Abstract
Shiga toxin-producing Escherichia coli isolates from two 2006 outbreaks were compared to other O157:H7 isolates for virulence genotype, biofilm formation, and stress responses. Spinach- and lettuce-related-outbreak strains had similar pulsed-field gel electrophoresis patterns, and all carried both stx2 and stx2c variant genes. Cooperative biofilm formation involving an E. coli O157:H7 strain and a non-O157:H7 strain was also demonstrated.
Hemorrhagic colitis, which occasionally progresses to hemolytic uremic syndrome (HUS), is a hallmark of human infection with Shiga toxin-producing Escherichia coli (STEC). Serotype O157:H7 is the serotype most commonly associated with clinical disease and food-associated outbreaks (18). However, other STEC serotypes have been associated with outbreaks and sporadic disease (4). There are two major types of Shiga toxin encoded by the stx1 and stx2 genes, and an increasing number of variants for both are being reported (5, 21). In addition to Shiga toxin, the locus of enterocyte effacement (LEE) and a 60-MDa, hemolysin-encoding plasmid are considered major STEC virulence determinants (21).
During 2006, there were three multistate, produce-associated outbreaks of E. coli O157:H7, two of which involved Pennsylvania. In September, 26 states reported illnesses linked to spinach which resulted in 205 confirmed cases of illness and three deaths (8, 10). Of 103 hospitalized patients, 31 (30%) developed HUS. In November, according to the Centers for Disease Control and Prevention, five states reported illnesses that were linked to lettuce (http://www.cdc.gov/ecoli/2006/december/121406.htm). Of 71 cases, 53 were hospitalized and 8 (15%) developed HUS.
Bacterial persistence on foods or in the production environment can be augmented by stress resistance genes and biofilm formation (9, 19). Acid resistance allows STEC to survive passage through the stomach and enhances survival in foods and in the processing environment (15, 26). Furthermore, sublethal acid conditions (acid adaptation) could increase the ability of cells to survive the severe acid challenge of gastric passage. Thermal processing is commonly used to reduce or eliminate pathogens from food. Although E. coli O157:H7 does not show an unusual tolerance to heat, variation in levels of heat resistance among strains has been demonstrated (3).
Biofilm formation can protect bacteria from environmental stress, increase their resistance to antimicrobials, and enhance persistence on foods and solid surfaces (6, 9, 17). We compared Pennsylvania STEC isolates from two 2006 outbreaks to serotype O157:H7 isolates from diverse sources for the presence of virulence genes, stress resistance characteristics, and biofilm formation.
Bacterial strains, media, and growth conditions.
The bacterial strains used in this study were propagated in brain heart infusion broth (Becton, Dickinson and Co., Sparks, MD) or on 1.5% brain heart infusion agar plates. Luria-Bertani (LB) broth (Becton, Dickinson and Co.) or agar (1.5%) made with no salt (LB-NS) and tryptic soy broth (Becton, Dickinson and Co.) or tryptic soy agar were used as noted below. Curli fiber expression was analyzed on Congo red indicator plates (24). Media for mixed-strain biofilm assays contained 100 μg/ml ampicillin (Sigma-Aldrich Corporation, St. Louis, MO).
Comparison of virulence genes, Shiga toxin sequencing, and pulsed-field gel electrophoresis (PFGE).
Seven strains submitted to Pennsylvania diagnostic laboratories from two 2006 outbreaks, including two sets of patient isolates matched with isolates from the spinach that the patients consumed (patient/food paired isolates), were compared to a diverse group of serotype O157:H7 strains by the use of PCR targeting of E. coli virulence genes (Table 1). Primers for the stx1 gene and/or the stx2 gene (11) amplified product from each of the 17 strains (Table 1). Primers targeting the STEC hly gene (12), the eae gene from O157 strains (Table 2), a 5′ conserved sequence of the eaeA gene in STEC (14), and the wzy gene in the O antigen gene cluster of O157 strains (Table 2) all amplified a product from each strain except for O6E01767 (O−:H4). Primers (not shown) targeting the cdt-I, cdt-III, cdt-IV, astA, bfp, cnf-1, cnf-2, fasA, faeG, fimF41a, fanA, fedA, elt, estIa, and estIb genes all failed to amplify a product from any of the 17 strains. These results indicate that the serotype O157:H7 strains from both 2006 outbreaks carried similar virulence genes and differed from the other O157:H7 strains only in the type of Shiga toxin gene(s). However, an O−:H4 strain isolated from a spinach bag was positive only for stx1 and contained none of the other genes considered important for STEC virulence.
TABLE 1.
E. coli strains and Shiga toxin amplification
| E. coli strain | Serotype | Source | stx1 | stx2 |
|---|---|---|---|---|
| Tarr1A | O157:H7 | Patient (24) | + | + |
| Tarr4A | O157:H7 | Patient (24) | + | + |
| 168-93 | O157:H7 | Beef brisket (FSIS) | + | |
| C7927 | O157:H7 | Patient (apple cider, CDC) | + | + |
| SEA13B88 | O157:H7 | Apple juice (FDA) | + | + |
| 180-93 | O157:H7 | Calf (FSIS) | + | |
| 194-93 | O157:H7 | “Downer” cow (FSIS) | + | + |
| 414-95 | O157:H7 | Ground beef (FSIS) | + | |
| ENT-C9490 | O157:H7 | Hamburger (CDC) | + | + |
| 380-94 | O157:H7 | Salami (CDC) | + | + |
| 06E01767 | O−:H4a | Spinach (patient A's home, coisolated with 06F00475) | + | |
| 06E02109 | O157:H7 | Stool, patient C (2006 shredded-lettuce outbreak) | + | |
| 06E02128 | O157:H7 | Stool, patient D (2006 shredded-lettuce outbreak) | + | |
| 06E01456 | O157:H7 | Stool, patient A (2006 spinach outbreak) | + | |
| 06F00475 | O157:H7 | Spinach (patient A's home, coisolated with 06E01767) | + | |
| 06F00480 | O157:H7 | Spinach (patient B's home) | + | |
| 06E01595 | O157:H7 | Stool, patient B (2006 spinach outbreak) | + |
The serotype of this strain was determined at the Gastroenteric Disease Center, Department of Veterinary and Biomedical Sciences, the Pennsylvania State University, University Park, PA. This strain did not react with any of the antisera used to type the somatic antigens of E. coli.
TABLE 2.
Primers
| Primer | Sequence | Purpose |
|---|---|---|
| O157wzy-F | CCTGTCAAAGGATAACCGTAATCC | O157 wzy PCR |
| O157wzy-R | TTGTTCTCCGTCTTGTCCTAAACT | O157 wzy PCR |
| Eae-2325-F | GTAAGTCTCAAACGCAAGCAACCAC | eae PCR |
| Eae-2491-R | AACCTTGTTGTCAATTTTCAGTTCATCA | eae PCR |
| stx1ABfor | ATGGTGCTCAAGGAGTATTGTG | stx1 PCR and sequencing |
| stx1ABrev | CATCTATTATCAGACCGGCAAC | stx1 PCR and sequencing |
| stx1seqfor | GTCTGGTGACAGTAGCTATACC | stx1 sequencing |
| stx1seqrev | CCTACACGAACAGAGTCTTGTC | stx1 sequencing |
| stx2ABfor | GGTCTGGTGCTGATTACTTCAG | stx2 PCR and sequencing |
| stx2ABrev | TCTGACAGGCAACTGTCAACTG | stx2 PCR and sequencing |
| stx2seqfor | TACGCTTCAGGCAGATACAGAG | stx2 sequencing |
| stx2seqrev | ATACTCCGGAAGCACATTGCTG | stx2 sequencing |
| 2851q | CCTCAATGCCTCGTTGTTTATG | stx2c PCR and sequencing |
| 933Wq | CGGTATGTTGAGCGTGAATTGC | stx2 PCR and sequencing |
To further characterize the seven 2006 outbreak strains, we sequenced the amplified stx operons using primers listed in Table 2. Strain O6E01767 (GenBank accession number EU273279) sequences shared the closest nucleotide identity (99%) with the stx1 variant gene reported by Asakura et al. (1) (GenBank accession number AB048235). The sequence also showed close identity to eight additional GenBank records for stx1c variants (GenBank accession numbers AY135685, Z36901, AJ413986, AJ314839, AJ314838, AJ312232, AB048231, and AB048234). All nine GenBank strains were isolated from either sheep (n = 6) or humans (n = 3), and all were of O serogroups other than O157. It is unknown what role, if any, strain 06E01767 (O−:H4) played in the 2006 outbreak, but past reports suggest that strains carrying stx1 variants may be associated with milder illness (16).
Analyses of amplified Shiga toxin DNA from each of the six serotype O157:H7 2006 outbreak strains suggested the presence of stx2 and a stx2c variant. Primers designed from the antitermination protein Q gene of phage 2851 and the antitermination protein Q gene (primer 933Wq) paired with primer stx2ABrev amplified two unambiguous sequences identical to a stx2c variant (22) and the stx2 gene of strain EDL933 (GenBank accession number AE005174) at the amino acid level. Freidrich et al. (13) showed that stx2 and stx2c are the two stx genes most likely to be associated with HUS and severe clinical disease, with stx2 being a greater risk factor than stx2c. It is unknown whether the presence of both these toxins within a single strain contributed to the strain virulence or the HUS associated with the outbreaks.
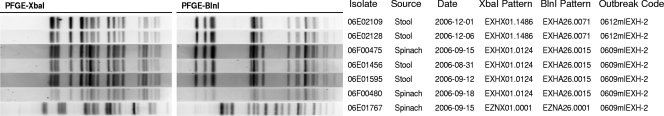
The perfect identity of virulence gene profiles and Shiga toxin sequences among the two sets of 2006 outbreak isolates prompted us to investigate strain relatedness using PFGE as described previously (20) (Fig. 1). The O157:H7 isolates from the spinach-associated outbreak had indistinguishable XbaI and BlnI restriction patterns. Likewise, the patterns from the shredded-lettuce-associated outbreak isolates were indistinguishable. Comparison of the serotype O157:H7 patterns of the spinach-associated outbreak to that of the lettuce-associated outbreak showed only minor band shifts or differences in band intensity, suggesting that the isolates from the two outbreaks could be closely related; further epidemiologic investigations may identify possible commonalities. The patterns for strain 06E01767 were unique from those of all other strains.
FIG. 1.
Comparison of PFGE patterns of spinach- and lettuce-associated-outbreak strains. The enzyme pattern names and outbreak codes were assigned by the PulseNet team of the CDC.
Thermal and acid tolerance.
Levels of thermal tolerance and tolerance to synthetic gastric fluid, pH 1.5, were compared among the strains in Table 1. The heat tolerance was determined at 60°C as described previously (25). Data (microbial counts versus time) were analyzed using linear regression (Excel 2002), and the D60 values were the slopes of the regression lines. The D values ranged from 1.72 ± 0.23 (strain Tarr4A) to 2.70 ± 0.25 (strain 414-95). There were no significant differences in D values (1.98 ± 0.10 to 2.32 ± 0.27) among the STEC strains associated with the two 2006 produce-associated outbreaks; however, the D value of strain 06E01767 (2.50 ± 0.43) was significantly different (P < 0.05) than that of strain Tarr4a. The results of the current study are comparable to those of Whiting and Golden (25), who also examined the D60 values of E. coli O157:H7 strains, including strains C7927, SEA13B88, and C9490.
The method of Buchanan and Edelson (7) was used to produce acid-adapted (pH ca. 4.7) and nonadapted (pH ca. 6.8) cell populations of the strains listed in Table 1. Portions (500 μl) of the cultures were inoculated into 50 ml of synthetic gastric fluid (pH 1.5), prepared as previously described (2), and incubated at 37°C without agitation. Samples taken at time zero and at 1, 2, 3, 4, and 5 h were enumerated by spread plating onto tryptic soy agar. Although there was some variation in the results, most strains showed a linear or quadratic decrease in population size with time, with certain strains, such as SEA13B88, 380-94, C9490, C7027, and Tarr4A, showing greater decreases than others (Table 3). Only serotype O157:H7 strain 180-93 showed a significant difference in inactivation from those of the other strains. Cubic polynomials were fit to the data over time, and for strains Tarr4A, 168-93, C7927, SEA13B88, 06E02109, 06E02128, 06E01456, 06F00475, 06F00480, and 06E01595, there was a significantly greater decline in the acid-adapted cell populations than in the nonadapted cell populations with exposure to synthetic gastric fluid. The acid tolerance of the spinach- and lettuce-associated-outbreak strains was not notably greater than that of most of the other E. coli O157:H7 strains tested. It is unknown why acid adaptation did not increase the ability of some strains to tolerate exposure to synthetic gastric fluid. Further studies to examine genetic differences among these strains, including the presence of mutations in stress response genes, are warranted.
TABLE 3.
Log numbers of CFU/ml of acid-adapted or non-acid-adapted cells following acid challenge with synthetic gastric fluid at pH 1.5
| E. coli strain | Cellsa | Mean log10 no. of CFU/ml ± SDb at:
|
|||||
|---|---|---|---|---|---|---|---|
| 0 h | 1 h | 2 h | 3 h | 4 h | 5 h | ||
| Tarr1A | AA | 6.75 ± 0.12 | 6.32 ± 0.18 | 6.25 ± 0.13 | 5.98 ± 0.21 | 5.76 ± 0.18 | 5.51 ± 0.40 |
| NA | 6.21 ± 0.27 | 5.74 ± 0.29 | 5.42 ± 0.19 | 5.16 ± 0.38 | 4.69 ± 0.47 | 4.20 ± 0.23 | |
| Tarr4A | AA | 6.82 ± 0.15 | 6.89 ± 0.04 | 6.63 ± 0.24 | 3.71 ± 0.58 | NDa | ND |
| NA | 6.52 ± 0.15 | 5.99 ± 0.16 | 5.79 ± 0.16 | 5.17 ± 0.26 | 4.59 ± 0.28 | 3.62 ± 0.44 | |
| 168-93 | AA | 7.05 ± 0.11 | 6.94 ± 0.08 | 6.81 ± 0.12 | 3.94 ± 0.27 | ND | ND |
| NA | 6.44 ± 0.03 | 5.95 ± 0.13 | 5.67 ± 0.14 | 5.36 ± 0.16 | 4.98 ± 0.42 | 4.26 ± 0.50 | |
| C7927 | AA | 6.93 ± 0.02 | 6.87 ± 0.14 | 6.47 ± 0.07 | 4.90 ± 1.29 | ND | ND |
| NA | 6.60 ± 0.10 | 5.98 ± 0.02 | 5.78.±0.50 | 4.74 ± 0.50 | 3.55 ± 0.74 | 1.89 ± 1.39 | |
| SEA13B88 | AA | 7.10 ± 0.07 | 6.70 ± 0.02 | 5.49 ± 0.67 | ND | ND | ND |
| NA | 6.51 ± 0.05 | 6.31 ± 0.26 | 5.78 ± 0.22 | 4.79 ± 0.79 | 4.10 ± 0.94 | 3.32 ± 1.40 | |
| 180-93 | AA | 6.52 ± 0.20 | ND | ND | ND | ND | ND |
| NA | 5.79 ± 0.39 | ND | ND | ND | ND | ND | |
| 194-93 | AA | 6.95 ± 0.10 | 6.68 ± 0.19 | 6.42 ± 0.26 | 6.18 ± 0.41 | 5.38 ± 0.48 | 3.61 ± 0.74 |
| NA | 6.62 ± 0.08 | 6.58 ± 0.14 | 6.23 ± 0.25 | 5.93 ± 0.63 | 5.11 ± 0.70 | 3.92 ± 0.86 | |
| 414-95 | AA | 6.83 ± 0.15 | 6.65 ± 0.26 | 6.32 ± 0.07 | 5.92 ± 0.03 | 3.63 ± 1.25 | ND |
| NA | 6.58 ± 0.17 | 6.44 ± 0.19 | 6.32 ± 0.27 | 5.40 ± 0.90 | 3.29 ± 1.41 | 0.63 ± 0.90 | |
| ENT | AA | 6.90 ± 0.24 | 6.80 ± 0.36 | 6.48 ± 0.62 | 4.21 ± 0.47 | ND | ND |
| C9490 | NA | 6.73 ± 0.10 | 6.47 ± 0.17 | 5.91 ± 0.38 | 4.52 ± 0.52 | 2.59 ± 0.62 | ND |
| 380-94 | AA | 7.01 ± 0.08 | 6.88 ± 0.07 | 6.79 ± 0.08 | 5.05 ± 1.27 | ND | ND |
| NA | 6.76 ± 0.13 | 6.52 ± 0.16 | 6.12 ± 0.33 | 5.28 ± 0.46 | 3.79 ± 0.99 | 2.33 ± 1.03 | |
| 06E01767 | AA | 6.83 ± 0.06 | 6.82 ± 0.09 | 6.63 ± 0.01 | 6.06 ± 0.40 | 4.72 ± 0.93 | 3.56 ± 1.10 |
| NA | 6.70 ± 0.09 | 6.63 ± 0.30 | 6.41 ± 0.35 | 6.25 ± 0.51 | 4.65 ± 0.52 | 2.95 ± 1.11 | |
| 06E02109 | AA | 6.93 ± 0.14 | 6.62 ± 0.10 | 6.78 ± 0.08 | 6.19 ± 0.47 | 3.20 ± 1.05 | ND |
| NA | 6.30 ± 0.13 | 6.04 ± 0.22 | 5.97 ± 0.23 | 5.74 ± 0.18 | 5.35 ± 0.32 | 4.92 ± 0.39 | |
| 06E02128 | AA | 6.85 ± 0.07 | 6.95 ± 0.09 | 6.86 ± 0.04 | 6.49 ± 0.11 | 2.93 ± 1.29 | ND |
| NA | 6.44 ± 0.34 | 5.55 ± 0.22 | 5.30 ± 0.19 | 5.10 ± 0.19 | 4.84 ± 0.02 | 4.65 ± 0.22 | |
| 06E01456 | AA | 6.93 ± 0.08 | 6.97 ± 0.07 | 6.87 ± 0.14 | 5.87 ± 0.66 | 2.45 ± 0.55 | ND |
| NA | 6.10 ± 0.06 | 5.59 ± 0.05 | 5.49 ± 0.31 | 5.31 ± 0.33 | 4.82 ± 0.26 | 4.64 ± 0.55 | |
| 06F00475 | AA | 6.69 ± 0.28 | 6.34 ± 0.46 | 6.05 ± 0.71 | 5.50 ± 0.91 | 3.38 ± 0.60 | ND |
| NA | 6.48 ± 0.19 | 6.01 ± 0.13 | 5.71 ± 0.11 | 5.33 ± 0.46 | 4.85 ± 0.75 | 4.63 ± 0.91 | |
| 06F00480 | AA | 6.96 ± 0.10 | 6.81 ± 0.16 | 6.72 ± 0.34 | 6.30 ± 0.52 | 3.75 ± 0.18 | ND |
| NA | 6.19 ± 0.16 | 5.81 ± 0.16 | 5.66 ± 0.09 | 5.49 ± 0.28 | 5.36 ± 0.40 | 4.43 ± 1.21 | |
| 06E01595 | AA | 6.98 ± 0.07 | 6.87 ± 0.14 | 6.75 ± 0.12 | 6.07 ± 0.97 | 3.98 ± 0.80 | ND |
| NA | 6.66 ± 0.10 | 6.14 ± 0.12 | 6.02 ± 0.10 | 5.86 ± 0.04 | 5.69 ± 0.38 | 5.39 ± 0.36 | |
AA, acid adapted; NA, non-acid adapted.
ND, not detected. The detection limit for plating was 1.32 log10 CFU. For the statistical analyses, random values were generated between 0 and 1.32 log10 CFU for samples in which no bacteria were detectable. The means were calculated with two independent samples of each strain.
Biofilm formation.
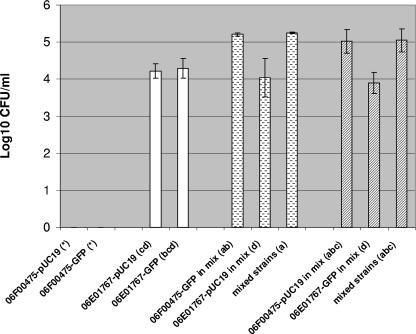
The formation of E. coli biofilm is often associated with the expression of Congo red-binding curli fimbriae and exopolysaccharides (27). Strain 06E01767 (O−:H4), but none of the 16 O157:H7 strains in this study, bound Congo red dye (results not shown). We compared all 17 strains for biofilm formation using crystal violet assays (optical density) or direct plate counts (CFU) and analyzed the results using analysis of variance with means separation by a Bonferroni least significant difference technique (23). Strain 06E01767 bound fivefold-greater amounts of crystal violet than the other 16 strains (P < 0.05), which were not different from the negative control (results not shown). Strains 06E01767, 06E02109 (an isolate from the lettuce outbreak), and 06F00475 (an isolate from the same spinach bag as 06E01767) were tested in a model DFR 110 drip flow biofilm reactor (BioSurface Technologies Corp., Bozeman, MT) by following the manufacturer's protocol. Strain 06F00475 produced only small amounts of patchy biofilm on glass, while strain 06E01767 generated a continuous, dense biofilm (not shown). These results agree with past findings (24) that the majority of serotype O157:H7 strains do not express curli fibers or form strong biofilms in the laboratory. However, as one of the serotype O157:H7 isolates resided in the same spinach bag as the strong-biofilm-forming strain 06E01767, we questioned whether strain 06F00475 could persist within a mixed-strain biofilm. We compared strains 06E01767 and 06F00475 and mixtures of both for biofilm formation on glass coupons in LB-NS medium, following 72 h of incubation by a previously described procedure with modifications (23). Biofilms were washed in phosphate-buffered saline (three times) by vortexing them for 30 s and transferred to tubes containing 0.1% peptone water and 0.3 g glass beads; the air-medium interface was scraped with a spatula. After being vortexed for 30 s and the removal of the slides, the tubes were vortexed again for 5 min and CFU were enumerated. Transformation with either pUC19 or pGFPuv (BD Biosciences Clontech, Palo Alto, CA) allowed for simultaneous quantification and differentiation of strains. While strain 06F00475 containing either plasmid could not be consistently recovered, strain 06E01767 was recovered at >4 log10 CFU/ml (Fig. 2). When cultured together, both strains were recovered at >4 log10 CFU/ml. When the hosts and plasmids were reversed, the results were similar. The total numbers of cells recovered from mixed-strain experiments were not different (P > 0.05), regardless of which strain carried green fluorescent protein. Likewise, the numbers of cells of strain 06F00475, when carrying either plasmid, were not different from each other or from the total number of mixed cells (P > 0.05). However, the recovered numbers of cells of strain 06E01767, carrying either plasmid, were lower (P < 0.05) than the total number of cells or the number of cells of strain 06F00475. These results indicate that the amount of biofilm produced by strain 06E01767 was constant whether the strain was incubated individually or in the presence of strain 06F00475. However, the non-biofilm-forming strain 06F00475, when grown together with strain 06E01767, persisted in numbers greater than those of strain 06E01767. It should be noted that strain 06E01767 biofilm could not be dislodged by vortexing it with glass beads; rather, it required physical abrasion. When slides were washed gently without vortexing, the numbers of cells recovered from 06F00475, 06E01767, and mixed-strain slides were similar, indicating that strain 06F00475 attached loosely to glass although it formed no biofilm (results not shown). Whether strain 06F00475 was an active or a passive participant in biofilm formation is being studied.
FIG. 2.
Mean log10 numbers of CFU/ml of strains recovered from biofilms formed on glass in LB-NS ± standard deviations. Strains 06F00475 (serotype O157:H7; spinach isolate) and 06E01767 (serotype O−:H4; spinach isolate), carrying either plasmid pGFPuv or plasmid pUC19, were tested in biofilms individually (plain bars) or as mixed strains (patterned bars). The two plasmids used in the experiment whose results are represented by the bars with broken horizontal lines are reversed from the plasmids used in the experiment whose results are represented by hatched bars. The means were calculated from three independent samples of each strain. Letters in parentheses following the strain designations represent the results of a Bonferroni least square difference means separation. Values for strains with the same letter are not statistically different from each other (P < 0.05). *, the number of CFU was below the limits of detection.
The results of this study suggest that the E. coli O157:H7 strains associated with spinach and lettuce in the outbreaks of 2006 are closely related. The finding of both the stx2 and stx2c variants in these strains supports the need for further research to investigate the role of the dual expression of stx2 and stx2c in STEC virulence and the development of HUS. Our studies with pairs of patient/food isolates suggest that differences in certain stress tolerances, virulence genotypes, or levels of biofilm formation did not develop following passage through the gastrointestinal tract. Moreover, the strains associated with the produce outbreaks did not display unusual stress resistance characteristics or biofilm-forming abilities compared to those of isolates from other sources, suggesting that the produce isolates did not undergo a drastic stress adaptation. However, broader and more-detailed studies need to be conducted to fully assess STEC adaptation to environmental growth. Finally, this study clearly demonstrates that in situations where environmental contamination with enteric bacteria results in the mixed-species contamination of food products, nonvirulent isolates could play an important role in the persistence of serotype O157:H7 on solid surfaces.
Acknowledgments
We gratefully acknowledge the work of James Tait, George Fraser, Barry Perry, Carol Sandt, Charles Cook, and Jonathan Sabo in serotyping and fingerprinting the human isolates and related food strains of E. coli. We also thank Giuseppina Esposito, Bryan Cottrell, Terence Strobaugh, Lori Bagi, Lisa Injaian, David Needleman, Peter Cooke, John Phillips, and Paul Pierlott for their technical assistance.
The mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 14 December 2007.
REFERENCES
- 1.Asakura, H., S.-I. Makino, T. Shirahata, T. Tsukamoto, H. Kurazono, T. Ikeda, and K. Takeshi. 1998. Detection and long-term existence of Shiga toxin (Stx)-producing Escherichia coli in sheep. Microbiol. Immunol. 42:683-688. [DOI] [PubMed] [Google Scholar]
- 2.Azizoglu, R. O., and M. Drake. 2007. Impact of antibiotic stress on acid and heat tolerance and virulence factor expression of Escherichia coli O157:H7. J. Food Prot. 70:194-199. [DOI] [PubMed] [Google Scholar]
- 3.Benito, A., G. Ventoura, M. Casadei, T. Robinson, and B. Mackey. 1999. Variation in resistance of natural isolates of Escherichia coli O157 to high hydrostatic pressure, mild heat, and other stresses. Appl. Environ. Microbiol. 65:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutin, L. 2006. Emerging enterohaemorrhagic Escherichia coli, causes and effects of the rise of a human pathogen. J. Vet. Med. B 53:299-305. [DOI] [PubMed] [Google Scholar]
- 5.Beutin, L., G. Krause, S. Zimmermann, S. Kaulfuss, and K. Gleier. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 42:1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandl, M. T. 2006. Fitness of human enteric pathogens on plants and implications for food safety. Annu. Rev. Phytopathol. 44:367-392. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan, R. L., and S. G. Edelson. 1996. Culturing enterohemorrhagic Escherichia coli in the presence and absence of glucose as a simple means of evaluating the acid tolerance of stationary-phase cells. Appl. Environ. Microbiol. 62:4009-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.California Food Emergency Response Team. 2007. Investigation of an Escherichia coli O157:H7 outbreak associated with Dole pre-packaged spinach. http://www.dhs.ca.gov/ps/fdb/local/PDF/2006%20Spinach%20Report%20Final%20redacted.PDF.
- 9.Carpentier, B., and O. Cerf. 1993. Biofilms and their consequences, with particular reference to hygiene in the food industry. J. Appl. Bacteriol. 75:499-511. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 2006. Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach—United States, September 2006. MMWR Morb. Mortal. Wkly. Rep. 55:1045-1046. [PubMed] [Google Scholar]
- 11.Fratamico, P. M., L. K. Bagi, E. J. Bush, and B. T. Solow. 2004. Prevalence and characterization of Shiga toxin-producing Escherichia coli in swine feces recovered in the National Animal Health Monitoring System's Swine 2000 study. Appl. Environ. Microbiol. 70:7173-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fratamico, P. M., S. K. Sackitey, M. Wiedmann, and M. Y. Deng. 1995. Detection of Escherichia coli O157:H7 by multiplex PCR. J. Clin. Microbiol. 33:2188-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich, A. W., M. Bielaszewska, W.-L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 14.Gannon, V. P. J., S. D'Souza, T. Graham, R. K. King, K. Rahn, and S. Read. 1997. Use of the flagellar H7 gene as a target in multiplex PCR assays and improved specificity in identification of enterohemorrhagic Escherichia coli strains. J. Clin. Microbiol. 35:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorden, J., and P. L. C. Small. 1993. Acid resistance in enteric bacteria. Infect. Immun. 61:364-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch, C., S. Hertwig, R. Lurz, B. Appel, and L. Beutin. 2001. Isolation of a lysogenic bacteriophage carrying the stx1ox3 gene, which is closely associated with Shiga toxin-producing Escherichia coli strains from sheep and humans. J. Clin. Microbiol. 39:3992-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar, C. G., and S. K. Anand. 1998. Significance of microbial biofilms in food industry: a review. Int. J. Food Microbiol. 42:9-27. [DOI] [PubMed] [Google Scholar]
- 18.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowbury, R. 2005. Stress responses of foodborne pathogens, with specific reference to the switching on of such responses, p. 77-97. In P. M. Fratamico, A. K. Bhunia, and J. L. Smith (ed.), Foodborne pathogens: microbiology and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 20.Sandt, C. H., D. A. Krouse, C. R. Cook, A. L. Hackman, W. A. Chmielecki, and N. G. Warren. 2006. The key role of pulsed-field gel electrophoresis in investigation of a large multiserotype and multistate food-borne outbreak of Salmonella infections centered in Pennsylvania. J. Clin. Microbiol. 44:3208-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, J. L., and P. M. Fratamico. 2005. Diarrhea-inducing Escherichia coli, p. 357-382. In P. M. Fratamico, A. K. Bhunia, and J. L. Smith (ed.), Foodborne pathogens: microbiology and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 22.Strauch, E., C. Schaudinn, and L. Beutin. 2004. First-time isolation and characterization of a bacteriophage encoding the Shiga toxin 2c variant, which is globally spread in strains of Escherichia coli O157. Infect. Immun. 72:7030-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhlich, G. A., P. H. Cooke, and E. B. Solomon. 2006. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl. Environ. Microbiol. 72:2564-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlich, G. A., J. E. Keen, and R. O. Elder. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:2367-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whiting, R. C., and M. H. Golden. 2002. Variation among Escherichia coli O157:H7 strains relative to their growth, survival, thermal inactivation, and toxin production in broth. Int. J. Food Microbiol. 75:127-133. [DOI] [PubMed] [Google Scholar]
- 26.Yokoigawa, K., A. Takikawa, Y. Okubo, and S. Umesako. 2003. Acid tolerance and gad mRNA levels of Escherichia coli O157:H7 grown in foods. Int. J. Food Microbiol. 82:203-211. [DOI] [PubMed] [Google Scholar]
- 27.Zogaj, X., W. Bokranz, M. Nimtz, and U. Römling. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]