Abstract
In this study, the genes involved in the initial attack on fluorene by Sphingomonas sp. strain LB126 were investigated. The α and β subunits of a dioxygenase complex (FlnA1-FlnA2), showing 63 and 51% sequence identity, respectively, to the subunits of an angular dioxygenase from the gram-positive dibenzofuran degrader Terrabacter sp. strain DBF63, were identified. When overexpressed in Escherichia coli, FlnA1-FlnA2 was responsible for the angular oxidation of fluorene, 9-hydroxyfluorene, 9-fluorenone, dibenzofuran, and dibenzo-p-dioxin. Moreover, FlnA1-FlnA2 was able to oxidize polycyclic aromatic hydrocarbons and heteroaromatics, some of which were not oxidized by the dioxygenase from Terrabacter sp. strain DBF63. The quantification of resulting oxidation products showed that fluorene and phenanthrene were the preferred substrates of FlnA1-FlnA2.
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental contaminants and are released during the burning, handling, or disposal of organic matter including coal tars, crude oil, and petroleum products. There are some natural origins, such as forest fires and natural oil seeps, but PAHs arise mainly from combustion- or oil-related anthropogenic activities. A number of organisms that are able to use PAHs as the sole source of carbon and energy have been isolated previously (6), and bioremediation strategies using these organisms have been proposed (17).
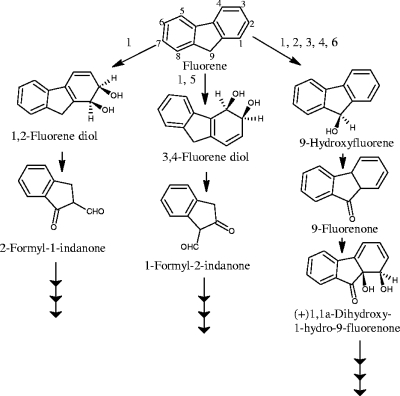
Fluorene, a tricyclic aromatic hydrocarbon containing a five-membered ring, offers a variety of possibilities for biochemical attack. Two of these pathways are initiated by dioxygenation at the 1,2 (5, 9) or 3,4 (5, 10, 28) positions (Fig. 1). The corresponding cis-dihydrodiols undergo dehydrogenation and then meta cleavage. The third route (38, 44) is initiated by monooxygenation at the C-9 position to give 9-hydroxyfluorene, which is then dehydrogenated into 9-fluorenone. This route is productive only if subsequent angular carbon dioxygenation forms 1-hydro-1,1a-dihydroxy-9-fluorenone, leading to phthalate, which is further degraded via protocatechuate (11, 28, 44) (Fig. 1).
FIG. 1.
Proposed pathways for fluorene degradation and the bacteria involved. 1, Arthrobacter sp. strain F101 (5, 9); 2, Terrabacter sp. strain DBF63 (28); 3, Brevibacterium sp. strain DPO1361 (44); 4, Pseudomonas sp. strain F274 (11); 5, Burkholderia cepacia F297 (12); and 6, Sphingomonas sp. strain LB126 (47).
Sphingomonads have been intensively studied for their ability to degrade a wide range of aromatic hydrocarbons (32, 34, 41, 42, 48, 49). The function and organization of PAH catabolic genes in Sphingomonas species often remain obscure since the genes involved in the degradation of aromatic compounds are not always arranged in discrete operons but are frequently dispersed throughout the genome. Sphingomonas sp. strain LB126 was isolated from PAH-contaminated soil and is capable of utilizing fluorene as the sole carbon source (3). Fluorene degradation by strain LB126 has been investigated previously (47), but the enzymes that govern the initial attack on fluorene were not identified.
In contrast, more genetic work done with gram-positive bacteria (13, 14) has shown that dibenzofuran-degrading Terrabacter sp. strain DBF63 can also oxidize fluorene, thanks to a cluster of plasmid-borne catabolic genes. The oxygenase component of an angular dioxygenase complex, encoded by dbfA1 and dbfA2, does not cluster with already known dioxygenases. Few data are available regarding genes involved in fluorene degradation by gram-negative bacteria. Although many PAH dioxygenases can oxidize fluorene, the respective host strains cannot use fluorene as the sole carbon source. Recently, the catabolic plasmid pCAR3 from Sphingomonas sp. strain KA1 was described (40). Genes homologous to dbfA1 and dbfA2, as well as all genes necessary for the complete degradation of fluorene, were found on pCAR3, but strain KA1 is unable to grow on fluorene as the sole source of carbon. We present here the first report, to our knowledge, of genes governing the angular attack on fluorene by the gram-negative Sphingomonas sp. strain LB126 using fluorene as the sole source of carbon and energy.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Sphingomonas sp. strain LB126 uses fluorene as the sole source of carbon and energy (3) and was kindly provided by Vlaamse Instelling voor Technologisch Onderzoek, Belgium. Escherichia coli Top10 was used as the recipient strain in all cloning experiments. E. coli BL21(DE3) was used for gene expression analysis. PCR amplicons were cloned into either pDrive (Qiagen, Valencia, CA), pGEM-T Easy vector (Promega, Madison, WI), or pGEM5Z vector (Promega, Madison, WI), while pET30f (Novagen, San Diego, CA) was used as an expression vector. MM284 minimal medium (27) was used for growing Sphingomonas sp. strain LB126 and was supplemented with phosphate buffer (50 mM; KH2PO4, K2HPO4, pH 7.2) instead of Tris buffer. Fluorene was provided as crystals in both solid and liquid media. Luria-Bertani (LB) broth (36) was used as a complete medium for growing E. coli strains. Solid medium contained 2% agar. When needed, ampicillin, streptomycin, or kanamycin was added to the medium at 100, 200, or 20 μg/ml, respectively. Sphingomonas sp. strain LB126 was grown at 30°C, and E. coli strains were grown at 37°C. Bacterial growth was determined by readings of the optical density at 600 nm (OD600).
DNA manipulations and molecular techniques.
Total DNA from pure cultures of Sphingomonas sp. strain LB126 was extracted using the UltraClean DNA isolation kit according to the recommendations of the manufacturer (MoBio, Carlsbad, CA) or using standard methods (36) when a higher DNA concentration was needed. Plasmid DNA extractions, restriction enzyme digestions, ligations, transformations, sequencing, and agarose gel electrophoresis were carried out using standard methods (36).
PCR.
Degenerate primers for amplifying conserved sequences of the gene encoding the angular dioxygenase were used as described elsewhere (15). PCR products were purified and cloned into either the pGEM-T or pDrive plasmid. The reverse transcriptase PCRs (RT-PCRs) were performed in 25 μl with 5 ng of total RNA and 20 pmol of each primer with the OneStep RT-PCR kit (Qiagen, Valencia, CA). Total RNA extractions were performed using the RNeasy kit (Qiagen, Valencia, CA), and the extracts were further purified by spin column and DNase I treatment according to the instructions of the kit manufacturer. The thermocycler program used for the RT-PCRs was as follows: 60°C for 30 min, 94°C for 15 min, 30 cycles (94°C for 30 s, 50°C for 30 s, and 72°C for 45 s), and 72°C for 7 min. The primer combinations AD1 (5′-CGTGACGGCGCGCTTC-3′) and AD2 (5′-CATGCGAAACTTCATC-3′), flnE-F (5′-CTGATCGCACTGGCG-3′) and flnE-R (5′-CGTTTCACCGACGTG-3′), and cirA-F (5′-GATCGGCTGGAAGTC-3′) and cirA-R (5′-CCGAGCTTCACGTCG-3′) were used to amplify internal fragments of flnA1 (252 bp), flnE (437 bp), and cirA (554 bp), respectively. The 16S rRNA gene-specific primers 63F (25) and 518R (30) were used as a control.
Southern blotting and cloning of catabolic genes.
Genomic DNA (2 μg) was digested with either BamHI, NotI, NsiI, or a combination of these enzymes, and the fragments were separated by gel electrophoresis and blotted onto a positively charged nylon membrane (Amersham, Buckinghamshire, United Kingdom) by using standard protocols (36). For Southern blot detection, a PCR-amplified digoxigenin-labeled probe was prepared according to the recommendations of the manufacturer (Roche Diagnostics, Mannheim, Germany) by using primers described by Iida et al. (15). Prehybridization and hybridization were carried out at 68°C. After hybridization, the membrane was washed twice with 2× SSC (20× SSC is 3 mol of NaCl/liter and 0.3 mol of sodium citrate/liter, pH 7.0) containing 0.1% (wt/vol) sodium dodecyl sulfate (SDS) for 5 min at room temperature and twice with 0.1× SSC containing 0.1% SDS for 15 min at 68°C. Detection was carried out according to standard protocols (36). To isolate catabolic genes, total DNA (10 μg) was digested with BamHI and NsiI and separated by gel electrophoresis and DNA fragments of about 7 kb were recovered from the agarose gel by using the UltraClean GelSpin DNA extraction kit (MoBio, Carlsbad, CA). The DNA obtained was cloned into pGEM5Z (Promega, Madison, WI), and E. coli Top10 cells (Invitrogen, Carlsbad, CA) were transformed with the resulting construct. Transformants were screened as pools of 10 clones by PCR using the above-mentioned primers (15). A 6.9-kb plasmid containing the angular dioxygenase gene was identified and sequenced by primer walking.
Construction of plasmids for protein overexpression.
The construction of the plasmids used in this study involved multiple PCR amplifications and cloning steps. The flnA1-flnA2 fragment (1,842 bp) was amplified by PCR with the primer pair 5′-CATATGGCCACAGCCCTCATGAACCACCC-3′ and 5′-AAGCTTGGCGCTCACAGGAACACCG-3′, introducing NdeI and HindIII sites (indicated by italics) at the ends of the amplicon. The PCR product was cloned into pDrive (Qiagen), sequenced, and then subcloned into the NdeI and HindIII sites of expression vector pET-30f (Novagen). E. coli BL21(DE3) was transformed with this construct for expression analysis.
SDS-PAGE.
Bacterial cells were pelleted by centrifugation and washed with 10 ml of ice-cold phosphate buffer (140 mM NaCl, 10 mM Na2HPO4, 2.7 mM KCl, 1.8 mM NaH2PO4, pH 7.4). One milliliter of ice-cold phosphate buffer was added to the pellet, and 550 μl of the suspension was subjected to sonication on ice for 20 s (5-s pulse interval; 40% of maximum amplitude). After centrifugation, the supernatant and the pellet were mixed with an equal volume of loading solution. SDS-polyacrylamide gel electrophoresis (PAGE) was performed on 13.3% polyacrylamide mini gels. After electrophoresis, protein staining was performed with Coomassie brilliant blue R-250.
Dioxygenase overexpression and in vivo assays.
Strain BL21(DE3)(pET30fflnA1A2) was grown overnight in 3 ml of LB medium with the suitable antibiotics. This culture was used to inoculate 25 ml of LB medium (at 0.1% by volume), which was incubated at 42°C up to an OD600 of 0.5. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.5 mM. The cells were further incubated for 7 h at 25°C. For in vivo assays, cells were centrifuged, washed, and resuspended to an OD600 of approximately 2 in M9 medium (36) containing 0.2% glucose. Cells (25 ml) were incubated for 48 h at 25°C with 5 ml of silicone oil containing each tested PAH at 0.1 g/liter.
GC-MS analysis of PAH oxidation products.
Water-soluble products resulting from PAH oxidation were extracted from the aqueous phase of the bacterial suspension by using columns filled with reverse phase-bonded silica (Upti-clean C18U, 0.5 g; Interchim, Montluçon, France). Columns were washed with 10 ml of water and then eluted with 1 ml of ethyl acetate. The solvent was dried over sodium sulfate and evaporated under nitrogen gas. The dried extracts were then dissolved in 100 or 200 μl of acetonitrile before being derivatized with N,O-bis(trimethylsilyl)trifluoroacetamide:trimethylchlorosilane (BSTFA) or n-butylboronate (NBB). In order to quantify the dihydrodiols formed upon the incubation of BL21(DE3)(pET30fflnA1A2) recombinant cells with PAHs, 2,3-dihydrobiphenyl (Sigma-Aldrich, St. Louis, MO) was added to a 0.1 μM final concentration in the aqueous phase prior to solid-phase extraction and was used as an internal standard. After derivatization and gas chromatography (GC)-mass spectrometry (MS) analysis, NBB dihydrodiol derivates were quantified on the basis of peak areas by using a calibration curve generated by analyzing known amounts of anthracene 1,2-dihydrodiol. GC-MS analysis of trimethylsilyl derivatives was carried out as previously described (18). NBB derivatives were separated on an MDN-12 capillary column (30 m in length, with a 0.25-mm internal diameter; Supelco) by using helium as a carrier gas at 1 ml/min. The oven temperature was held at 75°C for 1 min and then increased to 300°C at a rate of 14°C min−1 and held at 300°C for 8 min. The mass spectrometer was operated in the selected-ion-monitoring mode, selecting m/z values corresponding to the expected masses (M+) of the dihydrodiol derivatives (228 for naphthalene and 278 for anthracene and phenanthrene). The NBB derivative of trihydroxybiphenyl, the oxidation product of dibenzofuran, was monitored at an m/z of 268. The fluorene derivative was detected at an m/z of 196 (M+ − OBC4H9), because in contrast to that of other dihydrodiol derivatives, the abundance of the M+ ion was very low.
DNA and protein sequence analysis.
Sequence analysis was performed using the DNASTAR software package (Lasergene Inc., Madison, WI). The BLAST search tool was used for homology searches (1). Multiple alignments and phylogenetic trees were produced using the DNASTAR and MEGA3.1 software (23).
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the GenBank database under accession number EU024110.
RESULTS AND DISCUSSION
Cloning and sequence analysis of genes encoding a novel angular dioxygenase.
Sphingomonas strain LB126 has been studied for its ability to grow on fluorene and degrade phenanthrene, anthracene, and fluoranthene by cometabolism in the presence of pyruvate (46). In order to detect genes potentially involved in the initial attack of PAHs, a PCR strategy was chosen. The genes in strain LB126 involved in fluorene oxidation were expected to display some similarity to previously described counterparts in other PAH-degrading Sphingomonas strains. Several primer pairs corresponding to conserved domains of previously described PAH dioxygenases were tested (7, 19, 24, 29), but no amplification could be obtained (data not shown). Given the dearth of information regarding fluorene degradation genes in gram-negative bacteria, primers specific to angular dioxygenase genes from gram-positive bacteria were tested.
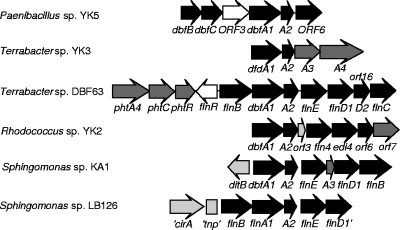
By using a set of such primers (15) and total DNA as a template, a 267-bp DNA fragment was amplified, and sequencing and translation revealed 57% protein sequence identity to a peptide in the dibenzofuran 4,4a-dioxygenase α subunit of Terrabacter sp. strain DBF63 (20). The 267-bp fragment was then used as a digoxigenin-labeled probe in Southern blot experiments with whole-genome extracts of strain LB126. A 6.9-kb fragment encompassing four entire open reading frames (ORFs; orf3 to orf6) and three truncated ones (orf1, orf2, and orf7) was cloned and sequenced as described in Materials and Methods (Table 1). The orf1 gene product did not show amino acid sequence similarities to previously described fluorene catabolic gene products but showed significant homology to the TonB-dependent receptor CirA from Sphingomonas wittichii strain RW1 (36%) and Novosphingobium aromaticivorans F199 (34%). orf2 encoded a truncated transposase, suggesting that the adjacent gene cluster was probably acquired by horizontal transfer, although no change in G+C content was noticed. orf3 to orf7 showed genetic organization similar to that of the dibenzofuran catabolic operon from Terrabacter sp. strain DBF63 (20) (Fig. 2). Nevertheless, the product of orf3, a putative dehydrogenase, did not show significant protein sequence similarity to its counterpart (FlnB) from strain DBF63. The highest degree of similarity was found with putative dehydrogenases from Mycobacterium strains MCS and KMS identified in whole-genome sequencing projects. orf4 and orf5 (flnA1 and flnA2) encoded the α and β subunits of a putative angular dioxygenase suggested to be involved in fluorene degradation (see below). The amino acid sequences of the subunits showed moderate identity (63 and 51%) to the sequences of DbfA1 and DbfA2 from strain DBF63. Phylogenetic analysis revealed that the orf4 product did not cluster with dioxygenase α subunits from other sphingomonads and was only distantly related to the angular dioxygenase from S. wittichii strain RW1 (4). The closest homologues within sphingomonads were the dioxygenase α subunits from the carbazole-degrading strains Sphingomonas sp. strain KA1 (36% amino acid identity) (40) and Sphingomonas sp. strain CB3 (35% amino acid identity) (39). Interestingly, in strain LB126, the genes encoding the lower pathway of fluorene degradation via protocatechuate (47) showed homology to the lig genes of Sphingomonas paucimobilis SYK-6 (26) and the fld genes of Sphingomonas sp. strain KA1 (40). The orf5 product showed moderate protein identity (48%) to the β subunit DbfA2 from gram-positive dibenzofuran-degrading Paenibacillus sp. strain YK5 (16). DbfA1 and DbfA2 from strain YK5 are the two subunits of a dioxygenase able to oxidize dibenzo-p-dioxin, dibenzothiophene, fluorene, and 9-fluorenone, compounds that, however, cannot be utilized as growth substrates by strain YK5 (16). The product of orf6, located downstream of flnA1 and flnA2, showed 42% protein identity to FlnE, a meta cleavage product hydrolase from strain DBF63 (14). The truncated orf7 gene product showed similarity to an extradiol dioxygenase α subunit (FlnD1) from strain DBF63. Since flnD1 from strain LB126 lacks a 3′ region, no conclusive homology search could be carried out. The genetic organization of the LB126 fragment was similar to the one found in gram-positive Terrabacter sp. strain DBF63 and is unusual among gram-negative bacteria. The presence of a truncated transposase gene indicates that this catabolic gene cluster present in strain LB126 may have been inherited by lateral transfer from other genera of PAH-degrading bacteria (Fig. 2) (33).
TABLE 1.
Homology search analyses of the recovered ORFs from fluorene-degrading Sphingomonas sp. strain LB126
| ORF | Gene | Probable function or product | Homologous protein | Source | % Identity | Accession no. |
|---|---|---|---|---|---|---|
| orf1 | Truncated cirA | TonB-dependent receptor | CirA | Sphingomonas wittichii RW1 | 36 | YP_001262040 |
| CirA | Novosphingobium aromaticivorans DSM 12444 | 34 | YP_001165948 | |||
| orf2 | Truncated tnp | Transposase | Transposase | Mesorhizobium loti MAFF303099 | 60 | NP_085624 |
| Transposase | Sinorhizobium medicae WSM419 | 57 | EAU08642 | |||
| orf3 | flnB | Probable dehydrogenase | Probable dehydrogenase | Mycobacterium sp. strain MCS | 40 | ABG07792 |
| Probable dehydrogenase | Mycobacterium sp. strain KMS | 40 | ZP_01286209 | |||
| Probable dehydrogenase | Rhodobacterales sp. strain HTCC2654 | 28 | ZP_01014534 | |||
| orf4 | flnA1 | Angular dioxygenase α subunit | DbfA1 | Terrabacter sp. strain DBF63 | 63 | BAC75993 |
| DbfA1 | Rhodococcus sp. strain YK2 | 54 | BAC00802 | |||
| DbfA1 | Paenibacillus sp. strain YK5 | 52 | BAE53401 | |||
| orf5 | flnA2 | Angular dioxygenase β subunit | DbfA2 | Terrabacter sp. strain DBF63 | 52 | BAC75994 |
| DbfA2 | Rhodococcus sp. strain YK2 | 51 | BAC00803 | |||
| DbfA2 | Paenibacillus sp. strain YK5 | 48 | BAE53402 | |||
| orf6 | flnE | Hydrolase | FlnE | Terrabacter sp. strain DBF63 | 42 | BAE45094 |
| ORF4 | Rhodococcus sp. strain YK2 | 42 | BAC00805 | |||
| a/b hydrolase 1 | Mycobacterium sp. strain MCS | 30 | YP_642596 | |||
| orf7 | Truncated flnD1 | Extradiol dioxygenase α subunit | FlnD1 | Terrabacter sp. strain DBF63 | 12 | BAC75996 |
| BphC6 | Rhodococcus rhodochrous | 12 | BAD10908 | |||
| Edi4 | Rhodococcus sp. strain YK2 | 12 | BAC00806 |
FIG. 2.
Genetic organization of the 6.9-kb DNA region containing fluorene catabolic genes in Sphingomonas sp. strain LB126 compared to that of corresponding clusters in Paenibacillus sp. strain YK5 (accession no. AB201843), Terrabacter sp. strain YK3 (accession no. AB075242), Rhodococcus sp. strain YK2 (accession no. AB070456), Sphingomonas sp. strain KA1 (accession no. NC_008308), and Terrabacter sp. strain DBF63 (accession no. AP008980). The arrows indicate the locations and the directions of transcription of the ORFs. Black arrows represent genes involved in the initial attack on fluorene, dark gray arrows indicate genes involved in the electron transport chain or phthalate degradation (pht), white arrows indicate regulatory genes, and light gray arrows represent genes not directly involved in fluorene oxidation. Apostrophes with gene designations indicate truncated ORFs. The truncated tnp gene of Sphingomonas sp. strain LB126 is represented by a box rather than an arrow because it is not involved in fluorene catabolism.
Transcriptional analysis by RT-PCR.
The transcriptional expression of cirA, flnA1, and flnE was studied by RT-PCR. Total RNA was extracted from cultures of Sphingomonas sp. strain LB126 grown on glucose or fluorene during the exponential growth phase (OD600 of 0.4). Results indicated that cirA, flnA1, and flnE transcripts were up-regulated in the presence of fluorene and were not or were only weakly detected when strain LB126 was grown on glucose (Fig. 3). These findings suggest that the cloned catabolic operon is involved in fluorene degradation by Sphingomonas sp. strain LB126.
FIG. 3.
RT-PCR transcriptional analysis of the expression of cirA, flnA1, and flnE in Sphingomonas sp. strain LB126 during growth on fluorene or glucose. RT-PCR products obtained from template RNA extracted from cells grown on glucose (lanes G) and fluorene (lanes F) were separated by gel electrophoresis. The positions of size markers (indicated in base pairs) (lane M) are shown on the left (1-kb Plus DNA ladder; Invitrogen, Carlsbad, CA). Differentially amplified RT-PCR fragments indicate differential gene expression in fluorene-grown Sphingomonas sp. strain LB126 cells. Lane −, negative control without RNA but with 16S rRNA gene primers. RNA quality was checked by amplifying 16S rRNA gene sequences with RT and Taq polymerase.
Functional expression of FlnA1-FlnA2 in E. coli.
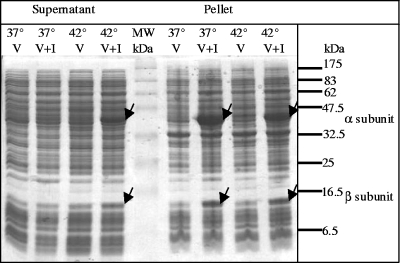
In order to study the enzymatic activity of FlnA1-FlnA2, the corresponding genes were introduced into pET30f and expressed in E. coli BL21(DE3). Protein extracts from IPTG-induced cells were separated by SDS-PAGE. The cells overproduced two polypeptides with Mrs of 45,000 and 14,000, which did not match exactly the predicted sizes of FlnA1 and FlnA2 as calculated from the deduced polypeptide sequences (49.5 and 19.4 kDa). Differences between the theoretical and apparent molecular masses upon SDS-PAGE gels were also observed for the DbfA1 and DbfA2 dioxygenase components from strain DBF63 (20). Significantly, it was found that the recombinant proteins were inactive and mostly insoluble (Fig. 4). When the recombinant strain was grown at 42°C up to an OD600 of 0.5 and subjected to IPTG induction at room temperature, a larger proportion of the FlnA1 and FlnA2 proteins was recovered in the soluble fraction (Fig. 4). In order to assess the enzymatic activity of FlnA1-FlnA2 in E. coli, biotransformation assays were carried out using induced cells incubated separately with fluorene, carbazole, dibenzofuran, dibenzothiophene, and dibenzo-p-dioxin, as well as with representative PAHs. Water-soluble oxidation products released into the culture medium were extracted and analyzed using GC-MS. The detection of fluorene oxidation products demonstrated that the recombinant enzyme was active in vivo (Table 2), suggesting that it recruited unspecific electron carriers from the host for function. When strain BL21(DE3)(pET30f), which lacked FlnA1-FlnA2, was incubated with the same PAHs under identical conditions, no oxidation product could be detected, demonstrating that FlnA1-FlnA2 was indeed responsible for fluorene transformation (Table 2).
FIG. 4.
Detection of FlnA1 and FlnA2 overproduced in E. coli BL21(DE3). Soluble (supernatant) and insoluble (pellet) proteins were analyzed by SDS-PAGE. E coli harboring pET30f lacking the flnA1-flnA2 insert (V) was used as a control. Protein extracts from cells overexpressing FlnA1 and FlnA2 (V+I) and grown at 37 and 42°C up to an OD600 of 0.5 prior to IPTG induction are shown. Arrows indicate the α and β subunits of the putative angular dioxygenase. A molecular mass ladder indicating masses in kilodaltons (prestained protein marker, broad range; New England Biolabs, Ipswich, MA) is included.
TABLE 2.
PAH selectivity of FlnA1-FlnA2 from Sphingomonas sp. strain LB126 as expressed in E. coli and comparison to DFDO (20) and CARDO (31)
| Substrate | Possible product | Principal fragment ionsa | Retention time (min) | Yield (%)b | Transformability byc:
|
|
|---|---|---|---|---|---|---|
| DFDO | CARDO | |||||
| Naphthalene | cis-1,2-Dihydroxy-1,2-dihydronaphthalened | 306 (8), 275 (5), 203 (32), 191 (100) | 13.5 | 93 | + | + |
| 1-Naphthold,g | 216 (86), 201 (100), 185 (46), 141 (24) | 12.4 | 7 | + | + | |
| Biphenyl | cis-2,3-Dihydroxy-2,3-dihydrobiphenyld | 332 (52), 243 (22), 227 (100), 211(18) | 15.1 | 84 | + | + |
| 2-Hydroxybiphenyld,g | 242 (48), 227 (76), 211 (100), 165 (7), 152 (20) | 12.9 | 8 | + | + | |
| 3-Hydroxybiphenyld,g | 242 (74), 227 (100), 211 (47), 165 (8), 152 (22) | 14.2 | 8 | + | + | |
| Phenanthrene | cis-9,10-Dihydroxy-9,10-dihydrophenanthrenee | 356 (16), 253, 191, 147 (100), 73 (99) | 16.7 | 95 | − | + |
| Monohydroxyphenanthreneg | 266 (100), 251 (65), 235 (27), 176 13) | 17.4 | 4 | − | + | |
| Anthracene | cis-1,2-Dihydroxy-1,2-dihydroanthracene,d,f | 356 (5), 266 (13), 253 (34), 191 (62), 147 (26), 73 (100) | 17.3 | 69 | − | + |
| Anthracenedihydrodiol | 356 (34), 266 (82), 253 (3), 191 (3), 147 (60), 73 (100) | 17.8 | 27 | − | − | |
| Monohydroxyanthraceneg | 266 (79), 251 (14), 235 (6), 191 (6), 165 (12), 73 (100) | 17.2 | 5 | − | + | |
| Fluorene | Dihydroxyfluorene | 342 (14), 253 (46), 152 (17), 73 (100) | 18.4 | 29 | − | + |
| 1-Hydro-1,1a dihydroxy-9-fluorenoned | 358 (65), 253 (59), 147 (36), 73 (100) | 16.7 | 64 | + | − | |
| Fluorenol-dihydrodiol | 360 (39), 270 (95), 242 (100), 181 (55), 165 (13) | 16.4 | 7 | − | − | |
| 9-Hydroxyfluorene | 1-Hydro-1,1a-dihydroxy-9-fluorenoned | 358 (40), 253 (41), 147 (39), 73 (100) | 16.8 | 82 | + | + |
| Fluorenol-dihydrodiol | 360 (34), 270 (90), 242 (100), 181 (78), 165 (23) | 16.4 | 18 | − | − | |
| 9-Fluorenone | 1-Hydro-1,1a-dihydroxy-9-fluorenoned | 358 (56), 253 (51), 147 (43), 73 (100) | 16.7 | 76 | + | + |
| Fluorenol-dihydrodiol | 360 (30), 270 (91), 242 (100), 181 (85), 165 (24) | 16.4 | 24 | − | − | |
| Fluoranthene | Monohydroxyfluorantheneg | 290 (81), 275 (54), 259 (47), 215 (76) | 20.0 | 100 | Not tested | + |
| Carbazole | Monohydroxycarbazoleg | 255 (100), 239 (51), 224 (47), 209 (22), 166 (11) | 17.1 | 56 | − | − |
| Dihydroxycarbazole | 343 (100), 327 (34), 252 (7), 164 (2) | 18.6 | 10 | − | − | |
| Monohydroxycarbazoleg | 327 (100), 312 (24), 165 (1), 73 (39) | 18.9 | 34 | − | − | |
| Dibenzofuran | 2,2′,3-Trihydroxybiphenyld | 418 (50), 403 (5), 315 (70), 73 (100) | 16.3 | 100 | + | + |
| Dibenzo-p-dioxin | 2,3,2′-Trihydroxydiphenyletherd | 434 (63), 419(11), 331 (77), 73 (100) | 16.9 | 100 | + | + |
| Dibenzothiophene | Dibenzothiophene sulfoned | 200 (7), 184 (100), 171 (7), 139 (18), 73 (4) | 17.7 | Traces | + | + |
| Dibenzothiophene sulfoxided | 216 (5), 200 (5), 184 (100), 147 (72), 73 (9) | 17.8 | Traces | + | + | |
Products were identified by GC-MS analysis (trimethylsilyl derivatization). Fragment ions are expressed as m/z values. Values in parentheses are relative abundances (in percentages) of major fragments.
In cases in which multiple oxidation products were detected, the relative abundance of each product is indicated as a percentage of the total product.
The ability or inability to transform each compound is shown by + or −, respectively.
Same mass spectrum as that of relevant PAH oxidation products generated by CARDO and DFDO and previously identified based on 1H and 13C nuclear magnetic resonance analyses (31).
Same retention time and mass spectrum as cis-9,10-dihydroxy-9,10-dihydrophenanthrene produced by Pdo1 (22).
Same retention time and mass spectrum as cis-1,2-dihydroxy-12-dihydroanthracene produced by Phn1 (7).
Monohydroxylated products are most probably formed by the spontaneous dehydration of the corresponding diols.
Substrate range of FlnA1-FlnA2.
The substrate range of FlnA1-FlnA2 was investigated and compared with those of the well-studied angular dioxygenases dibenzofuran 4,4a-dioxygenase (DFDO) from Terrabacter sp. strain DBF63 (20) and carbazole 1,9a-dioxygenase (CARDO) from Pseudomonas resinovorans CA10 (31, 37). When fluorene was used as a substrate, three oxidation products could be detected (Table 2). The major product was identified as 1-hydro-1,1a-dihydroxy-9-fluorenone (63%) based on the m/z fragment pattern of its mass spectrum, which was identical to that of the DFDO-mediated oxidation product of fluorene (20, 28) and the CARDO-mediated oxidation product of 9-hydroxyfluorene (43). Interestingly, CARDO does not yield the angular dioxygenation product 1-hydro-1,1a-dihydroxy-9-fluorenone when fluorene is used as a substrate (43). Fluorenol-dihydrodiol (7%) and dihydroxyfluorene (29%) were also produced by FlnA1-FlnA2 from strain LB126. The latter product was not formed by DFDO. Fluorenol-dihydrodiol probably resulted from the spontaneous transformation of 1-hydro-1,1a-dihydroxy-9-fluorenone, since this product was not detected after shorter incubations. 9-Hydroxyfluorene is likely oxidized into 9-fluorenone by a nonspecific dehydrogenase from E. coli. Indeed, we also observed such spontaneous oxidation upon the incubation of 9-hydroxyfluorene with the control strain BL21(DE3)(pET30f) lacking the flnA1-flnA2 construct. This result suggests that a dehydrogenase is not essential for the transformation of 9-hydroxyfluorene into 9-fluorenone but that it may be required in vivo to catalyze the reaction at a reasonable rate. 1-Hydro-1,1a-dihydroxy-9-fluorenone also accumulated when 9-hydroxyfluorene or 9-fluorenone was used as a substrate, showing that FlnA1-FlnA2 is involved in at least two steps in fluorene catabolism (Table 2). Given the low level of activity of FlnA1-FlnA2 and the necessity of monooxygenation before angular dioxygenation can occur, 9-hydroxyfluorene and 9-fluorenone are probably instantly consumed and are therefore not present for detection.
Three heteroatomic analogs of fluorene, i.e., dibenzofuran, carbazole, and dibenzothiophene, were tested as substrates for angular oxidation. Dibenzofuran was transformed into 2,2′,3-trihydroxybiphenyl by FlnA1-FlnA2, as previously found for DFDO (20) and CARDO (31). The initial attack occurred at the 4 and 4a carbon atoms, as put forward by Fortnagel et al. in 1989 (8). The dioxygenation of dibenzofuran produces a highly unstable hemiacetal product that could not be observed. Incubation with dibenzothiophene produced traces of dibenzothiophene sulfoxide and dibenzothiophene sulfone. These metabolites were previously identified as metabolic intermediates of dibenzothiophene degradation by Brevibacterium sp. strain DO (45), DFDO (28), and CARDO (31). Since FlnA1-FlnA2 was able to perform angular dioxygenation of fluorene and dibenzofuran, hydroxylation of dibenzothiophene sulfone at the angular position was expected. The level of activity of FlnA1-FlnA2 toward dibenzothiophene may have been too low to detect an angular dioxygenation product by GC-MS, although dibenzothiophene is degraded cometabolically by strain LB126 (46). No angular oxidation product was detected after incubation with carbazole, even though carbazole is a structural analogue of fluorene. Mono- and dihydroxycarbazole were the only oxidation products detected by GC-MS. DFDO from Terrabacter sp. strain DBF63 was not able to perform angular dioxygenation of carbazole either. The detection of monohydroxycarbazole suggests that FlnA1-FlnA2 transforms carbazole into the corresponding dihydrodiol by lateral dioxygenation. Resnick et al. reported that carbazole dihydrodiols are unstable and spontaneously form monohydroxycarbazole by dehydration (35). CARDO released 2′-aminobiphenyl-2,3-diol upon the angular oxidation of carbazole (31). Previously, the crystal structure of CARDO bound to carbazole was resolved and a molecular mechanism of angular dioxygenation for carbazole was proposed (2). Given the low level of protein sequence identity between CARDO and FlnA1 (16%), FlnA1-FlnA2 may be missing the amino acid sequence and hence the corresponding structural specificity for the angular dioxygenation of carbazole. The incubation of FlnA1-FlnA2 with dibenzo-p-dioxin yielded 2,3,2′-trihydroxydiphenylether via angular dioxygenation based on the described m/z fragments from DFDO and CARDO.
Since Sphingomonas sp. strain LB126 is able to use phenanthrene, fluoranthene, and anthracene in cometabolic degradation with pyruvate (46), we tested whether FlnA1-FlnA2 would attack these PAHs. cis-9,10-Dihydroxy-9,10-dihydrophenanthrene, which was previously identified as a product formed by pyrene dioxygenase from Mycobacterium sp. strain 6PY1 (22), was detected as the major oxidation product of phenanthrene. Interestingly, cis-3,4-dihydroxy-3,4-dihydrophenanthrene, which is produced in the catabolic pathway of known phenanthrene degraders, including sphingomonads (7, 34, 49), was not formed. Monohydroxyphenanthrene was detected in small amounts (4%) and may have resulted from the spontaneous dehydration of the corresponding dihydrodiol. In contrast, DFDO did not produce any metabolite when incubated in the presence of phenanthrene (21). When FlnA1-FlnA2 was incubated with fluoranthene, trace amounts of monohydroxyfluoranthene could be detected. Anthracene yielded three metabolites. The major compound could be identified as cis-1,2-dihydroxy-1,2-dihydroanthracene by comparison to the oxidation product formed by Phn1 from Sphingomonas sp. strain CHY-1 (7). Trace amounts of monohydroxyanthracene were also present. CARDO produced the same metabolites, but DFDO did not. Moreover, a second putative anthracene-diol was detected. Its mass spectrum was similar to that of cis-1,2-dihydroxy-12-dihydroanthracene, but the retention time was different. Since no angular attack on anthracene is possible without preliminary monooxygenation, we suggest that this compound may be cis-2,3-dihydroxy-2,3-dihydroanthracene. This metabolite has not been produced by any other enzyme reported so far. When incubated with biphenyl or naphthalene, FlnA1-FlnA2 produced the expected metabolites, which were also reported previously for DFDO and CARDO (20, 31). Our results show that FlnA1-FlnA2 from strain LB126 is unique in that it shares characteristics with both DFDO and CARDO.
Quantitative analysis of water-soluble dihydrodiols formed by FlnA1-FlnA2.
The catalytic activities of FlnA1-FlnA2 toward fluorene and other PAHs were compared by estimating the amount of di- or trihydroxylated products formed by strain BL21(DE3)(pET30fflnA1A2) after overnight incubation. Products were extracted and quantified as NBB derivatives by GC-MS analysis as described in Materials and Methods. Results showed that 1-hydro-1,1a-dihydroxy-9-fluorenone (97.5 μM) and 9,10-phenanthrene dihydrodiol (96.3 μM) accumulated at the highest concentrations, indicating that fluorene and phenanthrene were the preferred substrates (Table 3). GC-MS data from NBB derivatives confirmed that FlnA1-FlnA2 attacked fluorene in the angular position. The attack on phenanthrene generated 9,10-phenanthrene dihydrodiol instead of the more common 3,4-isomer. In this respect, the activity of FlnA1-FlnA2 is quite different from that of other known phenanthrene dioxygenases. In addition, FlnA1-FlnA2 showed a relatively low level of activity toward naphthalene (Table 3). Both dibenzofuran and dibenzo-p-dioxin yielded small amounts of products, essentially because the trihydroxylated compounds generated from these substrates reacted poorly with NBB (data not shown). The amounts of the trihydroxylated products were therefore tentatively calculated based on the peak areas of the trimethylsilyl derivatives determined using 2,3-dihydroxybiphenyl as the standard (Table 3). These results, together with the fact that neither phenanthrene nor dibenzofuran can support the growth of strain LB126, provide additional evidence that FlnA1-FlnA2 acts as an angular dioxygenase specifically dedicated to the initial attack of fluorene.
TABLE 3.
Comparison of the FlnA1-FlnA2 dioxygenase activities toward fluorene and other polycyclic substratesa
| Substrate | Product retention time (min) | Product m/z | Product concn (μM)b |
|---|---|---|---|
| Fluorene | 15.5 | 280 | 98 |
| Phenanthrene | 15.9 | 278 | 96 |
| Anthracene | 14.6 | 278 | 9 |
| 16.6 | 278 | 27 | |
| Naphthalene | 12.1 | 228 | 2 |
| Dibenzofuran | 13.5 | 418 | 10 |
| Dibenzo-p-dioxin | 14.1 | 434 | 10 |
Characteristics of the NBB derivatives, except for dibenzofuran and dibenzo-p-dioxin, which were analyzed as the trimethylsilyl derivatives, are shown. Anthracene yielded two isomers, one of which was identified as anthracene 1,2-dihydrodiol (retention time, 16.6 min).
Concentrations in the bacterial suspension were calculated after 23 h of incubation at 25°C. Values are means of results from duplicate experiments. The standard error was less than 10%.
In conclusion, the enzymes probably responsible for fluorene oxidation in strain LB126 were most closely related to proteins involved in the angular attack on dibenzofuran by gram-positive bacteria. The genes were likely acquired by lateral gene transfer, since a truncated transposase gene was identified upstream of the catabolic genes. RT-PCR analysis showed that cirA, flnA1, and flnE are expressed and induced when Sphingomonas sp. strain LB126 is grown on fluorene as opposed to glucose. The quantification of FlnA1-FlnA2 activity toward several PAHs showed that fluorene and phenanthrene are the preferred substrates, although phenanthrene cannot be used as a sole source of carbon and energy by Sphingomonas sp. strain LB126. Altogether, these data strongly suggest that the genes flnA1 and flnA2 are involved in fluorene degradation by Sphingomonas sp. strain LB126. The proteins encoded by flnA1 and flnA2 from Sphingomonas sp. strain LB126 appear to be quite unique, since no functional counterpart in gram-negative bacteria has been described to date. FlnA1-FlnA2 was shown to be capable of catalyzing monooxygenations and angular and lateral oxygenations of PAHs and heteroaromatics that are not oxidized by DFDO, the most closely related enzyme from Terrabacter sp. strain DBF63.
Acknowledgments
L.S. gratefully acknowledges the Fund for the Promotion of Research in Industry and Agriculture (FRIA), Belgium, for providing him with a doctoral fellowship.
L.S. also thanks the members of the Unit of Physiological Biochemistry (FYSA), Catholic University of Louvain, for their daily help and constructive remarks over many years. P.H. is a research associate at the Foundation for Scientific Research (FNRS), Belgium.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashikawa, Y., Z. Fujimoto, H. Noguchi, H. Habe, T. Omori, H. Yamane, and H. Nojiri. 2006. Electron transfer complex formation between oxygenase and ferredoxin components in Rieske nonheme iron oxygenase system. Structure 14:1779-1789. [DOI] [PubMed] [Google Scholar]
- 3.Bastiaens, L., D. Springael, P. Wattiau, H. Harms, R. deWachter, H. Verachtert, and L. Diels. 2000. Isolation of adherent polycyclic aromatic hydrocarbon (PAH)-degrading bacteria using PAH-sorbing carriers. Appl. Environ. Microbiol. 66:1834-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunz, P. V., and A. M. Cook. 1993. Dibenzofuran 4,4a-dioxygenase from Sphingomonas sp. strain RW1: angular dioxygenation by a three-component enzyme system. J. Bacteriol. 175:6467-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casellas, M., M. Grifoll, J. M. Bayona, and A. M. Solanas. 1997. New metabolites in the degradation of fluorene by Arthrobacter sp. strain F101. Appl. Environ. Microbiol. 63:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerniglia, C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351-368. [Google Scholar]
- 7.Demaneche, S., C. Meyer, J. Micoud, M. Louwagie, J. C. Willison, and Y. Jouanneau. 2004. Identification and functional analysis of two aromatic-ring-hydroxylating dioxygenases from a Sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70:6714-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortnagel, P., H. Harms, R. M. Wittich, W. Francke, S. Krohn, and H. Meyer. 1989. Cleavage of dibenzofuran and dibenzodioxin ring systems by a Pseudomonas bacterium. Naturwissenschaften 76:222-223. [DOI] [PubMed] [Google Scholar]
- 9.Grifoll, M., M. Casellas, J. M. Bayona, and A. M. Solanas. 1992. Isolation and characterization of a fluorene-degrading bacterium: identification of ring oxidation and ring fission products. Appl. Environ. Microbiol. 58:2910-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grifoll, M., S. Selifonov, and P. Chapman. 1995. Transformation of substituted fluorenes and fluorene analogs by Pseudomonas sp. strain F274. Appl. Environ. Microbiol. 61:3490-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grifoll, M., S. A. Selifonov, and P. J. Chapman. 1994. Evidence for a novel pathway in the degradation of fluorene by Pseudomonas sp. strain F274. Appl. Environ. Microbiol. 60:2438-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grifoll, M., S. A. Selifonov, C. V. Gatlin, and P. J. Chapman. 1995. Actions of a versatile fluorene-degrading bacterial isolate on polycyclic aromatic compounds. Appl. Environ. Microbiol. 61:3711-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habe, H., J.-S. Chung, A. Ishida, K. Kasuga, K. Ide, T. Takemura, H. Nojiri, H. Yamane, and T. Omori. 2005. The fluorene catabolic linear plasmid in Terrabacter sp. strain DBF63 carries the beta-ketoadipate pathway genes, pcaRHGBDCFIJ, also found in proteobacteria. Microbiology 151:3713-3722. [DOI] [PubMed] [Google Scholar]
- 14.Habe, H., J. S. Chung, H. Kato, Y. Ayabe, K. Kasuga, T. Yoshida, H. Nojiri, H. Yamane, and T. Omori. 2004. Characterization of the upper pathway genes for fluorene metabolism in Terrabacter sp. strain DBF63. J. Bacteriol. 186:5938-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iida, T., Y. Mukouzaka, K. Nakamura, and T. Kudo. 2002. Plasmid-borne genes code for an angular dioxygenase involved in dibenzofuran degradation by Terrabacter sp. strain YK3. Appl. Environ. Microbiol. 68:3716-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iida, T., K. Nakamura, A. Izumi, Y. Mukouzaka, and T. Kudo. 2006. Isolation and characterization of a gene cluster for dibenzofuran degradation in a new dibenzofuran-utilizing bacterium Paenibacillus sp. strain YK5. Arch. Microbiol. 184:305-315. [DOI] [PubMed] [Google Scholar]
- 17.Johnsen, A. R., L. Y. Wick, and H. Harms. 2005. Principles of microbial PAH-degradation in soil. Environ. Pollut. 133:71-84. [DOI] [PubMed] [Google Scholar]
- 18.Jouanneau, Y., C. Meyer, J. Jakoncic, V. Stojanoff, and J. Gaillard. 2006. Characterization of a naphthalene dioxygenase endowed with an exceptionally broad substrate specificity toward polycyclic aromatic hydrocarbons. Biochemistry 45:12380-12391. [DOI] [PubMed] [Google Scholar]
- 19.Kahng, H. Y., K. Nam, J. J. Kukor, B. J. Yoon, D. H. Lee, D. C. Oh, S. K. Kam, and K. H. Oh. 2002. PAH utilization by Pseudomonas rhodesiae KK1 isolated from a former manufactured-gas plant site. Appl. Microbiol. Biotechnol. 60:475-480. [DOI] [PubMed] [Google Scholar]
- 20.Kasuga, K., H. Habe, J. S. Chung, T. Yoshida, H. Nojiri, H. Yamane, and T. Omori. 2001. Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63. Biochem. Biophys. Res. Commun. 283:195-204. [DOI] [PubMed] [Google Scholar]
- 21.Kasuga, K., H. Nojiri, H. Yamane, and T. Omori. 1997. Genes of enzymes involved in the biodegradation of carbazole, dibenzofuran, fluorene, and dibenzo-p-dioxin by bacteria. Water Sci. Technol. 36:9-16. [Google Scholar]
- 22.Krivobok, S., S. Kuony, C. Meyer, M. Louwagie, J. C. Willison, and Y. Jouanneau. 2003. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: evidence for two ring-hydroxylating dioxygenases. J. Bacteriol. 185:3828-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 24.Larkin, M. J., C. C. Allen, L. A. Kulakov, and D. A. Lipscomb. 1999. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J. Bacteriol. 181:6200-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, D. Dymock, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masai, E., S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 1999. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J. Bacteriol. 181:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mergeay, M., D. Nies, H. G. Schlegel, J. Gerits, P. Charles, and F. Van Gijsegem. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monna, L., T. Omori, and T. Kodama. 1993. Microbial degradation of dibenzofuran, fluorene, and dibenzo-p-dioxin by Staphylococcus auriculans DBF63. Appl. Environ. Microbiol. 59:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moser, R., and U. Stahl. 2001. Insights into the genetic diversity of initial dioxygenases from PAH-degrading bacteria. Appl. Microbiol. Biotechnol. 55:609-618. [DOI] [PubMed] [Google Scholar]
- 30.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nojiri, H., J. Nam, M. Kosaka, K. Morii, T. Takemura, K. Furihata, H. Yamane, and T. Omori. 1999. Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase from Pseudomonas sp. strain CA10. J. Bacteriol. 181:3105-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinyakong, O., H. Habe, A. Kouzuma, H. Nojiri, H. Yamane, and T. Omori. 2004. Isolation and characterization of genes encoding polycyclic aromatic hydrocarbon dioxygenase from acenaphthene and acenaphthylene degrading Sphingomonas sp. strain A4. FEMS Microbiol. Lett. 238:297-305. [DOI] [PubMed] [Google Scholar]
- 33.Pinyakong, O., H. Habe, and T. Omori. 2003. The unique aromatic catabolic genes in sphingomonads degrading polycyclic aromatic hydrocarbons (PAHs). J. Gen. Appl. Microbiol. 49:1-19. [DOI] [PubMed] [Google Scholar]
- 34.Pinyakong, O., H. Habe, N. Supaka, P. Pinpanichkarn, K. Juntongjin, T. Yoshida, K. Furihata, H. Nojiri, H. Yamane, and T. Omori. 2000. Identification of novel metabolites in the degradation of phenanthrene by Sphingomonas sp. strain P2. FEMS Microbiol. Lett. 191:115-121. [DOI] [PubMed] [Google Scholar]
- 35.Resnick, S. M., D. S. Torok, and D. T. Gibson. 1993. Oxidation of carbazole to 3-hydroxycarbazole by naphthalene 1,2-dioxygenase and biphenyl 2,3-dioxygenase. FEMS Microbiol. Lett. 113:297-302. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1990. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Sato, S. I., J. W. Nam, K. Kasuga, H. Nojiri, H. Yamane, and T. Omori. 1997. Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA10. J. Bacteriol. 179:4850-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selifonov, S. A., M. Grifoll, J. E. Gurst, and P. J. Chapman. 1993. Isolation and characterization of (+)-1,1a-dihydroxy-1-hydrofluoren-9-one formed by angular dioxygenation in the bacterial catabolism of fluorene. Biochem. Biophys. Res. Commun. 193:67-76. [DOI] [PubMed] [Google Scholar]
- 39.Shepherd, J. M., and G. Lloyd-Jones. 1998. Novel carbazole degradation genes of Sphingomonas CB3: sequence analysis, transcription, and molecular ecology. Biochem. Biophys. Res. Commun. 247:129-135. [DOI] [PubMed] [Google Scholar]
- 40.Shintani, M., M. Urata, K. Inoue, K. Eto, H. Habe, T. Omori, H. Yamane, and H. Nojiri. 2007. The Sphingomonas plasmid pCAR3 is involved in complete mineralization of carbazole. J. Bacteriol. 189:2007-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Story, S. P., E. L. Kline, T. A. Hughes, M. B. Riley, and S. S. Hayasaka. 2004. Degradation of aromatic hydrocarbons by Sphingomonas paucimobilis strain EPA505. Arch. Environ. Contam. Toxicol. 47:168-176. [DOI] [PubMed] [Google Scholar]
- 42.Story, S. P., S. H. Parker, S. S. Hayasaka, M. B. Riley, and E. L. Kline. 2001. Convergent and divergent points in catabolic pathways involved in utilization of fluoranthene, naphthalene, anthracene, and phenanthrene by Sphingomonas paucimobilis var. EPA505. J. Ind. Microbiol. Biotechnol. 26:369-382. [DOI] [PubMed] [Google Scholar]
- 43.Takagi, T., H. Nojiri, T. Yoshida, H. Habe, and T. Omori. 2002. Detailed comparison between the substrate specificities of two angular dioxygenases, dibenzofuran 4,4a-dioxygenase from Terrabacter sp. and carbazole 1,9a-dioxygenase from Pseudomonas resinovorans. Biotechnol. Lett. 24:2099-2106. [Google Scholar]
- 44.Trenz, S. P., K. H. Engesser, P. Fischer, and H. J. Knackmuss. 1994. Degradation of fluorene by Brevibacterium sp. strain DPO 1361: a novel C-C bond cleavage mechanism via 1,10-dihydro-1,10-dihydroxyfluoren-9-one. J. Bacteriol. 176:789-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Afferden, M., S. Schacht, J. Klein, and H. G. Trüper. 1990. Degradation of dibenzothiophene by Brevibacterium sp. DO. Arch. Microbiol. 153:324-328. [Google Scholar]
- 46.van Herwijnen, R., P. Wattiau, L. Bastiaens, L. Daal, L. Jonker, D. Springael, H. A. Govers, and J. R. Parsons. 2003. Elucidation of the metabolic pathway of fluorene and cometabolic pathways of phenanthrene, fluoranthene, anthracene and dibenzothiophene by Sphingomonas sp. LB126. Res. Microbiol. 154:199-206. [DOI] [PubMed] [Google Scholar]
- 47.Wattiau, P., L. Bastiaens, R. van Herwijnen, L. Daal, J. R. Parsons, M. E. Renard, D. Springael, and G. R. Cornelis. 2001. Fluorene degradation by Sphingomonas sp. LB126 proceeds through protocatechuic acid: a genetic analysis. Res. Microbiol. 152:861-872. [DOI] [PubMed] [Google Scholar]
- 48.Willison, J. C. 2004. Isolation and characterization of a novel sphingomonad capable of growth with chrysene as sole carbon and energy source. FEMS Microbiol. Lett. 241:143-150. [DOI] [PubMed] [Google Scholar]
- 49.Zylstra, G. J., and E. Kim. 1997. Aromatic hydrocarbon degradation by Sphingomonas yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 19:408-414. [DOI] [PubMed] [Google Scholar]