehm, R. Kalscheuer, D. Baumeister, and A. Steinbüchel. 1999. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 171:73-80. [DOI] [PubMed] [Google Scholar]
- 40.Tseng, C. L., H. J. Chen, and G. C. Shaw. 2006. Identification and characterization of the Bacillus thuringiensis phaZ gene, encoding new intracellular poly-3-hydroxybutyrate depolymerase. J. Bacteriol. 188:7592-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uchino, K., T. Saito, B. Gebauer, and D. Jendrossek. 2007. Isolated poly(3-hydroxybutyrate) (PHB) granules are complex bacterial organelles catalyzing formation of PHB from acetyl coenzyme A (CoA) and degradation of PHB to acetyl-CoA. J. Bacteriol. 189:8250-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, L., W. Armbruster, and D. Jendrossek. 2007. Production of medium-chain-length hydroxyalkanoic acids from Pseudomonas putida in pH stat. Appl. Microbiol. Biotechnol. 74:1047-1053. [DOI] [PubMed] [Google Scholar]
- 43.York, G. M., J. Lupberger, J. Tian, A. G. Lawrence, J. Stubbe, and A. J. Sinskey. 2003. Ralstonia eutropha H16 encodes two and possibly three intracellular poly[d-(−)-3-hydroxybutyrate] depolymerase genes. J. Bacteriol. 185:3788-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]