Abstract
A strategy is described that enables the in situ detection of natural transformation in Acinetobacter baylyi BD413 by the expression of a green fluorescent protein. Microscale detection of bacterial transformants growing on plant tissues was shown by fluorescence microscopy and indicated that cultivation-based selection of transformants on antibiotic-containing agar plates underestimates transformation frequencies.
Horizontal gene transfer (HGT) plays a major role in shaping prokaryote genomes (1, 7, 14, 16, 17, 22, 24, 25). Of the three mechanisms involved in HGT, natural transformation has received limited attention due to the lack of knowledge of the factors regulating transformation in situ and the difficulties of reproducing in vitro the environmental conditions that are conducive to the process. Only a limited number of species have been demonstrated to be naturally transformable, and the majority of the evidence thereof is obtained from in vitro studies (6, 9, 19, 21).
Studies of HGT by natural transformation in complex environments have mostly been performed using bioreporter bacteria, i.e., naturally transformable strains carrying inserted DNA sequences facilitating homologous recombination with the donor DNA. Typically, the studies are based on monitoring the transfer of an antibiotic resistance gene. The antibiotic resistance gene can be either absent (12) or truncated (8) but is restored by homologous recombination with the functional form of the gene or its flanking regions present in the donor DNA. After exposure of the bacteria to the donor DNA, the cells are plated in the presence of the antibiotic, and only those cells that restore a functional antibiotic resistance gene by natural transformation grow in the medium.
A major limitation of this strategy is the agar-plating step required for the enumeration of transformants. The original material is destroyed during sampling, and no information can be obtained on the specific localization of the gene transfer event or on the interaction of the recipient cells with the material, including the donor DNA. Hence, it is not possible to localize hot spots of transformation in a complex environment and describe the microtopology. For example, it is interesting to know if natural transformation is modulated by nutrient levels released from different plant tissues (e.g., along the apical portion of the plant root) or at different depths of a bacterial biofilm. Moreover, conventional plating techniques are limited to culturable cells.
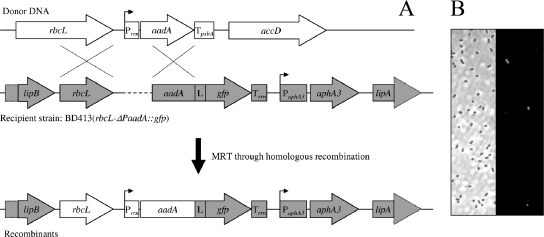
To overcome these limitations, we used a new strategy that allows the expression of a green fluorescent protein (GFP) in the naturally competent Acinetobacter baylyi strain BD413 (23) following a natural transformation event. A marker-reporter rescue cassette was designed to be used as donor DNA (12), and it was inserted into the chromosome of strain BD413, generating the recombinant strain BD413(rbcL-ΔPaadA::gfp) (Table 1 and Fig. 1A). The recipient reporter strain BD413 (rbcL-ΔPaadA::gfp) carries between the lipB gene and the lipA gene (13) an inactived aadA::gfp gene fusion downstream from the chloroplast gene rbcL. The aadA::gfp gene fusion is not expressed in the reporter strain since it lacks a functional promoter. Homologous recombination in the flanking rbcL and aadA loci by a double crossover event with the donor DNA and the recipient reporter strain gives rise to the insertion of the Prrn promoter, allowing the transcription of both the aadA and the gfp domain of the aadA::gfp gene fusion and the restoration of the spectinomycin resistance and fluorescence phenotypes. The detection of transformants can be accomplished by plating on a selective medium, as well as by direct cell fluorescence observation in situ. The recombination site is on the bacterial chromosome to maximize genetic stability and to reduce the likelihood of unintended transfer to other microorganisms.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli XL1-Blue | recA1 endA1 hsdR Tcr | Stratagene |
| A. baylyi BD413 (ADP1) | Rifr mutant of strain BD4 (ATCC 33305) | 11 |
| A. baylyi BD413 aadA::gfp | A BD413 derivative containing, in the lipBA operon in the chromosome, an aadA::gfp fusion gene cassette; Rifr Kmr Spcr; GFP positive | This study |
| A. baylyi BD413(rbcL-ΔPaadA::gfp) | A BD413 derivative containing, in the lipBA operon in the chromosome, the cassette rbcL-ΔP-ΦaadA::gfp, with a transcriptional aadA::gfp fusion inactivation by a Prrn deletion; Rifr Kmr Spcs; GFP negative | This study |
| Bacillus subtilis TS116 | A PB168 derivative containing, in the chromosome, an aadA gene cassette between the tobacco genes rbcL and accD; Cmr Spcr | 2 |
| Azospirillum brasilense SPF94 (pRK290TS) | A derivative of strain SPF94 containing, on plasmid pRK290TS, an aadA gene cassette between the tobacco genes rbcL and accD; Rifr Spcr | 2 |
| Agrobacterium tumefaciens LBA 4404 (pRK290TS) | A derivative of strain LBA4404 containing, on plasmid pRK290TS, an aadA gene cassette between the tobacco genes rbcL and accD; Rifr Spcr | 2 |
| Sinorhizobium meliloti SPF94 (pRK290TS) | A derivative of the USDA1002 mutant strain containing, on plasmid pRK290TS, an aadA gene cassette between the tobacco genes rbcL and accD; Rifr Spcr | 2 |
| Plasmids | ||
| pCLT (also named pLEP01) | Cloning vector, containing the aadA gene cassette between rbcL and accD; Ampr; 7.0 kb | 12 |
| pZR80 | Integrative vector in the chromosomal lipBA operon of strain BD413 (ADP1); Ampr Kmr; 6.4 kb | 13 |
| pZR80-2 | pZR80 derivative, integrative vector at lipBA operon site; Kmr; 4.9 kb | This study |
| pZR80-2 aadA::gfp | pZR80-2 containing the fusion aadA::gfp; Kmr Spcr; 7.2 kb | This study |
| pZR80-2 (rbcL-ΔPaadA::gfp) | pZR80-2 with Prrn upstream of aadA::gfp deleted and with a 1.1-kb fragment containing rbcL; Kmr Spcs; GFP negative; 7.8 kb | This study |
| pRK290TS | A derivative of the broad-host-range vector pRK290; contains an aadA gene cassette between rbcL and accD; Tcr Spcr | 2 |
Amp, ampicillin; Cm, chloramphenicol; Rif, rifampin; Spc, spectinomycin; Tc, tetracycline.
FIG. 1.
(A) Schematic representation (not drawn to scale) of the genetic composition of relevant regions in the donor DNA and in the recipient reporter strain A. baylyi BD413(rbcL-ΔPaadA::gfp). The recipient strain has been designed to recombine with donor DNA containing a functional aadA cassette inserted between the rbcL and the accD genes of the tobacco chloroplast genome. In the donor DNA, the transcription of the aadA gene is under the control of a modified plastid rRNA operon promoter, Prrn, and the 3′ region of the plastid psbA gene (TpsbA). The reporter strain contains a similar but promoter-free gene fusion, aadA::gfp, in which the aadA domain is physically linked to a gfp domain by an amino acid linker, L. A portion of the rbcL gene has been also cloned upstream from the aadA domain to provide a region for a second crossover event. The absence of a promoter (indicated by a dashed line upstream from the aadA domain) confers a spectomycin-susceptible and GFP-negative phenotype to the reporter strain. The region that is replaced by the double crossover event in the homologous recombination process is drawn in white. Arrows indicate genes. The relevant promoters and terminators are indicated by squares labeled with P and T, respectively. MRT, marker rescue transformation. (B) The expression of the aadA::gfp fusion in the bacterial transformants results in cell fluorescence. Transformant cells were added to a suspension of nontransformed cells of the reporter strain BD413(rbcL-ΔPaadA::gfp) and observed by phase-contrast (left) and fluorescence microscopy (right).
The functionality of the reporter strain was tested using the plasmid pCLT as donor DNA. This plasmid is not replicative in strain BD413 (plasmid pCLT is a pUC derivative that is not replicative in strain BD413 [18]) and contains the aadA gene inserted between the rbcL and accD plastid genes. Transformants could be grown on selective medium after exposure of the reporter strain to the DNA of pCLT and showed in all cases a fluorescent phenotype clearly distinguishable from the phenotype of the control strain (Fig. 1B). The transformation frequencies of strain BD413(rbcL-ΔPaadA::gfp) were quantified by standard in vitro assays (see the supplemental material for details) with purified bacterial or plant DNA, bacterial cell lysates, or plant tissue homogenates (see Tables S1 and S2 in the supplemental material). In general, the transformation frequencies were lower than those previously reported by some authors for double crossover homologous transformation with the aadA gene inserted into transplastomic tobacco (6, 12). These authors observed transformation frequencies of about 2 orders higher in magnitude. In contrast, we recorded transformation frequencies in the same order of magnitude as those measured by Gebhard and Smalla (8) by using the same filter transformation protocol. A factor that could influence transformation efficiency between the transformation systems is a possible slight toxic effect of GFP (4), while the chromosomal location of homologous recombination sites should guarantee the same frequency of the plasmid location (5).
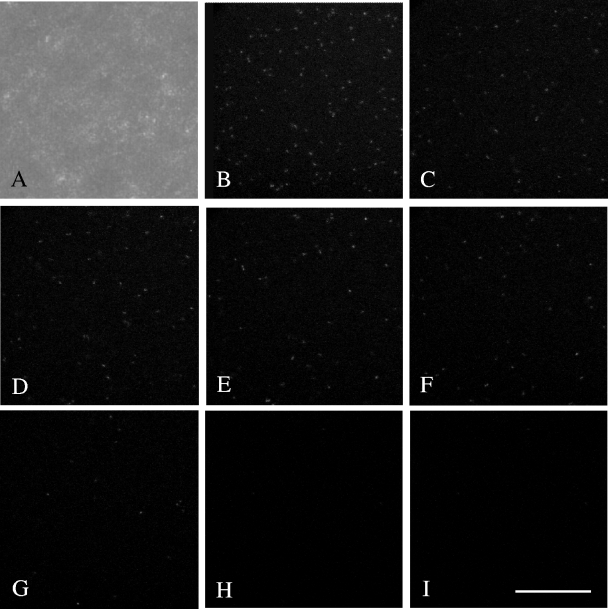
In situ detection of natural transformation experiments was carried out by exposing the reporter strain to lysates of Escherichia coli (pCLT) on filters placed on LB medium without antibiotics. The filters were observed after 2 days of incubation either by epifluorescence microscopy or by confocal laser scanning microscopy (CLSM), as explained in the supplemental material. CLSM imaging demonstrated green fluorescent cells in situ, indicating that GFP expression and, hence, the complementation of the aadA::gfp fusion with a functional promoter had occurred (Fig. 2A to I). Most of the transformants were localized on the top of the bacterial layer and did not seem to spread inside. This uneven distribution can be explained by the lack of oxygen inside the colony (20). To quantify the frequency of transformation observed in situ, 10 microscopic fields, randomly taken at different depths of the filter, were analyzed by CLSM as explained in detail in the supplemental material. A transformation frequency of 6.3 × 10−3 ± 1.0 × 10−3 (mean ± standard deviation) was measured, 2 orders of magnitude higher than the number determined by cultivation-based plating. A similar difference in the detection of the DNA transfer efficiency between culture and culture-independent methods has been reported for conjugal gene transfer in biofilms (20). One possible explanation of this remarkable difference could be that a proportion of the GFP-positive transformants are not readily culturable. Whatever the case, the data indicate that current cultivation-based methods for assessing natural transformation may underestimate the number of transformation events.
FIG. 2.
Visualization by CLSM of the results of a typical transformation experiment of A. baylyi BD413(rbcL-ΔPaadA::gfp) exposed to cell lysates of E. coli XL1-Blue(pCLT) and incubated for 2 days on a filter membrane. (A) Phase-contrast microscopy image. (B to I) Overlapping (B) of seven 5.8-μm sections (C to I) from the surface (C) to the depth (I) of the bacterial layer. The bar represents 30 μm. The total fluorescence in each layer was quantified by using the Histogram tool of Photoshop 6.0 software. The total green component of each layer was calculated as the average brightness level of the green channel for all the pixels of the image. The green-channel brightness, ranging from 0 to 256 units of brightness, was proportional to the richness in green pixels. The green-channel brightnesses of sections B to I were, respectively, 48.2, 13.2, 11.8, 11.3, 8.2, 4.2, 2.1, and 0.8.
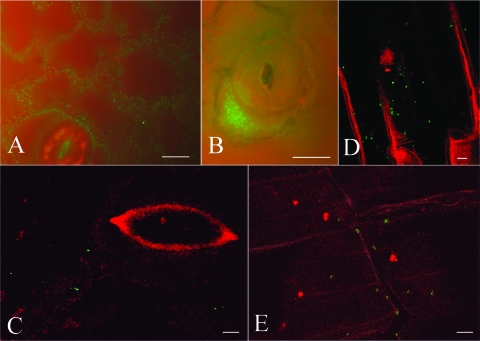
Natural transformation of the new reporter strain was also determined when it was present on defrosted and slightly abraded tobacco leaves, a simple system simulating natural plant decay, such as that occurring in the plant residuosphere (3; see Table S3 in the supplemental material). When purified DNA of transplastomic tobacco was externally supplemented, 10−9 transformants per recipient cell were obtained on defrosted leaf tissues placed on LB agar plates. In situ microscale localization of naturally transformed cells of the reporter strain on the surfaces of defrosted tobacco leaves and roots was observed by fluorescence microscopy and CLSM (Fig. 3). On the leaf tissue surface, fluorescent single cells and microcolonies of constitutively fluorescent A. baylyi BD413(rbcL-aadA::gfp) were localized in the interstices between epidermal cells and close to the stoma at the border with the other epidermal cells (Fig. 3A and B) (reference 20 and references therein). Transformants were detected on the leaf surface or close to a stoma (Fig. 3C) and on root surfaces (Fig. 3D and E) using either purified pCLT or E. coli XL1-Blue(pCLT) cell lysates as external donor DNA.
FIG. 3.
Visualization of fluorescent cells and natural transformation of the reporter strain A. baylyi BD413(rbcL-ΔPaadA::gfp) in situ on decaying tobacco tissues. To demonstrate fluorescence, A. baylyi strain BD413(rbcL-aadA::gfp), constitutively expressing GFP, was inoculated onto defrosted leaves and incubated for 5 days. GFP fluorescence showed single cells and microcolonies of A. baylyi in the interstices of epidermal cells (A) and between a stoma and the border of epidermal cells (B). (C to E) In situ detection by CLSM of natural transformation of the reporter strain BD413(rbcL-ΔPaadA::gfp) exposed to externally added DNA on decaying tissues of tobacco. In the experiment whose results are shown in panel C, leaf tissue was supplemented with purified pCLT DNA, while in the experiments whose results are shown in panels D and E, root tissue was supplemented with cell lysates of E. coli XL1-Blue(pCLT). The images show bacterial transformants on a leaf surface near a stoma (C) or scattered along the root tissues (D and E). Bars represent 10 μm.
Recently, nonculture-based strategies for the detection of natural transformation (21) have been tested in monocultures of strain BD413 growing in biofilms (10) and in bacteria from river water samples (15). The reporter strain BD413(rbcL-ΔPaadA:: gfp) presented here represents an additional tool for detailed studies of single-cell natural transformation events in the environment. Our approach has some limitations: the recipient cells have to be added to the studied environment, and low transformation frequencies will require the microscopic examination of a large number of microscopic fields. However, the presented strategy will expand our repertoire of methods available to better understand natural transformation processes, especially those occurring in environments considered “hot spots” for bacterial gene transfer activities (20, 24).
Supplementary Material
Acknowledgments
We thank D. Young and L. N. Ornston for providing the pZR80 plasmid and L. J. Halverson for plasmid pPnptII::gfp.
This work was supported by the EU TRANSBAC (QLK3-CT-2001-02242). D.D. and K.M.N. acknowledge financial support from the Consiglio dei Diritti Genetici, Italy, funded by the Cariplo Foundation, Italy.
Footnotes
Published ahead of print on 28 December 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bertolla, F., and P. Simonet. 1999. Horizontal gene transfers in the environment: natural transformation as a putative process for gene transfers between transgenic plants and microorganisms. Res. Microbiol. 150:375-384. [DOI] [PubMed] [Google Scholar]
- 2.Brusetti, L., A. Rizzi, A. Abruzzese, G. A. Sacchi, E. Ragg, M. Bazzicalupo, C. Sorlini, and D. Daffonchio. Effects of rhizodeposition of tobacco and transplastomic tobacco on the rhizosphere bacterial community. Environ. Biosafety Res., in press. [DOI] [PubMed]
- 3.Ceccherini, M. T., J. Poté, E. Kay, V. T. Van, J. Marechal, G. Pietramellara, P. Nannipieri, T. M. Vogel, and P. Simonet. 2003. Degradation and transformability of DNA from transgenic leaves. Appl. Environ. Microbiol. 69:673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dandie, C. E., S. M. Thomas, and N. C. McClure. 2001. Comparison of a range of green fluorescent protein-tagging vectors for monitoring a microbial inoculant in soil. Lett. Appl. Microbiol. 32:26-30. [DOI] [PubMed] [Google Scholar]
- 5.De Vries, J., and W. Wackernagel. 2003. Spread of recombinant DNA by roots and pollen of transgenic potato plants, identified by highly specific biomonitoring using natural transformation of an Acinetobacter sp. Appl. Environ. Microbiol. 69:4455-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Vries, J., and W. Wackernagel. 2004. Microbial horizontal gene transfer and the DNA release from transgenic crop plants. Plant Soil 266:91-104. [Google Scholar]
- 7.Doolittle, R. F. 1998. The case for gene transfers between very distantly related organisms, p. 311-320. In M. Syvanen and C. I. Kado (ed.), Horizontal gene transfer. Chapman & Hall, London, United Kingdom.
- 8.Gebhard, F., and K. Smalla. 1998. Transformation of Acinetobacter sp. strain BD413 by transgenic sugar beet DNA. Appl. Environ. Microbiol. 64:1550-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gogarten, J. P., and J. P. Townsend. 2005. Horizontal gene transfer, genome innovation and evolution. Nat. Microbiol. Rev. 3:679-687. [DOI] [PubMed] [Google Scholar]
- 10.Hendrickx, L., M. Hausner, and S. Wuertz. 2003. Natural genetic transformation in monoculture Acinetobacter sp. strain BD413 biofilms. Appl. Environ. Microbiol. 69:1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juni, E. 1972. Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J. Bacteriol. 112:917-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kay, E., T. M. Vogel, F. Bertolla, R. Nalin, and P. Simonet. 2002. In situ transfer of antibiotic resistance genes from transgenic (transplastomic) tobacco plants to bacteria. Appl. Environ. Microbiol. 68:3345-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kok, R. G., D. M. Young, and L. N. Ornston. 1999. Phenotypic expression of PCR-generated random mutations in a Pseudomonas putida gene after its introduction into an Acinetobacter chromosome by natural transformation. Appl. Environ. Microbiol. 65:1675-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koonin, E. V., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maruyama, F., K. Tani, T. Kenzaka, N. Yamaguchi, and M. Nasu. 2006. Quantitative determination of free-DNA uptake in river bacteria at single-cell level by in situ rolling-circle amplification. Appl. Environ. Microbiol. 72:6248-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen, K. M., A. M. Bone, K. Smalla, and J. D. Van Elsas. 1998. Horizontal gene transfer from transgenic plants to terrestrial bacteria—a rare event? FEMS Microbiol. Rev. 22:79-103. [DOI] [PubMed] [Google Scholar]
- 17.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 18.Palmen, R., and K. J. Hellingwerf. 1997. Uptake and processing of DNA by Acinetobacter calcoaceticus: a review. Gene 192:179-190. [DOI] [PubMed] [Google Scholar]
- 19.Smets, B. F., and T. Barkay. 2005. Horizontal gene transfer: perspectives at a crossroads of scientific disciplines. Nat. Microbiol. Rev. 3:675-678. [DOI] [PubMed] [Google Scholar]
- 20.Sørensen, S. J., M. Bailey, L. H. Hansen, N. Kroer, and S. Wuertz. 2005. Studying plasmid horizontal gene transfer in situ: a critical review. Nat. Microbiol. Rev. 3:700-710. [DOI] [PubMed] [Google Scholar]
- 21.Strätz, M., M. Mau, and K. N. Timmis. 1996. System to study horizontal gene exchange among microorganisms without cultivation of recipients. Mol. Microbiol. 22:207-215. [DOI] [PubMed] [Google Scholar]
- 22.Thomas, C. T., and K. M. Nielsen. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Microbiol. Rev. 3:711-721. [DOI] [PubMed] [Google Scholar]
- 23.Vaneechoutte, M., D. M. Young, L. N. Ornston, T. De Baere, A. Nemec, T. Van Der Reijden, E. Carr, I. Tjernberg, and L. Dijkshoorn. 2006. Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl. Environ. Microbiol. 72:932-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Elsas, J. D., S. Turner, and M. J. Bayley. 2003. Horizontal gene transfer in the phytosphere. New Phytol. 157:525-537. [DOI] [PubMed] [Google Scholar]
- 25.Wolf, Y. I., L. Aravind, N. V. Grishin, and E. V. Koonin. 1999. Evolution of aminoacyl-tRNA synthetases: analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 9:689-710. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.