Abstract
Minimization of chemical modifications during the production of proteins for pharmaceutical and medical applications is of fundamental and practical importance. The gluconoylation of heterologously expressed protein which is observed in Escherichia coli BL21(DE3) constitutes one such undesired posttranslational modification. We postulated that formation of gluconoylated/phosphogluconoylated products of heterologous proteins is caused by the accumulation of 6-phosphogluconolactone due to the absence of phosphogluconolactonase (PGL) in the pentose phosphate pathway. The results obtained demonstrate that overexpression of a heterologous PGL in BL21(DE3) suppresses the formation of the gluconoylated adducts in the therapeutic proteins studied. When this E. coli strain was grown in high-cell-density fed-batch cultures with an extra copy of the pgl gene, we found that the biomass yield and specific productivity of a heterologous 18-kDa protein increased simultaneously by 50 and 60%, respectively. The higher level of PGL expression allowed E. coli strain BL21(DE3) to satisfy the extra demand for precursors, as well as the energy requirements, in order to replicate plasmid DNA and express heterologous genes, as metabolic flux analysis showed by the higher precursor and NADPH fluxes through the oxidative branch of the pentose phosphate shunt. This work shows that E. coli strain BL21(DE3) can be used as a host to produce three different proteins, a heterodimer of liver X receptors, elongin C, and an 18-kDa protein. This is the first report describing a novel and general strategy for suppressing this nonenzymatic modification by metabolic pathway engineering.
Escherichia coli is a host commonly used for expression of proteins in research, diagnostic, therapeutic, and industrial applications. Current expression systems are capable of producing high yields of a wide variety of proteins (1). The E. coli strain used most frequently in industrial processes is BL21(DE3) owing to several important characteristics, including (i) fast growth, (ii) low acetate production even when it is grown with a high glucose concentration, and (iii) higher turnover of the tricarboxylic acid cycle (16, 17).
Nonenzymatic posttranslational modifications could be the result of unavoidable interference between bacterial metabolic pathways and the abundant heterologously expressed protein. Such modifications result in heterogeneity of the expressed protein. In the case of therapeutic proteins, efforts have to be made to correct such metabolic interference to help ensure the highest level of protein quality.
Nonenzymatic glycation is one type of posttranslational modification with important implications. This type of reaction has been extensively evaluated in higher eukaryotes (14, 21, 24) and in prokaryotes (4, 9, 13, 15, 26, 27).
Gluconoylation is a common posttranslational glycation that can adversely affect the quality of recombinant proteins. This modification is dependent on formation of 6-phosphogluconolactone (6-PGLac), an intermediate of the pentose phosphate (PP) pathway, which is produced by glucose-6-phosphate dehydrogenase. As a potent electrophile, 6-PGLac is expected to be involved in nonenzymatic glycation reactions in vivo. Due to the action of phosphogluconolactonase (PGL) (EC 3.1.1.31), 6-PGLac is converted to 6-phosphogluconic acid by hydrolysis. When metabolites such as 6-PGLac, gluconolactone, and glyceraldehyde begin to accumulate, they can become potent agents for covalent modification of proteins in eukaryotes and prokaryotes (19). In addition, spontaneous alpha-N-6-phosphogluconoylation has been detected in several recombinant proteins fused to hexahistidine affinity tags (9, 26, 27). All of these spontaneous modifications have been observed when heterologous proteins are expressed at high levels in E. coli (4, 15).
The implications of such posttranslational modifications can be significant. For example, glycation of alanine aminotransferase has been shown to markedly reduce its activity (2). Other researchers have shown that there is interference with crystallization of proteins (13). Due to the observation that E. coli host strain BL21(DE3) exhibits low endogenous PGL activity in vitro (23) and significant product gluconoylation, we hypothesized that overexpression of a PGL enzyme could suppress spontaneous gluconoylation. In addition, we reported that constitutive PGL overexpression increased the biomass yield, as well as the specific growth rate, of E. coli strain BL21(DE3) grown in glucose minimal medium and in fed-batch cultures (23). Furthermore, in the present work we describe the effect of overexpression of the PGL enzyme on the specific growth rate, the biomass yield, and protein productivity, as well as application of this approach to three unrelated heterologous proteins that exhibit gluconoylation adducts when they are expressed in E. coli BL21(DE3). One of the proteins analyzed corresponds to an 18-kDa polypeptide in which significant levels of the gluconoylated product were found for the first time (8). The second protein studied is associated with the liver X receptors (LXR and RXR) (12). In order to ascertain the general applicability of our findings, a third protein, elongin C, was also evaluated (25).
Results presented here demonstrate that gluconoylation in these proteins can be suppressed by PP pathway engineering by coexpressing a pgl gene in the BL21(DE3) production strain. The general applicability of our findings suggested that this approach could have a significant conceptual and economic impact on the commercial production of heterologous proteins.
MATERIALS AND METHODS
Materials.
Anhydrous glucose, magnesium sulfate 7-hydrate, anhydrous citric acid, ammonium hydroxide, potassium phosphate monobasic, ferric citrate, manganese chloride 4-hydrate, zinc acetate dehydrate, boric acid, sodium molybdate, calcium chloride, potassium chloride, cupric chloride dehydrate, cobalt chloride 6-hydrate, and EDTA disodium salt were obtained from Mallinckrodt-Baker, Inc. (Paris, KY). Ammonium phosphate dibasic was obtained from EMD Chemicals, Inc. (Gibbstown, NJ). Phytone-peptone and yeast extract were acquired from Becton Dickinson Biological Systems (Sparks, MD). Isopropyl-β-d-thiogalactopyranoside (IPTG) was purchased from Biovectra (Oxford, CT). Ampicillin, carbenicillin, kanamycin, chloramphenicol, glucose-6-phosphate, glucose-6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, morpholineethanesulfonic acid (MES) buffer, NADP+, and hexadecyltrimethylammonium bromide were all obtained from Sigma-Aldrich (St. Louis, MO).
Host strains and culture conditions.
A strain with a BL21(DE3) genetic background was purchased from Novagen (Madison, WI). The designation (DE3) indicates that the host is a lysogen of λDE3 and therefore carries a chromosomal copy of the T7 RNA polymerase gene under control of the lacUV5 promoter. This strain is suitable for production of proteins from target genes cloned in pET vectors.
Pseudomonas aeruginosa ATCC 27853was obtained from the American Type Culture Collection (Rockville, MD).
E. coli was grown under fed-batch culture conditions as described by Wang et al. (23), except that it was induced with IPTG for production of an 18-kDa protein. Also, the expression of either an LXRα/RXRα heterodimer or elongin C was IPTG inducible, but E. coli was simply grown in batch cultures (see the supplemental material).
Constructs and cloning vectors.
The pECO-1 plasmid can be used as a basic vector for coexpression in E. coli. The pECO-1 vector was constructed by digestion of pACYC184 with HindIII and HincII, and a 2,556-bp HindIII/HincII fragment containing the chloramphenicol resistance gene and origin of replication originally derived from p15A was isolated. The 2,556-bp fragment was then ligated with a synthetic DNA fragment with HindIII/HincII ends containing the tetracycline resistance gene promoter (tet promoter) and a multiple cloning site. The multiple cloning site contained NdeI, BamHI, SmaI, KpnI, SalI, SpeI, and HincII sites.
Plasmid pECO-1 was generated as the cloning vector for the 6-phosphogluconolactonase enzyme gene (pgl). The pgl sequence information was obtained from Hager et al. (10). The vector contained the p15A origin of replication and can coexist in E. coli cells with ColE1-bearing plasmids, such as the pET vectors and pGEX vectors. The pECO-1pgl plasmid contains the P. aeruginosa pgl gene in the XhoI site of pECO-1. 5′ and 3′ PCR amplification primers were designed and synthesized to direct PCR amplification of the entire pgl gene, including the region just upstream of the initiation codon, ATG (see the supplemental material).
XhoI sites were introduced into the gene to enable subsequent cloning steps. A 760-bp P. aeruginosa pgl sequence was PCR amplified. The amplified, gel-purified pgl DNA was eluted in water and cloned into the pCR2.1 vector using a TOPO TA cloning kit and was transformed into E. coli One Shot TOP10 (Invitrogen). Sequence-verified inserts were released from the cloning vector by XhoI digestion and ligated to SalI-cut pECO-1 expression vector DNA using T4 DNA ligase (New England Biolabs). Two microliters of the ligation reaction mixture was transformed into E. coli One Shot TOP10 cells. Colonies were screened for pgl inserts in the correct orientation by PCR.
The final construct, pECO-1pgl, was transformed into BL21(DE3) cells to generate a dual plasmid system in which PGL was constitutively expressed and the second plasmid expressed the heterologous protein of interest after IPTG addition.
pET16b.RXRaLBD.H6.LXRaLBD used was a bicistronic expression construct for coexpression of human RXR alpha ligand binding domain (LBD) (225- to 462-amino-acid sequence) and human LXR alpha LBD (205- to 447-amino-acid sequence) in E. coli (see the supplemental material).
pDEST.T7SUMO.ST.ElonginC was prepared by recombination of plasmid pENRT.tev.HumanTCEB1(ElonginC) DNA with pDEST.T7SUMO.ST by a ligation reaction to generate pDEST.T7H6.SUMO.att.tev.ElonginC (see the supplemental material).
In vitro assay of PGL activity.
The enzyme activity was determined by fluorimetric analysis at 340 nm and room temperature using a DU640 spectrophotometer (Beckman-Coulter, Fullerton, CA). Preparation of the cell extract and the enzymatic assay were performed as described by Sinha and Maitra (20) (see the supplemental material).
High-resolution RP-HPLC assay for analysis of 18-kDa protein variants.
The 18-kDa product-related variants and degradation products (i.e., sulfoxide-related species, inter- and intramolecular disulfide bonds) were monitored using a high-resolution reversed-phase HPLC (RP-HPLC) method. This analysis was performed with an Agilent 1100 system (Wilmington, DE), and samples (cell lysates) were injected onto a Bio-Wide Pore C8 reversed-phase column (2.1 by 150 mm; Supelco, Bellefonte, PA) maintained at 50°C with gradient separation consisting of 0.05% trifluoroacetic acid in water and 0.05% trifluoroacetic acid-80% acetonitrile and UV detection at 214 nm. Using a flow rate of 0.35 ml/min, protein was eluted with a 37.5 to 42.0% solvent-80% acetonitrile gradient in 0.1% formic acid in water from 0 to 38 min after injection.
Cell lysates were obtained from pellets obtained from 1 ml of whole-cell broth. Each cell pellet was resuspended in 5 ml of deionized water. Hexadecyltrimethylammonium bromide was added to a final concentration of 0.2% to induce cell lysis and aid in removal of DNA and host cell protein. Following 30 min of incubation, the sample pH was decreased to 4.5 by addition of acetic acid. Samples were again incubated for 30 min and then were centrifuged at 10,600 × g for 10 min. Supernatant was removed and filtered through a 0.45-μm filter for subsequent analysis by high-resolution RP-HPLC. The injection volume of cell lysate was adjusted to obtain a total peak area similar to that of the reference standard, which yielded a 3-μg sample load. The relative amounts of the product and its variants were estimated by using ChemStation software (Agilent, Wilmington, DE).
The identities of the peaks of product variants were confirmed by coupling RP-HPLC to electrospray ionization-mass spectrometry (ESI-MS). For identification of 18-kDa derivatives, samples from whole-cell lysates were first desalted by RP-HPLC using an acetonitrile gradient and UV absorbance at 214 nm. The flow was split postcolumn, and 0.1 ml/min was introduced into the ESI source of an LCT classic mass spectrometer (Waters, Milford, MA) (see the supplemental material).
Liquid chromatography (LC)/MS.
Eluates from highly purified LXR/RXR heterodimers, as well as from elongin C, were fractionated with an Alliance 2790 HPLC (Waters, Milford, MA). Samples (50 to 100 μl) were injected onto a POROS R1/10 column (2.1 by 30 mm) maintained at 60°C, which was equilibrated with 5% acetonitrile-0.1% formic acid in water. Protein was eluted using a 5 to 95% acetonitrile gradient in 0.1% formic acid in water at a rate of 0.75 ml/min from 0.3 to 1.8 min after injection. The flow was split postcolumn, yielding a total flow into the mass spectrometer of 0.3 ml/min. Mass analysis was performed using a time-of-flight mass analyzer (LCT Classic) coupled to an ESI mass spectrometer. Peaks were analyzed by ESI-MS in positive-ion (ES+) mode using Waters MassLynx, version 3.5. Mass was determined by using the MaxEnt Deconvolution software (version 1; Waters, Milford, MA).
Heterologous protein preparation.
After production by BL21(DE3) and the recombinant strain, heterologous proteins, such as LXR/RXR and elongin C, were purified from cell cultures using different methods. In the case of elongin C, cell lysates were obtained by sonication and centrifugation for 5 min at 34,000 × g. The soluble fraction was bound by the batch method to Ni-nitrilotriacetic acid (NTA) Superflow resin. The material was eluted in 250 mM imidazole-50 mM NaH2PO4. Eluates were then analyzed by the LC/MS method (see the supplemental material). For LXR/RXR protein preparation, bacterial cells were lysed with a high-pressure homogenizer in buffer A (50 mM Tris-HCl, 200 mM NaCl, 5% ethylene glycol, 1 mM dithiothreitol; pH 8.0) containing 25 mM imidazole. After centrifugation at 34,000 × g for 1 h, the supernatant was loaded onto Ni-NTA agarose (Qiagen). The Ni-NTA pool was loaded onto a Mono Q HR 10/10 column (Amersham Biosciences). Bound proteins were eluted with a linear 50 to 500 mM NaCl gradient (see the supplemental material).
Sampling cultures, measurements, and calculated parameters.
The E. coli batch and fed-batch cultures were sampled every 2 h after inoculation of the 15-liter bioreactor with either BL21(DE3) or the BL21(DE3)+PGL recombinant strain. The fermentations were performed in duplicate. At the indicated time points, pellets and supernatants were collected and stored at −70°C. These pellets were used to determine titers. Additionally, the glucose, lactate, acetate, glycerol, and ammonia contents of the corresponding supernatants were analyzed with a bioanalyzer (model Bioprofile 300B; NovaBiomedical, Waltham, MA).
Biomass concentration was monitored offline by determining the optical cell density at 550 nm and the cell dry weight.
The experimentally obtained specific growth rate was used for calculation of the fluxes through the PP pathway to generate the required anabolism, catabolism, and NAPDH to sustain the biomass accumulation rate with a particular macromolecular composition.
The molar biomass yield (Yx/s) was the experimentally determined Yx/s for E. coli strains BL21(DE3) and BL21(DE3)+PGL expressing the 18-kDa protein heterologously and growing in minimal medium on glucose as a sole carbon source. Yx/s was defined as the amount of biomass (in grams [dry weight]) synthesized from 1 mole of glucose (Tables 1 and 2).
TABLE 1.
Effect of PGL overexpression on the performance of glucose-grown cells during batch and fed-batch cultivation before IPTG-induction
| Strain | Batch cultivation
|
Fed-batch cultivation
|
||||
|---|---|---|---|---|---|---|
| Growth rate (1/h) | qGlc (mmol/h/g [dry wt]) | Yx/s (g/mol Glc) | Growth rate (1/h) | qGlc (mmol/h/g [dry wt]) | Yx/s (g/mol Glc) | |
| BL21 (PGL−) | 0.280 ± 0.001 | 4.59 ± 0.02 | 61.00 ± 0.06 | 0.140 ± 0.002 | 2.45 ± 0.03 | 57.26 ± 0.02 |
| BL21 (PGL+) | 0.310 ± 0.001 | 4.50 ± 0.05 | 68.89 ± 0.04 | 0.180 ± 0.001 | 2.29 ± 0.05 | 74.33 ± 0.05 |
TABLE 2.
Effect of PGL coexpression on BL21(DE3) growth and the heterologously expressed 18-kDa protein
| Strain | Production 0 to 12 after induction
|
Production 12 to 24 h after induction
|
Final titers 24 h after induction
|
|||||
|---|---|---|---|---|---|---|---|---|
| Growth rate (1/h) | qGlc (g/h/g [dry wt]) | Yx/s (g/mol Glc) | Growth rate (1/h) | qGlc (g/h/g [dry wt]) | Yx/s (g/mol Glc) | Volumetric productivity (g/liter) | Specific productivity (mg/h/g [dry wt]) | |
| BL21 (PGL−) | 0.030 ± 0.002 | 1.41 ± 0.02 | 21.4 ± 0.1 | 0.011 ± 0.001 | 0.52 ± 0.02 | 19.2 ± 0.1 | 3.9 ± 0.1 | 1.82 |
| BL21 (PGL+) | 0.038 ± 0.001 | 1.47 ± 0.07 | 29.8 ± 0.4 | 0.016 ± 0.001 | 0.42 ± 0.09 | 30.7 ± 0.1 | 6.7 ± 0.6 | 2.88 |
The specific glucose uptake rate (qGlc) was determined by determining the difference in carbon substrate in the extracellular medium between successive time points and was expressed in millimoles of glucose consumed per hour per gram (dry weight) (Tables 1 and 2).
Determination of E. coli macromolecular composition.
Proteins, nucleotides, and carbohydrates were quantified using the assays described by Cortassa et al. (5). The distributions of the monomers used as building blocks of the macromolecular fractions (e.g., amino acids for the protein fraction) were obtained from the study of Ingraham and coworkers performed in 1983 (11).
Metabolic flux analysis.
The rationale underlying evaluation of the quantitative flux in the presence and absence of PGL coexpression was to address the status of the redox intermediate NADPH, as well as the anabolic and catabolic fluxes in the E. coli PP pathway. The methodology proposed by Cortassa et al. (5) focuses on the instantaneous rates of synthesis of intermediate precursors and monomers for macromolecular synthesis necessary to sustain a certain growth rate in minimal medium.
Using the previously described E. coli metabolic pathway network (11), the estimated anabolic fluxes to the oxidative branch of the PP pathway were calculated as fluxes leading to the formation of key intermediates, including ribose-5-phosphate and erythrose-4-phosphate. According to the phosphorylation and NADPH requirements, the minimal catabolic fluxes needed to sustain experimentally determined growth rates were calculated by taking into consideration the theoretical aspects described by Cortassa et al. (5). The catabolic fluxes were also a function of the number of phosphorylation sites in the respiratory chain (P/O ratio) (5). The PP pathway had to be used to satisfy the constant demand of intermediates for the synthesis of amino acids and nucleotides. Additionally, the catabolic use of the PP pathway implied a cycling functioning mode generating NADPH with concomitant oxidation of glucose to CO2 to complete the NADPH demand at a P/O ratio of 1. All estimated metabolic fluxes were expressed in millimoles per hour per gram (dry weight) (Table 3).
TABLE 3.
Metabolic fluxes to the PP pathway exhibited by E. coli BL21(DE3) with and without PGL coexpression
| Strain | Period of induction (h) | Growth rate (1/h) | NADPH flux for anabolism (mmol/h/g [dry wt]) | Anabolic flux to PP pathway (mmol/h/g [dry wt]) | Catabolic flux to PP pathway (mmol/h/g [dry wt]) |
|---|---|---|---|---|---|
| BL21 (PGL+) | 0-12 | 0.038 | 0.618 (16)a | 0.044 (33) | 0.031 (41) |
| 12-24 | 0.016 | 0.301 (69) | 0.018 (63) | 0.001 | |
| BL21 (PGL−) | 0-12 | 0.030 | 0.534 | 0.033 | 0.022 |
| 12-24 | 0.011 | 0.178 | 0.011 | <0.001 |
The numbers in parentheses are percentages.
RESULTS
Comparison of PGL activities in P. aeruginosa, E. coli BL21(DE3), and the recombinant BL21(DE3)+PGL strain.
In the present work we hypothesized that overexpression of the PGL enzyme reduces the intracellular pool of 6-PGLac, thus suppressing spontaneous gluconoylation associated with highly expressed recombinant proteins in E. coli BL21(DE3).
PGL enzyme coexpression was achieved by cloning the P. aeruginosa pgl gene into the pECO-1 plasmid. The final construct, pECO-1pgl, was transformed into BL21(DE3) to generate the BL21(DE3)+PGL strains hosting a second plasmid to express the heterologous protein of interest (LXR/RXR heterodimer, elongin C, or the 18-kDa peptide).
Batch cultures were used to compare the endogenous PGL activity in P. aeruginosa and the activities in the parental BL21(DE3) and recombinant BL21(DE3)+PGL strains. The seed batch cultures were grown in 2.8-liter baffled shake flasks containing 1 liter of Luria-Bertani medium supplemented with 10 g of glucose and 25 μg of chloramphenicol as a selection marker for recombinant BL21(DE3)+PGL. Broth samples were collected at mid-exponential phase and processed to measure the in vitro specific PGL activity.
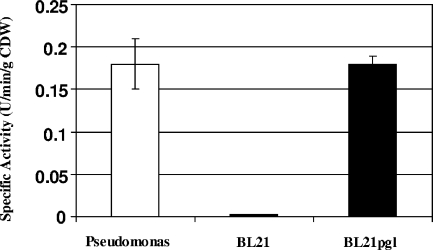
On average, the in vitro PGL specific activities exhibited by P. aeruginosa and the recombinant E. coli BL21(DE3)+PGL strain were 90-fold higher than the specific activity of the parental BL21(DE3) strain (Fig. 1). Accordingly, the large increases in PGL activity were achieved by expression of the heterologous PGL enzyme in BL21(DE3) at levels comparable to those shown by Pseudomonas under equivalent batch culture conditions. Then PGL was expressed in the fermentation culture under different conditions to produce the three unrelated recombinant proteins.
FIG. 1.
In vitro activities of the PGL enzyme from P. aeruginosa and E. coli batch cultures. Broth samples were collected at mid-exponential phase and processed to measure the in vitro PGL activities exhibited by P. aeruginosa and E. coli BL21(DE3). The enzyme activity is expressed in units (mmol NADPH/liter) per minute per gram (dry weight). CDW, cell dry weight.
In vitro PGL activity similar to the spontaneous hydrolysis rate was detected in the parental BL21(DE3) strain, confirming the absence of the PGL enzyme (see the supplemental material).
Effect of PGL overexpression on BL21(DE3) growth in batch and fed-batch cultures before IPTG induction.
When the PGL enzyme was constitutively overexpressed in BL21(DE3), qGlc and the specific growth rate were not significantly affected during the batch phase, but the Yx/s based on glucose consumption increased by 13% (Table 1). Once the fed-batch phase was initiated under glucose-limiting conditions until the cultures reached a critical cell density induced with IPTG, recombinant strain BL21(DE3)+PGL exhibited qGlc similar to those measured for the parental strain. However Table 1 shows that BL21(DE3)+PGL cells grew almost 29% faster and 30% more efficiently per mol of glucose assimilated than parental strain cells. These results suggested that the low PGL enzyme activity measured in the E. coli BL21(DE3) strain represented a bottleneck in the kinetics and efficiency (yield) of glucose utilization through the PP pathway to supply the required precursors for biosynthesis and redox metabolism.
PGL coexpression effect on the growth of strain BL21(DE3) and heterologous expression of the 18-kDa protein.
After addition of IPTG under glucose-limiting conditions during fed-batch culture, a general reduction in the three physiological parameters measured (specific growth rate, qGlc, and Yx/s) was observed in cultures of BL21(DE3) either with or without PGL coexpression (Table 2). However, growth, based on the specific growth rate and Yx/s of BL21(DE3)+PGL cultures during the first 12-h induction phase, was 27 and 50% greater, respectively, than that of the parental BL21(DE3) strain. In the second half of the induction period (12 to 24 h), the specific growth rate and Yx/s of BL21(DE3)+PGL were 45 and 50% greater than those of the parental strain. Furthermore, Table 2 shows that the final volumetric productivity of the recombinant BL21(DE3) strain increased by 70% and the specific productivity for the heterologous 18-kDa protein increased by 60% compared to cultures of parental strain BL21(DE3). On the other hand, the qGlc of BL21(DE3) and the BL21(DE3)+PGL recombinant were not significantly different throughout the production phase (Table 2). These results indicated that with similar qGlc, the overexpression of PGL allowed the cells to sustain higher growth rates and better yields through efficient distribution of carbon. Increases in both the growth rate and the Yx/s could result in increasing availability of key precursors for heterologous protein expression through the oxidative branch of the PP pathway (see Discussion).
Metabolic fluxes to the PP pathway exhibited by E. coli BL21(DE3) with and without PGL coexpression.
Before performing a metabolic flux analysis, we confirmed the carbon, nitrogen, and redox balances. Based on our knowledge of macromolecular composition and the monomer composition of each macromolecular fraction during E. coli balanced growth (i.e., constant growth rate and qGlc), carbon and redox equivalent requirements for biomass formation were calculated by adding all the phosphorylation, carbon, and NAD(P)H required by anabolic and minimum catabolic reactions, as previously evaluated for E. coli cells (11). Since the oxidation of glucose in the tricarboxylic acid cycle and respiration pathway did not produce enough NADPH (through the two different steps catalyzed by isocitrate dehydrogenase and malic enzyme), we considered ribose-5-phosphate and erythrose-4-phosphate to be produced through the oxidative branch of the PP pathway. Thus, three main fluxes ended up in our analysis, the NADPH flux for anabolism, the anabolic flux to the PP pathway, and the catabolic flux to the PP pathway (Table 3). The anabolic and catabolic fluxes to the PP pathway showed the total glucose diverted to this pathway to satisfy the demands of anabolism (erythrose-4-phosphate and ribose-5-phosphate) and catabolism (NADPH, NADH, and ATP) for biomass formation, and the NADPH flux for anabolism clearly indicated the net redox intermediate requirement.
Table 3 shows that in the first 12 h after IPTG induction of a BL21(DE3)+PGL culture there was a demand for an extra 16% NADPH compared with the estimated demand in the parental strain. Furthermore, between 12 and 24 h after induction the demand for NADPH became larger (69%) than the demand in parental cells. In addition, coexpression of the PGL enzyme induced higher anabolic (33%) and catabolic (41%) fluxes to the PP shunt than those estimated for the parental strain. The resulting estimates showed that BL21(DE3)+PGL had to sustain higher catabolic and anabolic fluxes, as well as NADPH generation, throughout the PP pathway because this strain had higher growth rates (Table 3). In addition, the biomass and product yields of BL21(DE3) (Table 2) could be higher because of the efficient functioning of the PP pathway, which not only satisfied the higher demand for NADPH and anabolic intermediates for biosynthesis but also provided surplus building blocks and redox intermediates for heterogeneously expressed protein, as reflected in higher volumetric and specific productivities (Table 2). These results clearly suggest that by redirecting the carbon flux through the PP pathway it might be possible to satisfy efficiently some of the extra requirements imposed by multicopy plasmid replication and high levels of expression of recombinant proteins.
Detection of adduct formation during production of different heterologous proteins.
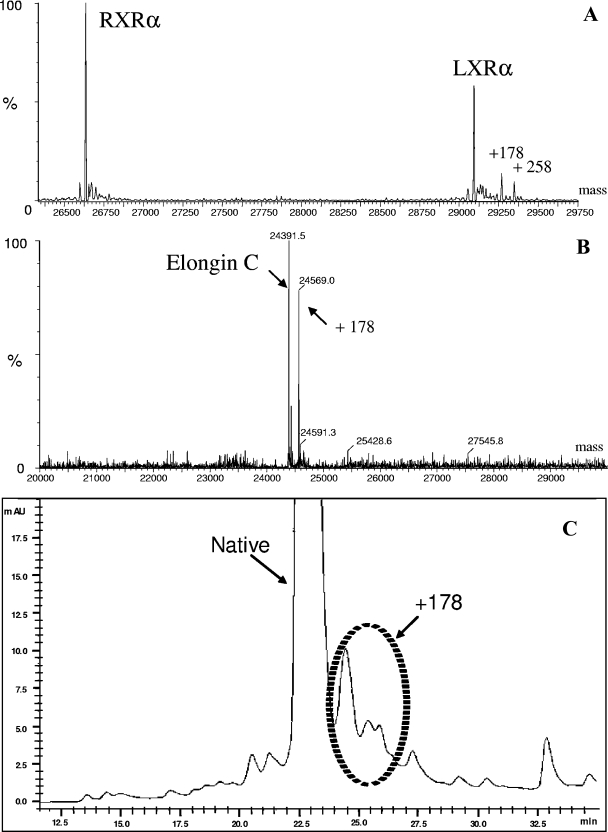
When the LC/MS method was used to analyze the proteins studied, a mass shift of 178 Da due to the gluconoylation of LXR (Fig. 2A) or elongin C (Fig. 2B) was detected. The other mass shift, a shift of 258 Da, was due to the phosphogluconoylation of LXR (Fig. 2A). These results are in close agreement with previous reports which showed that a nonenzymatic reaction of 6-PGLac with a protein could add 258 Da (phosphogluconoyl adduct). Subsequent dephosphorylation by a host cell phosphatase yielded a gluconoyl adduct with an expected mass shift 178 Da (9). Such autoxidative glycation reactions have been described for several hexahistidine-tagged proteins. Importantly, the 18-kDa protein investigated in this study does not contain a hexahistidine tag or a histidine-rich region but is still highly susceptible to this modification. Cell lysates analyzed by high-resolution RP-HPLC produced the main peak corresponding to the 18-kDa product, and some peaks corresponded to derivatives with different degrees of gluconoylation (Fig. 2C).
FIG. 2.
Evidence of adduct formation during production of LXR/RXR, elongin C, and the 18-kDa product. Broth samples were collected at the end of the fermentation and processed as described in Materials and Methods. The gluconoylated and phosphogluconoylated LXR derivatives (A) and the gluconoylated elongin C derivative (B) were evaluated by LC/MS analysis. In the case of the 18-kDa protein (C) the gluconoylated adduct was detected in whole-cell lysates using a high-resolution RP-HPLC method. The circled peaks are product derivatives exhibiting different extents of gluconoylation.
Removal of adducts from heterologous proteins by constitutive PGL coexpression.
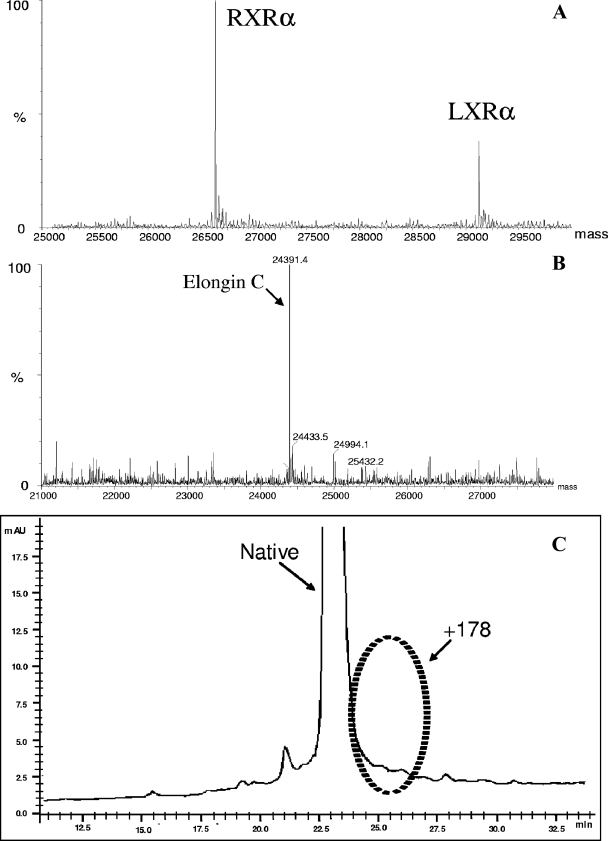
Batch and fed-batch cultures of recombinant E. coli overexpressing the pgl gene, BL21(DE3)+PGL, and parental strain BL21(DE3) were grown in parallel for production of the heterologous proteins studied. Figure 3 shows the absence of gluconoylated adducts for proteins expressed in E. coli strain BL21(DE3)+PGL (compare Fig. 2 and 3).
FIG. 3.
Evidence of adduct removal during production of LXR/RXR, elongin C, and the 18-kDa product. Cultures of the recombinant E. coli BL21(DE3)+PGL strain for production of LXR/RXR, elongin C, and 18-kDa protein were grown in parallel with corresponding “control” fermentations of the BL21(DE3) strain without PGL overexpression. The only difference between runs for product synthesis was the presence of the low-copy-number pECO-1pgl plasmid. As noted above, each comparative study included an assessment of the adduct levels by either LC/MS or high-resolution RP-HPLC analyses.
In the case of RXR/LXR, neither phosphogluconoylation nor gluconoylation was observed by MS analyses despite the fact that the yields ranged from 2.4 to 3.8 mg per g (dry weight) (compare Fig. 2A and 3A). Similarly, elongin C also did not exhibit these modifications when PGL was coexpressed despite an average yield of 1.67 mg per g (dry weight) (compare Fig. 2B and 3B). The magnitude of these increases in quality was substantial. In the case of the 18-kDa protein, the purity of the main product peak increased from 65 to 83% (compare Fig. 2C and 3C). The 18% increase in purity was attributed primarily to the removal of gluconoylation, as shown in Fig. 3C. The presence of the gluconoylated adducts in the controls (i.e., E. coli parental strain) grown under the same culture conditions is shown in Fig. 2.
Overall, the results obtained support the hypothesis that PGL overexpression hinders gluconoylation by reducing the available intracellular pool of 6-PGLac. It is worth noting that we observed both phosphogluconoylation and gluconoylation in the hexahistidine-tagged RXR/LXR heterodimer. Further, the elongin C and 18-kDa proteins exhibited only the gluconoyl adducts. Interestingly, while RXR/LXR and elongin C were both expressed as hexahistidine-tagged proteins, the 18-kDa protein studied did not contain either a hexahistidine tag or a histidine-rich region, which are usually associated with these modifications. Taken together, the results clearly show that all of the modifications were generated primarily by 6-PGLac because a reduction in the intracellular pool of this compound resulted in suppression of all of the nonenzymatic reactions mentioned above.
DISCUSSION
In support of heterologous protein production, extensive characterization studies are regularly performed to confirm structural integrity. Based on these characterizations, gluconoylation ranks high among the posttranslational modifications that adversely affect the quality of heterologous proteins. The present study describes the presence of significant gluconoylation in three heterologous proteins. The 18-kDa peptide was the first example of extensive gluconoylation found (8).
The main contribution of this work is the demonstration that the undesirable posttranslational modification by 6-PGLac of heterologous proteins can be suppressed by engineering the PP pathway through PGL coexpression. The general applicability of the pathway engineering strategy was assessed by studying the production of three unrelated heterologous proteins in E. coli host strain BL21(DE3). Additionally, as a consequence of the positive effect of constitutive PGL coexpression, E. coli strain B could satisfy the extra demand for precursors as well as the NADPH requirements to replicate plasmid DNA and express heterologous genes, as reflected by the 50% increase in the biomass yield and the 60% increase in specific productivity, as well as the increase in the specific growth rate (Table 2).
Different approaches have been taken to eliminate some posttranslational modifications, including methionine oxidation in human cystatin C (3) and glycation of human interferon gamma (15) and of gamma-B-crystallin (4) through a change in culture conditions and in the purification process or by site-directed mutagenesis of the N-terminal amino acids with a greater propensity to be modified. The N terminus phosphogluconoylation/gluconoylation in proteins could be eliminated by directly removing the hexahistidine tag, but this type of technology is very widely utilized. To our knowledge, this report describes a novel and general strategy for metabolically engineering the E. coli host cell toward efficient removal of posttranslational modifications by 6-PGLac.
Previous reports have documented the reactivity of lactones and the resulting nonenzymatic glycation reactions in biomolecules (19). In particular, it has been shown that reactive ketoaldehyde derivatives of glucose, like gluconolactone, can be involved in spontaneous in vitro and in vivo autooxidative glycation reactions of human and rat hemoglobin (14). It is known that the extent of these glycation reactions is dependent upon the number of reactive ɛ-amino groups of lysine residues in the protein (2, 21).
The probability of endogenous protein glycation by 6-PGLac and gluconolactone in higher eukaryotes is lower because (i) the concentration of glucose in the reactions is in the millimolar range, while the metabolites are typically present at substantially lower intracellular concentrations, and (ii) at least in human erythrocytes, the activity levels of the PGL enzyme are several times higher than those of glucose-6-phosphate dehydrogenase (18). It has been proposed that the role of the PGL enzyme in vivo is rapid and effective hydrolysis of lactones, preventing ester- and amide-forming reactions involving, e.g., basic amino acids, such as arginine and lysine (6). In the light of our results, there is complete agreement with the idea that the role of the PGL enzyme is to facilitate the flow of metabolic intermediates through the PP shunt. The increases in the specific growth rate and biomass yield in the E. coli BL21(DE3) strain are associated with a faster and efficient supply by the PP pathway of aromatic amino acids and nucleic acids, as well as NADPH, for biomass synthesis (23). Our studies showed that when PGL is overexpressed, it increases the biomass yield, as well as the yield of a heterologous product (e.g., the 18-kDa protein), in high-cell-density cultures. In both batch and fed-batch cultures there are increases in the biomass yield, and even after IPTG induction, cells coexpressing the PGL enzyme can sustain higher catabolic and anabolic fluxes through the PP pathway based on metabolic flux analysis estimates. Moreover, 13C-labeling and nuclear magnetic resonance studies have shown that when cells are growing aerobically in minimal medium with glucose as the carbon source, the main biosynthetic pathway in E. coli for ribose-5-phosphate and erythrose-4-phosphate, both of which are precursors for nucleotide and aromatic amino acid biosynthesis, is the oxidative branch of the PP pathway (22). It has been reported that 22% of the glucose transported into E. coli is driven by the glucose-6-phosphate dehydrogenase and that part is diverted to nucleotide, amino acid, and vitamin biosynthesis (7). Another important product of the PP oxidative branch is NADPH, which participates mostly in anabolic pathways (e.g., protein biosynthesis). Under the current conditions, where extra synthesis of nucleotides is required to maintain a multicopy plasmid and extra synthesis of amino acids is required to express a recombinant protein, by increasing the flux through the oxidative branch of the PP pathway it is possible to satisfy some of the extra requirements for production of a recombinant protein, as well as to increase the biomass yield and specific growth rate of the E. coli BL21(DE3) host (23). Expression of plasmid-encoded genes in bacteria is widely used for the production of recombinant proteins in biotechnological processes. However, the replication of plasmid DNA and the expression of plasmid-encoded proteins are a metabolic burden for the cells, as reflected by reduced growth rates and biomass yields.
In the studies described here, we hypothesized that the protein glycation occurred in response to three main conditions: (i) the high intracellular pool of 6-PGLac generated by the low PGL activity exhibited by the E. coli BL21(DE3) strain, (ii) the high intracellular expression level of the heterologous proteins, and (iii) the elevated levels of 6-PGLac and its dephosphorylated form, gluconolactone, which acted as potent glycating agents. Taking into consideration the necessity to remove undesired posttranslational modifications and the metabolic burden introduced by plasmid-encoded heterologous proteins to improve process productivity, engineering of E. coli central carbon metabolism definitely helps overcome these problems, as our results demonstrated. This approach could be quite important in the manufacture of high-quality therapeutic proteins.
Supplementary Material
Acknowledgments
We acknowledge GlaxoSmithKline for support of this research.
We thank A. Gardner, D. C. Johnson, Leanne Follweiler, J. Trulli, P. McCormick, G. F. Scott, C. Qiu, S. Solomon, and K. Bojczuk for suggestions and technical assistance.
Footnotes
Published ahead of print on 14 December 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baneyx, F. 1999. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10:411-421. [DOI] [PubMed] [Google Scholar]
- 2.Beranek, M., J. Drsata, and V. Palicka. 2001. Inhibitory effect of glycation on catalytic activity of alanine aminotransferase. Mol. Cell. Biochem. 218:35-39. [DOI] [PubMed] [Google Scholar]
- 3.Berti, P. J., I. Ekiel, P. Lindahl, M. Abrahamson, and A. C. Storer. 1997. Affinity purification and elimination of methionine oxidation in recombinant human cystatin C. Protein Expr. Purif. 11:111-118. [DOI] [PubMed] [Google Scholar]
- 4.Casey, E. B., H. R. Zhao, and E. C. Abraham. 1995. Role of glycine1 and lysine2 in the glycation of bovine gamma-B-crystallin. J. Biol. Chem. 270:20781-20786. [DOI] [PubMed] [Google Scholar]
- 5.Cortassa, S., J. C. Aon, and M. A. Aon. 1995. Fluxes of carbon, phosphorylation, and redox intermediates during growth of Saccharomyces cerevisiae on different carbon sources. Biotechnol. Bioeng. 47:193-208. [DOI] [PubMed] [Google Scholar]
- 6.Duffieux, F., J. van Roy, P. Michels, and F. R. Opperdoes. 2000. Molecular characterization of the first two enzymes of the pentose-phosphate pathway of Trypanosoma brucei. J. Biol. Chem. 275:27559-27565. [DOI] [PubMed] [Google Scholar]
- 7.Flores, S., G. Gosset, N. Flores, A. A. de Graaf, and F. Bolivar. 2002. Analysis of carbon metabolism in Escherichia coli strains with an inactive phosphotransferase system by 13C-labeling and NMR spectroscopy. Metab. Eng. 4:124-137. [DOI] [PubMed] [Google Scholar]
- 8.Gardner, A. R., T. D. Sweitzer, A. H. Taylor, and P. S. Patel. September 2004. Methods for preventing gluconoylation of proteins. U.S. patent 04006507.
- 9.Geoghegan, K. F., H. B. F. Dixon, P. J. Rosner, L. R. Hoth, A. J. Lanzetti, K. A. Borzilleri, E. S. Marr, L. H. Pezullo, L. B. Martin, P. K. LeMotte, A. S. McColl, A. V. Kamath, and J. G. Stroh. 1999. Spontaneous alpha-N-6-phosphogluconoylation of a “His tag” in Escherichia coli: the cause of extra mass of 258 or 178 Da in fusion proteins. Anal. Biochem. 267:169-184. [DOI] [PubMed] [Google Scholar]
- 10.Hager, P. W., M. W. Calfee, and P. Phibbs. 2000. The Pseudomonas aeruginosa devB/SOL homolog, pgl, is a member of the hex regulon and encodes 6-phosphogluconolactonase. J. Bacteriol. 182:3934-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingraham, J. L., O. Maaloe, and F. C. Neidhardt. 1983. Chemical synthesis of the bacterial cell: polymerization, biosynthesis, fueling reactions, and transport, p. 87-174. In Growth of the bacterial cell. Sinauer Associates, Sunderland, MA.
- 12.Joseph, S. B., A. Castrillo, B. A. Laffitte, D. J. Mangelsdorf, and P. Tontonoz. 2003. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat. Med. 9:213-219. [DOI] [PubMed] [Google Scholar]
- 13.Kim, K. M., E. C. Yi, D. Baker, and K. Y. J. Zhang. 2001. Post-translational modification of the N-terminal His tag interferes with the crystallization of the wild-type and mutant SH3 domains from chicken src tyrosine kinase. Acta Crystallogr. Sect. D 57:759-762. [DOI] [PubMed] [Google Scholar]
- 14.Lindsay, R. M., W. Smith, W. K. Lee, M. H. Dominiczak, and J. D. Baird. 1997. The effect of delta-gluconolactone, an oxidised analogue of glucose, on the nonenzymatic glycation of human and rat haemoglobin. Clin. Chim. Acta 263:239-247. [DOI] [PubMed] [Google Scholar]
- 15.Mironova, R., T. Niwa, R. Dimitrova, M. Bosanova, and I. Ivanov. 2003. Glycation and post-translational processing of human interferon-gamma expressed in Escherichia coli. J. Biol. Chem. 278:51068-51074. [DOI] [PubMed] [Google Scholar]
- 16.Noronha, S. B., H. J. C. Yeh, T. F. Spande, and J. Shiloach. 2000. Investigation of TCA cycle and glyoxylate shunt in Escherichia coli BL21 and JM109 using 13C-NMR/MS. Biotechnol. Bioeng. 68:316-327. [PubMed] [Google Scholar]
- 17.Phue, J., S. B. Noronha, R. Hattacharyya, A. J. Wolfe, and J. Shiloach. 2005. Glucose metabolism at high cell density growth of Escherichia coli B and Escherichia coli K. Biotechnol. Bioeng. 90:805-820. [DOI] [PubMed] [Google Scholar]
- 18.Rakitzis, E. T., and P. Papandreou. 1995. Reactivity of 6-phosphogluconolactone with hydroxylamine: the possible involvement of glucose-6-phosphate dehydrogenase in endogenous glycation reactions. Biochem. Int. 37:747-755. [DOI] [PubMed] [Google Scholar]
- 19.Rakitzis, E. T., and P. Papandreou. 1998. Kinetic analysis of 6-phosphogluconolactone hydrolysis in hemolysates. Chem.-Biol. Interact. 113:205-216. [PubMed] [Google Scholar]
- 20.Sinha, A., and P. K. Maitra. 1992. Induction of specific enzymes of the oxidative pentose pathway by glucono-delta-lactone in Saccharomyces cerevisiae. J. Gen. Microbiol. 138:1865-1873. [DOI] [PubMed] [Google Scholar]
- 21.Stitt, A. W. 2005. Maillard reaction in eye disease. Ann. N. Y. Acad. Sci. 1043:582-597. [DOI] [PubMed] [Google Scholar]
- 22.Szyperski, T. 1995. Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids: an efficient analytical tool to investigate intermediary metabolism. Eur. J. Biochem. 232:433-448. [DOI] [PubMed] [Google Scholar]
- 23.Wang, Y., S. L. Wu, W. S. Hancock, R. Trala, M. Kessler, A. H. Taylor, P. S. Patel, and J. C. Aon. 2005. Proteomic profiling of Escherichia coli proteins under high-cell density fed-batch cultivation with overexpression of phosphogluconolactonase. Biotechnol. Prog. 21:1401-1411. [DOI] [PubMed] [Google Scholar]
- 24.Wolff, S. P., and R. T. Dean. 1897. Glucose autooxidation and protein modification. The potential role of autoxidative glycosylation in diabetes. Biochem. J. 245:243-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan, Q., T. Kamura, Y. Cai, J. Jin, M. Ivan, A. Mushegian, R. C. Conaway, and J. W. Conaway. 2004. Identification of elongin C and Skp1 sequences that determine cullin selection. J. Biol. Chem. 279:43019-43026. [DOI] [PubMed] [Google Scholar]
- 26.Yan, Z., G. W. Caldwell, and P. A. McDonell. 1999. Identification of a gluconic acid derivative attached to the N-terminus of histidine-tagged proteins expressed in bacteria. Biochem. Biophys. Res. Commun. 262:793-800. [DOI] [PubMed] [Google Scholar]
- 27.Yan, Z., G. W. Caldwell, P. A. McDonell, W. J. Jones, A. August, and J. A. Masucci. 1999. Mass spectrometry determination of a novel modification of the N-terminus of histidine-tagged proteins expressed in bacteria. Biochem. Biophys. Res. Commun. 259:271-282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.