Abstract
Enantiopure sulfoxides are valuable asymmetric starting materials and are important chiral auxiliaries in organic synthesis. Toluene monooxygenases (TMOs) have been shown previously to catalyze regioselective hydroxylation of substituted benzenes and phenols. Here we show that TMOs are also capable of performing enantioselective oxidation reactions of aromatic sulfides. Mutagenesis of position V106 in the α-hydroxylase subunit of toluene ortho-monooxygenase (TOM) of Burkholderia cepacia G4 and the analogous position I100 in toluene 4-monooxygenase (T4MO) of Pseudomonas mendocina KR1 improved both rate and enantioselectivity. Variant TomA3 V106M of TOM oxidized methyl phenyl sulfide to the corresponding sulfoxide at a rate of 3.0 nmol/min/mg protein compared with 1.6 for the wild-type enzyme, and the enantiomeric excess (pro-S) increased from 51% for the wild type to 88% for this mutant. Similarly, T4MO variant TmoA I100G increased the wild-type oxidation rate by 1.7-fold, and the enantiomeric excess rose from 86% to 98% (pro-S). Both wild-type enzymes showed lower activity with methyl para-tolyl sulfide as a substrate, but the improvement in the activity and enantioselectivity of the mutants was more dramatic. For example, T4MO variant TmoA I100G oxidized methyl para-tolyl sulfide 11 times faster than the wild type did and changed the selectivity from 41% pro-R to 77% pro-S. A correlation between regioselectivity and enantioselectivity was shown for TMOs studied in this work. Using in silico homology modeling, it is shown that residue I100 in T4MO aids in steering the substrate into the active site at the end of the long entrance channel. It is further hypothesized that the main function of V106 in TOM is the proper positioning or docking of the substrate with respect to the diiron atoms. The results from this work suggest that when the substrate is not aligned correctly in the active site, the oxidation rate is decreased and enantioselectivity is impaired, resulting in products with both chiral configurations.
Biocatalysis has been gaining increasing recognition by pharmaceutical companies in recent years, especially in the synthesis of chiral molecules (3, 30, 43). Progress in the ability to access numerous genetic resources, high-throughput screening methodologies, and in vitro enzyme evolution have contributed to the growing use of biocatalysis in industry (24, 28). Chiral sulfoxides are of major importance in organic chemistry, as they are efficient chiral auxiliaries that lead to important asymmetric transformations. In addition, chiral sulfoxides possess a wide range of biological activities from flavor and aroma precursors to antimicrobial properties (14, 21). The world's best-selling antiulcer drug, (S)-omeprazole, is a chiral sulfoxide (14, 19). There have been numerous reports on chemical and biological methods for synthesizing chiral sulfoxides. The enantioselective oxidation of a prochiral sulfide is undoubtedly the most direct and economical approach for the synthesis of optically pure sulfoxides (9, 12).
Both isolated enzymes and whole cells have been used in the enantioselective oxidation of prochiral sulfides. Horseradish peroxidase and chloroperoxidase from Caldariomyces fumago, two isolated heme peroxidases, have been shown to catalyze a broad spectrum of stereoselective epoxidation and sulfoxidation reactions (8, 14, 52). The drawback of this approach is that most heme-containing enzymes utilize H2O2 as an oxidizing agent, which impairs their activity. Various flavin-containing Baeyer-Villiger monooxygenases have been used as biocatalysts for sulfoxidation reactions and have afforded high conversion rates and selectivity for the oxidation of numerous alkyl and aromatic sulfides (11, 12, 14, 21). A third class of enzymes used for obtaining chiral sulfoxides is toluene dioxygenase (TDO) from Pseudomonas putida F1 and naphthalene dioxygenase (NDO) from Pseudomonas sp. strain NCIB 9816-4 (23). Both purified enzymes and whole-cell biotransformations resulted in moderate to high yields with high enantioselectivity. For some alkyl aryl sulfides, the selectivity of these two enzymes was complementary (e.g., TDO favoring the R-enantiomer and NDO favoring the S-enantiomer) (2, 23). In addition to the aforementioned well-studied enzymes, various fungi and yeasts also oxidize sulfides into chiral sulfoxides with various degrees of selectivity (21, 31).
Toluene monooxygenases (TMOs) are soluble, nonheme, diiron-containing enzymes belonging to a group of four component alkene/aromatic monooxygenases (22). The hydroxylase is composed of two subunits in (αβγ)2 quaternary structure and is responsible for the regiospecificity of the enzyme (15, 29). An effector protein encoded binds to the hydroxylase complex, enhances the catalytic rate of the enzyme, and refines the product distribution (27, 38). A Rieske-type [2Fe-2S] ferredoxin transfers electrons from the NADH reductase to the hydroxylase (22). All four proteins encoded by six genes are required for efficient catalysis and high regiospecificity. The most extensively studied monooxygenases from this class are toluene ortho-monooxygenase (TOM) of Burkholderia cepacia G4, which hydroxylates toluene at the ortho position to form o-cresol (6, 33); toluene o-xylene monooxygenase (ToMO) of Pseudomonas stutzeri OX1, with a relaxed regiospecificity, producing a mixture of three isomers from toluene hydroxylation (49, 51); and toluene para-monooxygenase (TpMO) of Ralstonia pickettii PKO1 and toluene 4-monooxygenase (T4MO) of Pseudomonas mendocina KR1, both para-hydroxylating enzymes producing primarily p-cresol from toluene (18, 46).
TMOs have been shown to be effective bioremediating agents (capable of degrading trichloroethylene) as well as useful biocatalysts (6, 16, 49). Nitrohydroquinone (precursor for therapeutics of Parkinson's disease), 4-nitrocatechol (inhibitor of nitric oxide synthase), and 3-methoxycatechol (intermediate for an antivascular agent) are some of the compounds that were prepared using this class of enzymes. In addition, TOM, ToMO, T4MO, and TpMO are able to perform three successive hydroxylations, thus transforming benzene to phenol, catechol and 1,2,3-trihydroxybenzene (47). Recently, molecular evolution techniques were used by us to modify the regioselectivity of these enzymes through mutations at the hydroxylase gene (15-17). The advantages of TMOs are their high selectivity and their use of molecular oxygen as an inexpensive and safe oxidizing agent rather than H2O2. Lonza has exploited these advantages and used a similar enzyme, xylene monooxygenase from Pseudomonas putida ATCC 33015, to transform 2,5-dimethylpyrazine to 5-methylpyrazine-2-carboxylic acid, an intermediate for the production of an antilipolytic drug (40).
To this end, there is very limited knowledge on the enantioselectivity of TMOs and there are no reports on the utilization of TMOs for the synthesis of chiral sulfoxides. It was our objective to investigate the ability of TMOs to selectively oxidize alkyl aryl sulfides and to improve the activity and selectivity using saturation mutagenesis. An attempt to correlate structure and function of the mutants was performed using homology modeling with the known crystal structure of the ToMO hydroxylase (37) and the newly reported crystal structure of phenol hydroxylase (PH) (38), both from Pseudomonas stutzeri OX1.
MATERIALS AND METHODS
Chemicals.
Thioanisole (99%), methyl p-tolyl sulfide (99%), methyl phenyl sulfoxide (97%), and methyl p-tolyl sulfoxide (98%) were purchased from Aldrich (Sigma-Aldrich, Rehovot, Israel). All materials used were of the highest purity available and were used without further purification.
Bacterial strains and growth conditions.
Escherichia coli TG1 {supE hsdΔ5 thi Δ(lac-proAB) F′ [traD36 proAB+ lacIq lacZΔM15] } with the plasmid constructs was routinely cultivated at 37°C with shaking at 250 rpm on a TU-400 incubator shaker (Orbital shaker incubator; MRC, Holon, Israel) in Luria-Bertani (LB) medium (36) supplemented with kanamycin at 100 μg/ml to maintain the plasmids. To stably and constitutively express the TMO genes from the same promoter, the expression vectors pBS(Kan)TOM (6), pBS(Kan)TpMO (47), pBS(Kan)ToMO (50), and pBS(Kan)T4MO (47) were constructed as described earlier. All experiments were conducted by diluting overnight cells to an optical density (OD) at 600 nm of 0.1 and growing them to an OD of 1.3. The exponentially grown cells were centrifuged at 8,000 × g for 10 min at 25°C in a Sigma 4K15 centrifuge (Sigma, Osterode, Germany) and resuspended in potassium phosphate buffer (PB; 100 mM, pH 7.0). Expression of TMOs (wild-type [WT] and protein variants) by pBS(Kan) vectors within E. coli strains produced blue or brown cells on agar plates and in broth cultures. The color is indicative of indigoid compounds formed by oxidation of indole from tryptophan (13, 34).
Protein analysis and molecular techniques.
Protein samples of cells grown with and without 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) were analyzed on standard 12% Laemmli discontinuous sodium dodecyl sulfate-polyacrylamide gels (36). Plasmid DNA was isolated using a Midi kit (Promega, Madison, WI) or a Mini kit (Real Biotech Corp., Taipei, Taiwan), and DNA fragments were isolated from agarose gels using the RBC extraction kit (Real Biotech Corp., Taipei, Taiwan). E. coli strains were transformed by electroporation using a Micro-Pulser instrument (Bio-Rad, Richmond, CA) with the program Ec1 (1.8 kV, 1 pulse for an 0.1-cm cuvette).
Saturation mutagenesis.
A gene library encoding all possible amino acids at position 100 of T4MO tmoA in pBS(Kan)T4MO and the analogous position 106 of TOM tomA3 in pBS(Kan)TOM was constructed by replacing the target codon with NNN (N stands for A, T, G, or C) via overlap-extension PCR (35). Two primers, T4MO_100_Front and T4MO_100_Rear (Table 1), were designed to randomize position 100 of TmoA, and primers TOM3 and TOM4 were designed to randomize position 106 of TomA3. Two additional primers for cloning for each enzyme were T4MO_EcoRI_Front and T4MO_BglII_Rear_2 (for T4MO) and TOM1 and TOM2 (for TOM) (Table 1). In T4MO, the BglII site occurs naturally downstream from TmoA position 100 and the EcoRI site is upstream of tmoA in the multiple cloning site. In TOM, the BsiWI site is 206 bp upstream of position 106 and the SphI site is 127 bp downstream of position 106; both restriction sites occur naturally. Vent DNA polymerase (New England Biolabs, Ipswich, MA) was used in the PCR to minimize random point mutations, and pBS(Kan)T4MO or pBS(Kan)TOM was used as the template. For T4MO, the first 412-nucleotide degenerate fragment was amplified by PCR using primers T4MO_EcoRI_Front and T4MO_100_Rear, and the second degenerate fragment of 663 nucleotides was amplified by PCR using primers T4MO_100_Front and T4MO_BglII_Rear_2. After purification from agarose gels, the two fragments were combined at a 1:1 ratio as templates to obtain the full-length degenerate PCR product (1,039 bp) using T4MO_EcoRI_Front and T4MO_BglII_Rear as primers. For TOM, the 257-bp degenerate fragment was amplified using primers TOM1 and TOM3, and the 180-bp degenerate fragment was amplified using TOM4 and TOM2. After purification from agarose gels, the two fragments were combined at a 1:1 ratio as templates to obtain the full-length degenerate PCR product (375 bp) using primers TOM1 and TOM2. The PCR program for T4MO consisted of 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2.5 min, with a final extension at 72°C for 7 min, while the PCR program for TOM consisted of 30 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 2.25 min, with a final extension at 72°C for 7 min. The resulting PCR product of T4MO (1,039 bp) containing randomized nucleotides at TmoA position 100 was cloned into pBS(Kan)T4MO after double digestion with EcoRI and BglII, replacing the corresponding fragment in the original plasmid. Similarly, the resulting PCR product of TOM (375 bp), containing the randomized nucleotides at TomA3 position 106, was cloned into pBS(Kan)TOM after double digestion with BsiWI and SphI, replacing the corresponding fragment in the original plasmid. The resulting plasmid library was transformed into E. coli TG1 competent cells via electroporation.
TABLE 1.
Primers used for saturation mutagenesis and sequencing of the V106 region in the tomA3 gene in TG1/pBS(Kan)TOM and the I100 region in the tmoA gene in TG1/pBS(Kan)T4MO
| Primer | Nucleotide sequencea |
|---|---|
| Mutagenesis | |
| TOM1 | 5′-CCGATGGAGAAAGTGTTTCCGTACGAC-3′ |
| TOM2 | 5′-GTTGTAGTGCGAGAGAGCATGCATTTC-3′ |
| TOM3 | 5′-CCTCGGTGTGCCATATACTCCAACGGTGTNNNACCCTGG-3′ |
| TOM4 | 5′-GTCAACGCACTCAAGGTGTTCATCCAGGGTNNNACACCG-3′ |
| T4MO_EcoRI_Front | 5′-GGGAACAAAAGCTGGGTACCG-3′ |
| T4MO_BglII_Rear_2 | 5′-TCCAAGCCCAGATCTATCAACGAGCGTTCG-3′ |
| T4MO_100_Front | 5′-ACTTTGAAATCCCATTACGGCGCCNNNGCAGTTGG-3′ |
| T4MO_100_Rear | 5′-GCTGCATATTCACCAACTGCNNNGGCGCCGTAATGG-3′ |
| Sequencing | |
| T4_100_Check | 5′-GGATGCAGGTGCTTATTCGG-3′ |
| TOM1 | 5′-CCGATGGAGAAAGTGTTTCCGTACGAC-3′ |
Restriction enzyme sites are underlined.
DNA sequencing.
DNA sequencing was performed using the dideoxy chain termination technique (Multidisciplinary Laboratories, Technion, Haifa, Israel). Primer T4_100_Check (Table 1) was used for determining the sequence of saturation mutagenesis mutants of T4MO I100, and primer TOM1 (Table 1) was used for sequencing TOM V106 mutants. Analysis of DNA sequences was done with the Vector NTI program (Invitrogen, Carlsbad, CA).
Screening method.
Screening for mutants with improved activity was performed by growing them in 5 ml of LB medium supplemented with kanamycin (100 μg/ml). The cultures were grown for 20 h at 37°C with shaking at 250 rpm on a TU-400 incubator shaker (Orbital shaker incubator; MRC, Holon, Israel), and then cells were harvested at 8,000 × g for 10 min at 25°C using a Sigma 4K15 centrifuge (Sigma, Osterode, Germany). The cell pellets were resuspended with 2.5 ml PB, pH 7, 0.1 M, to a final OD at 600 nm of 4. The substrate thioanisole was added to 1 ml of cells in PB to a final concentration of 0.5 mM substrate (from a 50 mM stock solution in ethanol), and the biotransformation was carried out for 3 h in a 16-ml glass vial with shaking at 600 rpm using a Vibramax 100 shaker (Heidolph, Nuremberg, Germany) at room temperature. The reaction was stopped by adding 1 ml of ethyl acetate (1:1 volume) followed by vigorous vortexing. Phase separation was facilitated by a short centrifugation step (8,000 × g for 30 s), and the content of the organic phase was measured using a 6890N gas chromatograph (GC) (Agilent Technologies, Santa Clara, CA). The criteria for better performance were higher reaction rates and/or higher enantiomeric excess (EE) than the WT enzyme. In order to ensure the probability (99%) that all 64 possible outcomes from the single-site random mutagenesis had been sampled, 292 colonies needed to be screened (33).
Whole-cell enzymatic activity.
Whole-cell activity assays were performed in a manner similar to that for previous studies with TMOs (15, 46). Briefly, exponential-phase cultures were used in all experiments by diluting overnight cells to an OD (600 nm) of 0.1 and growing them to an OD of ∼1.3. The exponentially grown TG1 cells harboring the various pBS(Kan) vectors were centrifuged at 8,000 × g for 10 min at 25°C in a Sigma 4K15 centrifuge (Sigma, Osterode, Germany) and resuspended in PB to an OD of 11. The biotransformation was carried out in 16-ml glass vials with screw caps with 2 ml cells and 1 mM substrate (added from a 200 mM stock solution in ethanol). The vials were shaken at 600 rpm (Vibramax 100; Heidolph, Nuremberg, Germany) at room temperature. The reaction was stopped periodically (a vial was sacrificed) using 2 ml of ethyl acetate (1:1 volume) with vigorous vortexing. The product and remaining substrate were extracted into the organic phase and measured using the GC. The initial product formation rates were determined by sampling at 10-min intervals during the first hour. The specific activity (nmol/min/mg protein) was calculated as the ratio of the initial transformation rate and the total protein content. Total protein content was 0.22 (mg protein/ml/OD at 600 nm) for TOM and ToMO and 0.24 (mg protein/ml/OD at 600 nm) for T4MO and TpMO (16, 46, 51). Enantioselectivity was determined at 100% conversion, and in cases in which the activity was low and substrate was not depleted after 24 h, a longer period of time was used (48 h), with the substrate concentration reduced to 0.5 mM. Activity data reported in this paper are in the form of the mean ± 1 standard deviation (based on at least two independent results).
Measurement of apparent Vmax and Km values was performed similarly with 10-min intervals during the first hour of reaction. Substrate concentrations used were 0.1 to 2.0 mM with cell OD of 11. Kinetic constants were calculated using the Enzyme Kinetic Module for Sigma Plot (Systat Software Inc.).
Analytical methods.
Conversion of sulfides to sulfoxides was determined by GC with a 6890N GC (Agilent Technologies, Santa Clara, CA) using a 30-m × 0.32-mm × 0.25-μm capillary column packed with γ-cyclodextrin trifluoroacetyl (Chiraldex G-TA; Astec, Bellefonte, PA) and a flame ionization detector. The temperature for thioanisole was programmed as follows: T1 = 110°C; dT/dt = 10°C/min, T2 = 130°C; dT/dt = 20°C/min, T3 = 160°C, 13 min; split ratio, 1:3. Under these conditions, the retention times were 3.89 min for thioanisole, 10.35 min for (R)-methyl phenyl sulfoxide, and 14.15 min for (S)-methyl phenyl sulfoxide. For determination of methyl p-tolyl sulfide conversion, the temperature was programmed as follows: T1 = 110°C; dT/dt = 10°C/min, T2 = 130°C; dT/dt = 20°C/min, T3 = 160°C, 17.5 min; split ratio, 1:3. Under these conditions, the retention times were 4.45 min for methyl p-tolyl-sulfide, 12.56 min for (R)-methyl p-tolyl sulfoxide, and 13.46 min for (S)-methyl p-tolyl sulfoxide. The concentrations of the reactants were determined from calibration curves obtained with commercial standards. The identity of the sulfides and sulfoxides was confirmed by GC-mass spectrometry using a 6890N (Agilent Technologies, Santa Clara, CA) GC equipped with a capillary column (30 m × 0.32 mm × 0.25 μm) filled with HP-5 [(5%-phenyl)-methylpolysiloxane] (Agilent Technologies) and an HP-5975 mass spectrum detector (Agilent Technologies, Santa Clara, CA). As the HP-5 capillary column is not chiral, the R and S sulfoxides were seen as one peak. For determination of thioanisole and methyl p-tolyl-sulfide conversion, the temperature was programmed as follows: T1 = 90°C; dT/dt = 10°C/min, T2 = 190°C; split ratio, 1:10.
Homology modeling of TOM TomA3 and T4MO TmoA.
Parts of the amino acid sequences of the TOM α subunit (residues 1 to 300 of TomA3) and T4MO α subunit (residues 1 to 300 of TmoA) were modeled into the known three-dimensional structure of PH of P. stutzeri OX1 (for TOM) and ToMO TouA of P. stutzeri OX1 (for T4MO) (26, 37, 38). The three-dimensional model was obtained using the SWISS-MODEL server (http://swissmodel.expasy.org) (20, 42). The molecular visualization programs UCSF Chimera (http://www.cgl.ucsf.edu/chimera) and PyMOL (http://pymol.sourceforge.net) were utilized to visualize and manipulate the molecular model. The docking of methyl p-tolyl sulfoxide in ToMO diiron center was performed with the MEDock web server (http://bioinfo.mc.ntu.edu.tw/medock/index.html).
Protein structure accession numbers.
The Protein Data Bank accession codes of the enzymes that were used for the modeling are 2INP (2.30 resolution) for PH and 2INC (1.85 resolution) for ToMO TouA.
RESULTS
Evaluation of four WT TMOs.
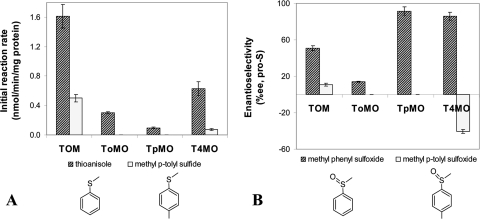
Four WT TMOs (TOM, ToMO, TpMO, and T4MO) were evaluated for their ability to oxidize two substrates, thioanisole and methyl p-tolyl sulfide, at a concentration of 1 mM. The results clearly indicated that TOM had the highest activity rate for both substrates: 1.6 ± 0.2 nmol/min/mg protein for thioanisole and 0.5 ± 0.05 nmol/min/mg protein for methyl p-tolyl sulfide (Fig. 1). TOM transformed thioanisole to the corresponding sulfoxides 2.5 times faster than T4MO, which had the second highest rate. ToMO exhibited a low transformation rate, while TpMO was a very poor catalyst (16 times lower than TOM). The transformation rate of methyl p-tolyl sulfide was dramatically lower for TOM and T4MO in comparison with thioanisole, while ToMO and TpMO had no activity at all (Fig. 1A). The enantiospecificity of the WT enzymes varied from 14% EE for ToMO to 91% EE for TpMO (Fig. 1B), with TOM exhibiting only moderate preference for the (S)-sulfoxide (51% EE). Surprisingly, the specificity of TOM was remarkably reduced in the sulfoxidation of methyl p-tolyl sulfide to a mere 11% EE, whereas T4MO completely changed its selectivity toward the R-enantiomer. It should be noted that in prolonged experiments (a few days) a small peak of sulfone was detected by GC-mass spectrometry, indicating further oxidation of the sulfoxide by the enzymes. However, within the short time limits of the experiments described in this work, oxidation of sulfoxides to sulfones was not detectable. As TOM showed the highest oxidation rate for both substrates and T4MO exhibited high enantioselectivity, these enzymes were further studied.
FIG. 1.
Initial sulfoxidation rate (A) and enantioselectivity (B) of TG1 cells expressing WT TMOs. Initial substrate concentration was 1 mM. Results represent an average of at least two independent experiments with the absolute measured error being less than 10%.
Apparent maximal reaction velocities (Vmax) and Michaelis-Menten constants (Km) were determined by whole-cell assays for TOM and T4MO with the two substrates (Table 2). The term apparent is used because the kinetic values were determined with whole cells. The Vmax values coincide well with the initial rates described in Fig. 1A, and therefore, a substrate concentration of 1 mM allows good comparison of maximal velocities for all enzymes studied in this work.
TABLE 2.
Apparent Vmax and Km values for E. coli TG1 expressing TOM and T4MO for oxidation of aryl alkyl sulfidesa
| Enzyme | Value for reaction:
|
|||||
|---|---|---|---|---|---|---|
| Thioanisole → methyl phenyl sulfoxide
|
Methyl-p-tolyl sulfide → methyl p-tolyl sulfoxide
|
|||||
| Vmax (nmol/min/mg protein) | Km (μM) | Vmax/Km | Vmax (nmol/min/mg protein) | Km (μM) | Vmax/Km | |
| TOM | 1.66 ± 0.49 | 129 ± 21 | 0.013 | 0.67 ± 019 | 68 ± 20 | 0.010 |
| T4MO | 0.85 ± 0.21 | 115 ± 28 | 0.007 | 0.070 ± 0.006 | 11 ± 4 | 0.006 |
Initial specific rates were determined for each reaction by monitoring the formation of the product using GC. Substrate concentrations were 0.1, 0.25, 0.50, 0.75, 1.0, 1.5, and 2.0 mM, and the cell OD was 11. Standard deviations are shown for two to three independent experiments.
Characterization of TOM mutants.
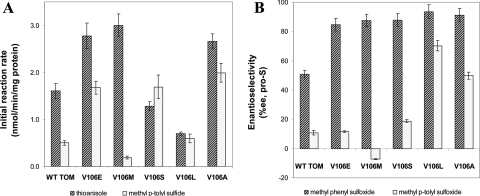
Saturation mutagenesis was performed at position TomA3 V106 since this residue was found to have influence on the regiospecificity of the enzyme in previous works (6, 34). From screening of approximately 300 mutant colonies on thioanisole, five improved variants appeared, TomA3 V106E, V106M, V106S, V106L, and V106A (Fig. 2). The initial thioanisole sulfoxidation rate of TG1 cells expressing WT TOM was improved by mutants V106E, V106M, and V106A (1.72-, 1.86-, and 1.65-fold, respectively), while V106S and V106L showed a decreased rate for thioanisole oxidation (0.8- and 0.43-fold, respectively). The enantioselectivity of all the V106 variants was improved compared to the WT, ranging from 85% EE for V106E up to 93% EE for the V106L (which had the lowest reaction rate). Mutants V106M and V106S exhibited the same percent EE, despite the big difference in rate.
FIG. 2.
Initial sulfoxidation rate (A) and enantioselectivity (B) of TG1 cells expressing WT TOM and TomA3 V106 mutants. Initial substrate concentration was 1 mM. Results represent an average of at least two independent experiments with the absolute measured error being less than 10%.
The results of the transformation of methyl p-tolyl sulfide to the corresponding sulfoxides were significantly different from those obtained for thioanisole (Fig. 2). The rates of V106E, V106S, and V106A were improved more than threefold, whereas mutant V106L showed only a minor improvement in rate; however, this was accompanied by an increase in selectivity from 11% EE (pro-S) to 70%. A most interesting result was obtained with the V106M mutant, which exhibited the highest transformation rate on thioanisole. With methyl p-tolyl sulfide, a substantial decrease in rate was measured accompanied by a reversal in the product configuration to 7% EE pro-R. Overall, with thioanisole there was a clear trend of improvement of the WT enantioselectivity by all of the mutants. However, large improvement in the percent EE of methyl p-tolyl sulfide oxidation was measured only for V106L and V106A. The only mutant that showed consistently better performance both in rate and in enantioselectivity for the two substrates was V106A.
Characterization of T4MO mutants.
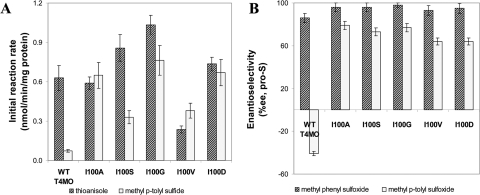
Screening of a library of T4MO TmoA I100 mutants resulted in five improved variants: TmoA I100A, I100S, I100G, I100V, and I100D. As seen in Fig. 3, the transformation rates of WT T4MO and TmoA mutants were lower than the rates of WT TOM and its mutants. T4MO mutants I100S, I100G, and I100D exhibited improved rates for thioanisole oxidation; mutant I100A had practically the same transformation rate as the WT; and mutant I100V exhibited less activity with thioanisole. The enantioselectivity of the T4MO mutants was remarkably improved from 86% EE to 98% EE for I100G. The methyl p-tolyl sulfide transformation rate was improved significantly by all of the mutants. The WT had a very low transformation rate with pro-R enantioselectivity (Fig. 3), while all the T4MO mutants switched their enantio preference from the R configuration to the S configuration. The transformation rates of methyl p-tolyl sulfide by mutants I100A, I100G, and I100D had a substantial improvement compared with WT T4MO by a factor of about 10, while mutants I100S and I100V had rates increased nearly fivefold. Variant TmoA I100V had a higher transformation rate for methyl p-tolyl sulfide than for thioanisole, which was exceptional even in comparison with the TOM mutants.
FIG. 3.
Initial sulfoxidation rate (A) and enantioselectivity (B) of TG1 cells expressing WT T4MO and TmoA I100 mutants. Initial substrate concentration was 1 mM. Results represent an average of at least two independent experiments with the absolute measured error being less than 10%.
Comparison of TOM and T4MO.
WT TOM showed a high rate and low enantioselectivity for thioanisole oxidation, while WT T4MO had a high percent EE and a lower rate (Fig. 1). When the methyl p-tolyl sulfide transformation was examined, the difference between the two enzymes became more evident with the transformation rate of TOM being almost an order of magnitude higher than that of T4MO, while the T4MO enantio preference was of the opposite enantiomer (pro-R selectivity). Two identical amino acid substitutions were found in the screen for both enzymes, Ser and Ala at position 100 in T4MO and 106 in TOM. Although T4MO variant TmoA I100V resembles WT TOM (both have Val at this position), there was no correlation in the oxidation rate or in percent EE for thioanisole transformation. However, the oxidation rates of methyl p-tolyl sulfide by the two (TmoA I100V and WT TOM) were similar. Both TOM and T4MO mutants had a comparable trend in results obtained with thioanisole sulfoxidation: the TOM variants had an overall lower percent EE on average compared with T4MO variants, but all of the variants showed an improvement in the percent EE with respect to WT. The best improvement in percent EE for thioanisole oxidation by a TOM variant was obtained by V106L (1.82-fold) while the best T4MO variant was I100G (1.14-fold). It is interesting that TomA3 V106G and TmoA I100L did not emerge from the screening process, as one would expect that a smaller residue such as Gly would improve the activity of TOM as well. Overall, the improvement in percent EE by TOM mutants for thioanisole oxidation was more dramatic, as the WT itself had a rather low selectivity. The presence of residues Ala and Ser had a more pronounced effect on the methyl p-tolyl sulfide oxidation than on thioanisole oxidation, for both TOM and T4MO. This implies that for the larger and less accepted methyl p-tolyl sulfide, a reduction in the residue size at position 106 (or 100 in T4MO) is crucial for better activity.
To verify that the increase in activity of the mutants derived from the amino acid substitutions rather than expression level changes, SDS-polyacrylamide gel electrophoresis was used to visualize two of the six subunits: TmoA (55 kDa) and a combined band from TmoE (35 kDa) and TmoF (36 kDa) in T4MO and the analogous TomA3 (61 kDa), TomA1 (37 kDa), and TomA5 (39 kDa) in TOM. All of the mutant and WT bands had similar intensities. As the cell growth and the biotransformation conditions were identical for the WT and mutants, the changes in activity appear to arise from the mutations at TmoA I100 for TOM or TomA3 V106 for T4MO and not from different expression levels.
Protein homology modeling.
In order to gain insight into the correlation between function and structure of the mutants, protein homology modeling was used. TOM TomA3 was modeled with the known crystal structure of PH of Pseudomonas stutzeri OX1 α-hydroxylase subunit (38) (74% identity), and T4MO TmoA was modeled with the known crystal structure of ToMO TouA (27, 37) (72% identity). The accuracy of the model was judged by the conservation of the positions of the diiron-coordinating residues in TOM (TomA3 E110, E140, H143, E201, E235, and H238) compared to PH and T4MO (TmoA E104, E134, H137, E197, E231, and H234) compared to ToMO (the average distance of the Cα carbons of the TOM TomA3 model to PH and T4MO TmoA to ToMO for the metal binding residues was less than 0.07 Å). The structural alignment of the template and model also showed conserved spatial configurations. Despite the limitations of homology-based modeling, it has become a common methodology in recent years for studying structure-function relationships in proteins (5, 16, 17, 20).
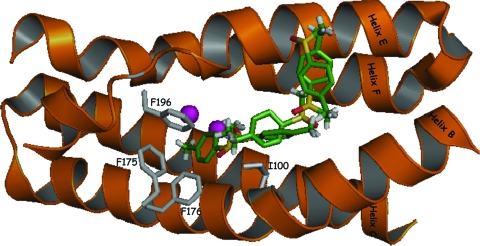
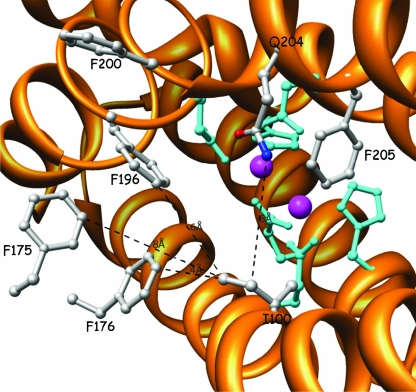
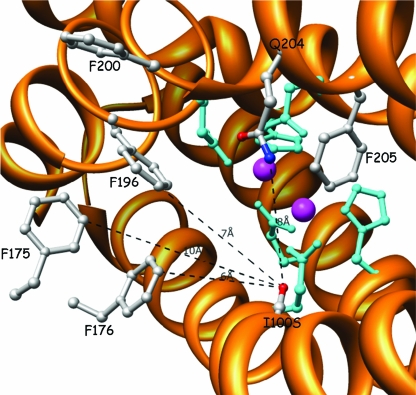
In order to visualize the route by which the substrate and product enter and leave the active site, there was use of maximum-entropy-based calculations of methyl p-tolyl sulfoxide docked in the diiron vicinity of ToMO, based on the location of 4-bromophenol in the original crystal structure (37) (Fig. 4). It is assumed that a similar pathway occurs in T4MO, which is closely related to ToMO. The substrate is facing the Phe residues (F175, F176, and F196) at the channel's end and is forced to turn into the diiron center, while residue I100 serves as an axis in this turn. The side chain of Ile-100 comprises the right side of the hydrophobic pocket and may cause a disturbance to the substrate entering the diiron center (Fig. 5). When changing Ile to Ser (Fig. 6), the access to the diiron center is more open and easier to approach. Consequently, a smaller side chain at position 100 makes the diiron center more accessible to the substrate. Similar results were obtained even with bulkier side chains like Asp, in which the carboxylic side chain, according to the model, had a bend to the far right side and therefore did not disturb the hydrophobic gate accessibility. This interpretation may explain the higher activity rate obtained with all five T4MO mutants: I100A, I100S, I100G, I100V, and I100D.
FIG. 4.
Maximum entropy-based docking of methyl p-tolyl sulfoxide in the ToMO active site. The four-helix bundle is colored in orange, Fe atoms are colored in magenta, and methyl p-tolyl sulfoxide carbon atoms are colored in green. Residues F175, F174, and F196 comprising the hydrophobic pocket are colored in gray. Position TouA I100 (analogous to TmoA I100 in T4MO) is colored in gray. Five sulfoxide molecules are seen at the hypothesized pathway leading to the active site. Graphics were prepared using PyMOL (http://pymol.sourceforge.net). Entropy/positioning calculations were made with the MEDock web server (http://bioinfo.mc.ntu.edu.tw/medock/index.html).
FIG. 5.
Active site of the α subunit of WT T4MO from the viewpoint of the substrate entering the diiron center. The Fe atoms (magenta) are coordinated by six residues (cyan). Residues I100, F175, F176, F196, and F200 (gray) comprise the hydrophobic gate (distances from I100 are presented in a dashed black line). All graphics here and below were prepared using UCSF Chimera unless noted otherwise (http://www.cgl.ucsf.edu/chimera).
FIG. 6.
Active site of the α subunit of T4MO variant TmoA I100S from the viewpoint of the substrate entering the diiron center. The Fe atoms (magenta) are coordinated by six residues (cyan). Residues I100S, F175, F176, F196, and F200 (gray) comprise the hydrophobic gate (distances from I100S are presented in a dashed black line).
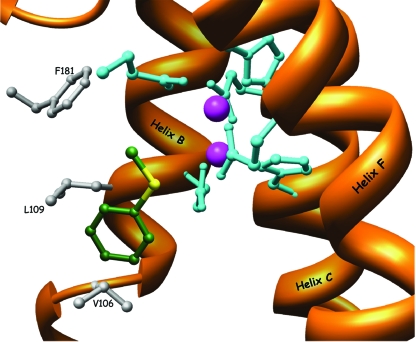
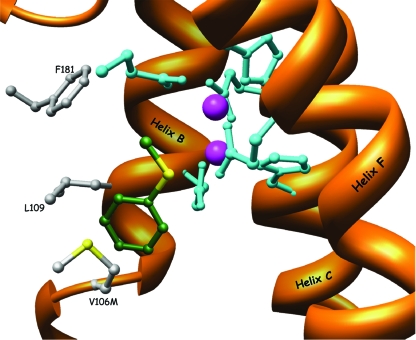
In the absence of structures of PH bound to a substrate or inhibitor, it is difficult to know the entrance path to the diiron center in TOM (38). However, the importance of residue V106 in TOM can be clarified from the hypothesized view of the orientation of the substrate within the diiron center between residues V106 and L109 (Fig. 7). The substrate hydroxylation presumably occurs at a bridging position between the two iron atoms (38). By decreasing the space between L109 and V106, as in the case of variant V106M (Fig. 8), the ability of the active site to accommodate the longer substrate (methyl p-tolyl sulfide) is damaged. However, the rate and enantioselectivity for thioanisole oxidation were improved, perhaps because of the closer proximity of the sulfur atom to the diiron atoms. The phenomenon may be explained by the differences in length between thioanisole and methyl p-tolyl sulfide (the latter is 1 Å longer). The increase in the size of the substrate corresponds to the distance that was shortened due to the presence of the side chain of Met in the V106M variant. Mutant TomA3 V106E, which has a large side chain, had such a positioning according to the model (not shown), which did not penetrate the crucial space needed for the proper alignment of the substrate in the active site.
FIG. 7.
Structure of the substrates' spatial positioning in the active-site pocket of WT TOM. The thioanisole molecule is manually docked within the active site and suggests a possible orientation of the substrate in the active site. The carbon atoms of thioanisole are colored in green. The aromatic ring is between the two residues L109 and V106 while the sulfur moiety of the molecule (yellow) points toward the diiron center. Substrate hydroxylation occurs at a bridging position between the two iron atoms (magenta).
FIG. 8.
Structure of the substrates' spatial positioning in the active-site pocket of TOM variant V106M. The thioanisole molecule is manually docked within the active site and suggests a possible orientation of the substrate in the active site. The carbon atoms of thioanisole are colored in green. The side chain of Met-106 enters the space between positions 109 and 106, which is important for the substrate alignment and steering within the active site.
DISCUSSION
Considering the great importance of chiral sulfoxides as functionalized substances in pharmaceuticals and as important chiral auxiliaries in organic synthesis (12, 14, 21, 44) and the ever-growing demand for green catalytic processes (41), TMOs were evaluated as potential biocatalysts in sulfoxidation reactions. Furthermore, saturation mutagenesis was applied to modify TOM and T4MO to increase the enantioselectivity and oxidation rate of two model substrates, namely, thioanisole and methyl p-tolyl sulfide.
All four WT TMOs (TOM, T4MO, TpMO, and ToMO) expressed in E. coli TG1 were able to oxidize thioanisole with variable degrees of enantioselectivities and rates. However, only TOM and T4MO showed substantial oxidation ability for methyl p-tolyl sulfide, albeit with a dramatic decrease in rate and enantioselectivity compared to the thioanisole results (T4MO actually changed its chiral preference from pro-S into pro-R) (Fig. 1). Results presented in this work regarding thioanisole oxidation disclose comparability between the regioselectivity of the enzymes and their enantioselectivity. ToMO, which has a relaxed regioselectivity and produces a mixture of three cresol isomers from toluene hydroxylation (16), had the lowest percent EE, making it the least enantioselective catalyst among the WT TMOs examined. TpMO and T4MO both hydroxylate toluene primarily at the para position (18, 46) and showed high enantioselectivity in the oxidation of thioanisole. TOM, which hydroxylates toluene specifically at the ortho position (34), showed moderate percent EE. The variability in enantioselectivity expressed by the WT TMOs implies that active-site structural differences that affect and control regioselectivity influence enantioselectivity as well.
It was previously shown that chiral sulfoxides can be synthesized with biocatalysts. Dai and Klibanov (10) described the asymmetric oxidation of thioanisole with H2O2 catalyzed by isolated horseradish peroxidase in various organic solvents. EE values of 60% (pro-S) in isopropyl alcohol and 33% in water were reported, suggesting that WT TOM and T4MO are more selective toward the same enantiomer.
In addition to purified enzymes, whole-cell systems with TDO or NDO expressed in P. putida were used for oxidation of thioanisole as well. TDO catalyzed with preferential formation of the (R)-sulfoxide (>98% EE), and NDO catalyzed with preferential formation of the (S)-sulfoxide (91% EE) (2). Some TMOs examined in this work catalyzed the formation of (S)-sulfoxide with similar or higher enantioselectivities: TpMO exhibited 91% EE and T4MO I100G exhibited 98% EE. Gibson and coworkers (23) reported that TDO oxidized thioanisole with >98% EE pro-R and methyl p-tolyl sulfide with 38% EE pro-S, similarly to cyclohexanone monooxygenase from Acinetobacter calcoaceticus (99% EE pro-R when oxidizing thioanisole but only 37% EE pro-S when oxidizing methyl p-tolyl sulfide) (7). T4MO exhibited the opposite trend in this work, a change in enantiospecificity from pro-S for thioanisole to pro-R for methyl p-tolyl sulfide, accompanied by a decrease in rate. Sheldon and coworkers reported lower yields for chloroperoxidase oxidation of methyl p-tolyl sulfide than for thioanisole (83% versus 100% conversion, respectively) (48). Analogous results were obtained with TMOs; Vmax dropped by 60% for WT TOM and by 90% for WT T4MO. Apparently, the addition of the methyl substituent at the para position on the benzyl ring results in a dramatic decrease in enzymatic activity and, in some cases, a change in enantioselectivity.
The only prior example regarding enantiospecificity of TMOs was reported by Fox and coworkers regarding the conversion of alkenes to chiral epoxides (25). WT T4MO formed (S)-butadiene epoxide with 34% EE, compared to 86% EE for (S)-methyl phenyl sulfoxide obtained in this work, suggesting higher selectivity toward the latter substrate.
TOM and T4MO were improved both in rate and in enantioselectivity via saturation mutagenesis at position I100 in T4MO and the analogous residue V106 in TOM. The fact that thioanisole was the substrate of choice resulted in variants more fit to oxidize this substrate selectively. Screening with methyl p-tolyl sulfide as a substrate may have resulted in different mutants. For example, variant TOM V106M would not have been chosen. Both TOM and T4MO mutants had notable improvement in percent EE and rate (Fig. 2 and 3). These improvements confirm the importance of position TomA3 V106 in TOM (and analogous TmoA I100 in T4MO) for enantioselective oxidation and show a correlation between regioselectivity and enantioselectivity of TMOs.
The improved variants selected following saturation mutagenesis and screening were TOM TomA3 V106E, V106M, V106S, V106L, and V106A and T4MO TmoA I100A, I100S, I100G, I100V, and I100D. The substituted amino acid residues were aliphatic (TmoA I100G, I100V, and I100A and TomA3 V106L and V106A), hydroxyl/sulfur-containing side chain (TmoA I100S and TomA3 V106S and V106M), or acidic (TmoA I100D and TomA3 V106E). In addition, the side chain was either small or big (for example, I100G versus I100D in TmoA and V106A versus V106M in TomA3). Three of the residues, Ser, Ala, and Asp/Glu, appeared in both enzymes, emphasizing their contribution to improvement in catalysis. It is also worth noting that some of the variants had been found beneficial in previous work (TOM V106A improved TOM activity for both chloroform degradation and naphthalene oxidation [33], T4MO TmoA I100A and I100S improved 4-nitrocatechol synthesis rate, and TmoA I100G enhanced the selectivity toward 2-naphthol formation from naphthalene [15, 45]). Subsequently, the current results as well as previous work by Fishman et al. (15), Pikus et al. (29), Tao et al. (45, 46), and Vardar and Wood (50) indicate together with the work of Sazinsky et al. (37) that the residue in this position need not be hydrophobic for TMOs to be efficient catalysts and suggest that a small side chain is not a prerequisite.
Comparison of TOM and T4MO variants reveals that the oxidation of thioanisole resulted in improvement in percent EE for all of the mutants. However, only two TOM variants showed improved enantioselectivity for methyl p-tolyl sulfoxide formation, and all of the T4MO variants had much better percent EE than did WT (with a change in the enantio preference toward the S configuration).
Vmax and Km values for T4MO were reported for nitrobenzene and m- and p-nitrophenols (15) and are similar to those reported here. However, the specificity constant (Vmax/Km) for nitrobenzene was 24-fold higher than that for thioanisole, suggesting a preference of the enzyme for ring hydroxylation. Additionally, the oxidation rate of thioanisole was lower than that of the natural substrate, toluene (16, 51). For example, T4MO oxidized 1 mM thioanisole at a rate of 0.63 ± 0.13 nmol/min/mg protein compared to 4.40 ± 0.30 for 109 μM toluene whereas TOM showed the same trend (1.61 ± 0.16 nmol/min/mg protein for thioanisole versus 2.5 ± 0.1 for toluene). It is worth emphasizing, however, that our experiments and the experiments on toluene were conducted using different substrate concentrations (1 mM using thioanisole and 109 μM using toluene). Kinetic constants for T4MO have been reported for the purified enzyme using toluene as a substrate (27); however, these values are not comparable to our system employing whole cells in which factors such as membrane permeability and cofactor availability affect the performance. A similar whole-cell system with a related enzyme, xylene monooxygenases of Pseudomonas putida expressed in E. coli JM101(pSPZ3), was used for oxidation of various substrates (4). The apparent Km values reported for pseudocumene and toluene were 202 ± 8 μM and 87 ± 17 μM, respectively, showing similarity to the values reported in Table 2 for T4MO and TOM. However, the rate constants reported by Buhler et al. (4) were 1 or 2 orders of magnitude higher for some of the substrates, suggesting that the TMOs described in this work need further improvement in rate before they can function in a commercial process.
Homology modeling was used to gain insight into the correlation between structure and function of the WT and mutants. Position TmoA I100 of T4MO and the analogous TomA3 V106 of TOM are part of the hydrophobic gate in the entrance to the active site (6) and a part of the hydrophobic cavity surrounding the diiron binding site (15, 33, 37, 38). A similar description of the active-site pocket was reported by Rosenzweig et al. for soluble methane monooxygenases (32). In their work, the analogous residue MmoX L110 was hypothesized to function as a gate, restricting the size of molecules entering and leaving the active site. It is expected that a larger channel or active-site entrance may be better suited for efficient movement of aromatic compounds to and from the diiron center. However, this finding was very recently questioned by Borodina et al. (1) regarding aromatic substrates. Mutations at L110 influenced the regiospecificity but did not extend activity to triaromatic hydrocarbons. Although soluble methane monooxygenases can hydroxylate aromatics, TMOs and PHs are 20 times more efficient in performing this reaction, perhaps because of easier substrate access and/or product egress (39). Enlargement of the hydrophobic gate in T4MO variants was acquired by mutants I100A, I100S, I100G, and I100V (TmoA I100S is presented in Fig. 6). For example, the rate of variant I100G was increased by 1.73-fold for thioanisole oxidation and by 11.4-fold for methyl p-tolyl sulfide oxidation, substantiating the role of position I100 as a gate. According to our model, even variant I100D of T4MO, which has a large side chain, made the entrance to the diiron center more accessible because of the side chain positioning.
The hydrophobic gate of TOM is less accessible for substrate entrance into the active site (compared with T4MO) because of the two Leu residues (L109 and L208) that do not exist in the T4MO hydrophobic gate. In addition, the active-site pocket of TOM is shallower than that of T4MO (38). Consequently, the activity rate of T4MO should have been higher if the “bottleneck” was indeed that gate. As the TOM α subunit (TomA3) is highly similar to the PH α subunit (74% identity), an alternative entrance to the diiron center may dominate and explain the higher activity of TOM. It is postulated that the entrance of the substrate may be directly from the protein surface through helices E and F of the four-helix bundle. The direct route to the diiron center, rather than passing through a ∼35-Å channel, can explain the substantially higher rate of WT TOM and variants compared with WT T4MO and variants, as well as the very low activity obtained by ToMO and TpMO. Direct entrance through helices E and F makes the hydrophobic gate hypothesis less relevant for TOM, which means that position V106 influences the activity of TOM by a second mechanism. In order to have an efficient oxidation of the substrate, it must align correctly in the active-site pocket. The space which the substrate supposedly occupies in the TOM active-site pocket lies between the two iron atoms and Leu-109 and Val-106 (Fig. 7). The sulfur moiety of the molecule which undergoes oxidation is positioned near the diiron atoms, whereas the aromatic ring is positioned in the space between Leu-109 and Val-106. The distance between Val-106 and the diiron center is 9 to 11 Å, and for methyl p-tolyl sulfide, which is ∼1 Å longer than thioanisole, the gap for proper alignment is marginal and therefore may be responsible for the lower activity obtained with the larger substrate. For TOM variant V106M this opening is even more limited because of the Met side chain, which penetrates the vital space between residues V106 and Leu-109, therefore restricting the space needed for proper substrate alignment (Fig. 8). Variant V106M possessed an improved activity rate and selectivity for oxidizing thioanisole perhaps because the sulfur atom is “pushed” by the V106M side chain and positioned closer to the diiron site, but the activity dropped dramatically for oxidizing methyl p-tolyl sulfide, possibly due to limited space for proper alignment within the active-site pocket. The T4MO I100 side chain distance from the diiron atoms is even less than that in variant TOM V106M (7 to 8 Å and 8 to 9 Å, respectively), thus resulting in very low activity accompanied by a small shift in enantio preference from S to R. Sazinsky et al. (38) suggested that the space occupied by residues I104 and L105 in the PH α subunit (analogous to TomA3 V106 and L109) is important for steering substrates, and thus, increasing the depth of the active-site pocket promotes hydroxylation of larger substrates. Indeed, the T4MO variants which showed significant activity with methyl p-tolyl sulfide had side chains positioned in such a manner that there was no penetration of the vital space in which the substrate is needed to align.
The enantio preference of the WT enzymes and their improved variants is pro-S for both thioanisole and methyl p-tolyl sulfide. However, both variants TOM TomA3 V106M and WT T4MO facilitated the pro-R configuration of methyl p-tolyl sulfide, accompanied by very low activity (Fig. 2 and 3). Thus, the results from this work suggest that when the substrate is not aligned correctly in the active site due to limited space, the oxidation rate is decreased and the enantioselectivity is impaired and can be of both chiral configurations. Whereas the main function of I100 in T4MO seems to be the steering of the substrate into the active site while passing through the entrance channel, the main function of V106 in TOM is hypothesized to be proper positioning or docking of the substrate with respect to the diiron atoms.
Acknowledgments
This research was supported by the Binational Science Foundation (BSF-2005173).
We thank Noam Adir from the Department of Chemistry at the Technion, Haifa, Israel, for assistance with the homology modeling and for fruitful discussions.
Footnotes
Published ahead of print on 11 January 2008.
REFERENCES
- 1.Borodina, E., T. Nichol, M. G. Dumont, T. J. Smith, and J. C. Murrell. 2007. Mutagenesis of the “leucine gate” to explore the basis of catalytic versatility in soluble methane monooxygenase. Appl. Environ. Microbiol. 73:6460-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, D. R., N. D. Sharma, S. A. Haughey, M. A. Kennedy, B. T. McMurray, G. N. Sheldrake, C. R. Allen, H. Dalton, and K. Sproule. 1998. Toluene and naphthalene dioxygenase-catalyzed sulfoxidation of alkyl aryl sulfides. J. Chem. Soc. Perkin Trans. 1:1929-1934. [Google Scholar]
- 3.Breuer, M., K. Ditrich, T. Habicher, B. Hauer, M. Kesseler, R. Sturmer, and T. Zelinski. 2004. Industrial methods for the production of optically active intermediates. Angew. Chem. Int. Ed. Engl. 43:788-824. [DOI] [PubMed] [Google Scholar]
- 4.Buhler, B., B. Witholt, B. Hauer, and A. Schmid. 2002. Characterization and application of xylene monooxygenase for multistep biocatalysis. Appl. Environ. Microbiol. 68:560-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulter, T., M. Alcalde, V. Sieber, P. Meinhold, C. Schlachtbauer, and F. H. Arnold. 2003. Functional expression of a fungal laccase in Saccharomyces cerevisiae by directed evolution. Appl. Environ. Microbiol. 69:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canada, K. A., S. Iwashita, H. Shim, and T. K. Wood. 2002. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J. Bacteriol. 184:344-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colonna, S., S. Del Sordo, N. Gaggero, G. Carrea, and P. Pasta. 2002. Enzyme mediated catalytic asymmetric oxidations. Heteroatome Chem. 13:467-473. [Google Scholar]
- 8.Colonna, S., N. Gaggero, C. Richelmi, and P. Pasta. 1999. Recent biotechnological developments in the use of peroxidases. Trends Biotechnol. 17:163-168. [DOI] [PubMed] [Google Scholar]
- 9.Cotton, H., T. Elebring, M. Larsson, L. Li, H. Sorensen, and S. von Unge. 2000. Asymmetric synthesis of esomeprazole. Tetrahedron Asymmetry 11:3819-3825. [Google Scholar]
- 10.Dai, L., and A. M. Klibanov. 2000. Peroxidase-catalyzed asymmetric sulfoxidation in organic solvents versus in water. Biotechnol. Bioeng. 70:353-357. [DOI] [PubMed] [Google Scholar]
- 11.de Gonzalo, G., G. Ottolina, F. Zambianchi, M. W. Fraaije, and G. Carrea. 2006. Biocatalytic properties of Baeyer-Villiger monooxygenases in aqueous and organic media. J. Mol. Catal. B Enzymatic 39:91-97. [Google Scholar]
- 12.de Gonzalo, G., D. E. Torres Pazmino, G. Ottolina, M. W. Fraaije, and G. Carrea. 2006. 4-Hydroxyacetophenone monooxygenase from Pseudomonas fluorescens ACB as an oxidative biocatalyst in the synthesis of optically active sulfoxides. Tetrahedron Asymmetry 17:130-135. [Google Scholar]
- 13.Eaton, R. W., and P. J. Chapman. 1995. Formation of indigo and related compounds from indolecarboxylic acids by aromatic acid-degrading bacteria: chromogenic reactions for cloning genes encoding dioxygenases that act on aromatic acids. J. Bacteriol. 177:6983-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez, I., and N. Khiar. 2003. Recent developments in the synthesis and utilization of chiral sulfoxides. Chem. Rev. 103:3651-3705. [DOI] [PubMed] [Google Scholar]
- 15.Fishman, A., Y. Tao, W. E. Bentley, and T. K. Wood. 2004. Protein engineering of toluene 4-monooxygenase of Pseudomonas mendocina KR1 for synthesizing 4-nitrocatechol from nitrobenzene. Biotechnol. Bioeng. 87:779-790. [DOI] [PubMed] [Google Scholar]
- 16.Fishman, A., Y. Tao, L. Rui, and T. K. Wood. 2005. Controlling the regiospecific oxidation of aromatics via active site engineering of toluene para-monooxygenase of Ralstonia pickettii PKO1. J. Biol. Chem. 280:506-514. [DOI] [PubMed] [Google Scholar]
- 17.Fishman, A., Y. Tao, G. Vardar, L. Rui, and T. K. Wood. 2006. Controlling regiospecific oxidation of aromatics and the degradation of chlorinated aliphatics via active site engineering of toluene monooxygenases, p. 237-286. In J.-L. Ramos and R. C. Levesque (ed.), Pseudomonas, vol. 4. Molecular biology of emerging issues. Springer US, New York, NY. [Google Scholar]
- 18.Fishman, A., Y. Tao, and T. K. Wood. 2004. Toluene 3-monooxygenase of Ralstonia pickettii PKO1 is a para-hydroxylating enzyme. J. Bacteriol. 186:3117-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimley, J. 2006. Pharma challenged. Chem. Eng. News 84:10-17. [Google Scholar]
- 20.Guex, N., A. Diemand, and M. C. Peitsch. 1999. Protein modeling for all. Trends Biochem. Sci. 24:364-367. [DOI] [PubMed] [Google Scholar]
- 21.Holland, H. L. 2001. Biotransformation of organic sulfides. Nat. Prod. Rep. 18:171-181. [DOI] [PubMed] [Google Scholar]
- 22.Leahy, J. G., P. J. Batchelor, and S. M. Morcomb. 2003. Evolution of the soluble diiron monooxygenases. FEMS Microbiol. Rev. 27:449-479. [DOI] [PubMed] [Google Scholar]
- 23.Lee, K., J. M. Brand, and D. T. Gibson. 1995. Stereospecific sulfoxidation by toluene and naphthalene dioxygenases. Biochem. Biophys. Res. Commun. 212:9-15. [DOI] [PubMed] [Google Scholar]
- 24.Lorenz, P., and J. Eck. 2005. Metagenomics and industrial applications. Nat. Rev. Microbiol. 3:510-516. [DOI] [PubMed] [Google Scholar]
- 25.McClay, K., B. G. Fox, and R. J. Steffan. 2000. Toluene monooxygenase-catalyzed epoxidation of alkenes. Appl. Environ. Microbiol. 66:1877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormick, M. S., M. H. Sazinsky, K. L. Condon, and S. J. Lippard. 2006. X-ray crystal structures of manganese(II)-reconstituted and native toluene/o-xylene monooxygenase hydroxylase reveal rotamer shifts in conserved residues and an enhanced view of the protein interior. J. Am. Chem. Soc. 128:15108-15110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell, K. H., J. M. Studts, and B. G. Fox. 2002. Combined participation of hydroxylase active site residues and effector protein binding in a para to ortho modulation of toluene 4-monooxygenase regiospecificity. Biochemistry 41:3176-3188. [DOI] [PubMed] [Google Scholar]
- 28.Panke, S., and M. Wubbolts. 2005. Advances in biocatalytic synthesis of pharmaceutical intermediates. Curr. Opin. Chem. Biol. 9:188-194. [DOI] [PubMed] [Google Scholar]
- 29.Pikus, J. D., J. M. Studts, K. McClay, R. J. Steffan, and B. G. Fox. 1997. Changes in the regiospecificity of aromatic hydroxylation produced by active site engineering in the diiron enzyme toluene 4-monooxygenase. Biochemistry 36:9283-9289. [DOI] [PubMed] [Google Scholar]
- 30.Pollard, D. J., and J. M. Woodley. 2007. Biocatalysis for pharmaceutical intermediates: the future is now. Trends Biotechnol. 25:66-73. [DOI] [PubMed] [Google Scholar]
- 31.Ricci, L. C., J. V. Comasseto, L. H. Andrade, M. Capelari, Q. B. Cass, and A. L. M. Porto. 2005. Biotransformations of aryl alkyl sulfides by whole cells of white-rot Basidiomycetes. Enzyme Microb. Technol. 36:937-946. [Google Scholar]
- 32.Rosenzweig, A. C., H. Brandstetter, D. A. Whittington, P. Nordlund, S. J. Lippard, and C. A. Frederick. 1997. Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions. Proteins Struct. Funct. Genet. 29:141-152. [PubMed] [Google Scholar]
- 33.Rui, L., Y. M. Kwon, A. Fishman, K. F. Reardon, and T. K. Wood. 2004. Saturation mutagenesis of toluene ortho-monooxygenase of Burkholderia cepacia G4 for enhanced 1-naphthol synthesis and chloroform degradation. Appl. Environ. Microbiol. 70:3246-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rui, L., K. F. Reardon, and T. K. Wood. 2005. Protein engineering of toluene-ortho-monooxygenase of Burkholderia cepacia G4 for regiospecific hydroxylation of indole to form various indigoid compounds. Appl. Microbiol. Biotechnol. 66:422. [DOI] [PubMed] [Google Scholar]
- 35.Sakamoto, T., J. M. Joern, A. Arisawa, and F. H. Arnold. 2001. Laboratory evolution of toluene dioxygenase to accept 4-picoline as a substrate. Appl. Environ. Microbiol. 67:3882-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Sazinsky, M. H., J. Bard, A. Di Donato, and S. J. Lippard. 2004. Crystal structure of the toluene/o-xylene monooxygenase hydroxylase from Pseudomonas stutzeri OX1. Insight into the substrate specificity, substrate channeling, and active site tuning of multicomponent monooxygenases. J. Biol. Chem. 279:30600-30610. [DOI] [PubMed] [Google Scholar]
- 38.Sazinsky, M. H., P. W. Dunten, M. S. McCormick, A. Didonato, and S. J. Lippard. 2006. X-ray structure of a hydroxylase-regulatory protein complex from a hydrocarbon-oxidizing multicomponent monooxygenase, Pseudomonas sp. OX1 phenol hydroxylase. Biochemistry 45:15392-15404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sazinsky, M. H., and S. J. Lippard. 2006. Correlating structure with function in bacterial multicomponent monooxygenases and related diiron proteins. Acc. Chem. Res. 39:558-566. [DOI] [PubMed] [Google Scholar]
- 40.Schmid, A., J. S. Dordick, B. Hauer, A. Kiener, M. Wubbolts, and B. Witholt. 2001. Industrial biocatalysis today and tomorrow. Nature 409:258-268. [DOI] [PubMed] [Google Scholar]
- 41.Schoemaker, H. E., D. Mink, and M. G. Wubbolts. 2003. Dispelling the myths—biocatalysis in industrial synthesis. Science 299:1694-1697. [DOI] [PubMed] [Google Scholar]
- 42.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Straathof, A. J., S. Panke, and A. Schmid. 2002. The production of fine chemicals by biotransformations. Curr. Opin. Biotechnol. 13:548-556. [DOI] [PubMed] [Google Scholar]
- 44.Tang, J., I. Brackenridge, S. M. Roberts, J. Beecher, and A. J. Willetts. 1995. Bakers' yeast oxidation of methyl para-tolyl sulfide: synthesis of a chiral intermediate in the preparation of the mevinic acid-type hypocholestemic agents. Tetrahedron 51:13217-13238. [Google Scholar]
- 45.Tao, Y., W. E. Bentley, and T. K. Wood. 2005. Regiospecific oxidation of naphthalene and fluorene by toluene monooxygenases and engineered toluene 4-monooxygenases of Pseudomonas mendocina KR1. Biotechnol. Bioeng. 90:85-94. [DOI] [PubMed] [Google Scholar]
- 46.Tao, Y., A. Fishman, W. E. Bentley, and T. K. Wood. 2004. Altering toluene 4-monooxygenase by active-site engineering for the synthesis of 3-methoxycatechol, methoxyhydroquinone, and methylhydroquinone. J. Bacteriol. 186:4705-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tao, Y., A. Fishman, W. E. Bentley, and T. K. Wood. 2004. Oxidation of benzene to phenol, catechol, and 1,2,3-trihydroxybenzene by toluene 4-monooxygenase of Pseudomonas mendocina KR1 and toluene 3-monooxygenase of Ralstonia pickettii PKO1. Appl. Environ. Microbiol. 70:3814-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Deurzen, M. P. J., F. van Rantwijk, and R. A. Sheldon. 1997. Selective oxidations catalyzed by peroxidases. Tetrahedron 53:13183-13220. [Google Scholar]
- 49.Vardar, G., K. Ryu, and T. K. Wood. 2005. Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for oxidizing nitrobenzene to 3-nitrocatechol, 4-nitrocatechol, and nitrohydroquinone. J. Biotechnol. 115:145-156. [DOI] [PubMed] [Google Scholar]
- 50.Vardar, G., and T. K. Wood. 2005. Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for enhanced chlorinated ethene degradation and o-xylene oxidation. Appl. Microbiol. Biotechnol. 68:510-517. [DOI] [PubMed] [Google Scholar]
- 51.Vardar, G., and T. K. Wood. 2004. Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for synthesizing 4-methylresorcinol, methylhydroquinone, and pyrogallol. Appl. Environ. Microbiol. 70:3253-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vargas, R. R., E. J. H. Bechara, L. Marzorati, and B. Wladislaw. 1999. Asymmetric sulfoxidation of a β-carbonyl sulfide series by chloroperoxidase. Tetrahedron Asymmetry 10:3219-3227. [Google Scholar]