Abstract
The genomic content of Enterobacter sakazakii strain ATCC BAA-894 was analyzed for variable-number tandem repeats (VNTRs). In this study we report the development of a multiple-locus VNTR analysis (MLVA) strategy for the subtyping of E. sakazakii. The method is based on a GeneScan analysis of four VNTR loci labeled with multiple fluorescent dyes. This approach was applied to a collection of 112 isolates representing all 16 of the currently defined E. sakazakii biogroups. MLVA successfully discriminated among these isolates and compared favorably with pulsed-field gel electrophoresis. The method was relatively fast and easy to perform. The potential value of MLVA as an epidemiological tool is discussed.
Enterobacter sakazakii is an emerging opportunistic pathogen that is the cause of rare cases of meningitis, necrotizing enterocolitis, and bacteremia in infants (1, 2, 9, 29, 40, 43). High mortality rates have been reported among those who develop meningitis, with the infants often presenting clinical complications, such as brain abscesses, seizures, and infarction (18). E. sakazakii, originally described as a “yellow-pigmented” coliform, first appeared in the literature in 1958 following a fatal case of infant meningitis in England (39). Since then, cases of infant infection have been reported from all regions of the developing world, with over 111 illness events and at least 26 deaths currently documented (27).
Based on genotyping and biotyping studies, E. sakazakii represents a diverse and taxonomically ill-defined species (11, 13, 14, 20). Iversen et al., (12) recently proposed the reclassification of the species into a new genus comprised of five species. Although clinical and domestic sources have been documented, the most frequently implicated mode of transmission of this bacterium is through consumption of contaminated powdered infant formula (4, 7, 26, 35, 40).
The ability to identify routes of transmission in cases of E. sakazakii infection relies on the availability of reproducible, highly discriminatory techniques for the purposes of surveillance and outbreak investigation. Strain subtyping by molecular methods is of particular interest. Ribotyping, random amplification of polymorphic DNA, and pulsed-field gel electrophoresis (PFGE) are methods that have been used to subtype E. sakazakii (5, 8, 28, 30, 37). In our laboratory, we showed that XbaI macrorestriction, followed by PFGE analysis, gives good discrimination and is highly reproducible (28). Although PFGE has been successfully applied to investigate E. sakazakii outbreaks (5, 37), the method has recognized limitations and may be superseded in the future by more modern approaches.
The availability of a complete E. sakazakii genome sequence (http://genome.wustl.edu/pub/organism/Microbes/Enteric_Bacteria/Enterobacter_sakazakii/assembly/Enterobacter_sakazakii-4.0/) provided the opportunity to search the genome for variable-number tandem-repeat (VNTR) motifs, which represent sources of genetic polymorphisms that, when suitably applied, can be used as a basis for subtyping. VNTRs consist of DNA elements that are repeated in tandem (41). The sequence element is often maintained within a bacterial species, with individual strains displaying different copy numbers. The length of a tandem repeat (TR) at a specific locus can vary as a consequence of DNA polymerase slippage during replication or unequal crossover events. These differences can be analyzed by amplification of the region and sizing of the resulting amplicons (15). The high degree of polymorphism at these loci has proven particularly useful as a target for strain discrimination of bacterial species. Multiple-locus variable-number TR analysis (MLVA) is a subtyping method that involves amplification and fragment size analysis of polymorphic VNTR regions. It has been successfully used to type other Enterobacteriaceae, including Salmonella (6, 23, 25, 33, 38), Escherichia coli (16, 21, 24, 32), and Yersinia pestis (17, 19). MLVA is a rapid method of high discriminatory power that is easily standardized between laboratories (22).
This study describes the development of an MLVA subtyping scheme and its application to a genotypically and phenotypically diverse collection of E. sakazakii isolates. To our knowledge, this is the first report of the application of MLVA to subtype E. sakazakii.
MATERIALS AND METHODS
Bacterial strains.
A summary of the E. sakazakii strains used in the present study is shown in Table 1. A global collection of 112 isolates was selected to represent the phenotypic, genetic, and epidemiologic diversity of E. sakazakii. The isolates included both domestic strains (35) and strains from the United Kingdom, France, Switzerland, Denmark, The Netherlands, the Czech Republic, the United States, Canada, Malaysia, Pakistan, Indonesia, Korea, and New Zealand. These strains represent all currently known biogroups (13). All isolates were confirmed as E. sakazakii by real-time PCR using a primer set and probe targeting the dnaG gene on the macromolecular synthesis operon (34). All bacteria were cultured on nutrient agar (Oxoid, Basingstoke, United Kingdom), and for long-term storage, the isolates were maintained in cryopreservation fluid at −80°C (Technical Service Consultants Ltd., Lancashire, England).
TABLE 1.
Characteristics of the E. sakazakii isolates used in this study
| Strain | Origin
|
Biotype | |
|---|---|---|---|
| Country | Source | ||
| CFS01 | Ireland | Milk-processing facilitya | 1 |
| CFS06 | Ireland | Milk-processing facilitya | 1 |
| CFS07 | Ireland | Milk-processing facilitya | 1 |
| CFS09 | Ireland | Milk-processing facilitya | 1 |
| CFS10 | Ireland | Milk-processing facilitya | 5 |
| CFS101 | Ireland | PIF facilityb | 2 |
| CFS104 | Ireland | PIF facilityc | 2 |
| CFS108 | Ireland | PIF facilityb | 2 |
| CFS110 | Ireland | PIF facilityb | 2 |
| CFS115 | Ireland | PIF facilityb | 2 |
| CFS116 | Ireland | PIF facilityb | 8 |
| CFS122 | Ireland | PIF facilityb | 2 |
| CFS127 | Ireland | PIF facilitya | 2 |
| CFS128 | Ireland | PIF facilityc | 2 |
| CFS129 | Ireland | PIF facilityb | 5 |
| CFS130 | Ireland | PIF facilitya | 8 |
| CFS131 | Ireland | PIF facilitya | 1 |
| CFS135 | Ireland | PIF facilityb | 8A |
| CFS136 | Ireland | PIF facilityb | 8 |
| CFS138 | Ireland | PIF facilityb | 2 |
| CFS139 | Ireland | PIF facilityb | 2 |
| CFS148 | Ireland | PIF facilityb | 8 |
| CFS150 | Ireland | PIF facilityb | 8A |
| CFS152 | Ireland | PIF facilityb | 2 |
| CFS155 | Ireland | PIF facilityb | 2 |
| CFS156 | Ireland | PIF facilityb | 1 |
| CFS158 | Ireland | PIF facilityb | 2 |
| CFS167 | Ireland | PIF facilityb | 8A |
| CFS170 | Ireland | PIF facilityb | 2 |
| CFS173 | Ireland | PIF facilitya | 8 |
| CFS174 | Ireland | PIF facilityb | 3 |
| CFS175 | Ireland | PIF facilityb | 1 |
| CFS176 | Ireland | PIF facilityb | 1 |
| CFS237 | Ireland | PIF facilityb | 12 |
| CFS1001 | Ireland | PIF facilitya | 2 |
| 206N | New Zealand | Milk powder | 1 |
| 207N | New Zealand | Milk powder | 1 |
| 228N | New Zealand | Milk powder | 1 |
| 229N | New Zealand | Milk powder | 1 |
| 254N | New Zealand | Milk powder | 2 |
| 302N | New Zealand | Milk powder | 1 |
| 305N | New Zealand | Milk powder | 2 |
| 37 | Unknown | Unknown | 1 |
| 44 | Unknown | Unknown | 1 |
| 50 | Unknown | Unknown | 1 |
| 53 | Unknown | Unknown | 1 |
| 71 | Unknown | Unknown | 1 |
| 73 | Unknown | Unknown | 1 |
| 80 | Unknown | Unknown | 1 |
| 82 | Unknown | Unknown | 1 |
| 88 | Unknown | Unknown | 2 |
| 90 | Unknown | Unknown | 2 |
| 93 | Unknown | Unknown | 1 |
| 102 | Unknown | Unknown | 2 |
| 107 | Unknown | Unknown | 1 |
| 109 | Unknown | Unknown | 1 |
| 112 | Unknown | Unknown | 1 |
| 195 | Unknown | Unknown | 1 |
| 336 | Unknown | Unknown | 2 |
| 338 | Unknown | Unknown | 5 |
| 343 | Unknown | Unknown | 1 |
| 344 | Unknown | Unknown | 1 |
| E265 | Malaysia | Milk powder | 5 |
| E290 | Switzerland | PIF facilityb | 2 |
| E465 | Pakistan | PIF facilityb | 6 |
| E515 | Switzerland | Water | 10 |
| E604 | Canada | Clinical | 1 |
| E616 | Unknown | Food | 15 |
| E625 | Korea | Infant food | 16 |
| E626 | Korea | Infant food | 16 |
| E632 | United States | Infant food | 2 or 4 |
| E681 | Unknown | Infant food | 16 |
| E685 | Unknown | Food | 14 |
| E694 | Unknown | Food | 16 |
| E760 | Czech Republic | Clinical | 3 |
| E765 | Czech Republic | Clinical | 9 |
| E770 | Denmark | Milk powder | 2 |
| E772 | France | Milk powder | 5 |
| E775 | Russia | Milk powder | 13 |
| E777 | Netherlands | Milk powder | 2 |
| E784 | Netherlands | Neonate | 2 |
| E786 | Netherlands | Neonate | 1 |
| E787 | Netherlands | Neonate | 1 |
| E789 | Netherlands | Neonate | 2 |
| E808 | Indonesia | Milk powder | 2 |
| E809 | Indonesia | Milk powder | 1 |
| E825 (CDC 1058-77) | United States | Human (breast abscess) | 9 |
| E827 (CDC 407-77) | United States | Human (septum) | 4 |
| E828 (CDC 3128-77) | United States | Human (septum) | 11 |
| E829 (CDC 1716-77) | United States | Human (blood) | 9 |
| E830 (CDC 9369-75) | United States | Unknown | 7 |
| E834 | France | Neonate (feces) | 1 |
| E837 | France | Neonate (feces) | 4 |
| E844 | France | Neonate (feces) | 4 |
| E846 | France | Neonate (feces) | 1 |
| E866 | Switzerland | Neonate | 16 |
| E888 | Denmark | Milk powder | 15 |
| E892 | United States | Clinical | 3 |
| E893 | United States | Clinical | 8 |
| E899 | United States | Clinical | 1 |
| E900 | United States | Clinical | 1 |
| E901 | United States | Clinical | 3 |
| NCTC 29004 | United States | Unknown | 1 |
| NCTC 11467 | United States | Human (throat) | 1 |
| NCTC 8155 | United Kingdom | Milk powder | 1 |
| ATCC 12868 | Unknown | Unknown | 1 |
| ATCC 51329 | Unknown | Unknown | 15 |
| NCTC 9529 | United Kingdom | Water | 16 |
| NCTC 9238 | United Kingdom | Human (abdominal pus) | 1 |
| NCTC 9844 | United Kingdom | Unknown | 10 |
| ATCC BAA 893 | United States | Milk powder | 2 |
| ATCC BAA 894 | United States | Neonate | 2 |
Final product.
Environmental.
Intermediate product; PIF, powdered infant formula.
Genomic-DNA extraction.
Briefly, all bacteria were cultured on nutrient agar at 37°C for 18 h, and an isolated colony was then inoculated into 10 ml tryptone soy broth (Oxoid). One milliliter of overnight culture was recovered by centrifugation at 13,000 to 16,000 × g for 2 min. Total DNA was prepared using the Wizard Genomic DNA purification kit (Promega, Madison, WI) and quantified using spectrophotometry (NanoDrop). The integrity of the purified template DNA was assessed by conventional agarose gel (1% [wt/vol]) electrophoresis, and the DNA preparations were stored at 4°C.
Identification of TRs.
The TR DNA motifs were identified from version 4 of a 6.4 covered draft genome sequence of E. sakazakii ATCC BAA-894 using Tandem Repeats Finder software (3; http://tandem.bu.edu/trf/trf.html). A large number of TRs were identified, and a selection of these were initially assessed for their potential value in a typing scheme. Loci were chosen based on the following features: copy number and repeat units that were both practical in size and free of nucleotide errors. PCR primers flanking the selected TRs were designed using the Primer 3 website (http://frodo.wi.mit.edu/). Care was taken to generate primers with similar or identical melting temperatures that provided a single distinct amplicon at each locus. Initially, the size variation and utility of particular TR loci were assessed across a subset of E. sakazakii isolates and analyzed by conventional agarose gel electrophoresis. Using this approach, four novel loci were selected and taken forward for full assessment (Table 2). All four of these VNTRs (denoted Enterobacter sakazakii tandem repeat 1 [ESTR-1] through -4) were localized in intergenic regions of the E. sakazakii genome and contained repeat motifs ranging from 7 to 15 base pairs.
TABLE 2.
Selected VNTR loci in the E. sakazakii BAA-894 genomea
| Locus | TR sequence (5′-3′) | Gene | % GC content | Contig no. | Location on contig (nt) |
|---|---|---|---|---|---|
| ESTR-1 | GTTGAGGTGGCGGGT | Intergenic region | 67 | 1.19 | 86316-86346 |
| ESTR-2 | TTTGAGGCGAACG | Intergenic region | 54 | 0b | 501708-501744 |
| ESTR-3 | GACCGCA | Intergenic region | 71 | 0b | 1207159-1207222 |
| ESTR-4 | GGCTGCGGTTTTTCG | Intergenic region | 60 | 1b | 153842-153872 |
TR from sequence information from supercontigs.
MLVA PCR amplification.
All four selected MLVA loci were amplified individually by PCR. The reaction mixtures consisted of 1× PCR buffer containing 1.5 mM MgCl2 (New England Biolabs, United Kingdom), 100 mM deoxynucleoside triphosphates, 0.1 μM dye-labeled forward primer (MWG Biotech AG, Ebersberg, Germany), 0.1 μM nonlabeled reverse primer, 2.5 U Taq DNA polymerase (New England Biolabs, United Kingdom), 100 ng template DNA, and PCR grade water (Invitrogen, California) in a volume of 50 μl. Forward primers were fluorescently labeled with 4,7,2′,7′-tetrachloro-[3′,6′-dipivaloylfluoresceinyl]-6- carboxamidohexyl]-1-O-[2-cyanoethyl]-[N,N-diisopropyl]-phosphoramidite] (TET), 3′, 6′-dipivaloylfluoresceinyl-6-carboxamidohexyl-1-O-2-cyanoethyl-N,N-diisopropyl- phosphoramidite (FAM), or 4,7,2′,4′,5′,7′-hexachloro-3′,6′-dipivaloylfluoresceinyl-6-carboxamidohexyl-1-O-2-cyanoethyl-N,N-diisopropyl-phosphoramidite (HEX) (Table 3). The thermal amplification conditions were 95°C for 1 min, followed by 35 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min, with a final extension of 72°C for 5 min. Following amplification, equal volumes (2 μl) from each of the four individual PCR mixtures (ESTR-1 to −4) were electrophoresed on a 1% (wt/vol) agarose gel at 100 V for 90 min in 1× Tris-borate-EDTA. The band intensity of each amplicon was examined prior to GeneScan analysis. All four loci were amplified two or three times from each isolate to ensure reproducibility.
TABLE 3.
Primers for PCR amplification of selected VNTR used in this study
| Locus | Primer | Primer sequence (5′-3′) | Product size (bp)a | Annealing temp (oC) |
|---|---|---|---|---|
| ESTR-1 | ESTR1-F | 5′ TET-TCCGGCCATTTCGCTCTG | 183 | 52 |
| ESTR1-R | 5′ TTACAGCGGCCCGAATACTC | |||
| ESTR-2 | ESTR2-F | 5′ FAM-AGGATAAAAATGCCGAGGGAATGT | 309 | 52 |
| ESTR2-R | 5′ TGGGCAACGATAAATCAAACACT | |||
| ESTR-3 | ESTR3-F | 5′ HEX-CGAAGACGGGTGATGAAAAA | 155 | 52 |
| ESTR3-R | 5′ CAGGCGAATTCCTCTCTGTT | |||
| ESTR-4 | ESTR4-F | 5′ HEX-GTGATACCGGCGTAATCCTG | 384 | 52 |
| ESTR4-R | 5′ GGCACCGCATAAAGAGAATG |
Size of product in absence of TR.
MLVA.
Prior to GeneScan analysis, amplicons of uniform intensity were diluted 1:100 in distilled water. For each individual isolate, 1 μl of the diluted amplified product for each locus was added to a fresh tube. One microliter of this pooled mixture was added to the following: 2.5 μl deionized formamide, 0.5 μl of EDTA (50 mM; pH 8.0)/blue dextran (50 mg/ml) loading dye (Applied Biosystems, Foster City, CA), and 1 μl of GeneFLO tetramethylcarboxyrhodamine (TAMRA)-labeled 625 DNA Ladder (Chimerx, Madison, WI). This mixture was denatured at 92°C for 5 min and immediately placed on ice. An aliquot of the final mixture (3 μl) was loaded and electrophoresed at 3,000 V, 60 mA, and 200 W for 1.5 h at 52°C on an ABI 377 DNA analyzer (Applied Biosystems, Foster City, CA) using a 5% (wt/vol) LongRanger denaturing acrylamide gel (Cambrex, Nottingham, United Kingdom). The dyes were detected using filter set C, a filter set suitable for the detection of the dyes HEX, TAMRA, FAM, and TET. Following electrophoresis, sample lanes were tracked and extracted. Size values were assigned to the GeneFlo TAMRA-labeled 625 DNA ladder. Other dye-labeled fragments were sized relative to these, using GeneScan analysis software, version 3.1.2.
The analysis parameters on the GeneScan analysis software settings were set as follows. The data point range was set from 1,250 to 10,000. The baseline line and multicomponent data-processing tools were selected, and the peak amplitude thresholds were set as follows: green, 50; blue, 50; black, 50;, and red, 5. The peak half-width was set at two points, all sizes were selected in the size call range, the local Southern size-calling method was selected, and finally, the split-peak correction was set at the leftmost peak, with a correction limit of 30 data points. Following analysis, correct sizing of the doublets contained within the GeneFLO TAMRA-labeled 625 DNA ladder was checked.
Data analysis.
A four-digit allele string based on the number of repeats was assigned to all isolates in the following order: ESTR-1, ESTR-2, ESTR-3, and ESTR-4. One hundred and seven allele strings were imported into a Bionumerics software package (version 4.5; Applied Maths, Sint-Martens-Latem, Belgium) and a minimum-spanning tree (MST) was generated based on the categorical and the priority rule highest number of single-locus variants (where two types were at equal distances from the tree, the type with the highest number of single-locus variants was linked first).
PFGE and analysis.
All isolates were typed by PFGE using methods previously described by Mullane et al. (28). DNA fingerprints were stored as tagged image format files and imported into BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium), where a dendrogram was created using the DICE coefficient and the unweighted pair group method with arithmetic mean. A position tolerance of 1.5% and optimization of 1.5% were applied. The genetic diversity and relatedness of the E. sakazakii isolates were compared at 80% similarity.
Sequence verification and diversity.
To confirm that length polymorphisms were the result of repeat copy number variation, a subset of amplicons were purified with a Qiagen MiniElute gel extraction kit (Qiagen, Hilden, Germany), and DNA sequencing was performed on PCR products representing at least two alternative alleles for all four novel VNTR loci. Each PCR product was sequenced in both directions (GATC, Constance, Germany), and sequence information was analyzed with DNAStar (DNAStar, Madison, WI).
The genetic diversity of TRs for each locus was calculated using Simpson's index, while the discriminatory index of PFGE and MLVA was assessed using Simpson's index of diversity (10, 36).
RESULTS
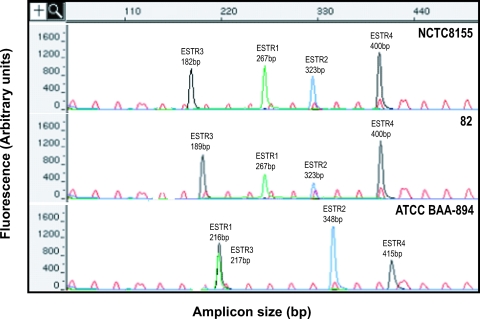
For the development of this MLVA method, primers flanking each locus were designed to amplify all four TR-containing regions (ESTR-1, ESTR-2, ESTR-3, and ESTR-4). The amplified products were pooled for each isolate and then resolved on a 377 DNA sequencer and sized using GeneScan analysis software. The electropherogram of the pooled amplicons showed a clear sizing pattern that was easily interpreted (Fig. 1). The VNTR loci from three isolates are shown, and the internal size standard is displayed. The internal size standard gave uniform peak spacing at 25-bp intervals up to 625 bp (Fig. 1). All loci were identified automatically by the software according to their sizes and fluorescences. The use of multiple dyes allowed ease of interpretation of the electopherogram in cases where amplicons overlapped. An example of this is shown in Fig. 1, where ESTR-1 and ESTR-3 of ATCC BAA-894 differ by a single base pair. Isolate 80 and NCTC 8155 shared three loci but differed by a single repeat motif at ESTR-3, while ATCC BAA-894 differed at all four loci (Fig. 1).
FIG. 1.
E. sakazakii GeneScan traces generated from four dye-labeled VNTR loci showing three different profiles. The red peaks are the GenFLO TAMRA-labeled internal size standards. ESTR-2 was labeled with FAM (blue peak), ESTR-3 and ESTR-4 loci were labeled with HEX (black peaks), and the ESTR-1 locus was labeled with TET (green peak). The size of each locus is indicated in the three windows of isolates NCTC 8155 (top), 80 (middle), and ATCC BAA-894 (bottom).
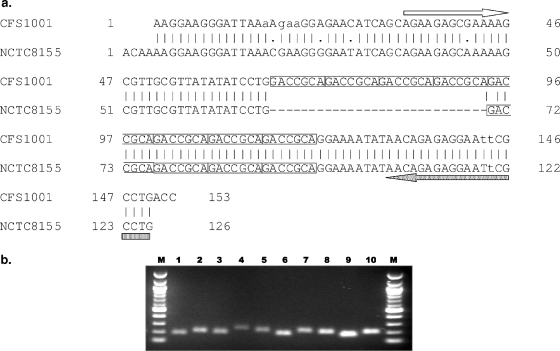
Nucleotide sequencing of each of the four amplified loci from two isolates confirmed that size polymorphisms were due to varying repeat units. An example of this is shown in Fig. 2a, where the size variation in the amplicons from the ESTR-3 loci in CFS1001 and NCTC 8155 can be attributed to the presence of eight and four TRs, respectively. Figure 2b shows a selection of 10 alleles varying in size from 168 bp containing 2 repeats to 245 bp containing 13 repeats.
FIG. 2.
(a) Sequencing alignment of ESTR-3 loci from isolates CFS1001 and NCTC8155. The result from sequencing shows that the amplified fragment size variation at the loci between isolates CFS1001 (top) and NCTC8155 (bottom) is due to different numbers of -GACCGCA- repeats alone. The arrows indicate primer binding sites. (b) Agarose gel image of ESTR-3 from strains NCTC 8155 (182-bp product) (lane 1), CFS1001 (210-bp product) (lane 2), 82 (196-bp product) (lane 3), 102 (245-bp product) (lane 4), ATCC BAA-894 (217-bp product) (lane 5), 73 (182-bp product) (lane 6), 338 (217-bp product) (lane 7), 336 (203-bp product) (lane 8), 344 (168-bp product) (lane 9), and 109 (203-bp product) (lane 10). Lanes M, 100-bp molecular size marker (New England Biolabs, England).
MLVA was used to genotype a geographically diverse and previously characterized collection of 112 E. sakazakii isolates from clinical, food, and environmental origins representing all 16 currently identified biogroups. Excluding null-allele types, a range of 2 to 14 alleles was found for the four loci, with VNTRs repeating from 0 times (absence of a repeat unit) at some loci up to 19 times at others (ESTR-3). ESTR-3 was the most variable locus, displaying 14 alleles with repeat copy numbers ranging from 1 to 19. A summary of the allelic variability of the loci is given in Table 4. The diversity index, which reflects the value of a TR locus for the purpose of typing, was calculated in each case using Simpson's index. These values ranged from 61.0 to 89.7% (Table 4). Five isolates (E888, E866, E515, E616, and E694) failed to produce amplicons at any of the four loci. These isolates were excluded from further study but were included in PFGE analysis.
TABLE 4.
MLVA locus characteristics
| Locus | No. of repeats
|
No. of alleles | % Missing amplicons | Locus diversity index (%) | |
|---|---|---|---|---|---|
| Minimum | Maximum | ||||
| ESTR-1 | 0 | 9 | 9 | 32 | 75.8 |
| ESTR-2 | 0 | 3 | 4 | 12 | 74.7 |
| ESTR-3 | 1 | 19 | 14 | 16 | 89.7 |
| ESTR-4 | 1 | 2 | 2 | 11 | 61.0 |
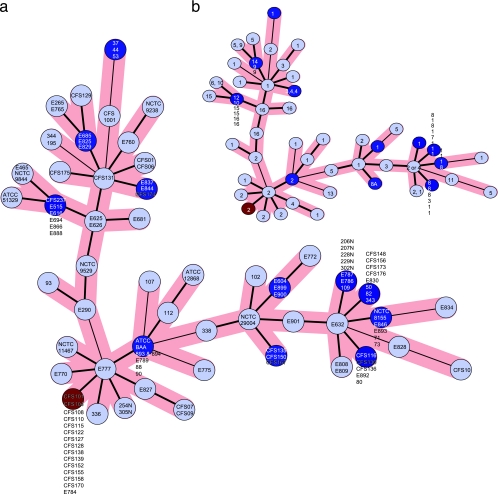
The genetic relationships of 107 of the 112 isolates were deduced by the construction of an MST (Fig. 3). Forty-nine unique allele strings were observed among these isolates. The MST highlights the diversity among the tested isolates. A number of branched clusters were visible, but no association could be established between the isolate type, geographical origin, or biogroup (Fig. 3).
FIG. 3.
(a) MST of MLVA (where the weight of the edge between each pair of points is the distance between those two points) analysis of 107 isolates of E. sakazakii. (b) MST of MLVA analysis showing biogroup identities of each cluster.
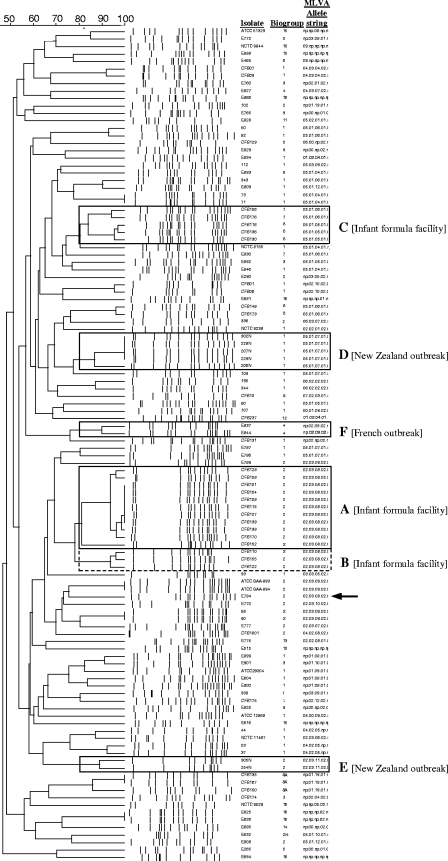
Figure 4 shows a PFGE dendrogram of all isolates with corresponding MLVA allele strings. A total of 49 MLVA and 64 PFGE types were identified among the 107 isolates analyzed (Fig. 3 and 4). The discriminatory indices of the two methods, calculated using Simpson's diversity index, were 0.960 and 0.981 for MLVA and PFGE, respectively. This indicates that if two isolates are sampled randomly from a collection and analyzed by MLVA, then 96% of the samples would fall into different types.
FIG. 4.
Dendrogram of the 112 isolates that were previously typed by XbaI PFGE. Allele strings and biogroups are also included. The arrow indicates isolate E784.
The PFGE dendrogram identified 23 clusters containing from 2 to 11 isolates that displayed >80% similarity (Fig. 4). The same allele string was assigned to 16 of these clusters, 7 of which were represented by unique allele strings. The allele strings assigned to the remaining nine clusters were shared with isolates of different PFGE types. Conversely, seven PGFE clusters were subdivided into different MLVA types. The largest cluster, cluster A (n = 11), consisting of isolates from an infant milk powder production facility and clustering at 80% similarity by PFGE, shared the same allele identifier (02.03.08.02) by MLVA. The conservation of the allele string in these isolates sampled over a 1-year period demonstrates the stability of each locus over time. Cluster B shared 77% similarity with cluster A; however, MLVA failed to make a distinction between the two groups. Furthermore, isolate E784, which was associated with neonatal infection in The Netherlands, was also assigned the allele string 02.03.08.02, although it shared only 64% similarity with the isolates from the production factory. Two different allele codes were assigned to isolates in cluster C (n = 5), which were also obtained from the same infant milk powder production facility. Of these, the allele string 05.01.05.01 was shared with isolates E892 and 80, which are represented by different PFGE pulse types. Similarly, allele code 05.01.06.01 was shared with isolates CFS148, CFS173, E830, 50, 82, and 343, which are associated with four different PFGE types. The PFGE types of these isolates showed between 58 and 74% similarity with cluster C. PFGE cluster D contained isolates that were epidemiologically linked to cases of meningitis in 2004 (31). These were all given the same allele identifier by MLVA. This identifier was also shared with isolate 109 of unknown origin and the clonally related isolates E787 and E786, which showed 78 and 52% similarity to the cluster, respectively. The other subtype implicated in the New Zealand 2004 case (cluster E) was successfully grouped by this method. The two clonal isolates, E837 and E844, involved in an outbreak in France (cluster F) displayed the same allele string, while the other two implicated isolates (E834 and E846), in agreement with PFGE, gave different allele identifiers.
DISCUSSION
A greater understanding of the routes of E. sakazakii contamination of powdered infant formula facilities would significantly reduce the risk of infection among vulnerable groups. The availability of rapid and accurate surveillance tools would enable the development of more targeted and effective control strategies to provide a safer powdered product. This approach would also be of value when applied in clinical and care settings to support epidemiological investigations. Controlling the processing environment by exclusion of E. sakazakii remains a significant challenge for those producing infant formula powder. Carefully developed Hazard Analysis and Critical Control Point plans could benefit from a clearer description of the epidemiology of the organism in the powdered infant formula production environment, as previously shown by Mullane et al., (28). Molecular typing remains an important tool for surveillance, outbreak investigation, and tracing of bacteria through the food chain. The ability to trace E. sakazakii is of epidemiological importance because of the implications for infant health and of economic value to infant formula producers. No standardized protocol to investigate the dissemination of E. sakazakii has been agreed on. PFGE, along with other subtyping strategies, has been applied (5, 8, 28, 30, 37). Despite the apparent advantages of PFGE, the approach will not evolve technically in the future. Therefore, molecular biologists have begun to assess the utility of second-generation subtyping methods. MLVA is one such approach. In this paper, we have described the development and application of an MLVA typing protocol for E. sakazakii. A set of four discriminatory TR markers were identified and used in the development of the typing scheme.
Our method was applied to 112 previously characterized E. sakazakii isolates representing all 16 biogroups. The study collection was cultured from clinical, food, and environmental sources from diverse geographical origins. In addition, a number of strains from a narrower population, all originating from the one infant formula production facility, were included to assess the method's ability to identify clonal populations (28). A virtually continuous range of repeats was observed across the four loci, suggesting a well-balanced diversity among the isolates in the study. Stability of a given MLVA type over a 1-year period was shown. Although the Simpson's indices calculated for PFGE and MLVA were similar, more subtypes could be identified by PFGE, indicating the higher discriminatory power of this method when applied to this collection of isolates. Nonetheless, MLVA grouped together all of the clonal isolates identified by PFGE in cluster A. Failure to discriminate between the clonally related isolates in cluster A and cluster B could possibly be due to incomplete XbaI digestion, resulting in one- or two-band differences. MLVA clustered isolates obtained from a recent outbreak in New Zealand and clearly distinguished them from isolates from an unrelated French outbreak that occurred later. However, identical MLVA types from the New Zealand outbreak were also found among nonrelated isolates, and one nonrelated isolate in cluster A had the same allele code as isolates in this cluster.
Including all 112 isolates, we noted that TR amplicons were absent for 31% of the strains at ESTR-1, 11% at ESTR-2, 15% at ESTR-3, and 10% at ESTR-4. Failure of amplification of TR loci is mainly attributable to polymorphisms in the primer-annealing site(s) that flank the TR or to absence of the corresponding locus in some strains. Bearing in mind that E. sakazakii is poorly defined taxonomically and may represent more than one species, amplification failure of all loci may reflect the diversity of the species being typed. Recently, it has been proposed that E. sakazakii be reclassified as four species, one genomospecies, and two subspecies in a new genus “Cronobacter” within the family Enterobacteriaceae (12). Using this classification system, “C. sakazakii” comprises biogroups 1 to 4, 7, 8, 11, and 13. The other species are represented by biogroups 5, 6, 9 10, 12, and 14 to 16. Excluding all strains (n = 20) belonging to these biogroups from MLVA analysis, fewer amplification failures were noted at all loci: 10% at ESTR-1, 0% at ESTR-2, 2% at ESTR-3, and 3% at ESTR-4. As our primers were designed based on ATCC BAA-894 (the only available genome sequence), which belongs to “C. sakazakii,” these findings support the reclassification of the species as described by Iversen et al. (12). However, use of TR motifs to examine taxonomy in bacteria is open to criticism, largely due to concerns in relation to rapid evolution and possible homoplasty that could potentially result in misleading conclusions (42).
In this study, no relationship between the geographical origin and conservation of particular allele motifs could be established. Allelic relationships could perhaps develop following analysis of other collections and when a reference set of variable loci has been established.
In conclusion, we have described the development and application of an MLVA method to subtype E. sakazakii. Although this method has less discriminatory power than PGFE, it could potentially be used to rapidly monitor clonal outbreaks of the organism in food production facilities. It is realistic to assume that the inclusion of additional polymorphic loci would further increase the discriminatory power of the method.
Acknowledgments
We acknowledge the financial support provided through the Irish government's Food Institutional Research Measure (FIRM) grant no. 05/R&D/D/363.
We thank The Nestlé Research Centre and The University of Zurich for the donation of strains.
Footnotes
Published ahead of print on 14 December 2007.
REFERENCES
- 1.Arseni, A., E. Malamou-Ladas, C. Koutsia, M. Xanthou, and E. Trikka. 1987. Outbreak of colonization of neonates with Enterobacter sakazakii. J. Hosp. Infect. 9:143-150. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Oz, B., A. Preminger, O. Peleg, C. Block, and I. Arad. 2001. Enterobacter sakazakii infection in the newborn. Acta Paediatr. 90:356-358. [PubMed] [Google Scholar]
- 3.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biering, G., S. Karlsson, N. C. Clark, K. E. Jonsdottir, P. Ludvigsson, and O. Steingrimsson. 1989. Three cases of neonatal meningitis caused by Enterobacter sakazakii in powdered milk. J. Clin. Microbiol. 27:2054-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Block, C., O. Peleg, N. Minster, B. Bar-Oz, A. Simhon, I. Arad, and M. Shapiro. 2002. Cluster of neonatal infections in Jerusalem due to unusual biochemical variant of Enterobacter sakazakii. Eur. J. Clin. Microbiol. Infect. Dis. 21:613-616. [DOI] [PubMed] [Google Scholar]
- 6.Boxrud, D., K. Pederson-Gulrud, J. Wotton, C. Medus, E. Lyszkowicz, J. Besser, and J. M. Bartkus. 2007. Comparison of multiple-locus variable-number tandem repeat analysis, pulsed-field gel electrophoresis, and phage typing for subtype analysis of Salmonella enterica serotype Enteritidis. J. Clin. Microbiol. 45:536-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, N. C., B. C. Hill, C. M. O'Hara, O. Steingrimsson, and R. C. Cooksey. 1990. Epidemiologic typing of Enterobacter sakazakii in two neonatal nosocomial outbreaks. Diagn. Microbiol. Infect. Dis. 13:467-472. [DOI] [PubMed] [Google Scholar]
- 8.Drudy, D., M. O'Rourke, M. Murphy, N. R. Mullane, R. O'Mahony, L. Kelly, M. Fischer, S. Sanjaq, P. Shannon, P. Wall, M. O'Mahony, P. Whyte, and S. Fanning. 2006. Characterization of a collection of Enterobacter sakazakii isolates from environmental and food sources. Int. J. Food Microbiol. 110:127-134. [DOI] [PubMed] [Google Scholar]
- 9.Gallagher, P. G., and W. S. Ball. 1991. Cerebral infarctions due to CNS infection with Enterobacter sakazakii. Pediatr. Radiol. 21:135-136. [DOI] [PubMed] [Google Scholar]
- 10.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iversen, C., L. Lancashire, M. Waddington, S. Forsythe, and G. Ball. 2006. Identification of Enterobacter sakazakii from closely related species: The use of Artificial Neural Networks in the analysis of biochemical and 16S rDNA data. BMC Microbiol. 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iversen, C., A. Lehner, N. Mullane, E. Bidlas, I. Cleenwerck, J. Marugg, S. Fanning, R. Stephan, and H. Joosten. 2007. The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov., Cronobacter sakazakii subsp. sakazakii comb. nov., Cronobacter sakazakii subsp. malonaticus,subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies 1. BMC Evol. Biol. 7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iversen, C., M. Waddington, J. J. Farmer III, and S. J. Forsythe. 2006. The biochemical differentiation of Enterobacter sakazakii genotypes. BMC Microbiol. 6:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iversen, C., M. Waddington, S. L. On, and S. Forsythe. 2004. Identification and phylogeny of Enterobacter sakazakii relative to Enterobacter and Citrobacter species. J. Clin. Microbiol. 42:5368-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem-repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keys, C., S. Kemper, and P. Keim. 2005. Highly diverse variable number tandem repeat loci in the E. coli O157:H7 and O55:H7 genomes for high-resolution molecular typing. J. Appl. Microbiol. 98:928-940. [DOI] [PubMed] [Google Scholar]
- 17.Klevytska, A. M., L. B. Price, J. M. Schupp, P. L. Worsham, J. Wong, and P. Keim. 2001. Identification and characterization of variable-number tandem repeats in the Yersinia pestis genome. J. Clin. Microbiol. 39:3179-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai, K. K. 2001. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine (Baltimore) 80:113-122. [DOI] [PubMed] [Google Scholar]
- 19.Le Fleche, P., Y. Hauck, L. Onteniente, A. Prieur, F. Denoeud, V. Ramisse, P. Sylvestre, G. Benson, F. Ramisse, and G. Vergnaud. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehner, A., T. Tasara, and R. Stephan. 2004. 16S rRNA gene based analysis of Enterobacter sakazakii strains from different sources and development of a PCR assay for identification. BMC Microbiol. 4:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindstedt, B. A., E. Heir, E. Gjernes, T. Vardund, and G. Kapperud. 2003. DNA fingerprinting of Shiga-toxin producing Escherichia coli O157 based on multiple-locus variable-number tandem-repeats analysis (MLVA). Ann. Clin. Microbiol. Antimicrob. 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindstedt, B. A., M. Torpdahl, E. M. Nielsen, T. Vardund, L. Aas, and G. Kapperud. 2007. Harmonization of the multiple-locus variable-number tandem repeat analysis method between Denmark and Norway for typing Salmonella typhimurium isolates and closer examination of the VNTR loci. J. Appl. Microbiol. 102:728-735. [DOI] [PubMed] [Google Scholar]
- 23.Lindstedt, B. A., T. Vardund, L. Aas, and G. Kapperud. 2004. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J. Microbiol. Methods 59:163-172. [DOI] [PubMed] [Google Scholar]
- 24.Lindstedt, B. A., T. Vardund, and G. Kapperud. 2004. Multiple-locus variable-number tandem-repeats analysis of Escherichia coli O157 using PCR multiplexing and multi-colored capillary electrophoresis. J. Microbiol. Methods 58:213-222. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Y., M. A. Lee, E. E. Ooi, Y. Mavis, A. L. Tan, and H. H. Quek. 2003. Molecular typing of Salmonella enterica serovar Typhi isolates from various countries in Asia by a multiplex PCR assay on variable-number tandem repeats. J. Clin. Microbiol. 41:4388-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullane, N. R., R. Drudy, P. Whyte, M. O'Mahony, A. G. M. Scannell, P. G. Wall, and S. Fanning. 2006. Enterobacter sakazakii: biological properties and significance in dried infant milk formula (IMF) powder. Int. J. Dairy Technol. 59:102-111. [Google Scholar]
- 27.Mullane, N. R., C. Iversen, B. Healy, C. Walsh, P. Whyte, P. G. Wall, T. Quinn, and S. Fanning. 2007. Enterobacter sakazakii, an emerging bacterial pathogen with implications for infant health. Minerva Pediatr. 59:137-148. [PubMed] [Google Scholar]
- 28.Mullane, N. R., P. Whyte, P. G. Wall, T. Quinn, and S. Fanning. 2007. Application of pulsed-field gel electrophoresis to characterise and trace the prevalence of Enterobacter sakazakii in an infant formula processing facility. Int. J. Food Microbiol. 116:73-81. [DOI] [PubMed] [Google Scholar]
- 29.Muytjens, H. L., H. C. Zanen, H. J. Sonderkamp, L. A. Kollee, I. K. Wachsmuth, and J. J. Farmer III. 1983. Analysis of eight cases of neonatal meningitis and sepsis due to Enterobacter sakazakii. J. Clin. Microbiol. 18:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nazarowec-White, M., and J. M. Farber. 1999. Phenotypic and genotypic typing of food and clinical isolates of Enterobacter sakazakii. J. Med. Microbiol. 48:559-567. [DOI] [PubMed] [Google Scholar]
- 31.New Zealand Ministry of Health. July 2005. Enterobacter sakazakii (E. sakazakii) invasive disease became notifiable on 21 July 2005. http://www.moh.govt.nz/moh.nsf/0/526cb4a064b49a31cc257042007c4ef3?OpenDocument. Accessed 11 October 2007.
- 32.Noller, A. C., M. C. McEllistrem, A. G. Pacheco, D. J. Boxrud, and L. H. Harrison. 2003. Multilocus variable-number tandem-repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J. Clin. Microbiol. 41:5389-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramisse, V., P. Houssu, E. Hernandez, F. Denoeud, V. Hilaire, O. Lisanti, F. Ramisse, J. D. Cavallo, and G. Vergnaud. 2004. Variable number of tandem repeats in Salmonella enterica subsp. enterica for typing purposes. J. Clin. Microbiol. 42:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo, K. H., and R. E. Brackett. 2005. Rapid, specific detection of Enterobacter sakazakii in infant formula using a real-time PCR assay. J. Food Prot. 68:59-63. [DOI] [PubMed] [Google Scholar]
- 35.Simmons, B. P., M. S. Gelfand, M. Haas, L. Metts, and J. Ferguson. 1989. Enterobacter sakazakii infections in neonates associated with intrinsic contamination of a powdered infant formula. Infect. Control Hosp. Epidemiol. 10:398-401. [DOI] [PubMed] [Google Scholar]
- 36.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 37.Smeets, L. C., A. Voss, H. L. Muytjens, J. F. G. M. Meis, and W. J. G. Melchers. 1998. Genetische karakterisatie van Enterobacter sakazakii-isolaten van Nederlandse patiënten met neonatale meningitis. Ned. Tijdschr. Med. Microbiol. 6:113-115. [Google Scholar]
- 38.Torpdahl, M., G. Sorensen, S. Ethelberg, G. Sando, K. Gammelgard, and L. J. Porsbo. 2006. A regional outbreak of S. typhimurium in Denmark and identification of the source using MLVA typing. Euro. Surveill. 11:134-136. [PubMed] [Google Scholar]
- 39.Urmenyi, A. M., and A. W. Franklin. 1961. Neonatal death from pigmented coliform infection. Lancet i:313-315. [DOI] [PubMed] [Google Scholar]
- 40.van Acker, J., F. de Smet, G. Muyldermans, A. Bougatef, A. Naessens, and S. Lauwers. 2001. Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J. Clin. Microbiol. 39:293-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Belkum, A. 2007. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA). FEMS Immunol. Med. Microbiol. 49:22-27. [DOI] [PubMed] [Google Scholar]
- 42.Whatmore, A. M., S. J. Shankster, L. L. Perrett, T. J. Murphy, S. D. Brew, R. E. Thirlwall, S. J. Cutler, and A. P. MacMillan. 2006. Identification and characterization of variable-number tandem-repeat markers for typing of Brucella spp. J. Clin. Microbiol. 44:1982-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willis, J., and J. E. Robinson. 1988. Enterobacter sakazakii meningitis in neonates. Pediatr. Infect. Dis. J. 7:196-199. [DOI] [PubMed] [Google Scholar]