Abstract
We present the development of a simple, high-throughput screen for identifying bacterial strains capable of l-tyrosine production. Through the introduction of a heterologous gene encoding a tyrosinase, we were able to link l-tyrosine production in Escherichia coli with the synthesis of the black and diffusible pigment melanin. Although melanin was initially produced only at low levels in morpholinepropanesulfonic acid (MOPS) minimal medium, phosphate supplementation was found to be sufficient for increasing both the rates of synthesis and the final titers of melanin. Furthermore, a strong linear correlation between extracellular l-tyrosine content and melanin formation was observed by use of this new medium formulation. A selection strategy that utilizes these findings has been developed and has been shown to be effective in screening large combinatorial libraries in the search for l-tyrosine-overproducing strains.
Traditional metabolic engineering has often focused on the rational design of metabolic pathways, relying on extensive a priori knowledge of cellular mechanisms in order to redirect metabolite flow, revise metabolic regulation, or introduce new pathways to achieve a particular phenotype (3). In recent years, however, considerable advances in molecular biology and the growing availability of annotated genome sequences have made combinatorial methods of metabolic engineering an increasingly attractive approach for strain improvement. With these search strategies, random, traceable genetic-level perturbations are introduced into a cell to yield a new population of strains with a diverse range of properties. A screen is then implemented in order to probe these mutant libraries for strains exhibiting enhancements in the trait of interest. Although the potential of the combinatorial approach has already been demonstrated for a number of genetic tools, including transposon mutagenesis, genomic complementation, and global transcription machinery engineering (1, 12), each of these examples has dealt with easily accessible phenotypes. In the case of lycopene production in Escherichia coli, desirable clones assumed a red pigmentation and could therefore be selected by visual inspection. For solvent (sodium dodecyl sulfate, ethanol) tolerance in E. coli and Saccharomyces cerevisiae, simple growth competition assays were used to heavily enrich a population with the best performers. For most other systems of interest, however, the widespread use of these combinatorial approaches is hampered by the absence of a high-throughput method for selecting strains with the desired cellular properties.
Although l-tyrosine has received far less attention than the other aromatic amino acids, l-tryptophan and l-phenylalanine, it remains a valuable target compound for microbial production. Apart from its use as a dietary supplement, l-tyrosine also serves as a precursor for 3,4-dihydroxy-l-phenylalanine (l-DOPA, or levodopa), an important drug for the treatment of Parkinson's disease (5). Additionally, l-tyrosine is involved in the synthesis of p-hydroxycinnamic acid and p-hydroxystyrene, both of which serve as starting materials for a variety of novel polymers, adhesives and coatings, pharmaceuticals, biocosmetics, and health and nutrition products (20, 23).
Most prior work on the microbial production of aromatic amino acids has focused largely on two main goals: (i) alleviating the feedback regulation of the product-forming pathway and (ii) altering central carbon metabolism in order to increase the supply of the two main precursors, erythrose-4-phosphate and phosphoenolpyruvate (4, 5, 11, 24). Although these approaches have certainly led to significant increases in aromatic amino acid production, further gains in yield and productivity may require the modulation of factors that are not directly involved in the biosynthetic pathway or the related precursor-forming/utilization reactions. Implementation of the combinatorial metabolic engineering approaches discussed earlier would allow for the identification of these more obscure targets, which may act through unknown or poorly understood mechanisms. A high-throughput screen capable of selecting l-tyrosine-producing mutants from a large, diverse population thus becomes an important tool for the future engineering of these production strains.
Here we present the development of a high-throughput screen for l-tyrosine production based on the synthesis of the black, diffusible pigment melanin. This is accomplished through the heterologous expression of a bacterial tyrosinase in the production strain of interest. Tyrosinases, which contain a pair of cupric ions in their active site, use molecular oxygen to catalyze the ortho-hydroxylation of l-tyrosine to l-DOPA, followed by its oxidation to dopachrome. The reactive quinones that are generated then polymerize nonenzymatically to form melanin (8). Since melanin is a black pigment with a characteristic absorbance profile, the production of melanin can easily be detected by both visual and spectrophotometric means. The coupling of l-tyrosine production and melanin synthesis thus enables a simple method for identifying high l-tyrosine producers within a mixed population of cells. In the present study, we have introduced the melA gene from Rhizobium etli (7, 13) into a series of E. coli l-tyrosine production strains. Strains that either produced or were exposed to greater amounts of l-tyrosine could be distinguished by the unique pigmentation imparted by the synthesis of melanin.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
R. etli CFN42 was kindly provided by G. Dávila (10) and was cultured in peptone-yeast extract (PY) medium at 30°C (6). E. coli DH5α (Invitrogen) was used for routine transformations as described in the protocol and was cultivated in Luria-Bertani (LB) medium. The following plasmids were transformed into E. coli K-12 ΔpheA ΔtyrR (T. Lütke-Eversloh and G. Stephanopoulos, unpublished data) and/or E. coli K-12 ΔpheA ΔtyrR ΔwecB (C. N. S. Santos and G. Stephanopoulos, unpublished data) and used for l-tyrosine and melanin production experiments: pCL1920::tyrAWTaroGWT, pCL1920::tyrAfbraroGWT, pCL1920::tyrAWTaroGfbr, pCL1920:: tyrAfbraroGfbr, pTrcmelA, and pTrcmelAmut1(15). l-Tyrosine production experiments were performed at 37°C with 225-rpm orbital shaking in 50 ml morpholinepropanesulfonic acid (MOPS) minimal medium (Teknova) (17) cultures supplemented with 5 g/liter glucose and an additional 4 g/liter NH4Cl. Liquid melanin production experiments were performed at 30°C with 225-rpm orbital shaking in 50 ml M9 (22) or MOPS minimal medium cultures supplemented with 5 g/liter glucose, an additional 4 g/liter NH4Cl, 40 μg/ml CuSO4, and l-tyrosine at the indicated concentrations. All liquid cultivations were conducted at least in triplicate. Solid melanin production experiments were performed at 30°C in MOPS minimal medium supplemented with 5 g/liter glucose, an additional 4 g/liter NH4Cl, 0.4 μg/ml CuSO4, 15 g/liter Bacto agar (BD Diagnostics), and l-tyrosine at the indicated concentrations. When appropriate, antibiotics were added at the following concentrations: 100 μg/ml carbenicillin for maintenance of pTrcmelA and 50 μg/ml spectinomycin for maintenance of pCL1920-derived plasmids. Carbenicillin was chosen in place of ampicillin due to its improved stability during the longer cultivations (>48 h) required for the synthesis of melanin. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added at concentrations of 1 mM for the induction of pTrcmelA and 3 mM for the induction of both pTrcmelA and pCL1920-derived plasmids. All chemicals, including those used in the supplementation experiments—CaCl2, NaCl, Na2HPO4, NaH2PO4, and K2HPO4—were purchased from Sigma, J. T. Baker, or Mallinckrodt Chemicals.
Construction of pTrcmelA.
R. etli CFN42 genomic DNA was extracted with the Wizard genomic DNA purification kit (Promega) and used as a template for the amplification of melA with Pfu Turbo DNA polymerase (Stratagene) and the following primers: melA sense NcoI (5′-TAA ACC ATG GCG TGG CTG GTC GGC A-3′) and melA anti Hind III (5′-ACG AAG CTT TTA GGC GGA CAC TAT GGC TAT TTC TAG CTT-3′). In order to introduce an NcoI restriction site for cloning, the start codon was changed from TTG to ATG, and the second codon was changed from CCG to GCG. This second alteration resulted in a proline-to-alanine substitution in the second amino acid. The melA PCR product was digested with NcoI and HindIII and then ligated into the NcoI/HindIII-digested plasmid pTrcHis2B (Invitrogen) for 1 h at room temperature. The plasmid was transformed into chemically competent E. coli DH5α cells (Invitrogen) and plated onto LB agar plates containing 100 μg/ml ampicillin, 1 mM IPTG, 500 mg/liter l-tyrosine, and 0.4 μg/ml CuSO4. The latter step was designed to facilitate the selection of clones with correct plasmids, which should synthesize melanin and form dark colonies. Plasmid constructs were isolated and verified by sequencing. All enzymes used in the cloning procedure were purchased from New England Biolabs.
Analytical methods.
For the quantification of l-tyrosine, cell-free culture supernatants were filtered through 0.2-μm-pore-size polytetrafluoroethylene membrane syringe filters (VWR International) and used for high-performance liquid chromatography (HPLC) analysis with a Waters 2690 Separations module connected with a Waters 996 photodiode array detector (Waters) set to a wavelength of 278 nm. The samples were separated on a Waters Resolve C18 column with 0.1% (vol/vol) trifluoroacetic acid in water (solvent A) and 0.1% (vol/vol) trifluoroacetic acid in acetonitrile (solvent B) as the mobile phase. The following gradient was used at a flow rate of 1 ml/min: at 0 min, 95% solvent A plus 5% solvent B; at 8 min, 20% solvent A plus 80% solvent B; at 10 min, 80% solvent A plus 20% solvent B; at 11 min, 95% solvent A plus 5% solvent B. For the quantification of melanin, the optical densities of cell-free culture supernatants at 400 nm were determined with an Ultrospec 2100 pro UV/visible spectrophotometer (Amersham Biosciences) and compared to a synthetic melanin standard (Sigma). For cell density determinations, the optical densities of cultures and cell-free culture supernatants were measured at 600 nm. Since melanin affects the absorbance measurements at this wavelength, the cell density is calculated by taking the difference between these two values. pH measurements were taken with a SympHony SP20 pH meter and electrode (VWR International).
Library construction and screening.
Transposon mutagenesis (random knockout) libraries for K-12 ΔpheA ΔtyrR/pCL1920::tyrAfbraroGfbr/pTrcmelAmut1 were generated by transformation with 1,000 to 1,300 ng of the pJA1 vector (2). After an initial 1-h outgrowth at 37°C, cells were centrifuged at 2,000 × g and resuspended in 1 ml MOPS minimal medium. Cells were then plated on 150- by 15-mm petri dishes containing MOPS minimal medium with 5 g/liter glucose, an additional 4 g/liter NH4Cl, 40 μg/ml CuSO4, and 20 mM Na2HPO4. Additionally, the medium was supplemented with 10 μg/ml kanamycin to select for strains with transposon-mediated chromosomal integrations. After an incubation period of 120 to 144 h at 30°C, 165 of the darkest colonies (representing 7.9% of the total population) were chosen and restreaked onto a fresh set of MOPS agar plates. Thirty colonies exhibiting the greatest melanin production after an additional 120 to 144 h of incubation were used to inoculate 200 μl of LB medium containing 1 mM IPTG and 50 μg/ml spectinomycin. After four rounds of subculturing, with each round lasting at least 5 to 6 h, individual colonies were isolated and tested for the loss of pTrcmelAmut1 by streaking onto LB plates with and without ampicillin (Amp+ and Amp−, respectively). Ampicillin-sensitive colonies were then analyzed for l-tyrosine production under the cultivation conditions described above. A modified thermal asymmetric interlaced PCR protocol was used to sequence and identify promising transposon targets (1).
RESULTS
Isolation of a melA variant with an enhanced capacity for melanin synthesis.
The melA gene was amplified from R. etli genomic DNA and introduced into the pTrcHis2B vector under the control of the IPTG-inducible promoter Ptrc. During sequence verification of the plasmid construct, a variant was discovered containing a C→T base pair substitution at the 1,000th nucleotide of the melA gene, a change that results in a proline-to-serine switch in the 334th amino acid. This single amino acid substitution led to a significant reduction in the lag time before the onset of melanin production, with the mutant showing signs of melanin synthesis 12 h ahead of the wild type (Fig. 1). The mutated plasmid variant, named pTrcmelAmut1, was therefore selected for use in subsequent melanin production experiments.
FIG. 1.
Growth and melanin production of K-12 ΔpheA ΔtyrR expressing two versions of the R. etli melA gene. Cultures were grown in M9 minimal medium with l-tyrosine supplementation at 500 mg/liter. Squares, pTrcmelA; triangles, pTrcmelAmut1; open symbols, growth (expressed as optical density at 600 nm [OD600]); solid symbols, melanin production (in milligrams per liter).
Melanin production in M9 and MOPS minimal media.
In order to demonstrate the feasibility of probing l-tyrosine concentrations through melanin production, K-12 ΔpheA ΔtyrR/pTrcmelAmut1 was cultured in liquid M9 minimal medium supplied with varying amounts of l-tyrosine (0 to 500 mg/liter in 50-mg/liter increments). As expected, a positive linear trend was observed between extracellular l-tyrosine supplementation and melanin production after 48 h of cultivation, with a linear regression R2 value of 0.922. Cultures grown with higher concentrations of l-tyrosine (>250 mg/liter) continued to produce melanin after this period, leading to even greater resolution and higher R2 values after 72 and 96 h (Fig. 2A).
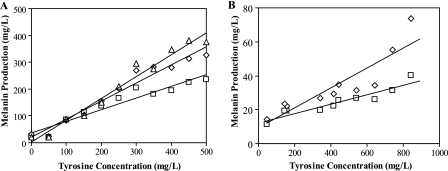
FIG. 2.
Correlation between melanin production and external l-tyrosine concentrations in different medium formulations. (A) M9 minimal medium. K-12 ΔpheA ΔtyrR/pTrcmelAmut1 was cultivated in 0 to 500 mg/liter l-tyrosine in 50-mg/liter increments. Melanin measurements were taken after 48 h (□), 72 h (⋄), and 96 h (▵) of cultivation. R2 values for the linear regressions were 0.922, 0.956, and 0.968, respectively. (B) MOPS minimal medium. Five l-tyrosine production strains (Table 1, strains A to E) were transformed with pTrcmelAmut1 and assayed for melanin production in medium without l-tyrosine supplementation. In order to probe a wider l-tyrosine concentration range, strain D was also cultivated in medium containing 100, 200, 300, 400, or 500 mg/liter l-tyrosine. For these data points, l-tyrosine concentration was calculated as the sum of strain D's l-tyrosine production level after 24 h (347 mg/liter) and the amount of l-tyrosine that was externally supplemented. Melanin measurements are shown after 313 h (□) and 410 h (⋄) of growth. R2 values for the linear regressions were 0.875 and 0.797, respectively.
Since l-tyrosine production in MOPS minimal medium is typically enhanced as much as 15-fold over that in M9 minimal medium (data not shown), steps were taken to determine whether these initial results could be reproduced in this alternate medium formulation. For these experiments, five l-tyrosine production strains (Table 1) were transformed with pTrcmelAmut1 and cultivated in media without l-tyrosine supplementation. Additionally, K-12 ΔpheA ΔtyrR/pCL1920:: tyrAfbraroGfbr/pTrcmelAmut1 (strain D) was cultured in media containing 100 to 500 mg/liter l-tyrosine (in 100-mg/liter increments) to extend the range of l-tyrosine concentrations tested. Under these conditions, two significant drawbacks with the use of MOPS minimal medium were encountered: (i) the poor resolving power of the assay due to the low levels of melanin produced and (ii) the inordinate length of time needed for melanin synthesis to occur. Although a weaker linear correlation between l-tyrosine and melanin concentrations was still observed after 313 and 410 h (Fig. 2B), the highest melanin titers were fivefold lower than those produced in M9 minimal medium (74 mg/liter versus 375 mg/liter). Furthermore, a sixfold-longer incubation period (313 h versus 48 h) was required for this trend to develop.
TABLE 1.
Production strains and l-tyrosine titers after 24 h
| Strain | Genotype | l-Tyrosine production (mg/liter) |
|---|---|---|
| A | K-12 ΔpheA ΔtyrR/pCL1920::tyrAWTaroGWT | 7 |
| B | K-12 ΔpheA ΔtyrR/pCL1920::tyrAWTaroGfbr | 156 |
| C | K-12 ΔpheA ΔtyrR/pCL1920::tyrAfbraroGWT | 175 |
| D | K-12 ΔpheA ΔtyrR/pCL1920::tyrAfbraroGfbr | 347 |
| E | K-12 ΔpheA ΔtyrR ΔwecB/pCL1920::tyrAfbraroGfbr | 433 |
Optimizing melanin production in MOPS minimal medium.
Supplementation experiments were conducted in order to determine which M9 component, if any, could improve tyrosinase enzyme activity in MOPS minimal medium. A comparison of medium compositions revealed that M9 minimal medium contains 180-fold more calcium chloride (CaCl2) and 32-fold more hydrogen phosphate (HPO42−) than MOPS minimal medium (17); hence, the effects of CaCl2 and sodium phosphate (dibasic, Na2HPO4) supplementation were examined. Although the addition of 0.09 mM CaCl2 had a slightly detrimental effect on melanin synthesis, both concentrations of Na2HPO4 tested (40 and 60 mM) were sufficient to restore melanin production in MOPS minimal medium (Table 2). Melanin concentrations measured after 72 and 96 h were comparable to those previously observed for M9 minimal medium. In order to minimize the deviation from the standard recipe for MOPS minimal medium, lower concentrations of Na2HPO4 were also tested for their effects on melanin synthesis. Further optimization of the Na2HPO4 concentration revealed that only 20 mM was necessary to achieve adequate levels of melanin production (Fig. 3).
TABLE 2.
Melanin production by K-12 ΔpheA ΔtyrR/pTrcmelAmut1 in MOPS minimal medium with supplementationa
| Supplementation | Melanin production (mg/liter)
|
|
|---|---|---|
| 72 h | 96 h | |
| None | 72 | 97 |
| CaCl2 (0.09 mM) | 28 | 41 |
| Na2HPO4 | ||
| 40 mM | 319 | 425 |
| 60 mM | 338 | 349 |
All cultures were supplemented with 500 mg/liter l-tyrosine.
FIG. 3.
Melanin production by K-12 ΔpheA ΔtyrR/pTrcmelAmut1 in MOPS minimal medium with different amounts of Na2HPO4 supplementation. All cultures were additionally supplemented with 500 mg/liter l-tyrosine. Melanin measurements were taken after 72 h (open bars) and 96 h (solid bars).
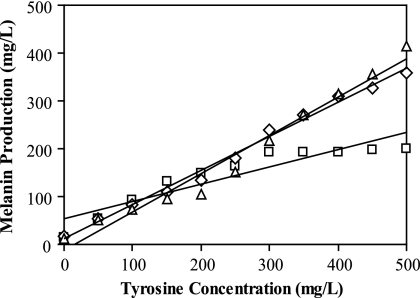
To determine whether a correlation between melanin production and l-tyrosine concentration could be established with the optimized medium formulation, K-12 ΔpheA ΔtyrR/ pTrcmelAmut1 was once again cultured in varying concentrations of l-tyrosine. In stark contrast to the original MOPS minimal medium experiment, supplementation with 20 mM Na2HPO4 led to significant increases in the rates of melanin synthesis, as well as in the final titers of melanin. A linear trend was observed up to 300 mg/liter l-tyrosine after 72 h. After a cultivation period of 96 h, a strong correlation was seen for the entire range of l-tyrosine concentrations tested, exhibiting a linear regression R2 value of 0.992 (Fig. 4).
FIG. 4.
Correlation between melanin production and l-tyrosine supplementation in MOPS minimal medium with 20 mM Na2HPO4. K-12 ΔpheA ΔtyrR/pTrcmelAmut1 was cultivated in 0 to 500 mg/liter l-tyrosine in 50-mg/liter increments. Melanin measurements were taken after 72 h (□), 96 h (⋄), and 120 h (▵) of cultivation. R2 values for the linear regressions were 0.853, 0.992, and 0.970, respectively.
Since the high-throughput implementation of this assay will likely require use in a solid-medium format, a series of experiments was conducted to determine whether incremental differences in melanin production could also be distinguished by visual inspection on agar plates. K-12 ΔpheA ΔtyrR/pTrcmelAmut1 colonies were streaked onto l-tyrosine-supplemented MOPS agar plates and incubated at 30°C for the specified periods. After 72 h, plates with l-tyrosine concentrations differing by as little as 50 mg/liter were easily differentiated based on both the intensity of pigmentation and the radial diffusion of melanin (Fig. 5A). The visual contrast between colonies became even more pronounced with increasing incubation times. These favorable trends were not limited to externally supplied l-tyrosine; a similar pigmentation pattern was observed among strains capable of different levels of l-tyrosine production (Table 1, strains A to E), with the highest l-tyrosine producer exhibiting the darkest coloration (Fig. 5B).
FIG. 5.
Melanin production on MOPS agar plates with 20 mM Na2HPO4. (A) Melanin production by K-12 ΔpheA ΔtyrR/pTrcmelAmut1 with l-tyrosine supplementation. (B) Melanin production by five l-tyrosine production strains, A to E, as listed in Table 1. Image brightness and contrast were adjusted with Adobe Photoshop CS2 (brightness, +25; contrast, +45).
Na2HPO4 supplementation increases the buffering capacity of MOPS minimal medium.
To gain a better understanding of how Na2HPO4 exerts its effect on melanin synthesis, additional supplementation experiments were performed with the following chemicals: sodium chloride (NaCl), potassium phosphate (dibasic, K2HPO4), and sodium phosphate (monobasic, NaH2PO4). A proper basis for comparison was established by maintaining equivalent concentrations of sodium (Na+) or phosphate (HPO42− or H2PO4−) ions in all medium formulations. The addition of NaCl to MOPS minimal medium had a slightly beneficial effect on melanin production but was able to increase titers only to 24 to 30% of the values observed with 20 mM Na2HPO4 (Table 3). Thus, the Na+ concentration has only a marginal impact on melanin synthesis, and the bulk of the improvement in tyrosinase enzyme activity relies on the increase in HPO42− availability. In accordance with this hypothesis, high melanin titers were once again attained when K2HPO4, another source of HPO42−, was added to the medium. Interestingly, the addition of NaH2PO4 did not have a significant effect on melanin production, suggesting that Na2HPO4 supplementation is needed simply to provide the medium with extra buffering capacity. Further evidence arises from an apparent correlation between culture pH and melanin titers, with the highest pH values (6.55 to 6.67) resulting in the greatest melanin titers. When a 50:50 mixture of NaH2PO4 and Na2HPO4 was added to the medium, intermediate values for both pH and melanin concentrations were observed (Table 3). These findings are consistent with other recent reports on R. etli MelA activity. Using a partially purified MelA tyrosinase, Cabrera-Valladares et al. found that the pH optimum for the enzyme's l-DOPA oxidase activity lies in the range of 6.5 to 7.5 (7). Subsequent optimization of melanin production in stationary-phase E. coli cultures revealed a pH optimum of 7.5 (13).
TABLE 3.
Melanin production and pH of K-12 ΔpheA ΔtyrR/pTrcmelAmut1 in MOPS minimal medium with supplementation
| Supplementationa | Melanin production (mg/liter)
|
Culture pH
|
||
|---|---|---|---|---|
| 72 h | 96 h | 72 h | 96 h | |
| None | 25 | 37 | 4.60 | 4.57 |
| Sodium phosphate, dibasic (Na2HPO4) | 305 | 401 | 6.57 | 6.55 |
| Sodium chloride (NaCl) | 74 | 117 | 4.91 | 4.91 |
| Potassium phosphate, dibasic (K2HPO4) | 303 | 392 | 6.65 | 6.67 |
| Sodium phosphate, monobasic (NaH2PO4) | 22 | 34 | 4.74 | 4.76 |
| Sodium phosphate, monobasic and dibasic (NaH2PO4 + Na2HPO4)b | 85 | 136 | 5.04 | 5.04 |
All cultures were supplemented with 500 mg/liter l-tyrosine. Where used, Na2HPO4 was added at a final concentration of 20 mM. For all other medium formulations, supplemental components were added such that the sodium (Na+) or phosphate (HPO42− or H2PO4−) ion concentrations were equivalent to those found in 20 mM Na2HPO4.
NaH2PO4 and Na2HPO4 contributed equal amounts of HPO42− or H2PO4−.
Effects of Na2HPO4 supplementation and pTrcmelAmut1 on l-tyrosine production.
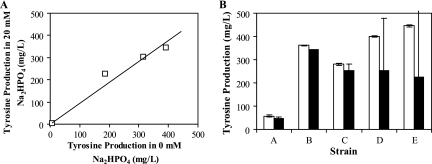
During the course of developing this assay, two major genotypic and environmental perturbations were introduced. Both the heterologous expression of the R. etli melA gene and the change in medium formulation (as necessitated by the Na2HPO4 requirement for melanin synthesis) have the potential to interfere with l-tyrosine production in the strains of interest. Thus, studies were conducted to elucidate what effects, if any, such alterations have on the behavior of these strains. To ensure that Na2HPO4 supplementation does not have a negative effect on l-tyrosine synthesis, the production levels of four strains (Table 1, strains A, C, D, and E) were characterized in MOPS minimal medium without Na2HPO4 or with 20 mM Na2HPO4. A plot of the concentrations achieved by these strains reveals that the overall trends in l-tyrosine production are maintained (Fig. 6A). It is therefore reasonable to assume that the strains exhibiting the greatest capacity for l-tyrosine production in 20 mM Na2HPO4 will remain the top performers under standard cultivation conditions. To determine the impact of expression of a heterologous tyrosinase, the production levels of five strains (Table 1, strains A to E) were measured both in the presence and in the absence of pTrcmelAmut1. As seen in Fig. 6B, the presence of the reporter plasmid had an unfavorable effect on the final l-tyrosine titers for the top two production strains, D and E, when they were cultivated at 37°C in liquid MOPS minimal medium. The l-tyrosine concentrations measured for these strains exhibited wide standard deviations, even for biological replicates within the same experiment. This result demonstrates the importance of removing pTrcmelAmut1 from strains identified through the melanin screen prior to a more rigorous quantification of l-tyrosine production through liquid cultivation and HPLC analysis.
FIG. 6.
Effects of cellular and cultivation perturbations on l-tyrosine production. (A) l-Tyrosine production by strains A, C, D, and E (Table 1) in 0 and 20 mM Na2HPO4 after 24 h. Each data point represents one strain. (B) l-Tyrosine production of strains A to E (Table 1) with (solid bars) and without (open bars) pTrcmelAmut1 after 24 h.
Screening of a random knockout library.
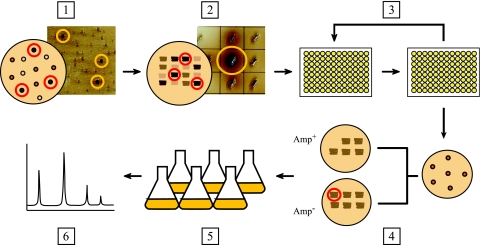
Taking these results together, we have developed a selection strategy for utilizing this assay to screen large combinatorial libraries of mutant strains (Fig. 7). In our initial application of the screen, a random knockout library was generated by the transposon-mediated mutagenesis of the parental strain K-12 ΔpheA ΔtyrR/pCL1920::tyrAfbraroGfbr/pTrcmelAmut1 (2). Following transformation with the pJA1 vector, the library was plated directly onto MOPS agar supplemented with 20 mM Na2HPO4 and 0.4 μg/ml CuSO4, providing the optimum conditions for the synthesis of melanin. The plates were incubated at 30°C for a period of 120 to 144 h, during which colonies of noticeably different pigmentation intensities were detected. To achieve greater resolving power, the darkest colonies from this stage (165 colonies) were streaked out on a fresh set of MOPS agar plates and incubated for an additional 120 to 144 h. Subjecting strains to this second round of selection allowed us to more clearly differentiate between the levels of melanin produced by these isolates, as well as to limit further selection of false positives. Following this second incubation period, 30 strains exhibiting the most intense coloration underwent repeated rounds of subculturing in Amp− medium to facilitate the loss of pTrcmelAmut1, which was shown earlier to have a detrimental effect on final l-tyrosine titers. In most cases, the plasmid was easily lost after four rounds of reinoculation, with each round lasting at least 5 to 6 h (data not shown). Individual clones were isolated and tested for growth on both Amp+ and Amp− media to verify the loss of the plasmid, and ampicillin-sensitive mutants were then cultivated under standard l-tyrosine production conditions and analyzed by HPLC.
FIG. 7.
Strategy for screening libraries on solid media. (Step 1) Plate the library of mutants on MOPS agar and incubate strains at 30°C until clear differences in melanin pigmentation are observed (120 to 144 h). Select the darkest colonies from this first round of screening. (Step 2) Streak out the selected colonies on a fresh set of MOPS agar plates. Incubate the plates for an additional 120 to 144 h to allow for melanin synthesis. Select the darkest streaks from this round. (Step 3) Proceed to the plasmid-curing step. This is achieved by subculturing mutants in Amp− medium at 37°C to facilitate the loss of pTrcmelAmut1. (Step 4) To verify the loss of the plasmid, isolate single colonies and check for growth on Amp− and Amp+ plates. (Step 5) Strains that now exhibit ampicillin sensitivity are cultivated under conditions appropriate for l-tyrosine production (MOPS minimal medium, 37°C). (Step 6) The cell-free culture supernatant is collected and analyzed by HPLC to quantify the l-tyrosine content of the medium. Image brightness and contrast were adjusted with Adobe Photoshop CS2 (brightness, +30; contrast, +30).
Out of an initial library of 21,000 viable colonies, 30 mutants were chosen for a rigorous quantification of l-tyrosine production. Of these isolated strains, two mutants were found to possess l-tyrosine titers 57 to 71% above that of the parental strain (Table 4). The regions of chromosomal integration were sequenced, and it was verified that the integrations had occurred in the C-terminal region of the epsilon subunit of DNA polymerase III, encoded by dnaQ (25), and in a small, 48-amino-acid hypothetical protein encoded by ygdT (http://ecocyc.org/).
TABLE 4.
l-Tyrosine production by strains isolated from a random knockout library (24 h)
| Straina | Genotype | l-Tyrosine production (mg/liter) | % Increase above parental production level |
|---|---|---|---|
| D (parental) | K-12ΔpheA ΔtyrR/pCL1920::tyrAfbraroGfbr | 347 | |
| dnaQ KO | K-12 ΔpheA ΔtyrR dnaQ::kan/pCL1920::tyrAfbraroGfbr | 545 | 57 |
| ygdT KO | K-12 ΔpheA ΔtyrR ygdT::kan/pCL1920::tyrAfbraroGfbr | 594 | 71 |
KO, knockout.
DISCUSSION
Although the microbial production of aromatic amino acids, such as l-tyrosine, has been extensively studied and reviewed in recent years (4, 5, 9, 11, 14, 24), strategies for strain improvement have typically exhibited a narrow focus on well-characterized biochemical pathways and regulatory mechanisms. Combinatorial methods for metabolic engineering were previously of limited use due to the absence of a high-throughput screen for assessing the phenotype of interest. In this study, we have described the development of a simple high-throughput screen for the microbial production of l-tyrosine based on the synthesis of the dark pigment melanin. Although proof-of-concept experiments were carried out with E. coli, such a screen can be readily applied to any microorganism capable of expressing a tyrosinase enzyme with high activity towards l-tyrosine. Many bacteria, including several species of Rhizobium, Streptomyces, Pseudomonas, and Bacillus, naturally express these enzymes and produce melanin for protection against UV damage and for increased virulence and pathogenesis (7, 8, 18, 21, 27). Hence, such strains possess the innate ability to act as sensors for the production of l-tyrosine. Additionally, in cases where the host strain lacks an endogenous tyrosinase, the appropriate gene(s) can easily be introduced on a plasmid or integrated into the bacterial chromosome.
Screening for l-tyrosine production by monitoring melanin synthesis is a versatile technique that can be implemented in a variety of formats. Although the screening strategy presented here focuses on a solid-medium implementation, liquid culture experiments can also be carried out in 96-well microtiter plates with individual strains from a combinatorial library inoculated into separate wells. After cultivation in a medium conducive to melanin production, the absorbance at 400 nm is measured, and desirable mutants are selected for further characterization. In addition, although the method described suggests the use of the same strain for both l-tyrosine production and detection, it is possible to decouple these two steps by creating a strain exclusively for detection. In such a screening strategy, mutants from a combinatorial library are first individually cultured in 96-well microtiter plates. After a set period of time, the culture supernatants, which contain different amounts of microbially produced l-tyrosine, are used as a growth medium component for a separate reporter strain expressing melA. Detection strains grown in the highest l-tyrosine concentrations will synthesize the most melanin and exhibit the highest absorbances at 400 nm, allowing for the identification of the best-performing mutants. This strategy bears similarity to a recently published method for mevalonate detection with a green fluorescent protein-expressing mevalonate auxotroph (19). This “biosensor,” as it was termed, allows one to measure the mevalonate content of a culture by monitoring the growth or fluorescence of the auxotrophic reporter strain. Although such a technique can also be used for l-tyrosine production through the construction of the appropriate auxotroph, the coupling of l-tyrosine production with melanin synthesis offers the added convenience of requiring only one culturing step for simultaneous production and detection of the compound of interest. This important feature also allows for the simple execution of this screen in a solid-medium format, which can be used to further enhance the high-throughput nature of the assay. As described above, with this approach, combinatorial libraries are plated directly onto MOPS agar medium, and colonies that exhibit the darkest pigmentation are selected for further analysis. Such a technique precludes the need for expensive robotics to automate the selection and subsequent inoculation of colonies into microtiter plates and for multiplate scanners to increase the throughput of the absorbance measurements. Through this method, libraries on the order of 106 strains can be probed with relative ease. Although the utility of the mevalonate biosensor strain was also demonstrated in a solid-medium format by utilizing a plate-spraying technique, this method was shown to distinguish only between mevalonate-producing and non-mevalonate-producing colonies (19). This approach would be particularly difficult to implement on agar plates in the case of l-tyrosine production, since the parental strain that is used to generate the combinatorial libraries already produces an elevated level of l-tyrosine. It is therefore likely that the most severe growth-limiting factor for the auxotrophic strain will be the depletion of a carbon source rather than l-tyrosine.
More recently, an alternative assay for l-tyrosine production has been described which utilizes a chemical reaction between 1-nitroso-2-naphthol and l-tyrosine to produce a yellow, fluorescent product. This method, which was originally developed for the determination of l-tyrosine levels in blood plasma (26), was adapted for microbial l-tyrosine production in microtiter plates (16). Again, however, the high-throughput implementation of this assay relies heavily on the availability of expensive robotics to automate sampling, reaction preparations, and fluorescence measurements.
Screening of a random knockout library by this melanin-based selection strategy has led to the discovery of two targets that were successful in eliciting significant increases in l-tyrosine production. A dnaQ::kan mutation in the background of the parental strain K-12 ΔpheA ΔtyrR/pCL1920::tyrAfbraroGfbr led to a 57% increase in l-tyrosine production; a ygdT::kan mutation resulted in even further increases (71%). It should be noted that rational design approaches would not have been capable of predicting either of these genetic changes, particularly the deletion of ygdT, which codes for an as yet unidentified hypothetical protein. Certainly, for both mutant strains identified, further work must be conducted to elucidate the complex relationship between genotype and cellular phenotype. This example, however, serves to illustrate the great potential that can be unlocked by such a screening strategy. Indeed, the application of this simple assay for probing a variety of combinatorial libraries will likely lead to the discovery of additional targets that were previously unreachable through traditional methods of metabolic engineering.
Acknowledgments
This work was supported by the Dupont-MIT Alliance and a National Science Foundation Graduate Fellowship (CNSS).
We thank Guillermo Dávila for providing R. etli strain CFN42.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Alper, H., K. Miyaoku, and G. Stephanopoulos. 2005. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechnol. 23:612-616. [DOI] [PubMed] [Google Scholar]
- 2.Badarinarayana, V., P. W. Estep III, J. Shendure, J. Edwards, S. Tavazoie, F. Lam, and G. M. Church. 2001. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat. Biotechnol. 19:1060-1065. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, J. E. 1991. Toward a science of metabolic engineering. Science 252:1668-1675. [DOI] [PubMed] [Google Scholar]
- 4.Berry, A. 1996. Improving production of aromatic compounds in Escherichia coli by metabolic engineering. Trends Biotechnol. 14:250-256. [DOI] [PubMed] [Google Scholar]
- 5.Bongaerts, J., M. Kramer, U. Muller, L. Raeven, and M. Wubbolts. 2001. Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab. Eng. 3:289-300. [DOI] [PubMed] [Google Scholar]
- 6.Bravo, A., and J. Mora. 1988. Ammonium assimilation in Rhizobium phaseoli by the glutamine synthetase-glutamate synthase pathway. J. Bacteriol. 170:980-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabrera-Valladares, N., A. Martinez, S. Pinero, V. H. Lagunas-Muñoz, R. Tinoco, R. de Anda, R. Vazquez-Duhalt, F. Bolivar, and G. Gosset. 2006. Expression of the melA gene from Rhizobium etli CFN42 in Escherichia coli and characterization of the encoded tyrosinase. Enzyme Microb. Technol. 38:772-779. [Google Scholar]
- 8.Claus, H., and H. Decker. 2006. Bacterial tyrosinases. Syst. Appl. Microbiol. 29:3-14. [DOI] [PubMed] [Google Scholar]
- 9.Flores, N., J. Xiao, A. Berry, F. Bolivar, and F. Valle. 1996. Pathway engineering for the production of aromatic compounds in Escherichia coli. Nat. Biotechnol. 14:620-623. [DOI] [PubMed] [Google Scholar]
- 10.González, V., P. Bustos, M. A. Ramirez-Romero, A. Medrano-Soto, H. Salgado, I. Hernandez-Gonzalez, J. C. Hernandez-Celis, V. Quintero, G. Moreno-Hagelsieb, L. Girard, O. Rodriguez, M. Flores, M. A. Cevallos, J. Collado-Vides, D. Romero, and G. Davila. 2003. The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments. Genome Biol. 4:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda, M. 2006. Towards bacterial strains overproducing l-tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 69:615-626. [DOI] [PubMed] [Google Scholar]
- 12.Jin, Y. S., and G. Stephanopoulos. 2007. Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli. Metab. Eng. 9:337-347. [DOI] [PubMed] [Google Scholar]
- 13.Lagunas-Muñoz, V. H., N. Cabrera-Valladares, F. Bolivar, G. Gosset, and A. Martinez. 2006. Optimum melanin production using recombinant Escherichia coli. J. Appl. Microbiol. 101:1002-1008. [DOI] [PubMed] [Google Scholar]
- 14.Lütke-Eversloh, T., C. N. S. Santos, and G. Stephanopoulos. 2007. Perspectives of biotechnological production of l-tyrosine and its applications. Appl. Microbiol. Biotechnol. 77:751-762. [DOI] [PubMed] [Google Scholar]
- 15.Lütke-Eversloh, T., and G. Stephanopoulos. 2007. l-Tyrosine production by deregulated strains of Escherichia coli. Appl. Microbiol. Biotechnol. 75:103-110. [DOI] [PubMed] [Google Scholar]
- 16.Lütke-Eversloh, T., and G. Stephanopoulos. 2007. A semi-quantitative high-throughput screening method for microbial l-tyrosine production in microtiter plates. J. Ind. Microbiol. Biotechnol. 34:807-811. [DOI] [PubMed] [Google Scholar]
- 17.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nosanchuk, J. D., and A. Casadevall. 2003. The contribution of melanin to microbial pathogenesis. Cell. Microbiol. 5:203-223. [DOI] [PubMed] [Google Scholar]
- 19.Pfleger, B. F., D. J. Pitera, J. D. Newman, V. J. Martin, and J. D. Keasling. 2007. Microbial sensors for small molecules: development of a mevalonate biosensor. Metab. Eng. 9:30-38. [DOI] [PubMed] [Google Scholar]
- 20.Qi, W. W., T. Vannelli, S. Breinig, A. Ben-Bassat, A. A. Gatenby, S. L. Haynie, and F. S. Sariaslani. 2007. Functional expression of prokaryotic and eukaryotic genes in Escherichia coli for conversion of glucose to p-hydroxystyrene. Metab. Eng. 9:268-276. [DOI] [PubMed] [Google Scholar]
- 21.Ruan, L., W. He, J. He, M. Sun, and Z. Yu. 2005. Cloning and expression of mel gene from Bacillus thuringiensis in Escherichia coli. Antonie van Leeuwenhoek 87:283-288. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Sariaslani, F. S. 2007. Development of a combined biological and chemical process for production of industrial aromatics from renewable resources. Annu. Rev. Microbiol. 61:51-69. [DOI] [PubMed] [Google Scholar]
- 24.Sprenger, G. A. 2007. From scratch to value: engineering Escherichia coli wild type cells to the production of l-phenylalanine and other fine chemicals derived from chorismate. Appl. Microbiol. Biotechnol. 75:739-749. [DOI] [PubMed] [Google Scholar]
- 25.Taft-Benz, S. A., and R. M. Schaaper. 1999. The C-terminal domain of DnaQ contains the polymerase binding site. J. Bacteriol. 181:2963-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waalkes, T. P., and S. Udenfriend. 1957. A fluorometric method for the estimation of tyrosine in plasma and tissues. J. Lab. Clin. Med. 50:733-736. [PubMed] [Google Scholar]
- 27.Wang, G., A. Aazaz, Z. Peng, and P. Shen. 2000. Cloning and overexpression of a tyrosinase gene mel from Pseudomonas maltophila. FEMS Microbiol. Lett. 185:23-27. [DOI] [PubMed] [Google Scholar]