Abstract
Natural remediation of oil spills is catalyzed by complex microbial consortia. Here we took a whole-community approach to investigate bacterial incorporation of petroleum hydrocarbons from a simulated oil spill. We utilized the natural difference in carbon isotopic abundance between a salt marsh ecosystem supported by the 13C-enriched C4 grass Spartina alterniflora and 13C-depleted petroleum to monitor changes in the 13C content of biomass. Magnetic bead capture methods for selective recovery of bacterial RNA were used to monitor the 13C content of bacterial biomass during a 2-week experiment. The data show that by the end of the experiment, up to 26% of bacterial biomass was derived from consumption of the freshly spilled oil. The results contrast with the inertness of a nearby relict spill, which occurred in 1969 in West Falmouth, MA. Sequences of 16S rRNA genes from our experimental samples also were consistent with previous reports suggesting the importance of Gamma- and Deltaproteobacteria and Firmicutes in the remineralization of hydrocarbons. The magnetic bead capture approach makes it possible to quantify uptake of petroleum hydrocarbons by microbes in situ. Although employed here at the domain level, RNA capture procedures can be highly specific. The same strategy could be used with genus-level specificity, something which is not currently possible using the 13C content of biomarker lipids.
Coastal environments are threatened by petroleum spills ranging from low-level discharges to catastrophic accidents. Large spills commonly are followed by clean-up efforts, but complete containment is rare. In all cases, remediation ultimately depends on microbial degradation. The rate of this natural bioremediation varies with physical and biological factors (temperature, wind and wave action, macroecology, and microbial community diversity), all of which have been extensively studied and reviewed (3, 4, 23, 27, 42, 69).
The bacterial genera most commonly identified as hydrocarbon degraders include Pseudomonas, Alcanivorax, Marinobacter, and Cycloclasticus (gammaproteobacteria) (5, 14, 19, 74); Sphingomonas (alphaproteobacteria) (18); gram-positive bacterial genera such as Staphylococcus and Geobacillus (43, 76); and deltaproteobacterial genera such as Desulfobacter, Desulfococcus, Desulfosarcina, and Desulfatibacillum (1, 12, 24, 51, 63). However, because many of the methods for measuring bacterial degradation of petroleum rely on enrichment cultures or isolates (58, 68, 72), the rates at which hydrocarbons are consumed in situ can be difficult to determine. Studies of the degradation of model hydrocarbons by species in pure or enrichment cultures cannot reflect the rates of conversion of oil into biomass by complex microbial communities in their native environments. Since petroleum is a mixture of many compounds with different degradation potentials, estimates based on the uptake of radiolabeled substrates (usually 14C-labeled n-alkanes) (69) likely do not represent the utilization rate for the total oil. This is especially evident in gas chromatograms of contaminated sediments, where typically an “unresolved complex mixture” (UCM) is a dominant feature in addition to n-alkanes (54). Newer methods of stable-isotope labeling (8) or probing (52) combined with molecular analyses of community diversity by PCR-denaturing gradient gel electrophoresis (35, 56) are promising, but they can require long incubation times and the results are difficult to convert to estimates of oil utilization rates. These methods also are subject to the challenges described above, namely, selection of a single molecule as a proxy for a complex, heterogeneous mixture.
To quantify the microbial incorporation of oil in coastal salt marsh habitats, isotope labeling experiments may not be necessary. It is possible to use whole oil and thus avoid many of the concerns raised above. Variations in the natural abundance of the stable isotopes of carbon, 12C and 13C, can be used to trace the carbon sources used by heterotrophic organisms (7, 13). These ratios are expressed as values of δ13C:
 |
Coffin and colleagues (11) demonstrated that the natural 13C abundance of total RNA extracted from a coastal salt marsh could be used to determine the sources of carbon utilized by the active biological community. Spartina alterniflora, a common grass found in salt marshes, has a δ13C value of approximately −13‰ (48). The values of δ13C measured for heterotrophic bacteria living on salt marsh detritus are similar to the values for Spartina, as the marsh grass is the primary producer in these ecosystems. In contrast, fossil fuels, which are derived mainly from marine products, have a lower natural abundance of 13C, resulting in more negative values of δ13C. Although petroleum mixtures contain a wide variety of hydrocarbon structures, the δ13C value of most oils is near −27‰ (55). In this work, our goal was to utilize the difference between the natural isotopic compositions of these substrates (13C-enriched Spartina and 13C-depleted petroleum) to monitor uptake of oil-derived carbon into salt marsh bacteria.
We studied whole-community uptake of hydrocarbons using natural-abundance 13C while simultaneously investigating the microbes likely to be responsible for the uptake. The 16S rRNA molecule (rRNA) is a logical “biomarker” that can link isotopic composition and taxonomy (39, 40, 46). Specifically, we combined (i) phylogenetic approaches to obtain sequences of 16S rRNA genes with (ii) carbon isotopic analyses of native rRNA molecules. Since rRNA in actively growing bacteria can account for 20% of the cellular mass (dry weight), it is possible to purify sufficient quantities of total bacterial or group-specific rRNA molecules to subject the material directly to isotopic analysis.
To this end, we used a moving-wire interface that allows 13C analysis of complex, nonvolatile organic samples, such as nucleic acids (9). Recent improvements in the sensitivity of the device, now termed “spooling-wire microcombustion” (SWiM) (15, 60), made this work feasible by providing sample requirements compatible with RNA purification protocols. Typical elemental analyzers require several orders of magnitude more material. The rRNA-SWiM approach is likely to have many future applications in investigations of carbon metabolism in heterogeneous microbial communities.
In September 1969, the oil barge Florida spilled 700,000 liters of diesel fuel (no. 2 fuel oil) in Buzzards Bay near West Falmouth, MA (22). This event has become one of the most well-studied oil spills in history. Thirty years after the spill, sediments in nearby Wild Harbor contain remains of degraded oil. The abundance and composition of the residue, known as a UCM, changed only slightly between 1973 and 2000, and the residue remains confined to a narrow 2- to 3-cm horizon now located approximately 10 cm deep in the marsh sediments (54). In the current work, Wild Harbor served as a control. Specifically, we expected to detect no petroleum-related signal in the δ13C values of rRNA, since active biodegradation is now undetectable in the contaminated sediment (62). We then used natural-abundance 13C-rRNA-SWiM measurement to quantify the microbial assimilation of hydrocarbons from a modern, simulated spill in a similar environment. We selected nearby Woodneck Marsh, an uncontaminated site, for the simulated spill. This location is a tidally flushed salt marsh similar to Wild Harbor and is also adjacent to Sippewissett Marsh, where community microbial diversity and metabolism have been studied extensively (21, 25, 61). Here we describe both the microbial diversity enriched by addition of no. 2 fuel oil and the use of carbon isotopic signatures of rRNA to quantify incorporation of oil-derived carbon into microbial biomass.
MATERIALS AND METHODS
Wild Harbor samples.
A 32-cm-long core was taken from Wild Harbor marsh, West Falmouth, MA, at a temperature of 5°C (early April 2003) using a 10-cm-diameter polyvinyl chloride tube. This core was frozen at −70°C immediately and returned to the laboratory, where it was later cut in situ into 1-cm slices using a power saw (Ryobi TS230). Each slice was divided into two halves; one half was used for analysis of hydrocarbons by gas chromatography-mass spectrometry, and the other half was used for RNA extraction.
Woodneck Marsh bulk samples.
Bulk samples of Spartina and wild rose were collected from Woodneck Beach Salt Marsh, Falmouth, MA. In addition, bulk biomass was skimmed from the surface of a shallow tidal pool, and a mussel was collected from the same location. These samples were frozen in N2(l) and homogenized with a mortar and pestle. A portion of each bulk sample was prepared for analysis of δ13C of total organic carbon using standard sealed-tube combustion procedures (44). Values of δ13C were determined using purified CO2 by dual-inlet isotope ratio mass spectrometry (VG Prism).
Woodneck Marsh incubation.
Twelve shallow cores (diameter, 14 cm; depth, 5 cm) were harvested in midsummer and immediately incubated in sterile (baked in air at 450°C for 8 h) Pyrex crystallizing dishes for 0, 1, or 2 weeks under ambient temperature and sunlight conditions; 8 of these cores were incubated with no. 2 fuel oil (Mobil diesel), and 4 were incubated without fuel oil. An excess of no. 2 fuel oil was supplied as an irregular layer of oil-coated sand on top of the eight inoculated samples. The sand was used to keep the oil from forming a continuous film and thus to allow gas exchange. Oil-free zones were observed throughout the experiment, and some areas of the sediment surface remained exposed. To simulate tidal flushing and prevent desiccation, ∼20 ml of sterile-filtered marsh water was added to the preparations daily. Four of the samples were harvested after 1 week, and the other four were harvested after 2 weeks. The samples were frozen at −80°C for extraction of hydrocarbons and nucleic acids.
Purification of bacterial rRNA. (i) RNA extraction.
Total RNA was extracted using methods described previously (46). Some samples were lysed in 50 mM sodium acetate (pH 5.2) containing 0.5% sodium dodecyl sulfate and 0.1 mg/ml proteinase K (Life Technologies), followed by extraction with phenol-chloroform-isoamyl alcohol (25:24:1) and 100% chloroform, and some samples were extracted with RNAWiz (Ambion) used according to the manufacturer's instructions. Nucleic acids were precipitated with isopropanol and washed three times with 70% ethanol, and the pellets were resuspended in Nanopure H2O. Visual inspection of electrophoresis gels confirmed that the total extracts were enriched in RNA but that some samples also contained small amounts of DNA. Humic acids were removed effectively by the RNAWiz method in particular, and significant A320 generally was not observed for these samples.
(ii) Magnetic bead capture of small-subunit rRNA.
Small-subunit rRNA (16S and 23S rRNA) of bacteria was captured from the total RNA extracts using an Ambion MICROBExpress bacterial mRNA enrichment kit, with minor modifications to the manufacturer's protocol. The MICROBExpress kit contains a probe cocktail designed to bind, capture, and remove bacterial 16S and 23S rRNA molecules to enrich the mRNA in the remaining solution (compatible genera are listed at http://www.ambion.com/techlib/misc/microbe.html). Here, the kit was employed to recover the bacterial rRNA fraction from mixed environmental RNA, and the uncaptured (mRNA) fraction was discarded. The total RNA, capture probe cocktail, and binding solution were hybridized and combined with the included paramagnetic beads, as instructed. After capture of the bead-probe-rRNA complex using a magnetic particle separator as described previously (46), the complex was washed and eluted in Nanopure H2O at 90°C for 60 s. Recovered rRNA was precipitated in 500 μl of isopropanol plus 10 μl of 5 M NaCl, centrifuged (10,000 × g, 15 min), and washed with 70% ethanol.
To remove residual carbon from buffers used in the capture procedure, samples were cleaned using an Ambion MEGAclear RNA purification kit. RNA recovered from the MEGAclear elution step was precipitated in 500 μl of isopropanol plus 10 μl of 5 M NaCl, centrifuged, and washed as described above. Recovered pellets were redissolved in 400 ml of Nanopure H2O and reprecipitated using high-performance liquid chromatography-grade isopropanol (Fisher). The final pellets were washed three times in 70% ethanol, dried at 60C, and redissolved in 10 μl of Nanopure H2O for isotopic analysis. Method blanks were prepared by processing aliquots of Nanopure water that were the same size using the entire hybridization, capture, precipitation, and clean-up procedure.
Isotopic analysis using a SWiM interface.
The SWiM system was described by Sessions et al. (60) and is a modification of the original device built by Brand and Dobberstein (9). Briefly, samples in ∼1-μl droplets of water are applied to a wire (0.25-mm nickel wire moving at a rate of 0.8 cm/s), which passes sequentially through a cleaning oven (950°C), a drying oven (120°C), and then a combustion oven (750°C) containing wireform CuO. The atmosphere is excluded from the combustion furnace by positive pressure of the helium carrier gas. A portion of the combustion gases flows through a countercurrent Nafion membrane to remove H2O, through an open split, and then to a ThermoFinnigan 252 isotope ratio mass spectrometer. CO2 reference gas for calibration of isotopic ratios was supplied via a custom-built interface similar to the Conflo III device, and samples were analyzed as sets of five to seven injections spaced at 45-s intervals. The carbon content and δ13C values were calculated from the peak areas of m/z 44 and 45 ion chromatograms. The precision of δ13C values attainable with this system is roughly 1‰ for samples containing as little as 10 ng C and 0.2‰ for samples containing 100 ng C (60).
Analytical blanks were estimated by repeatedly measuring 1-μl aliquots of sample-free extracts (described above). They typically contained <10% of the typical carbon concentration in processed RNA samples and had δ13C values of −23 to −24‰. The contributions to the overall analytical blank arising from the SWiM system are estimated to be <0.2 ng C (55), with the balance of C presumably derived from RNA extraction procedures. Blank contributions were subtracted from all reported sample δ13C values, and the error was propagated to the stated uncertainties using equations given by Hayes (26).
Analysis of hydrocarbons.
Aliquots of homogenized material were extracted by vortexing wet sediment in a 1:1 mixture of CH2Cl2 and H2O in 50-ml Teflon tubes. The tubes were centrifuged at 5,000 × g, and the CH2Cl2 layer was moved to a separatory funnel, where it was washed with H2O. The total organic extract was passed over anhydrous Na2SO4, reacted with HCl-activated Cu to remove elemental sulfur, and fractionated over SiO2 gel in hexane-CH2Cl2 (95:5). Hydrocarbons were analyzed by gas chromatography-mass spectrometry using an Agilent 6890N gas chromatograph interfaced to a 5973 MSD equipped with an HP-5MS column (30 m by 0.25 mm [inside diameter]; film thickness, 0.25 μm).The gas chromatograph temperature program was 65°C for 2 min, increasing to 130°C (0 min) at a rate of 20°C/min, to 280°C (0 min) at a rate of 6°C/min, and finally to 320°C (25 min) at a rate of 3°C/min.
Phylogenetic characterization.
Total DNA was extracted using an UltraClean Mega soil DNA kit from MoBio Labs and was concentrated and purified using a Qiagen QIAquick kit. Partial 16S rRNA genes were amplified from Woodneck Marsh samples using universal bacterial primers (0.4 μM each) 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1100R (5′-AGGGTTGCGCTCGTTG-3′), a total DNA template (20 ng), and Sigma JumpStart RedTaq Ready Mix for PCR. The thermal cycling conditions were 2 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1.5 min at 72°C and then a final extension step of 10 min at 72°C. Duplicates were run for each sample, and 2.5-μl portions of the duplicates were combined in one reaction mixture and amplified for an additional five cycles using a modified “low bias and reconditioning” protocol (67). PCR products were visualized on a 1.0% agarose gel using SYBR green. All reactions were performed using 25-μl (final volume) mixtures. Negative controls (reagents, lab water) were included with every PCR.
PCR products from all samples were cloned using an Invitrogen TOPO TA cloning kit for sequencing with E. coli One Shot TOP10 chemically competent cells. Clones were incubated overnight at 37°C on plates containing LB medium with ampicillin (50 μg/ml), inoculated into 5 ml of liquid medium, and incubated for 24 h at 37°C. Plasmids were purified using a Qiagen QIAprep spin miniprep kit. Sequencing was done at the Dana-Farber/Harvard DNA Resource Core (http://dnaseq.med.harvard.edu/).
Preliminary 16S rRNA sequence alignments were obtained using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and then were manually inspected for realignment and trimmed to ∼700 bp. A check for chimeric sequences suggested that there were no chimeras (Bellerophon server) (30). Maximum likelihood trees were created from the clone sequences using PHYML (20; http://atgc.lirmm.fr/phyml/). The substitution model was HKY, and 100 bootstrap replicates were calculated. Sequences from related species and from similar environmental clones were downloaded from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/) and included in the alignments and trees. Aquifex aeolicus served as the outgroup.
Rarefaction curves were calculated using distance-based operational taxonomic unit (OTU) and richness determination (DOTUR) (59). Distance matrices for the DNA sequences of the clones were calculated using default parameters of dnadist in PHYLIP (http://evolution.genetics.washington.edu/phylip.html). Sequences were then assigned to OTUs based on the unweighted-pair group method using average linkages (average neighbor) clustering algorithm implemented in DOTUR with default parameters for precision (0.01) and bootstrapping (1,000).
RESULTS
Wild Harbor relict oil spill.
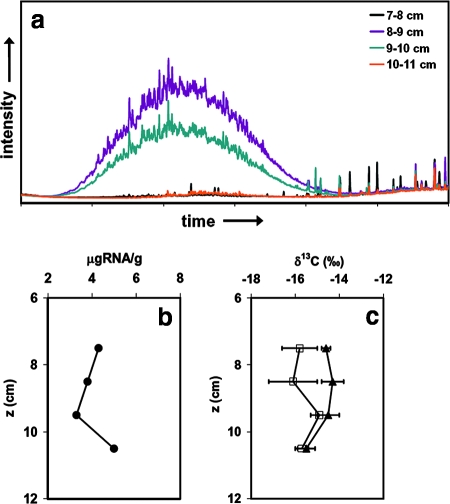
The residue of the no. 2 fuel oil spilled from the barge Florida was detected at a depth of 8 to 10 cm in the sediment core from Wild Harbor (Fig. 1a). This residue appeared to be a UCM of hydrocarbons, consistent with previous reports (54). The horizons immediately above and below did not contain significant UCM (Fig. 1a).
FIG. 1.
(a) Chromatograms of the sediment core from Wild Harbor showing a degraded UCM and no clear signature of n-alkanes. (b) Total RNA yield (in μg RNA/g [wet weight] of sediment). (c) δ13C values for total RNA (▴) and captured bacterial rRNA (□).
The RNA yields from this core were low (3.3 to 5.0 μg RNA/g [wet weight]) (Fig. 1b). Assuming that the RNA content of a typical bacterial cell is 6 × 10−14 g and using a conversion based on the estimated growth rate (6), this RNA concentration is consistent with the presence of 2 × 108 to 4 × 108 cells/g of sediment if all of the RNA was from bacterial biomass. This represents an upper boundary for the microbial cell density, as some of the RNA may have been derived from microaerophilic eukaryotes and/or from the roots of the Spartina grass itself. The results are also consistent with estimates derived from phospholipid fatty acid concentrations (62). The extractable RNA showed distinct bands representing the 16S and 23S rRNA molecules by gel electrophoresis; this indicated that there were intact ribosomes in the total community and presumably reflected the potential for metabolic activity.
The values of δ13C for the total RNA extracts ranged from −15.5 to −14.3‰ (Table 1). The differences between the values for extracts obtained in the region of the UCM and the values for the horizons above and below the residue of the spill were insignificant (Fig. 1c). The values of δ13C for the captured fraction of bacterial rRNA were systematically more negative by ∼1‰ than the values for the total RNA. Three of the four values were in the range from −16.1 to −15.7‰, while one sample from within the oil spill horizon (9 to 10 cm) had a slightly more positive δ13C value than the other samples (−14.9‰). The difference is not significant given the analytical uncertainties, and the direction of the change is opposite what would be expected if the bacteria in this horizon were using oil as a substrate (the δ13C for oil is approximately −27‰).
TABLE 1.
RNA yields from environmental samples
| Sample | RNA concn (μg/g)a |
|---|---|
| Wild Harbor core | |
| 7-8 cm | 4.3 |
| 8-9 cm | 3.8 |
| 9-10 cm | 3.3 |
| 10-11 cm | 5.0 |
| Woodneck Marsh surface sediment | |
| Control | 55 ± 13 |
| Incubation with oil for 1 wk | 40 ± 14 |
| Incubation with oil for 2 wk | 37 ± 10 |
The data are data for single measurements for Wild Harbor and for three measurements for Woodneck Marsh.
Woodneck Marsh fuel oil incubation experiment.
Eight samples of surface sediment from nearby Woodneck Marsh were amended with no. 2 fuel oil and harvested over a 2-week period. Four additional samples were reserved as controls (no oil). Four of the amended samples were harvested after 1 week, and four were harvested after 2 weeks. The controls were harvested simultaneously with the samples (two at each time point).
(i) Composition and degradation of oil.
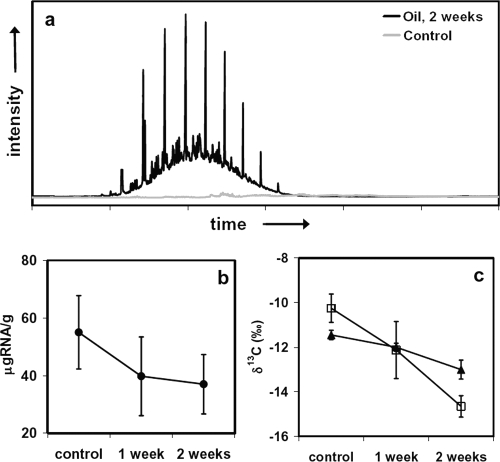
The control samples contained no detectable hydrocarbon-rich UCM and therefore had an insignificant petroleum component. All of the oil-inoculated samples still contained readily detectable petroleum (Fig. 2a). The chromatograms included a UCM similar to that found in the deeper, Wild Harbor sediments. As a result of the quantity of fresh oil added, n-alkanes were also visible in the chromatograms (Fig. 2a). We examined the composition of this oil before and after incubation by using gas chromatography-mass spectrometry. The net uptake of oil represented a very small fraction of the total amount added, and the composition remained unchanged; we thus presumed that the isotopic composition of the residual oil did not differ from the original composition.
FIG. 2.
(a) Chromatograms of the sediment cores from Woodneck Marsh showing, for incubation with oil, a regular series of n-alkanes centered around n-C20, which is absent in the control. (b) Total RNA yield (in μg RNA/g [wet weight] of sediment). (c) δ13C values for total RNA (▴) and captured bacterial rRNA (□).
(ii) Total RNA.
The RNA yields from the Woodneck Marsh samples were 1 order of magnitude higher than those from the Wild Harbor samples (37 to 55 μg RNA/g [wet weight]) (Table 1), indicating that the surface samples had a greater biomass density than the deeper sediments of the Wild Harbor core. The estimates in both cases could have been influenced by a small amount of residual DNA in the extracts (they were not treated with DNase), although visualization on an agarose gel suggested that the majority of the nucleic acid fractions obtained from both Woodneck Marsh and Wild Harbor was RNA. The Woodneck Marsh data suggest that the concentration was approximately 4 × 109 to 6 × 109 cells/g of sediment if all of the RNA was from bacterial biomass. The total RNA content—and therefore probably the total biomass—of the treatment samples decreased relative to the content of the control samples over the duration of the experiment (Fig. 2b), but 16S rRNA sequence data suggested that the total microbial diversities were similar in the two samples (see below).
The values of δ13C for the total RNA extracts ranged from −11.5 to −13.0‰ (Table 1). These values also decreased moderately over the course of the experiment (Fig. 2c), and the difference, ∼1.5‰, was greater than the isotopic heterogeneity among the replicate samples (the sample standard deviations were ±0.2 to 0.4‰). All treatment samples displayed isotopic depletion relative to the control (no-oil) samples.
(iii) Bacterial rRNA.
The values of δ13C for the captured fraction of bacterial rRNA ranged from −10.3 to −14.7‰. These values reflect a 4.4‰ difference between the control samples and the oil incubation samples at 2 weeks (Fig. 2c). The results are consistent with the incorporation of oil-derived carbon into bacterial biomass. The 4.4‰ decrease in the value of δ13C of captured bacterial rRNA could be used to estimate the fraction of new bacterial biomass (fnew) in the samples that was oil derived, either directly (heterotrophically) or indirectly (autotrophic incorporation of respired CO2).
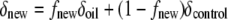 |
Using the control bacterial rRNA end member value of −10.3 ‰ ± 0.6‰, an assumed composition for oil of −27.0‰ ± 0.5‰, and the final value of −14.7‰ ± 0.5‰, the isotopic mass balance suggested that at 2 weeks, 26% ± 4% of the carbon in the captured bacterial rRNA originated from the oil.
Woodneck Marsh microbial community.
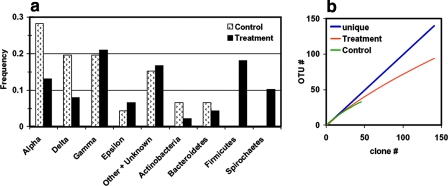
Partial gene sequences of 16S rRNA genes were obtained from three of the 2-week incubation samples and from one of the control samples (also after 2 weeks of incubation). Forty-six clones were sequenced from control sample SM-10, and 138 clones were sequenced from samples SM-3, SM-6, and SM-9 (treatments). The clones sequenced from the control sample included Alpha-, Delta-, and Gammaproteobacteria at the highest frequencies (Fig. 3a). In contrast, the treatment samples contained primarily Gammaproteobacteria, Firmicutes, Spirochaetes, and other or unknown organisms, while the relative number of Deltaproteobacteria clones decreased (Fig. 3a). The number of unique OTUs (the OTU threshold was set at 3% divergence) detected at similar sampling levels remained approximately the same, regardless of the presence of oil. Calculations using DOTUR (58) yielded 94 unique OTUs among the 138 sequences in the treatment samples (Fig. 3b) and 33 OTUs among the 46 sequences in the control sample (Fig. 3b). These numbers should be interpreted with caution, however, as the numbers of sequences examined were not the same for the two samples (46 sequences versus 138 sequences) and an estimate of community diversity obtained using the set of only 46 control sequences, in particular, was not likely to be very robust. The individual treatment samples had a diversity similar to that of the control sample; SM-3 contained 36 OTUs, SM-6 contained 43 OTUs, and SM-9 contained 35 OTUs. These numbers are consistent with similar levels of total diversity in all samples.
FIG. 3.
(a) Frequency of detection of 16S rRNA clones in the control and 2-week treatment samples. Control plot clones were designated SM-10 (46 clones were sequenced), and treatment plot clones were designated SM-3, SM-6, and SM-9 (138 clones were sequenced). (b) Rarefaction curves obtained by DOTUR for the treatment and control samples, using a cutoff value of 97% sequence identity.
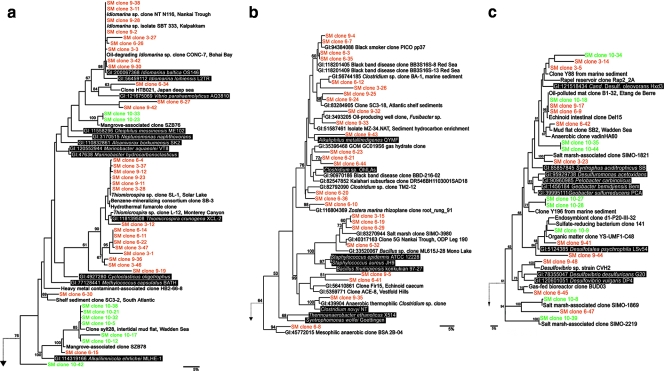
As expected, the vast majority of clones were closely related to previously reported marine or salt marsh microbial sequences. Many also were closely related to bacteria that grow in limited-oxygen environments, such as Clostridium, Methylococcus, and Desulfovibrio spp. (Fig. 4a, b, and c). Several of the sequences identified in the treatment samples shared 16S rRNA sequence similarity with other clones that are associated with hydrocarbon-contaminated environments (29, 49, 75). These clones include clones obtained from gas hydrates, oil wells, and other sediments.
FIG. 4.
Maximum likelihood tree of all sequenced Gammaproteobacteria (a), Firmicutes (b), and Deltaproteobacteria (c) clones calculated using PHYML as described in the text. Bootstrap support values are indicated for major nodes having values of >50%. Green, clones from the control samples; orange, clones from the treatment plots. Reference species are indicated by white letters on a black background. No Firmicutes were detected among the 45 clones from the control plot.
Although Gammaproteobacteria comprised about 20% of the clones detected in both the control and treatment samples (Fig. 3a), the distributions differed markedly (Fig. 4a). In the control samples, the clones were related to Methylococcus, Marinobacter, and Alkalilimnicola. Additional groups were detected in the treatment samples. Most of the new clones aligned with Idiomarina spp. or Thiomicrospira spp. and were similar to other oil-mineralizing consortia. Idiomarina spp. are heterotrophic aerobes (31), and Thiomicrospira spp. are microaerophilic sulfur oxidizers that fix CO2 (37).
No Firmicutes were identified in the control sample. However, 18% of the clones detected in the treatment samples were related to Firmicutes. The species detected were putative anaerobes closely related to Clostridium and Alkaliphilus (Fig. 4b) and were similar to other clones from gas hydrates, oil wells, and anaerobic hydrocarbon enrichments (53, 75).
Many of the new Deltaproteobacteria in the treatment library were similar to clones from hydrocarbon-contaminated systems. These Deltaproteobacteria sequences, however, were more evenly distributed among both the control and treatment samples (Fig. 4c). In both types of samples, clones were found that were related to alkane-degrading species, such as “Candidatus Desulfococcus oleovorans” Hxd3 (1, 63). Clones whose sequences were 100% identical to the sequence of a clone from an oil-degrading hydrocarbon mat from Etang de Berre (29) were also found in both the treatment and control samples; this group also clustered more closely with Desulfococcus spp. than with other sulfate-reducing Deltaproteobacteria (Fig. 4c) The preponderance of the remaining clones appeared to be related to sulfate-reducing species of Deltaproteobacteria and included Desulfovibrio and Desulfotalea spp. No clones closely related to Geobacter or Pelobacter spp. were detected.
Many of the clones in the other major categories (Fig. 3a) also were closely related to clones from hydrocarbon-rich environments, consistent with the overall observation that the experimental clone library reflected many species that have previously been shown to be involved, directly or indirectly, in the metabolism of hydrocarbons (29, 64, 66, 70).
Additional measurements.
The SWiM device was used to obtain δ13C data for RNA extracts of pure cultures for method development (Table 2). Concurrent analyses of nucleotide standards, bulk RNA standards, and total nucleic acid extracts produced δ13C results that had an analytical precision (based on three to five injections) of around ± 0.2‰ and that were accurate to within ±1.0‰ for samples containing 100 nmol of carbon (60). Values of δ13C measured for RNA using the SWiM device were consistently 1 to 2‰ enriched in 13C relative to the biomass from which they were derived (shown for E. coli in Table 2; also measured for Ferroplasma acidarmanus [data not shown]). The results agree with previous data which established that the 13C content of RNA is influenced by the ribosugar component (7), because sugars tend to be enriched in 13C relative to total biomass.
TABLE 2.
Isotopic compositions of samples from Wild Harbor and Woodneck Marsh
| Sample | Type | δ13C (‰) | Error (1σ) (‰) |
|---|---|---|---|
| Wild Harbora | |||
| 7.5 cm | Total RNA | −14.6 | 0.2 |
| 8.5 cm | Total RNA | −14.3 | 0.5 |
| 9.5 cm | Total RNA | −14.5 | 0.5 |
| 10.5 cm | Total RNA | −15.5 | 0.4 |
| 7.5 cm | Captured rRNA | −15.8 | 0.8 |
| 8.5 cm | Captured rRNA | −16.1 | 1.1 |
| 9.5 cm | Captured rRNA | −14.9 | 0.4 |
| 10.5 cm | Captured rRNA | −15.7 | 0.3 |
| Woodneck Marshb | |||
| Control | Total RNA | −11.5 | 0.2 |
| Oil incubation, 1 wk | Total RNA | −12.0 | 0.2 |
| Oil incubation, 2 wk | Total RNA | −13.0 | 0.4 |
| Control | Captured RNA | −10.3 | 0.6 |
| Oil incubation, 1 wk | Captured rRNA | −12.1 | 1.3 |
| Oil incubation, 2 wk | Captured rRNA | −14.7 | 0.5 |
| Bulk samples from Woodneck Marsh | |||
| Grass (S. alterniflora) | Emerged grass | −13.9 | 0.2c |
| Wild rose | Whole leaf | −27.4 | 0.2c |
| Mussel (G. demissa) | Whole tissue | −14.8 | 0.2c |
| Marsh microbial mat total organic carbon | Whole mat & sediment | −15.6 | 0.2c |
| Blanks | |||
| Total | Total RNA blank | −23.4 | 1.2d |
| Captured | Captured RNA blank | −24.2 | 1.2d |
| E. coli controls | |||
| E. coli whole | Whole cells | −22.8 | 0.2c |
| E. coli total | Total RNA | −21.5 | 0.1 |
| E. coli captured | Captured RNA | −20.6 | 0.6 |
The values of δ13C for Wild Harbor are based on triplicate measurements of individual samples.
The values of δ13C for Woodneck Marsh are based on triplicate measurements for four replicate samples; the errors were calculated by determining sample standard deviations (1σ).
Estimated error for closed-tube combustion.
The error for the captured sample was measured; the error for the total sample was assumed to be equivalent to the error for the captured sample.
Other carbon isotopic data were obtained for bulk biomass of major components of the Woodneck Marsh salt marsh community. The data include samples of the major primary producer, the C4 grass S. alterniflora, as well as a C3 plant (wild rose). A marsh mud-dwelling mussel, Geukensia demissa, and a mixed sample of whole marsh mud plus bacteria (mat total organic carbon) also were examined.
DISCUSSION
Residual no. 2 fuel oil remains in the sediment in Wild Harbor (54, 71), where it spilled from the barge Florida in 1969. Active biodegradation of this residue by the bacterial community is undetectable, as determined previously using natural-abundance 14C measurements of intact phospholipids extracted from the sediments (62). Our data agree with this previous conclusion; the microbial incorporation of highly degraded oil is insignificant. The amount of 13C in bacterial rRNA was uniform, regardless of whether it was purified from samples obtained within or outside the relict, spill-contaminated horizons (Fig. 1c). However, this environment also is different from Woodneck Marsh in several ways that could significantly affect (reduce) the microbial metabolism of hydrocarbons. It is buried deep within the anoxic zone of the marsh sediment and has no contact with oxygenated substrates other than possible irrigation via the Spartina roots. The total carbon environment and total microbial community in this anoxic setting also must be different from those at Woodneck Marsh. This is reflected in the δ13C value for total RNA in Wild Harbor, which is 3‰ less isotopically (approximately −14.5‰) than the value for untreated total RNA from the surface community in Woodneck Marsh (−11.5‰). This effect could be the result of differences in the carbon substrates and assimilation pathways used by the microbial communities in the two locations.
In the Woodneck Marsh sediments, both the phylogenetic distribution and the 13C content of bacterial rRNA were affected by the addition of fresh fuel oil. The data demonstrate that the salt marsh microbial community responded rapidly to the introduction of hydrocarbon contaminants. The observation that this carbon entered the active metabolic pool—as detected using the labile 16S and 23S rRNA molecules—is consistent with the incorporation of carbon from petroleum into microbial biomass. Biodegradation of hydrocarbons proceeds first through the more labile n-alkanes to more recalcitrant molecules, such as aromatic hydrocarbons and branched alkanes (54, 58, 69). The duration of the experiment captured only the initial degradation of the labile fraction of oil, and the experiment was not sensitive enough to detect differential utilization of structurally or isotopically distinct components; a longer experiment is needed to determine changes in the microbial community after the depletion of n-alkanes.
However, in this short experiment we estimated that after 2 weeks, up to ∼26% of the bacterial carbon was derived from the oil. Stable isotope labeling and analysis of captured rRNA have been used previously to assess microbial uptake of a variety of “tracer” carbon compounds (40). Other studies have examined the catabolic conversion of hydrocarbon substrates into CO2 (2), while more recent approaches have included isotope labeling methods (DNA stable isotope probing or lipids) to detect the flow of oil-derived carbon into active microbes (8, 41, 47). Many of these studies utilized labeled substrates and/or examined only a single compound. To our knowledge, the study presented here is the first study to use the natural abundance of 13C in rRNA to trace the microbial incorporation of carbon from total oil as a substrate. Conversely, we could not detect if some components of the oil were preferentially consumed relative to other components, other than to note that the total composition of the mixture did not appear to change with time. Regardless, in general, this experiment expanded the potential for using non-isotope-labeled nucleic acids as tracers for natural biogeochemical processes. In the case of petroleum biodegradation, this approach may be particularly well suited.
Oil spills affect sediment-associated microbial consortia by stimulating the growth of species involved directly and indirectly in metabolism of the complex hydrocarbon mixtures (28, 32, 57, 75). In the Woodneck Marsh incubations we observed differences in the community composition between the controls and the treatment experiments, both within groups (Gammaproteobacteria) and with the appearance of new groups (Firmicutes). The frequency of our clone phylogenies shifted from primarily Alpha-, Gamma-, and Deltaproteobacteria in the control samples to mainly Gammaproteobacteria, Firmicutes, and Spirochaetes in the oil-treated samples. Sulfate-reducing Deltaproteobacteria were present in both types of samples, but the relative number of clones decreased and the diversities were comparable in the control and treatment samples. Also, we did not find examples of frequently isolated oil-degrading genera, such as Azoarcus, Cycloclasticus, or Alcanivorax. Alcanivorax and Cycloclasticus are two of the best-known genera of marine hydrocarbon degraders (14, 33, 34, 56). The failure to detect clones related to these genera may have been due to environmental differences between the Woodneck Marsh salt marsh sediments and other, more open, oceanic sites or could have been due to the relatively short incubation time of the experiment (14 days). The overall biodiversity did not decrease, consistent with some previous reports for hydrocarbon-contaminated marine systems (29, 56).
Many genera of Gammaproteobacteria degrade hydrocarbons in pure or enrichment cultures, and clones associated with Gammaproteobacteria commonly are detected in environmental incubations and in hydrocarbon-contaminated sediment samples (16, 29, 68, 74, 75). Our enrichment of a set of clones with high similarity to Idiomarina spp. (Fig. 4a) also is consistent with the previous detection of this genus in association with oil-rich microbial mats and sediments, (29, 36, 57), although the role that Idiomarina plays in the degradation of hydrocarbons is not known.
The sediment microbial community changed visibly when the oil was added, and gradually white filaments appeared to replace pink and purple filaments. This observation is consistent with the results of our clone library sampling analysis; decreased numbers of Alphaproteobacteria clone representatives and increased numbers of clones that are related to sulfur oxidizers (among the total clones of Gammaproteobacteria) were detected in the library generated for the treatment samples. Many of the new clones observed in the oil-treated samples were associated with Thiomicrospira (73) and other sulfur-oxidizing environmental clones (49, 65). Local anoxia also was indicated by the results for the Woodneck Marsh treatment samples. Microbial consortia associated with hydrocarbon degradation in marine sediments frequently contain sulfate reducers (Deltaproteobacteria) (49, 57, 66), and sulfate-reducing Deltaproteobacteria clones were detected throughout the experiment. Microaerophilic or anoxic conditions also were suggested by the presence of clones affiliated with the genus Clostridium, which were absent from the controls.
While we found that the phylogenetic distributions of clones were different for the treatment and control samples, our results do not quantitatively reflect the population changes induced by the mock oil spill. Biases can be introduced by differential copy numbers of 16S rRNA genes or through artifacts of extraction or PCR (10, 17, 38, 50). The results described here thus provide an overview of differences between the oil-treated and control samples. Future work should combine quantitative PCR with the greater phylogenetic specificity of the 13C-rRNA-SWiM approach in order to obtain a more detailed understanding of the quantitative role that specific microbial groups play in oil degradation.
This work provides a foundation for studies of the metabolic pathways of carbon assimilation in heterogeneous microbial consortia. Here, measurements obtained using the natural isotopic contrast between 13C-depleted oil and a 13C-enriched salt marsh ecosystem showed that hydrocarbons serve as a carbon source at the same time that their oxidation serves as a source of energy. The 13C content of purified bacterial rRNA showed the formation of 13C-depleted biomass over a period of 2 weeks. Most of the isotopically depleted signal was presumed to reflect carbon assimilated from the hydrocarbon amendment, and clonal enrichment of organotrophs is consistent with this observation.
Further study should allow us to investigate hydrocarbon uptake by environmental species using rRNA capture probes having greater specificity. Genus-specific 13C-rRNA-SWiM should be able to quantify the clones detected in the treatment samples, including Idiomarina spp., that reflect carbon assimilated directly from oil. We also noted that while the suite of RNA capture probes used here recovers most genera of bacteria, it is not universally compatible. Some excluded organisms of potential environmental interest are Pirellula (and presumably other Planctomycetales), Chloroflexus (and presumably other Chloroflexales), and Thermatogales (presumably including any of their low-temperature relatives). These exceptions highlight the need for group-targeted probes in future work.
The 13C-rRNA-SWiM approach offers several advantages over traditional substrate incubations. Such approaches provide a valuable new method for studying hydrocarbon assimilation by specific organisms and complement other methods, such as DNA isotope probing (52), labeled-substrate RNA capture (40), and analysis of labeled phospholipid fatty acids (8, 47). Additionally, the investment in technology and labor is arguably less resource-intensive than ion probe (secondary ion mass spectrometry) (45) techniques, although to date our laboratories house the only SWiM interfaces available. The next steps toward broader application of this approach, therefore, include development of new capture probes and broader collaborations between microbiologists and isotope geochemists at the interface of these analytical approaches. Routine δ13C analyses of rRNA now require <50 ng C, which is equivalent to the RNA content of 104 to 106 cells, depending on the growth rate. This should open new windows for understanding carbon flows and pathways in complex microbial consortia.
Acknowledgments
We are indebted to John M. Hayes for his enduring guidance and support. The manuscript was greatly improved by the detailed comments of two anonymous reviewers. Helen White, Bob Nelson, and Chris Reddy provided access to samples from Wild Harbor. Helen White also gave helpful comments on an early manuscript draft. Sean Sylva, Dan Rogers, Steve Petsch, and Susan Carter are thanked for general laboratory assistance. Al Gagnon provided the analyses of δ13C by closed-tube combustion.
This work was supported by a grant from the Reinhart Coastal Research Center of Woods Hole Oceanographic Institution (to A.P. and K.J.E.) and by grant NSF-EAR-0311937 (to A.P.).
Footnotes
Published ahead of print on 14 December 2007.
REFERENCES
- 1.Aeckersberg, F., F. Bak, and F. Widdel. 1991. Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch. Microbiol. 156:5-14. [Google Scholar]
- 2.Aggarwal, P. K., M. E. Fuller, M. M. Gurgas, J. F. Manning, and M. A. Dillon. 1997. Use of stable oxygen and carbon isotope analyses for monitoring the pathways and rates of intrinsic and enhanced in situ biodegradation. Environ. Sci. Technol. 31:590-596. [Google Scholar]
- 3.Atlas, R. M., and R. Bartha. 1973. Stimulated biodegradation of oil slicks using oleophilic fertilizers. Environ. Sci. Technol. 7:538-541. [DOI] [PubMed] [Google Scholar]
- 4.Atlas, R. M. 1981. Microbial degradation of petroleum hydrocarbons—an environmental perspective. Microbiol. Rev. 45:180-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartha, R. 1977. The microbiology of aquatic oil spills. Adv. Appl. Microbiol. 22:225-266. [DOI] [PubMed] [Google Scholar]
- 6.Binder, B. J., and Y. C. Liu. 1998. Growth rate regulation of rRNA content of a marine Synechococcus (cyanobacterium) strain. Appl. Environ. Microbiol. 64:3346-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blair, N., A. Leu, E. Muñoz, J. Olsen, E. Kwong, and D. DesMarais. 1985. Carbon isotopic fractionation in heterotrophic microbial metabolism. Appl. Environ. Microbiol. 50:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. deGraaf, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by C-13-labelling of biomarkers. Nature 392:801-805. [Google Scholar]
- 9.Brand, W. A., and P. Dobberstein. 1996. Isotope-ratio-monitoring liquid chromatography mass spectrometry (IRM-LCMS). First results from a moving wire interface system. Isot. Environ. Health Stud. 32:275-283. [DOI] [PubMed] [Google Scholar]
- 10.Chandler, D. P., J. K. Fredrickson, and F. J. Brockman. 1997. Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol. Ecol. 6:475-482. [DOI] [PubMed] [Google Scholar]
- 11.Coffin, R. B., D. J. Velinsky, R. Devereux, W. A. Price, and L. A. Cifuentes. 1990. Stable carbon isotope analysis of nucleic acids to trace sources of dissolved substances used by estuarine bacteria. Appl. Environ. Microbiol. 56:2012-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cravo-Laureau, C., R. Matheron, J.-L. Cayol, C. Joulian, and A. Hirschler-Rea. 2004. Desulfatibacillum aliphaticivorans gen. nov., sp. nov., an n-alkane- and n-alkene-degrading, sulfate-reducing bacterium. Int. J. Syst. Evol. Microbiol. 54:77-83. [DOI] [PubMed] [Google Scholar]
- 13.DeNiro, M. J., and S. Epstein. 1978. Influence of diet on distribution of carbon isotopes in animals. Geochim. Cosmochim. Acta 42:495-506. [Google Scholar]
- 14.Dyksterhouse, S. E., J. P. Gray, R. P. Herwig, J. C. Lara, and J. T. Staley. 1995. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int. J. Syst. Bacteriol. 45:116-123. [DOI] [PubMed] [Google Scholar]
- 15.Eek, K. M., A. L. Sessions, and D. P. Lies. 2007. Carbon-isotopic analysis of microbial cells sorted by flow cytometry. Geobiology 5:85-95. [DOI] [PubMed] [Google Scholar]
- 16.Engelhardt, M. A., K. Daly, R. P. Swannell, and I. M. Head. 2001. Isolation and characterization of a novel hydrocarbon-degrading, Gram-positive bacterium, isolated from intertidal beach sediment, and description of Planococcus alkanoclasticus sp. nov. J. Appl. Microbiol. 90:237-247. [DOI] [PubMed] [Google Scholar]
- 17.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fredrickson, J. K., D. L. Balkwill, G. R. Drake, M. F. Romine, B. Ringelberg, and D. C. White. 1995. Aromatic-degrading Sphingomonas isolates from the deep subsurface. Appl. Environ. Microbiol. 61:1917-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauthier, M. J., B. LaFay, R. Christen, L. Fernandez, M. Acquaviva, P. Bonin, and J. C. Bertrand. 1992. Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int. J. Syst. Bacteriol. 42:568-576. [DOI] [PubMed] [Google Scholar]
- 20.Guindon, S., and O. Gascuel. 2003. PhyML—a simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 21.Hamlett, N. V. 1986. Alteration of a salt-marsh bacterial community by fertilization with sewage sludge. Appl. Environ. Microbiol. 52:915-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampson, G. R., and H. L. Sanders. 1969. Local oil spill. Oceanus 15:8. [Google Scholar]
- 23.Harayama, S., Y. Kasai, and A. Hara. 2004. Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 15:205-214. [DOI] [PubMed] [Google Scholar]
- 24.Harms, G., K. Zengler, R. Rabus, F. Aeckersberg, D. Minz, R. Rosselló-Mora, and F. Widdel. 1999. Anaerobic oxidation of o-xylene, m-xylene, and homologous alkylbenzenes by new types of sulfate-reducing bacteria. Appl. Environ. Microbiol. 65:999-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harvey, R. W., and L. Y. Young. 1980. Enrichment and association of bacteria and particulates in salt marsh surface water. Appl. Environ. Microbiol. 39:894-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes, J. M. 1983. Practice and principles of isotopic measurements in organic geochemistry, p 5.1-5.31. In W. G. Meinschein (ed.), Organic geochemistry of contemporaneous and ancient sediments. Great Lakes Section, Society of Economic Paleontologists and Mineralogists, Bloomington, IN.
- 27.Head, I. M., and R. P. J. Swannell. 1999. Bioremediation of petroleum hydrocarbon contaminants in marine habitats. Curr. Opin. Biotechnol. 10:234-239. [DOI] [PubMed] [Google Scholar]
- 28.Hedlund, B. P., A. D. Geiselbrecht, T. J. Bair, and J. T. Staley. 1999. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium, Neptunomonas naphthovorans gen. nov., sp. nov. Appl. Environ. Microbiol. 65:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez-Raquet, G., H. Budzinski, P. Caumette, P. Dabert, K. Le Menach, G. Muyzer, and R. Duran. 2006. Molecular diversity studies of bacterial communities of oil polluted microbial mats from the Etang de Berre (France). FEMS Microbiol. Ecol. 58:550-562. [DOI] [PubMed] [Google Scholar]
- 30.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 31.Ivanova, E. P., L. A. Romanenko, J. Chun, M. H. Matte, G. R. Matte, V. V. Mikhailov, V. I. Svetashev, A. Huq, T. Maugel, and R. R. Colwell. 2000. Idiomarina gen. nov., comprising novel indigenous deep-sea bacteria from the Pacific Ocean, including descriptions of two species, Idiomarina abyssalis sp. nov. and Idiomarina zobellii sp. nov. Int. J. Syst. Evol. Microbiol. 50:901-907. [DOI] [PubMed] [Google Scholar]
- 32.Kasai, Y., H. Kishira, K. Syutsubo, and S. Harayama. 2001. Molecular detection of marine bacterial populations on beaches contaminated by the Nakhodka tanker oil-spill accident. Environ. Microbiol. 3:246-255. [DOI] [PubMed] [Google Scholar]
- 33.Kasai, Y., H. Kishira, and S. Harayama. 2002. Bacteria belonging to the genus Cycloclasticus play a primary role in the degradation of aromatic hydrocarbons released in a marine environment. Appl. Environ. Microbiol. 68:5625-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasai, Y., H. Kishira, T. Sasaki, K. Syutsubo, K. Watanabe, and S. Harayama. 2002. Predominant growth of Alcanivorax strains in oil-contaminated and nutrient-supplemented sea water. Environ. Microbiol. 4:141-147. [DOI] [PubMed] [Google Scholar]
- 35.Kasai, Y., Y. Takahata, M. Manefield, and K. Watanabe. 2006. RNA-based stable isotope probing and isolation of anaerobic benzene-degrading bacteria from gasoline-contaminated groundwater. Appl. Environ. Microbiol. 72:3586-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleinsteuber, S., V. Riis, I. Fetzer, H. Harms, and S. Muller. 2006. Population dynamics within a microbial consortium during growth on diesel fuel in saline environments. Appl. Environ. Microbiol. 72:3531-3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuenen, J. G., and R. F. Beudeker. 1982. Microbiology of thiobacilli and other sulphur-oxidizing autotrophs, mixotrophs and heterotrophs. Phil. Trans. R. Soc. London 298:473-497. [DOI] [PubMed] [Google Scholar]
- 38.Luna, G. M., A. Dell'Anno, and R. Danovaro. 2006. DNA extraction procedure: a critical issue for bacterial diversity assessment in marine sediments. Environ. Microbiol. 8:308-320. [DOI] [PubMed] [Google Scholar]
- 39.MacGregor, B. J., V. Brüchert, S. Fleischer, and R. Amann. 2002. Isolation of small-subunit rRNA for stable isotopic characterization. Environ. Microbiol. 4:451-464. [DOI] [PubMed] [Google Scholar]
- 40.MacGregor, B. J., H. T. S. Boschker, and R. Amann. 2006. Comparison of rRNA and polar-lipid-derived fatty acid biomarkers for assessment of 13C-substrate incorporation by microorganisms in marine sediments. Appl. Environ. Microbiol. 72:5246-5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madsen, E. L. 2006. The use of stable isotope probing techniques in bioreactor and field studies on bioremediation. Curr. Opin. Biotechnol. 17:92-97. [DOI] [PubMed] [Google Scholar]
- 42.Margesin, R., M. Hammerle, and D. Tscherko. 2007. Microbial activity and community composition during bioremediation of diesel-oil-contaminated soil: effects of hydrocarbon concentration, fertilizers, and incubation time. Microb. Ecol. 53:259-269. [DOI] [PubMed] [Google Scholar]
- 43.Maugeri, T. L., C. Gugliandolo, D. Caccamo, and E. Stackebrandt. 2002. Three novel halotolerant and thermophilic Geobacillus strains from shallow marine vents. Syst. Appl. Microbiol. 25:450-455. [DOI] [PubMed] [Google Scholar]
- 44.McNichol, A. P., E. A. Osborne, A. R. Gagnon, B. Fry, and G. A. Jones. 1994. TIC, TOC, DIC, DOC, PIC, POC—unique aspects in the preparation of oceanographic samples for 14C-AMS. Nucl. Instr. Methods Phys. Res. B 92:162-165. [Google Scholar]
- 45.Orphan, V. J., C. H. House, K. U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Coupled isotopic and phylogenetic characterization of single cells: direct evidence for a methane-consuming archaeal/bacterial consortium. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 46.Pearson, A., A. L. Sessions, K. J. Edwards, and J. M. Hayes. 2004. Phylogenetically specific separation of rRNA from prokaryotes for isotopic analysis. Mar. Chem. 92:295-306. [Google Scholar]
- 47.Pelz, O., N. Chatzinotas, N. Andersen, S. M. Bernasconi, C. Hesse, J. Abraham, and W.-R. Zeyer. 2001. Use of isotopic and molecular techniques to link toluene degradation in denitrifying aquifer microcosms to specific microbial populations. Arch. Microbiol. 175:270-281. [DOI] [PubMed] [Google Scholar]
- 48.Peterson, B. J., R. W. Howarth, and R. H. Garritt. 1985. Multiple stable isotopes used to trace the flow of organic matter in estuarine food webs. Science 227:1361-1363. [DOI] [PubMed] [Google Scholar]
- 49.Phelps, C. D., L. J. Kerkhof, and L. Y. Young. 1998. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol. Ecol. 27:269-279. [Google Scholar]
- 50.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rabus, R., R. Nordhaus, W. Ludwig, and F. Widdel. 1993. Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl. Environ. Microbiol. 59:1444-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 53.Ravot, G., M. Magot, M. L. Fardeau, B. K. Patel, P. Thomas, J. L. Garcia, and B. Ollivier. 1999. Fusibacter paucivorans gen. nov., sp. nov., an anaerobic, thiosulfate-reducing bacterium from an oil-producing well. Int. J. Syst. Bacteriol. 49:1141-1147. [DOI] [PubMed] [Google Scholar]
- 54.Reddy, C. M., T. I. Eglinton, A. Hounshell, H. K. White, L. Xu, R. B. Gaines, and G. S. Frysinger. 2002. The West Falmouth oil spill after thirty years: the persistence of petroleum hydrocarbons in marsh sediments. Environ. Sci. Technol. 36:4754-4760. [DOI] [PubMed] [Google Scholar]
- 55.Reddy, C. M., L. Xu, J. G. Quinn, and P. C. Hartmann. 2002. Investigating the radiocarbon content of the unresolved complex mixture. Abstr. Am. Chem. Soc. Div. Environ. Chem. 224:32. [Google Scholar]
- 56.Röling, W. F. M., M. G. Milner, D. M. Jones, K. Lee, F. Daniel, R. J. Swannell, and I. M. Head. 2002. Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl. Environ. Microbiol. 68:5537-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Röling, W. F. M., M. G. Milner, D. M. Jones, F. Fratepietro, R. P. Swannell, F. Daniel, and I. M. Head. 2004. Bacterial community dynamics and hydrocarbon degradation during a field-scale evaluation of bioremediation on a mudflat beach contaminated with buried oil. Appl. Environ. Microbiol. 70:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rueter, P., R. Rabus, H. Wilkes, F. Aeckersberg, F. A. Rainey, H. W. Jannasch, and F. Widdel. 1994. Anaerobic oxidation of hydrocarbons in crude oil by new types of sulphate-reducing bacteria. Nature 372:455-458. [DOI] [PubMed] [Google Scholar]
- 59.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sessions, A. L., S. P. Sylva, and J. M. Hayes. 2005. Moving-wire device for carbon isotopic analyses of nanogram quantities of nonvolatile organic carbon. Anal. Chem. 77:6519-6527. [DOI] [PubMed] [Google Scholar]
- 61.Simmons, S. L., and K. J. Edwards. 2007. Unexpected diversity in populations of the many-celled magnetotactic prokaryote. Environ. Microbiol. 9:206-215. [DOI] [PubMed] [Google Scholar]
- 62.Slater, G. F., H. K. White, T. I. Eglinton, and C. M. Reddy. 2005. Determination of microbial carbon sources in petroleum contaminated sediments using molecular C-14 analysis. Environ. Sci. Technol. 39:2552-2558. [DOI] [PubMed] [Google Scholar]
- 63.So, C. M., and L. Y. Young. 1999. Isolation and characterization of a sulfate-reducing bacterium that anaerobically degrades alkanes. Appl. Environ. Microbiol. 65:2969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki, Y., F. Inagaki, K. Takai, K. H. Nealson, and K. Horikoshi. 2004. Microbial diversity in inactive chimney structures from deep-sea hydrothermal systems. Microb. Ecol. 47:186-196. [DOI] [PubMed] [Google Scholar]
- 65.Takai, K., H. Hirayama, T. Nakagawa, Y. Suzuki, K. H. Nealson, and K. Horikoshi. 2004. Thiomicrospira thermophila sp. nov., a novel microaerobic, thermotolerant, sulfur-oxidizing chemolithomixotroph isolated from a deep-sea hydrothermal fumarole in the TOTO caldera, Mariana Arc, Western Pacific. Int. J. Syst. Evol. Microbiol. 54:2325-2333. [DOI] [PubMed] [Google Scholar]
- 66.Teske, A., K.-U. Hinrichs, V. Edgcomb, A. D. Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson, J. R., L. A. Marcelino, and M. F. Polz. 2002. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR.’ Nucleic Acids Res. 30:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, Y., P. C. Lau, and D. K. Button. 1996. A marine oligobacterium harboring genes known to be part of aromatic hydrocarbon degradation pathways of soil pseudomonads. Appl. Environ. Microbiol. 62:2169-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ward, D. W., R. M. Atlas, P. D. Boehm, and J. A. Calder. 1980. Microbial biodegradation and chemical evolution of oil from the Amoco spill. AMBIO 9:277-283. [Google Scholar]
- 70.Werne, J. P., R. R. Haese, T. Zitter, G. Aloisi, I. Bouloubassi, S. Heijs, A. Fiala-Medioni, R. D. Pancost, J. S. Sinninghe Damste, G. de Lange L. J. Forney, J. C. Gottschal, J.-P. Foucher, J. Mascle, and J. Woodside. 2004. Life at cold seeps: a synthesis of biogeochemical and ecological data from Kazan mud volcano, eastern Mediterranean Sea. Chem. Geol. 205:367-390. [Google Scholar]
- 71.White, H. K., C. M. Reddy, and T. I. Eglinton. 2005. Isotopic constraints on the fate of petroleum residues sequestered in salt marsh sediments. Environ. Sci. Technol. 39:2545-2551. [DOI] [PubMed] [Google Scholar]
- 72.Whyte, L. G., L. Bourbonniere, and C. W. Greer. 1997. Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways. Appl. Environ. Microbiol. 63:3719-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wirsen, C. O., T. Brinkhoff, J. Kuever, G. Muyzer, S. Molyneaux, and H. W. Jannasch. 1998. Comparison of a new Thiomicrospira strain from the mid-Atlantic ridge with known hydrothermal vent isolates. Appl. Environ. Microbiol. 64:4057-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yakimov, M. M., P. N. Golyshin, S. Lang, E. R. Moore, W. R. Abraham, H. Lunsdorf, and K. N. Timmis. 1998. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Bacteriol. 48:339-348. [DOI] [PubMed] [Google Scholar]
- 75.Yakimov, M. M., R. Denaro, M. Genovese, S. Cappello, G. D'Auria, T. N. Chernikova, K. N. Timmis, P. N. Golyshin, and L. Giluliano. 2005. Natural microbial diversity in superficial sediments of Milazzo Harbor (Sicily) and community successions during microcosm enrichment with various hydrocarbons. Environ. Microbiol. 7:1426-1441. [DOI] [PubMed] [Google Scholar]
- 76.Zhuang, W. Q., J. H. Tay, A. M. Maszenan, and S. T. Tay. 2003. Isolation of naphthalene-degrading bacteria from tropical marine sediments. Water Sci. Technol. 47:303-308. [PubMed] [Google Scholar]