Abstract
Lactobacillus sakei is extensively used as functional starter culture in fermented meat products. One of the safety criteria of a starter culture is the absence of potentially transferable antibiotic resistance determinants. However, tetracycline-resistant L. sakei strains have already been observed. In this paper, we show that tetracycline resistance in L. sakei Rits 9, a strain isolated from Italian Sola cheese made from raw milk, is mediated by a transposon-associated tet(M) gene coding for a ribosomal protection protein and a plasmid-carried tet(L) gene coding for a tetracycline efflux pump. pLS55, the 5-kb plasmid carrying the tet(L) gene, is highly similar to the pMA67 plasmid recently described for Paenibacillus larvae, a species pathogenic to honeybees. pLS55 could be transferred by electroporation into the laboratory strain L. sakei 23K. While the L. sakei 23K transformant containing pLS55 displayed an intermediate tetracycline resistance level (MIC, <32 μg/ml), L. sakei Rits 9, containing both tetracycline-resistant determinants, had a MIC of <256 μg/ml, suggesting that Tet L and Tet M confer different levels of resistance in L. sakei. Remarkably, in the absence of tetracycline, a basal expression of both genes was detected for L. sakei Rits 9. In addition, subinhibitory concentrations of tetracycline affected the expression patterns of tet(M) and tet(L) in different ways: the expression of tet(M) was induced only at high tetracycline concentrations, whereas the expression of tet(L) was up-regulated at lower concentrations. This is the first time that two different mechanisms conferring resistance to tetracycline are characterized for the same strain of a lactic acid bacterium.
Lactobacillus sakei is a facultative heterofermentative psychrotrophic lactic acid bacterium (LAB) that has been isolated from several raw fermented food products of plant and animal origin. It is found in kimchi, silage, cheese, sauerkraut, sourdough, and smoked fish but is mainly found in meat products (4, 7, 8). Though some L. sakei strains have been identified as responsible for the spoilage of vacuum-packaged meat products, this bacterium is widely used as a starter culture for the production of fermented sausages and has biotechnological potential for biopreservation and food safety (6). Lactobacilli are generally recognized as safe and they are not responsible for human infections in healthy people (46). However, they might act as reservoirs of transmissible antibiotic resistance genes that under certain conditions could be transferred to food or gut microbiota (27). In addition, the emergence of antibiotic-resistant food-borne pathogens originating from meat products (14) raises the question of the possibility of gene transfer between industrial bacterial species and food-borne pathogens. Therefore, a consensus criterion has been issued for which strains to be used in food systems should be free of potentially transferable antibiotic resistance traits (15).
Tetracyclines are a group of broad-spectrum antibiotics whose general usefulness has been reduced with the onset of bacterial resistance. Tetracycline resistance (Tcr) is the most frequent bacterial antibiotic resistance found in nature and is mostly acquired by horizontal gene transfer. Nowadays, 39 acquired tetracycline determinants are known for bacteria (37). Usually, these genes code for energy-dependent efflux systems or for proteins that protect the bacterial ribosomes from the blockage of protein synthesis (9, 10, 37). In rare cases, Tcr is mediated through direct inactivation of the antibiotic (40) or by mutations in the 16S rRNA that prevent the binding of tetracycline to the ribosome (38).
Currently, data on antibiotic resistance in lactobacilli are relatively scarce. However, in recent years a number of studies have correlated atypically high phenotypic resistances with the presence of tet genes (11, 17, 18, 19, 20, 26). Tetracycline resistance in Lactobacillus has commonly been associated with the presence of tet(M) (19, 20), but recently the gene coding for the efflux transporter Tet L was also described for some cloacal isolates (5). However, data about the functionality of both genes when they coexist in the same bacterium were not available until now. In this context, this study reports the isolation of a Tcr L. sakei strain from Italian Sola cheese and the molecular characterization of both ribosomal protection- and efflux pump-encoding genes, tet(M) and tet(L), responsible for Tcr in this strain.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. sakei Rits 9 was isolated on MRS agar (Oxoid Limited, Hampshire, United Kingdom) containing 16 μg/ml tetracycline (Sigma, St. Louis, MO) from an Italian Sola cheese made from raw cow's milk according to International Dairy Federation (IDF) standard 122C:1996. An internal 474-bp fragment of the 16S rRNA gene and an internal 424-bp fragment of the katA gene (encoding the L. sakei heme-dependent catalase) were amplified using primers Y1 (45) and R518 and primers 702-F and 310-R (2), respectively (Table 1). The resulting nucleotide sequences showed to be identical to the corresponding partial sequences in L. sakei 23K (6). L. sakei 23K, a laboratory strain originally isolated from sausage and cured of plasmids (3), was used as the recipient strain for genetic constructions. L. sakei 23K electrocompetent cells were prepared and transformed with pLS55 as described previously (3). After an incubation period of 2 h following electroporation, bacterial suspensions were plated on MRS medium containing 4, 8, 16, or 32 mg/liter tetracycline and incubated for 48 h at 30°C.
TABLE 1.
Primers used in this study
| Primer | R/Fa | Sequence (5′-3′) | Specificity | Amplicon size | Reference or source |
|---|---|---|---|---|---|
| Y1 | F | GACAAGGTGCGCCATATGT | 16S rRNA | 474 bp | 44 |
| R518 | R | GCTTATCTTTTGGTCTTTGG | This study | ||
| 702-F | F | AATTGCCTTCTTCCGTGTA | katA | 424 bp | 2 |
| 310-R | R | AGTTGCGCACAATTATTTTC | |||
| TetL-FW3 | F | GTMGTTGCGCGCTATATTCC | tet(L) | 0.7 kb | J. M. Collard, personal communication |
| TetL-RV3 | R | GTGAAMGRWAGCCCACCTAA | |||
| DI | F | GAYACNCCNGGNCAYRTNGAYTT | tet(M) | 1.5 kb | 9 |
| TetM-R | R | CACCGAGCAGGGATTTCTCCAC | |||
| tetM-revF | F | GTTACCACTGGCGAACCTG | tet(M) flanking regions | 2.5 kb | This study |
| tetM-revR | R | GTCCACGCTTCCTAATTCTG | |||
| tetM-tn | F | CTCGTCAAAATGAACGGACTAC | Tn916-like | 4.2 kb | This study |
| tetM-revR | R | GTCCACGCTTCCTAATTCTG | tet(M) flanking regions | ||
| TetL-FW-RT | F | TTTCCAGCACTCGTGATGGTT | tet(L) | 70 bp | This study |
| TetL-RV-RT | R | GACCAAACGCTTTACCCCTATTTT | |||
| TetM-FW-RT | F | AAATGGGCTTAGTGTTTGTTTAGCA | tet(M) | 78 bp | This study |
| TetM-RV-RT | R | CGACGGGTCTGGCAAACAG | |||
| F_alllact_IS | F | TGGATGCCTTGGCACTAGGA | Lactobacillus ITSb | 90 bp | 21 |
| R_alllact_IS | R | AAATCTCCGGATCAAAGCTTACTTAT |
R, reverse; F, forward.
ITS, internal transcribed spacer.
Bacterial strains were stored at −80°C and routinely cultured on MRS agar. All incubations were performed aerobically at 30°C for 48 h.
Determination of the MICs of tetracycline.
The MICs of tetracycline for the different strains were determined by microdilution. Briefly, colonies obtained after growth on solid media were picked up and incubated overnight at 30°C in LSM broth (29). The optical density at 625 nm (OD625) of the cultures was adjusted to 0.2 in LSM broth, and the suspension was diluted 500-fold in the same medium. One hundred microliters of this dilution was then transferred to 100 μl of LSM containing the appropriate amount of tetracycline in serial twofold dilutions, and the microtiter plates were incubated at 30°C for 24 h. The growth was recorded with a Benchmark plus microplate spectrophotometer (Bio-Rad, Hercules, CA). All the experiments were carried out in triplicate.
DNA and RNA techniques. (i) Nucleic acids extractions and labeling.
Genomic DNA was isolated using the GenElute bacterial genomic DNA kit (Sigma). Plasmid DNA was isolated using either the large-scale Qiagen kit (Qiagen Inc. Valencia, CA) or the procedure of O'Sullivan and Klaenhammer (33). Total RNA was extracted from cells grown up to an OD600 of about 1.6 by use of an RNeasy mini kit (Qiagen) following the manufacturer's instructions with the following modifications: the lysis buffer was supplemented with 30 μg/ml lysozyme (Sigma) and 100 U/ml mutanolysin (Sigma) and the samples were incubated for 30 min under gentle stirring. DNA was removed by on-column digestion using an RNase-free DNase set (Qiagen). Four microliters of RNA (about 3 μg) was reverse transcribed into cDNA by use of a cDNA archive kit (Applied Biosystems, Foster City, CA). The cDNA was stored at −80°C until use.
(ii) Microarray hybridization.
DNA microarrays contained 327 oligonucleotides (50 to 60 base pairs long), including control probes and oligonucleotides specific for 250 antibiotic resistance genes, including 28 tet genes (1). Spotting of the oligonucleotides, hybridization conditions, and analysis of the results were as previously described (43).
(iii) Real-time PCR conditions.
Real-time PCR was used to assess the influence of different subinhibitory concentrations of tetracycline (16, 32, and 64 μg/ml) on the expression levels of tet(L) and tet(M) in L. sakei Rits 9. All the primers used in this study are listed in Table 1. Primers TetL-FW-RT and TetL-RV-RT and TetM-FW-RT and TetM-RV-RT were designed to amplify internal fragments of 70 and 78 bp, respectively. The rRNA 16S-to-23S intergenic region was used as the endogenous control by using Lactobacillus-specific primers (24). PCR was performed in an ABI Prism 7500 fast real-time PCR system (Applied Biosystems), and SYBR green I fluorophore was used to correlate the amount of PCR product with the fluorescent signal. Amplification was carried out in a 25-μl final volume containing 1 μl of cDNA as a template, 200 nM of each primer, and 12.5 μl of SYBR green PCR master mix (Applied Biosystems). Thermal cycling consisted of an initial cycle of 95°C for 10 min followed by 35 cycles of 95°C for 15 s and 60°C for 1 min. The expression levels in the presence of antibiotic were refereed to those obtained for the control culture (absence of antibiotic). Two independent experiments were carried out and each sample was analyzed in duplicate in two independent PCR runs. Negative controls, including all the elements of the reaction mixture except the template cDNA, were also included.
(iv) Pulsed-field gel electrophoresis (PFGE) and Southern hybridization conditions.
The genetic location of tet(L) and tet(M) was assessed by hybridization using as probes 0.7- and 1.5-kb internal segments of the genes obtained by PCR and labeled with digoxigenin (Roche Applied Science, Basel, Switzerland). The tet(L) and tet(M) fragments were amplified using primer pairs TetL-FW3/TetL-RV3 and DI/TetM-R (9), respectively. Total and plasmid DNAs digested with the restriction enzymes EcoRI, HindIII, AscI, and PstI (Takara Bio Inc., Shiga, Japan) were hybridized using high-stringency standard conditions at 68°C.
For PFGE analysis, the strain was inoculated in 10 ml MRS supplemented with 20 mM dl-threonine and incubated at 30°C until the OD600 was 0.5 to 1.0 or above. The cells were harvested by centrifugation, washed in 10 ml 50 mM EDTA, and resuspended in 50 mM EDTA (300 μl × OD600). A 125-μl cell suspension was mixed gently with 750 μl 1% low-melting-point agarose (prepared in 50 mM EDTA). The cell-agarose suspension was pipetted into the Bio-Rad plug mold. The agarose plugs were incubated at 37°C overnight in a lysozyme solution (2 mg/ml lysozyme, 20 units/ml mutanolysin, 0.05% N-lauroyl sarcosine in 50 mM EDTA). The lysozyme solution was replaced by a sodium dodecyl sulfate-proteinase solution (10 mM Tris, pH 8.0, 1% sodium dodecyl sulfate, 2 mg/ml proteinase K in 0.5 M EDTA, pH 8.5) and incubated at 50°C overnight. The agarose plugs were washed six times for 30 min in 50 mM EDTA and stored at 4°C in 50 mM EDTA. Slices of 1 to 2 mm of the agarose plugs were incubated in 200 μl of restriction enzyme buffer for 1 to 4 h at 4°C. The buffer was replaced with 200 μl fresh restriction enzyme buffer, 2 μl acetylated bovine serum albumin (10 mg/ml stock), and 20 to 40 units of AscI. The agarose plugs were incubated for 30 to 45 min at 4°C and then at 37°C overnight. The samples were loaded on a 1.1% agarose gel prepared in 0.5× Tris-borate-EDTA buffer. The DNA fragments were resolved on a Bio-Rad contour-clamped homogeneous electric field mapper using a 24-h program with a linear ramp factor, an initial switch time of 2 s, and a final switch time of 30 s. The gel was stained in ethidium bromide and destained in 0.5× Tris-borate-EDTA buffer.
Southern blotting of PFGE gels was performed with DNA probes labeled with horseradish peroxidase with the ECL direct nucleic acid labeling kit (Amersham Biosciences, Buckinghamshire, United Kingdom) according to the manufacturer's instructions.
(v) Sequencing strategy for the tet genes and sequence analysis.
Plasmid DNA was sequenced after serial runs using the first-round primers, which consisted of the complementary sequences of TetL-FW3 and TetL-RV3 and then primers designed from the DNA sequence newly obtained. The plasmid was thereafter resequenced on the other strand in order to check for sequence accuracy.
For sequencing the tet(M) region, a pair of primers was designed from the tet(M) sequence of Staphylococcus aureus subsp. aureus Mu50 and served for the amplification of L. sakei Rits 9 tet(M). Primers tetM-revF and tetM-revR (Table 1) were used to amplify regions upstream and downstream of the tet(M) genes. The sequencing of the flanking regions of tet(M) was carried out using inverse PCR as described elsewhere (16). In short, total genomic DNA was digested with HindIII and self-ligated overnight. The ligated DNA was precipitated, centrifuged, dried, and resuspended in 100 μl Tris-EDTA prior to use as the template for PCR amplification. Purified PCR products were sequenced by cycle extension in an ABI 370 DNA sequencer (Applied Biosystems).
Phylogenetic analyses were performed on sequences available in the GenBank database, using the Treetop software (http://www.genebee.msu.su/services/phtree_full.html).
Nucleotide sequence accession numbers.
The nucleotide sequences described in this paper have been deposited in the GenBank database with the following accession numbers: for L. sakei Rits 9 plasmid pLS55, EF605268; and for L. sakei Rits 9 tet(M) and flanking regions, EF605269.
RESULTS
L. sakei Rits 9 possesses tet(L) and tet(M) resistance genes.
L. sakei Rits 9 was isolated from an Italian Sola cheese as spontaneously resistant to tetracycline. The presence of genes responsible for such resistance was searched by hybridization with DNA microarrays containing oligonucleotides characteristic of 28 known tetracycline resistance genes. The results showed the strain to harbor both tet(M) and tet(L). Hybridization signals were quite strong for both 50- and 60-mer oligonucleotides used for identifying the respective Tcr genes. Except for positive signals obtained with control probes targeting lactobacillus tuf genes, no other positive signals were found with any of the remaining spots, indicating the absence of other antibiotic resistance determinants. This shows either that other resistance genes are absent or that similar genes may be present but with a homology too low to get a hybridization signal (data not shown). To verify the presence of both genes, primers derived from known tet gene sequences were used. Amplification of internal fragments of tet(L) and tet(M) with the primers TetL-FW3 and TetL-RV3 and DI and TetM-R, respectively, resulted in amplicons of about 0.7 kb and 1.5 kb, confirming that L. sakei Rits 9 possesses both genes. L. sakei Rits 9 harbors one small plasmid of 5 kb, as revealed by a plasmid profile analysis using the O'Sullivan and Klaenhammer method (Fig. 1A), and at least one large plasmid, as revealed by PFGE (Fig. 1B). Southern blots showed tet(L) to be located on the 5-kb small plasmid (data not shown) and tet(M) on a large AscI PFGE chromosomal fragment (>450 kb) (Fig. 1C). In order to determine the involvement of those two genes in the resistance phenotype of L. sakei Rits 9, the 5-kb plasmid containing tet(L) was totally sequenced, as was the chromosomal region encompassing tet(M).
FIG. 1.
(A) Plasmid profile of L. sakei Rits 9 undigested (lane 1) and digested with PstI (lane 2). (B) PFGE analysis of total DNA from L. sakei Rits 9 undigested (lane 1) and digested with AscI (lane 2). (C) Southern blot analysis of the PFGE gel with the internal tet(M) probe. MWM, molecular weight marker.
The tet(L) gene is contained by a plasmid, and the tet(M) gene is flanked by transposon-like regions.
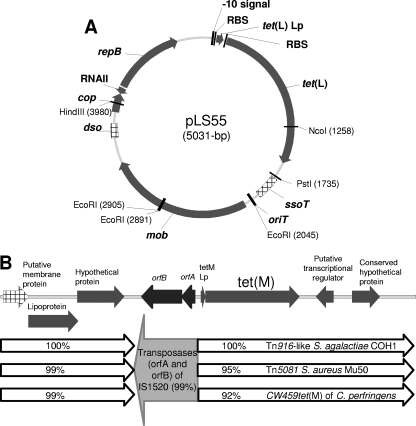
The 5-kb plasmid containing tet(L), named pLS55, was sequenced. It was found to be composed of 5,031 bp, consistent with its predicted size. The plasmid was almost 100% identical to pMA67, a plasmid recently described for the gram-positive bacterial pathogen of honeybees Paenibacillus larvae (32). Indeed, only seven of the base pairs were found to be different, four of them located in the tet(L) structural gene (positions 1, 287, 859, and 1197), and the plasmids differ in size by only one nucleotide (5,030 bp for pMA67). Remarkably, a different initiation codon was found for tet(L) in pLS55 (ATG instead of GTG), which could suggest a more efficient translation of the gene in L. sakei (30). The expression of tet(L) seems to depend on the synthesis of a 20-amino-acid leader peptide encoded 22 bp upstream of the tet(L) ribosome binding site, which is typical of inducible tet genes (25). A phylogenetic analysis performed on all complete tet(L) sequences available in the GenBank database showed that both P. larvae and L. sakei Rits 9 tet(L) genes are different from all previously described tet(L) genes and form an independent branch associated with a very strong bootstrap value (Fig. 2A).
FIG. 2.
Phylogenetic tree of homologs of the deduced Tet(L) and Tet(M) proteins (A and B, respectively). Protein accession numbers are given in brackets. Trees were constructed by the neighbor-joining algorithm and clustered by the unweighted-pair group method using average linkages, and bootstrap values (100 replicates) are given at the branch points. The distances refer to the percentages of different residues. Abbreviations: E. faecium, Enterococcus faecium; B. cereus, Bacillus cereus; M. haemolytica, Mannheimia haemolytica; A. pleuropneumoniae, Actinobacillus pleuropneumoniae; C. difficile, Clostridium difficile; E. coli, Escherichia coli; N. meningitidis, Neisseria meningitidis.
Apart from tet(L), pLS55 contains all the elements for replication control (12, 13, 22, 28) (Fig. 3A). Interestingly, the Rep protein is 80% identical to the Rep proteins of L. sakei plasmid pLS141-1 and of pLC2 identified for Lactobacillus curvatus, a lactobacillus species closely related to L. sakei (GenBank accession no. AB109041 and CAA78602, respectively). It can be deduced that pLS55 would likely be a mobilizable rolling-circle replication plasmid in the group II family (also called the pMV158 family).
FIG. 3.
Genetic structure of the tet(L)-containing plasmid pLS55 (A) and L. sakei Rits 9 tet(M) and flanking regions (B). Arrows show the direction of transcription of the open reading frames. Relevant restriction sites and their locations are indicated. The genes which matched the highest homology scores and the homologies with the partial sequences of different transposons are indicated.
The sequence of a region encompassing 8,524 bp around the tet(M) gene was obtained by several PCR and sequencing steps. The nucleotide sequence of the L. sakei Rits 9 tet(M) gene was shown to be identical to the one described for S. aureus subsp. aureus Mu50 and for Streptococcus agalactiae COH1 (Fig. 2B). The tet(M) gene was flanked downstream and upstream by regions with high similarity to the tet(M)-surrounding regions of several gram-positive bacteria (31, 36, 41), corresponding to transposon-like sequences (Fig. 3B). Upstream of tet(M), a 181-bp region mainly features a sequence corresponding to a 28-amino-acid leader peptide. Immediately upstream the leader peptide sequence, we found a 1,305-bp sequence that shares more than 99% identity with L. sakei IS1520, encompassing the transposase subunits A and B of an IS element present in five copies in the L. sakei 23K chromosome (6).
Regulation and expression levels of tet(M) and tet(L).
Real-time PCR was used to assess the influence of different subinhibitory concentrations of tetracycline (16, 32, and 64 μg/ml) on the expression levels of tet(M) and tet(L) in L. sakei Rits 9. Concentrations higher than 64 μg/ml affected the growth rate of the strain and therefore were not included in the study. A basal constitutive expression of both genes was observed independent of the presence of tetracycline. Remarkably, we noticed that tet(M) expression was gradually induced by exposure to increasing amounts of tetracycline. Indeed, tet(M) induction was about 13% increased at low tetracycline concentration (16 μg/ml) and up to 100% (relative induction was 2.095 ± 0.215) after exposure to 64 μg/ml compared to the control conditions (absence of antibiotic) (Fig. 4). On the contrary, the tet(L) gene was induced up to 2.74-fold at the lower tetracycline concentration, and its relative expression remained similar at higher tetracycline concentrations (between 2.74 ± 0.40- and 3.07 ± 0.44-fold increases) (Fig. 4).
FIG. 4.
Relative expression levels of tet(L) and tet(M) in L. sakei Rits 9 grown in the presence of different tetracycline concentrations refereed to those obtained for the control culture (absence of antibiotic).
pLS55 is able to replicate in L. sakei 23K.
To determine whether pLS55 replication is possible in another L. sakei strain, and to assess the functionality of the tet(L) gene, the transformation of the plasmid into L. sakei 23K was attempted, and transformants were plated with different tetracycline concentrations (4, 8, 16, and 32 μg/ml). When 4 μg/ml was used, a background was quite visible, but the background disappeared when 8-, 16-, and 32-μg/ml concentrations of tetracycline were used. Several transformants were obtained on plates with 8 and 16 μg/ml of tetracycline. No transformants were obtained at 32 μg/ml. Plasmid preparations of four clones confirmed the presence of a 5-kb plasmid in all of them. Then, one of them, named L. sakei 23K-TL, was selected to analyze its MIC to tetracycline in comparison with the control L. sakei 23K and L. sakei Rits 9. While the MIC of the Rits 9 strain was found to be <256 μg/ml and that of the 23K strain <1 μg/ml of tetracycline, the MIC of L. sakei 23K-TL was <32 μg/ml.
DISCUSSION
Tetracyclines have been extensively used in the prophylaxis and treatment of human and animal infections. Furthermore, they have been administered at subtherapeutic concentrations as growth promoters in animal feeds (34, 44). This intensive and extensive use has caused Tcr to spread to a large number of commensal bacteria (9, 37). In fact, different Tcr genes are present in the fecal microbiota of babies not previously exposed to the antibiotic (23). At present, there is great concern that animal and human commensal bacteria, such as LAB, could act as a reservoir for antibiotic resistance genes. These microorganisms may subsequently contaminate the raw milk and meat produced from these animals, and the foods prepared from those raw materials can therefore be considered as potential vehicles for the spread of antibiotic-resistant LAB along the food chain to the consumer (42). Resistances could ultimately be transferred to human pathogenic and opportunistic bacteria, hampering the treatment of infections (27).
Several Tcr LAB have been isolated from raw milk dairy products, e.g., Lactobacillus fermentum ROT1 (21) and Lactococcus lactis subsp. lactis K214 (34), and from raw meat-based fermented products, such as L. alimentarius, L. curvatus, L. plantarum, and L. sakei (19). The Tcr has been found to be mediated mainly by tet(M), which could be plasmid encoded and transferred through interspecies and intergenus conjugation mechanisms (17, 27). In this study, we show that L. sakei Rits 9, a Tcr strain isolated from a dairy product, harbors two Tcr genes, namely, the ribosomal protection tet(M) gene frequently encountered in lactobacilli and the efflux pump-encoding tet(L) gene. This combination of tet(L) and tet(M) genes is very frequently found for Streptococcus spp. and Enterococcus sp. strains (35, 39) and also for cloacal Lactobacillus salivarius subsp. salivarius isolates (5). However, to the best of our knowledge this is the first report on the coexistence of two genes encoding different mechanisms of Tcr in the same L. sakei strain.
The gene tet(L) was found to be associated with the plasmid pLS55, which is highly similar to pMA67, a plasmid described for the honeybee-pathogenic species P. larvae (32). As L. sakei and P. larvae are not known to share a common ecological niche, it is therefore plausible that such a plasmid has been horizontally transferred in these two hosts through different microorganisms. The presence of a Mob protein encoded by pLS55 and the 80% identity between the Rep protein of pLS55 and some Rep proteins described for other L. sakei or L. curvatus plasmids suggest that pLS55 can be transferred and stably maintained in L. sakei. Indeed, we could electroporate it in the plasmid-free laboratory strain L. sakei 23K, in which it autonomously replicated.
On the other hand, tet(M) was shown to be located on a transposon-like region. Upstream of tet(M), a fragment of 1,305 bp identical to L. sakei IS1520 was also present. This suggests that the acquisition of tet(M) by L. sakei Rits 9 occurred through an insertion event, although a more detailed study is necessary to corroborate this.
The high Tcr level in L. sakei Rits 9 and the absence of positive hybridization results other than the ones obtained with the tet(L) and tet(M) oligonucleotides in the microarray analysis suggest that Tcr in this strain is linked to the presence of one or both genes. In order to ascertain the functionalities of both genes and the partial contribution of each to the Tcr phenotype, we have transformed the plasmid-free laboratory strain L. sakei 23K with the tet(L)-containing plasmid pLS55. The resulting strain displayed an intermediate Tcr level compared with the Rits 9 strain, which displayed a much higher MIC. Thus, the higher Tcr level of L. sakei Rits 9 could be due to the presence of tet(M) or to a synergistic effect of both genes. Furthermore, these data indicate that both tet genes are functional in L. sakei, with tet(L) conferring a moderated resistance level, whereas tet(M) confers a high Tcr level to this bacteria. In relation to this, it has been shown that tet(L) and tet(M) can contribute differently to the Tcr phenotype depending on the Enterococcus or Streptococcus strain (39). It is also likely that the resistance level conferred by these two genes is species dependent and probably strain dependent.
Finally, expression studies were carried out to go more deeply into the functionality of tet(L) and tet(M) in L. sakei Rits 9. The fact that tet(M) expression was mainly induced at high Tcr levels, whereas tet(L) induction was achieved at lower concentrations, sheds some light onto the physiological function of both genes. These data indicate that, at a low tetracycline concentration, the activity of the efflux pump Tet L is enough for L. sakei Rits 9 to cope with antibiotic challenge; however, at concentrations higher than 16 μg/ml, the cells need an extra input, which is supplied by a higher amount of the ribosomal protection protein Tet M. These findings also support the previous results just discussed above, indicating that Tet M is responsible, to a larger extent than Tet L, for the high Tcr phenotype of L. sakei Rits 9.
In conclusion, the results of the current study indicate that Lactobacillus species from raw milk cheese can harbor acquired Tcr determinants associated with mobile elements, potentially enabling them to spread to other LAB or potentially pathogenic bacteria. We also demonstrated, for the first time, that two different Tcr mechanisms, active efflux and ribosomal protection, are functional when they are together in the same strain. Remarkably, our data suggest that the two genes are dedicated to cope with two different physiological conditions, low and high tetracycline concentrations. This functional complementarity of both mechanisms and their involvement in the physiology of L. sakei under tetracycline challenge will contribute to an understanding of how a bacterium makes use of different resistance determinants and of how they are engaged to fight against the deleterious action of antimicrobials.
Acknowledgments
Work on antibiotic resistance at our laboratories was supported by an EU project within the Sixth Framework Programme (ACE-ART, reference no. FP6-506214). M. S. Ammor was awarded a postdoctoral fellowship from the Secretaría de Estado de Universidades e Investigación of the Spanish Ministry of Education and Science (reference no. SB2004-0165). Miguel Gueimonde is the recipient of a Juan de la Cierva contract from the Spanish Ministry of Education and Science.
Footnotes
Published ahead of print on 11 January 2008.
REFERENCES
- 1.Ammor, M. S., A. B. Flórez, A. H. A. M. van Hoek, C. G. de los Reyes-Gavilán, H. J. M. Aarts, A. Margolles, and B. Mayo. 2008. Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and bifidobacteria. J. Mol. Microbiol. Biotechnol. 14:6-15. [DOI] [PubMed] [Google Scholar]
- 2.Ammor, M. S., E. Dufour, M. Zagorec, S. Chaillou, and I. Chevallier. 2005. Characterization and selection of Lactobacillus sake [sic] strains isolated from traditional dry sausage for their potential use as starter cultures. Food Microbiol. 22:529-538. [Google Scholar]
- 3.Berthier, F., M. Zagorec, M. Champomier-Vergès, S. D. Ehrlich, and F. Morel-Deville. 1996. Efficient transformation of Lactobacillus sakei by electroporation. Microbiology 142:1273-1279. [DOI] [PubMed] [Google Scholar]
- 4.Cappello, M. S., B. Laddomada, P. Poltronieri, and G. Zacheo. 2001. Characterisation of lab in typical Salento Pecorino cheese. Meded. Rijksuniv. Gent Fak. Landbouwkd. Toegep. Biol. Wet. 66:569-572. [PubMed] [Google Scholar]
- 5.Cauwerts, K., F. Pasmans, L. A. Devriese, F. Haesebrouck, and A. Decostere. 2006. Cloacal Lactobacillus isolates from broilers often display resistance toward tetracycline antibiotics. Microb. Drug Resist. 12:284-288. [DOI] [PubMed] [Google Scholar]
- 6.Chaillou, S., M. C. Champomier-Verges, M. Cornet, A. M. Crutz-Le Coq, A. M. Dudez, V. Martin, S. Beaufils, E. Darbon-Rongere, R. Bossy, V. Loux, and M. Zagorec. 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 23:1527-1533. [DOI] [PubMed] [Google Scholar]
- 7.Champomier-Vergès, M. C., S. Chaillou, M. Cornet, and M. Zagorec. 2002. Lactobacillus sakei: recent developments and future prospects. Res. Microbiol. 153:115-123. [DOI] [PubMed] [Google Scholar]
- 8.Choi, I. K., S. H. Jung, B. J. Kim, S. Y. Park, J. Kim, and H. U. Han. 2003. Novel Leuconostoc citreum starter culture system for the fermentation of kimchi, a fermented cabbage product. Antonie van Leeuwenhoek 84:247-253. [DOI] [PubMed] [Google Scholar]
- 9.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clermont, D., O. Chesneau, G. De Cespedes, and T. Horaud. 1997. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob. Agents Chemother. 41:112-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danielsen, M. 2002. Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid 48:98-103. [DOI] [PubMed] [Google Scholar]
- 12.del Solar, G., P. Acebo, and M. Espinosa. 1995. Replication control of plasmid pLS1: efficient regulation of plasmid copy number is exerted by the combined action of two plasmid components, CopG and RNA II. Mol. Microbiol. 18:913-924. [DOI] [PubMed] [Google Scholar]
- 13.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle, M. P., and M. C. Erickson. 2006. Emerging microbiological food safety issues related to meat. Meat Sci. 74:98-112. [DOI] [PubMed] [Google Scholar]
- 15.European Commission. 2001. Opinion of the Scientific Committee on Animal Nutrition on the criteria for assessing the safety of micro-organisms resistant to antibiotics of human clinical and veterinary importance. http://www.europa.eu.int/comm/food/fs/sc/scan/out64_en.pdf. Revised 18 April 2001.
- 16.Florez, A. B., M. S. Ammor, S. Delgado, and B. Mayo. 2006. Molecular analysis of a chromosomally encoded erm(B) gene and its flanking insertion points in Lactobacillus johnsonii G41. Antimicrob. Agents Chemother. 50:4189-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gevers, D., G. Huys, and J. Swings. 2003. In vitro conjugal transfer of tetracycline resistance from Lactobacillus isolates to other Gram-positive bacteria. FEMS Microbiol. Lett. 225:125-130. [DOI] [PubMed] [Google Scholar]
- 18.Gevers, D., G. Huys, F. Devlieghere, M. Uyttendaele, J. Debevere, and J. Swings. 2000. Isolation and identification of tetracycline resistant lactic acid bacteria from pre-packed sliced meat products. Syst. Appl. Microbiol. 23:279-284. [DOI] [PubMed] [Google Scholar]
- 19.Gevers, D., L. Masco, L. Baert, G. Huys, J. Debevere, and J. Swings. 2003. Prevalence and diversity of tetracycline resistant lactic acid bacteria and their tet genes along the process line of fermented dry sausages. Syst. Appl. Microbiol. 26:277-283. [DOI] [PubMed] [Google Scholar]
- 20.Gevers, D., M. Danielsen, G. Huys, and J. Swings. 2003. Molecular characterization of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl. Environ. Microbiol. 69:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gfeller, K. Y., M. Roth, L. Meile, and M. Teuber. 2003. Sequence and genetic organization of the 19.3-kb erythromycin- and dalfopristin-resistance plasmid pLME300 from Lactobacillus fermentum ROT1. Plasmid 50:190-201. [DOI] [PubMed] [Google Scholar]
- 22.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gueimonde, M., S. Salminen, and E. Isolauri. 2006. Presence of specific antibiotic (tet) resistance genes in infant faecal microbiota. FEMS Immunol. Med. Microbiol. 48:21-25. [DOI] [PubMed] [Google Scholar]
- 24.Haarman, M., and J. Knol. 2006. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 72:2359-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoshino, T., T. Ikeda, N. Tomizuka, and K. Furukawa. 1985. Nucleotide sequence of the tetracycline resistance gene of pTHT15, a thermophilic Bacillus plasmid: comparison with staphylococcal TcR controls. Gene 37:131-138. [DOI] [PubMed] [Google Scholar]
- 26.Huys, G., K. D'Haene, and J. Swings. 2006. Genetic basis of tetracycline and minocycline resistance in potentially probiotic Lactobacillus plantarum strain CCUG 43738. Antimicrob. Agents Chemother. 50:1550-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobsen, L., A. Wilcks, K. Hammer, G. Huys, D. Gevers, and S. R. Andersen. 2007. Horizontal transfer of tet(M) and erm(B) resistance plasmids from food strains of Lactobacillus plantarum to Enterococcus faecalis JH2-2 in the gastrointestinal tract of gnotobiotic rats. FEMS Microbiol. Ecol. 59:158-166. [DOI] [PubMed] [Google Scholar]
- 28.Khan, S. A. 1997. Rolling-circle replication of bacterial plasmids. Microbiol. Mol. Biol. Rev. 61:442-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klare, I., C. Konstabel, S. Müller-Bertling, R. Reissbrodt, G. Huys, M. Vancanneyt, J. Swings, H. Goossens, and W. Witte. 2005. Evaluation of new broth media for microdilution antibiotic susceptibility testing of lactobacilli, pediococci, lactococci, and bifidobacteria. Appl. Environ. Microbiol. 71:8982-8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak, M. 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361:13-37. [DOI] [PubMed] [Google Scholar]
- 31.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 32.Murray, K. D., K. A. Aronstein, and J. H. de León. 2007. Analysis of pMA67, a predicted rolling-circle replicating, mobilizable, tetracycline-resistance plasmid from the honey bee pathogen, Paenibacillus larvae. Plasmid 58:89-100. [DOI] [PubMed] [Google Scholar]
- 33.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 59:2730-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perreten, V., F. Schwarz, L. Cresta, M. Boeglin, G. Dasen, and M. Teuber. 1997. Antibiotic resistance spread in food. Nature 389:801-802. [DOI] [PubMed] [Google Scholar]
- 35.Petsaris, O., F. Miszczak, M. Gicquel-Bruneau, A. Perrin-Guyomard, F. Humbert, P. Sanders, and R. Leclercq. 2005. Combined antimicrobial resistance in Enterococcus faecium isolated from chickens. Appl. Environ. Microbiol. 71:2796-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts, A. P., P. A. Johanesen, D. Lyras, P. Mullany, and J. I. Rood. 2001. Comparison of Tn5397 from Clostridium difficile, Tn916 from Enterococcus faecalis and the CW459tet(M) element from Clostridium perfringens shows that they have similar conjugation regions but different insertion and excision modules. Microbiology 147:1243-1251. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, M. C. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245:195-203. [DOI] [PubMed] [Google Scholar]
- 38.Ross, J. I., E. A. Eady, J. H. Cove, and W. J. Cunliffe. 1998. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob. Agents Chemother. 42:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sapkota, A. R., K. K. Ojo, M. C. Roberts, and K. J. Schwab. 2006. Antibiotic resistance genes in multidrug-resistant Enterococcus spp. and Streptococcus spp. recovered from the indoor air of a large-scale swine-feeding operation. Lett. Appl. Microbiol. 43:534-540. [DOI] [PubMed] [Google Scholar]
- 40.Speer, B. S., L. Bedzyk, and A. A. Salyers. 1991. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J. Bacteriol. 173:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, Y. Margarit, I. Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. B. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ‘pan-genome. ’ Proc. Natl. Acad. Sci. USA 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teuber, M., L. Meile, and F. Schwarz. 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie van Leeuwenhoek 76:115-137. [PubMed] [Google Scholar]
- 43.van Hoek, A. H. A. M., I. M. Scholtens, A. Cloeckaert, and H. J. M. Aarts. 2005. Detection of antibiotic resistance genes in different Salmonella serovars by oligonucleotide microarray analysis. J. Microbiol. Methods 62:13-23. [DOI] [PubMed] [Google Scholar]
- 44.Wegener, H. C. 2003. Antibiotics in animal feed and their role in resistance development. Curr. Opin. Microbiol. 6:439-445. [DOI] [PubMed] [Google Scholar]
- 45.Young, J. P., H. L. Downer, and B. D. Eardly. 1991. Phylogeny of the phototrophic rhizobium strain BTAi1 by polymerase chain reaction-based sequencing of a 16S rRNA gene segment. J. Bacteriol. 173:2271-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Z'Graggen, W. J., H. Fankhauser, F. Lammer, T. Bregenzer, and D. Conen. 2005. Pancreatic necrosis infection due to Lactobacillus paracasei in an immunocompetent patient. Pancreatology 5:108-109. [DOI] [PubMed] [Google Scholar]