Abstract
A sensitive method for specific detection of viable Escherichia coli O157:H7 cells, including viable but nonculturable (VBNC) cells, in water samples was developed. This method involved capture of the bacterial cells on a low-protein-binding membrane and direct extraction and purification of RNA followed by reverse transcription-PCR and electronic microarray detection of the rfbE and fliC genes of E. coli O157:H7. It detected as few as 1 CFU of E. coli O157:H7 in diluted cultures, 3 to 4 CFU/liter in tap water, 7 CFU/liter in river water, and 50 VBNC cells in 1 liter of river water, demonstrating the best limit of detection reported to date for VBNC cells in environmental water samples.
Escherichia coli O157:H7 is a human pathogen of animal origin, and as few as 10 cells can cause serious human illness and even death (22, 32). Outbreaks due to E. coli O157:H7 contamination in water and food still occur (1, 14), making it important to monitor this pathogen in water and food.
E. coli O157:H7 can readily adapt to and survive under a wide range of environmental conditions, including changes in temperature, low pH, and desiccation (1, 16). This bacterium can live in laboratory media mimicking environmental conditions as well as in environmental samples, such as water, food, soil, and manure (41). Several outbreaks are attributed to the presence of E. coli O157:H7 in ground water, surface water, and private drinking water supplies (9, 20). Recent microbiological studies demonstrated that E. coli O157:H7 and other bacteria may become viable but nonculturable (VBNC) when exposed to sublethal stresses, such as changes in temperature, available nutrients, or pH or exposure to chlorine (17, 18, 25, 26, 29, 30). Under simple starvation conditions, E. coli O157:H7 can also enter into a VBNC state (37). However, expression of the toxin gene stx1 can still occur in the VBNC cells of E. coli O157:H7 (41), and three strains of this bacterium in the VBNC state can become culturable again in the presence of Oxyrase antioxidant, enterobacterial autoinducer, or sodium pyruvate (2, 33), suggesting the potential health concerns regarding the VBNC cells. Therefore, it is particularly important to monitor the VBNC cells of E. coli O157:H7 in drinking water and source water because of the potential of water distribution to transmit this pathogen.
A number of methods have been developed for detection of E. coli O157:H7, but they often do not differentiate viable cells from dead ones (7, 43). Specific detection of viable pathogens is important because only the viable cells have the potential to cause infections and, thus, pose a significant health risk. Culture-based methods are still widely used for positive identification of E. coli O157:H7 in a sample. However, these methods do not detect VBNC cells and may give false-negative results. They are also time-consuming and impractical for tracking outbreaks and routine monitoring. Immunoassays are based on antibody and antigen recognition. Antigens may be present in both viable and dead cells, and cross-reaction with matrices may result in false-positive results (43). PCR-based methods can sensitively and specifically detect E. coli O157:H7 but cannot distinguish viable from dead cells (7). A recombinant bacteriophage-based assay (3) using a combination of fluorescence intensity and nutrient uptake analysis can differentiate viable from dead bacteria. However, it is not capable of analyzing environmental samples because of the interference from auto-fluorescing or light-quenching particles in the samples.
Most mRNA species have a short half-life of only a few minutes; thus, mRNA is a better viability marker than DNA (19). Our studies and those of others demonstrate that the rfbE gene is a good viability marker for E. coli O157:H7 (23, 41). The rfbE gene-based reverse transcription-PCR (RT-PCR) method reported by Yaron and Matthews (41) has a detection limit of 106 CFU, which does not provide sufficient sensitivity for monitoring culturable and/or VBNC cells of E. coli O157:H7 in water. In practice, only a few cells may be present in a large volume of water, and so detection of the pathogenic bacteria is difficult.
Using mRNAs encoding the E. coli O157 lipopolysaccharide gene (rfbE) and the H7 flagellin gene (fliC) of E. coli O157:H7 as the markers, we have developed a combined RT-PCR and electronic microarray technique for specific detection of viable E. coli O157:H7 (23). Microarray-based bacterial detection with 16S rRNA, various functional genes, or whole genomic DNA have been reported for a wide range of bacteria (5, 10, 11, 13, 34, 38-40, 42, 44, 45) because of the high-throughput nature of the microarrays. Some microarray techniques have been shown to be useful for quantitative analysis of particular genes of interest (34, 42) as well as of whole microbial genomes (39, 40). A specific type of microarray, namely, the electronic microarray, uses an electric field to deliver DNA probes or targets to defined positions on the microarray chip (15), which enables concentration of the negatively charged DNA molecules on the chip, resulting in an improvement in detection sensitivity and specificity. For these reasons, an electronic microarray is used in the present study.
The objective of this study was to develop a method that could specifically detect a small number of culturable and VBNC E. coli O157:H7 cells in 1 liter of tap water as well as in river water. To achieve this, a set of procedures was developed for efficient cell capture and RNA extraction and purification from a small number of culturable bacterial cells in 1 liter of tap water or river water that contained complex matrices, including many organisms, compounds, and some debris. The captured target bacteria were detected using the rfbE and fliC genes as the markers with the combined RT-PCR and electronic DNA microarray method. This method was further demonstrated to be capable of detecting VBNC E. coli O157:H7 cells in 1 liter of the river water.
MATERIALS AND METHODS
Materials.
A sample of E. coli O157:H7 culture (strain ADRI V241) was provided by the Agri-Food Laboratories Branch, Alberta Agriculture and Food (Edmonton, Alberta, Canada). This strain, producing both Shiga toxin 1 (Stx1) and Stx2, was isolated from cattle.
The researchers involved in this study were certified for handling biosafety level 2 pathogens. Experiments were performed in a biosafety level 2 laboratory. The waste materials were autoclaved and disposed of according to the biosafety guidelines throughout the study.
TRIzol reagents for RNA extraction and PCR and RT-PCR reagents were obtained from Invitrogen (Carlsbad, CA). QIAquick PCR purification kits used for PCR product purification were purchased from Qiagen (Valencia, CA). Short DNA labeled with either biotin (captures) or fluorescent Cy3 (reporters) and PCR primers were custom synthesized by Integrated DNA Technologies (Coralville, IA). Low-protein-binding polyvinylidene difluoride hydrophilic membranes (HVLP) (pore size, 0.45 μm; diameter, 47 mm) were purchased from Millipore (Bedford, MA).
Microarray instrumentation.
A NanoChip molecular biology workstation (Nanogen, San Diego, CA) was used as the platform to facilitate microarray analysis. The NanoChip workstation consisted of a microchip cartridge, a loader, and a reader. The microchip was a semiconductor device with 100 test spots on an active surface with dimensions of 3 mm by 3 mm and was coated with an agarose permeation layer containing streptavidin. A Horiba twin compact B-173 conductivity meter (Horiba, Irvine, CA) was used to measure the conductivity of the 50 mM histidine buffer prior to each microarray analysis.
Bacterial cultures, RNA extraction, and RT-PCR.
E. coli O157:H7 cultures were grown in tryptic soy broth at 37°C with shaking (125 rpm). Bacterial growth was monitored by determining the optical density at 600 nm using a SmartSpec 3000 spectrophotometer (Bio-Rad, Hercules, CA). To count the number of bacterial cells, cultures were serially diluted with tryptic soy broth, and 100 μl of the chosen dilution was mixed with warm tryptic soy agar (TSA) and poured into plates in triplicate. Plates were incubated at 37°C for 24 h. For calculation of the numbers of CFU per milliliter, dilutions showing between 30 and 300 colonies on TSA were used. In parallel, the numbers of cells in cultures were determined by direct microscopic counting. The CFU and microscopic counts were in agreement.
Late-log-phase (optical density at 600 nm = 0.9) cells were collected by 10 min of centrifugation at 12,000 × g at 4°C using a Micromax RF centrifuge (Thermo IEC, Waltham, MA). The supernatant was removed, and the total RNA amount was extracted from the cell pellets by use of TRIzol reagent and following the manufacturer's specifications with the following modifications. The cells were mixed with 1 ml TRIzol reagent and incubated for 5 min at room temperature. For samples containing less than 104 cells, 5 to 10 μg of RNase-free glycogen was added into the mixture for coprecipitation of RNA. Then, chloroform (0.4 ml) was added and mixed. The rest of the extraction was done according to the manufacturer's instructions. The RNA extracts obtained were immediately frozen at −80°C prior to the RT-PCR assays.
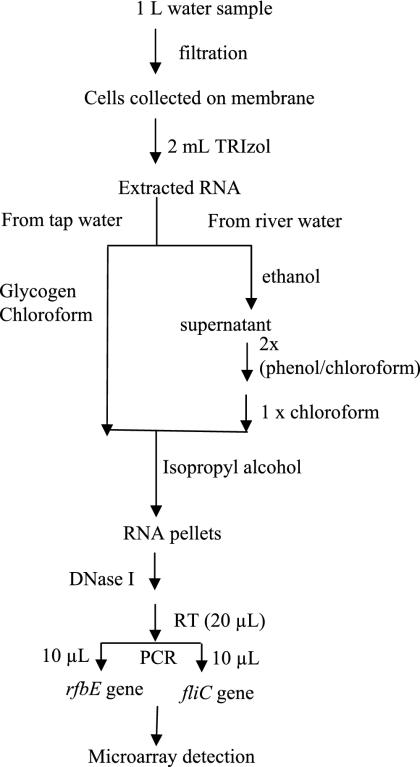
The RT-PCRs were performed under conditions that were previously described in detail (27). These procedures are briefly described in Fig. 1. PCR was carried out using a GeneAmp PCR 2700 system (Applied Biosystems, Foster City, CA), using an initial denaturation step of 3 min at 94°C, amplification for 50 cycles with denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s followed by a final extension at 72°C for 10 min.
FIG. 1.
Flow chart for concentration and RT-PCR electronic microarray detection of E. coli O157:H7 from spiked tap water and river water samples.
Electronic microarray detection.
The principle of electronic microarray detection has been described in detail elsewhere (23). Briefly, three major steps are involved in this technique: immobilization of captures to individual spots on the chip; hybridization with targets; and detection of reporter fluorescence. Specific captures were designed to be complementary to the targets of interest, and fluorescently labeled reporters were complementary to the targets and sat adjacent to the captures.
Capture and reporter sets for the E. coli O157:H7 rfbE and fliC genes were designed based on GenBank sequence data, including 17 available sequences of the rfbE gene for serotype O157 O antigen from 17 E. coli strains and 28 available sequences of the fliC gene for serotype H7 flagellin from 28 E. coli strains. For the rfbE gene, a 100% conserved fragment in all 17 available sequences was identified and used to design the capture and the reporter. Similarly, a 100% conserved fragment in all 28 available sequences was identified and used to design the capture and reporter sets for the fliC gene. The capture and reporter sets for the rfbE and fliC genes were tested to specifically detect E. coli O157:H7 and not other bacteria, as described in our previous study (23). The electronic microarray analyses were also performed according to procedures that were previously described (23).
Analysis of the fluorescence intensity for individual test sites was performed using built-in NanoChip configuration software. In each set of sample analyses, the average fluorescence intensity of three negative controls (50 mM histidine) was used as the background. The fluorescence intensity values from three spots addressed with the same capture and target set were analyzed for statistical agreement. The Q-test was used to reject values that were not in agreement with the others in the set. The signal-to-background (S/B) ratio represents the average of the fluorescence intensity values from three spots loaded with the same capture and target set relative to the background value. The threshold for a positive identification was an S/B ratio higher than 5 (23).
Preparation of 1-CFU samples.
To prepare representative samples containing 1 CFU of the bacterial cell, the following procedures were used. A 1-ml bacterial sample, which contained 20 CFU based on consistent plate counting and microscopic counting results, was divided into 10 aliquots, and the RNA was extracted separately from each aliquot. Due to Poisson distribution effects, each aliquot may not have contained the same frequency of 2 CFU. Therefore, the resulting 10 RNA samples were combined and then the RNA mixture was again divided into 10 aliquots. Each aliquot of the RNA sample was presumed to contain RNA equivalent to 2 CFU. This RNA sample was treated with DNase and used to generate cDNA. Then, the resulting cDNA was divided into two subparts: one for PCR-electronic microarray detection of the rfbE gene and the other for detection of the fliC gene. Therefore, the S/B ratio from microarray detection using the rfbE or fliC gene corresponded to 1 CFU. All the data were obtained from triplicate samples.
Generation of E. coli O157:H7 in a VBNC state.
E. coli O157:H7 cells in late log phase were harvested by centrifugation at 10,000 × g for 12 min and were washed three times with sterilized deionized water to prevent the carryover of nutrients upon inoculation of the microcosm. Cells were diluted with sterilized deionized water to a cell density of 106 CFU/ml and were maintained in three sterile screw-cap bottles at 25°C in the dark without shaking. Every 7 days, after being mixed well, samples were taken from each bottle to determine the total cell number, viability, and culturability. The total numbers of bacterial cells and the numbers of viable cells were determined using a Live/Dead BacLight bacterium viability kit (Molecular Probes Europe, Leiden, The Netherlands) as described in the supplier's instructions. This kit has been used to determine the numbers of total and viable cells of several other bacterial species (6, 28). It uses SYTO 9 stain and propidium iodide to measure membrane integrity to distinguish live cells (intact cell membrane) from dead cells (lacking an intact cell membrane). Samples stained with these reagents were filtered through 0.22-μm-pore-size polycarbonate black filters (Osmonics, Livermore, CA). An epifluorescence microscope (Zeiss Axioskop 2 FS) was used to view live and dead cells. A series of 20 images was taken from each sample in duplicate. The bacteria were then counted and calibrated to give total and live counts per milliliter.
To assess the culturability of E. coli O157:H7 cells, samples were taken once a week from the microcosms prepared as described above, inoculated on TSA plates in triplicate, and incubated for 48 h at 37°C before the CFU numbers were assessed. Upon prolonged incubation (5 days), no further colony development could be observed. To determine whether culturable cells persisted at the end of the incubation period, an aliquot (10 ml) of sample from the microcosm was filtered through a 0.22-μm-pore-size membrane filter (Millipore, Bedford, MA), and the filtrate was then placed on solid medium and observed for growth. When less than 0.1 CFU/ml of microcosm was culturable, the cells were considered to be in the nonculturable state.
Concentration of bacteria from tap water and river water samples.
To prepare tap water or river water samples containing different numbers of bacterial CFU or VBNC cells, the E. coli O157:H7 normal culture or VBNC cells were serially diluted and the appropriate number of CFU or VBNC cells was spiked in 1 liter of tap water (Edmonton, Alberta, Canada) or river water (North Saskatchewan River, Edmonton, Alberta, Canada) in triplicate. The numbers of cells in the spiked samples were determined by direct plate counting for culturable cells, and the numbers of the VBNC cells were determined using fluorescence microscopy with the Live/Dead kit.
To concentrate the bacterial cells, the spiked water samples were filtered using a low-protein-binding HVLP membrane (pore size, 0.45 μm; diameter, 47 mm). Controls consisting of 1 liter of unspiked tap water and river water samples were prepared in triplicate using the same experimental procedures. For the spiked and unspiked tap water samples, the total RNA was directly extracted from the cells captured on the membrane filters by use of 2 ml of TRIzol reagent per sample following the procedures described above.
For the spiked and unspiked river water samples, the procedures for the total RNA extraction were modified. The cells on the membrane filter were mixed with 2 ml of TRIzol reagent followed by 5 min of incubation at room temperature. An aliquot of 144 μl of 100% ethanol was slowly added into the mixture and mixed thoroughly, followed by a 30-min incubation at −20°C. Following centrifugation at 12,000 × g for 10 min at 4°C, the supernatant was transferred into a fresh tube. Then, 0.8 ml of chloroform was added, mixed, and centrifuged at 12,000 × g for 15 min at 4°C. The upper aqueous phase was collected and extracted with an equal volume of phenol-chloroform (1:1) and centrifuged at 12,000 × g for 15 min at 4°C. This step was repeated one more time followed by addition of an equal volume of chloroform to remove residual phenol by use of the same centrifugation conditions. The rest of the extraction was performed according to the TRIzol reagent manufacturer's instructions. The RNA samples obtained were used for the RT-PCR electronic microarray detection according to the procedure described above. The protocol for concentration and RT-PCR electronic microarray detection of E. coli O157:H7 from spiked tap water and river water samples is schematically shown in Fig. 1. Because all RT products were separated into two portions to separately detect the rfbE gene and fliC gene, each S/B ratio of the microarray detection corresponds to half of the initial volume of cells used.
Statistical analysis.
All experiments were conducted in triplicate, and each sample was analyzed in triplicate. Standard deviations of the means (SD) were calculated from the data of the triplicate samples.
RESULTS AND DISCUSSION
Detecting 1 CFU of E. coli O157:H7.
The minimum infectious dose of E. coli O157:H7 is as few as 10 cells (32). It is difficult to specifically detect a few viable cells of this pathogen due to the interference from dead cells and other materials. To address this limitation, a new method that combined highly efficient RNA extraction with highly sensitive detection by RT-PCR and the electronic microarray technique was developed and used in this study.
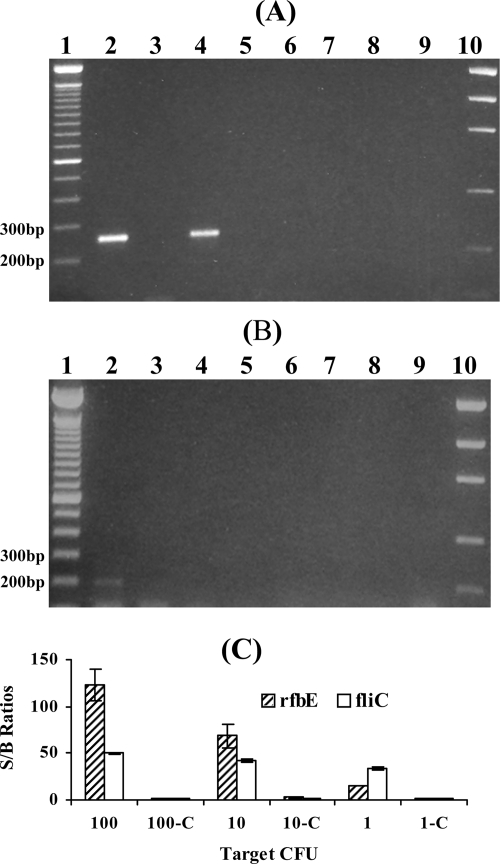
The samples containing 1, 10, and 100 CFU (equivalent to the numbers of the cells) were analyzed using this method. The numbers of cells in the samples were confirmed by microscopic counting and by plating the cultures in parallel experiments. The results obtained with the E. coli O157:H7 rfbE and fliC genes in these samples as detected by conventional gel electrophoresis are shown in Fig. 2A and B; the results as detected by the electronic DNA microarray technique are shown in Fig. 2C. With the gel electrophoresis assay, visible bands were detected from the samples containing 100 and 10 CFU for the rfbE gene (lanes 2 and 4 in Fig. 2A) but only from the samples containing 100 CFU for fliC (lane 2 in Fig. 2B). RT-PCR products from the samples containing 1 CFU of the rfbE or fliC genes were not detectable on the gel (lane 6, Fig. 2A, and lane 6, Fig. 2B). However, the electronic DNA microarray detection provided positive S/B ratios (i.e., values greater than 5) when the targets of the E. coli O157:H7 rfbE and fliC genes from the samples containing 1 CFU were used. A 106-fold improvement in the sensitivity of detection of culturable E. coli O157:H7 cells in the results for the rfbE gene and a 107-fold improvement in the results for the fliC gene were obtained compared to the results previously reported (41). These improvements were due to the efficient extraction and detection methods we developed here. Two steps in the extraction procedure were important for the successful detection seen with the results obtained using the samples containing 1 CFU. First, a volume of 400 μl (instead of 200 μl) of chloroform was used for RNA extraction to ensure complete denaturation of RNase and to improve the efficiency of mRNA extraction (35). Second, RNase-free glycogen was used to coprecipitate RNA so that the RNA pellet became visible, minimizing the loss of RNA. Detection of 1 CFU per reaction by use of DNA as a template by real-time PCR (4) and by use of antibodies as a target by immunoassay (43) was reported. However, these methods did not differentiate viable cells from dead ones. The results of the present study, together with previous results (23), demonstrate that the RT-PCR/electronic microarray method could specifically detect as few as 1 CFU of viable E. coli O157:H7 (as distinguished from dead cells) without cultural enrichment. Although quantification of this method is limited due to PCR bias in template-to-product ratios (15), the detection of a low number of E. coli O157:H7 is useful for surveillance of pathogen contamination in water or food.
FIG. 2.
(A and B) Agarose gel electrophoresis analysis of the E. coli O157:H7 samples for the rfbE gene (A) and fliC gene (B). Lanes: 1, 100-bp DNA ladder; 2, RT-PCR product from sample containing 100 CFU; 3, DNase I control from 100 CFU; 4, RT-PCR product from 10 CFU; 5, DNase I control from 10 CFU; 6, RT-PCR product from 1 CFU; 7, DNase I control from 1 CFU; 8, RT negative control; 9, PCR negative control; 10, low-mass DNA marker. (C) Electronic microarray analysis of dilutions of E. coli O157:H7 cultures for the rfbE and fliC genes. Electronic microarray S/B ratios obtained from the E. coli O157:H7 rfbE and fliC targets amplified from 100, 10, and 1 CFU were positive. The S/B ratios from 100-C, 10-C, and 1-C (DNase I control samples) were negative. The error bars of S/B ratios represent one SD of the S/B ratio from triplicate samples.
Analysis of E. coli O157:H7 in tap water.
One important transmission route for E. coli O157:H7 outbreaks is through drinking water (9, 20). Tap water is generally characterized as being highly oligotrophic and is oxidatively stressful to bacterial cells, and the bacteria can be induced to enter a VBNC state (30). In practice, it is likely that a few culturable and/or VBNC cells can be present in a large volume of water. Routine culture-based methods do not detect very small numbers of bacterial cells in the VBNC state. The challenge is to capture the few viable cells, including VBNC cells, in a large volume of water. In this study, we first developed a method to capture a small number of bacterial cells from 1 liter of tap water.
Conventional concentration methods, including centrifugation and filtration, were initially tested; however, several problems were encountered. It was not practical to use centrifugation for the concentration of a few bacterial cells from a large-volume water sample, as others have reported (36). Filtration is feasible for handling a large volume of water. With currently available filtration methods using electropositively charged filters, bacterial cells could be efficiently captured due to a net negative charge on the surface of the bacteria. The problem was the poor recovery of the cells from the filters due to strong cell attachment to the filter (36). To overcome this problem, a capture method combining cell capture and RNA extraction into one step using zero-charged and low-protein-binding membrane filters (HVLP pore size, 0.45 μm) was developed here. A low background of extractable components and low adsorption of nucleic acids were obtained with the zero-charged HVLP filters, which was consistent with a previous study (31). The pore size of this membrane, suitable for trapping E. coli O157:H7, was also one of the reasons for its use in this method. The results from the detection of 3 to 4, 7, and 70 CFU of E. coli O157:H7 in 1-liter tap water samples are summarized in Table 1 and support the hypothesis that a small number of bacterial cells were captured from 1-liter tap water samples by use of the HVLP membranes and that the cells captured on the membranes were efficiently extracted for RNA by use of TRIzol reagent. The combination of this one-step capture-extraction method with the RT-PCR electronic DNA microarray detection technique was demonstrated to be capable of detecting 3 to 4 CFU of E. coli O157:H7 in 1 liter of tap water without cultural enrichment. Compared to the detection limits of 1,000 CFU/liter in drinking water (with DNA as a target) after 21 h of culture enrichment (7) and of 105 CFU/liter (based on antibody determinations) in tap water (12), our method represents a substantial improvement for specific detection of viable E. coli O157:H7 bacteria. This method needs only minimum sample preparation and does not require culture enrichment.
TABLE 1.
Signal-to-background (S/B) ratios from the electronic microarray analysis of spiked tap and river water samples for the rfbE and fliC genes
| Sample category | No. of bacterial cells spiked (CFU) | S/B ratio (SD)a
|
|||
|---|---|---|---|---|---|
|
rfbE gene
|
fliC gene
|
||||
| Spiked sample | DNase I control | Spiked sample | DNase I control | ||
| Tap water | 70 | 22 (0.6) | 1.8 (0.1) | 34 (2) | 1.7 (0.2) |
| 7 | 20 (2) | 1.8 (0.3) | 29 (5) | 1.7 (0.3) | |
| 3-4 | 12 (1) | 1.7 (0.2) | 13 (1) | 0.7 (0.5) | |
| Negative controlb | 2.1 (0.2) | 2.0 (0.3) | |||
| River water | 70 | 12 (0.4) | 1.8 (0.1) | 33 (4) | 1.3 (0.1) |
| 7 | 7.2 (0.6) | 1.5 (0.1) | 13 (0.2) | 1.9 (0.2) | |
| Negative controlc | 1.4 (0.03) | 2.3 (0.3) | |||
An S/B ratio of >5 was the basis for positive identification; SD values are presented in parentheses.
Tap water without spiking with E. coli O157:H7.
River water without spiking with E. coli O157:H7.
Analysis of E. coli O157:H7 in river water.
Having established the method for detecting a few CFU of this bacterium in 1 liter of tap water, the following experiments were carried out to detect small numbers of CFU of E. coli O157:H7 in complex environmental water samples. A set of river water samples (1 liter each) from the North Saskatchewan River was spiked with different numbers of E. coli O157:H7 bacteria. Unlike the tap water, which contained a relatively low amount of organic matter, the river water from the North Saskatchewan River contained a high content of complex matrices, for example, microorganisms, algae, organic matter, and other components. When the TRIzol reagent was used to extract RNA from bacteria in the river water samples according to the manufacturer's instructions, a brown pellet was obtained, indicating interference due to polysaccharides and other matrices. The subsequent RT-PCR amplification was not successful; thus, no bacteria were detected in the spiked samples. This was suspected to have been due to inhibition of enzymatic PCRs by organic components in the sample; therefore, we first attempted to improve the results by removing these inhibitory compounds. The RNA extraction procedures were modified. A low concentration of ethanol was used to precipitate inhibiting components (possibly polysaccharides) while leaving the RNA in the supernatant (21). The supernatant was extracted with phenol-chloroform (1:1) twice followed by chloroform alone once to remove residual polysaccharides and other inhibitory compounds. After these modified procedures were used, a white RNA pellet was obtained, indicating the removal of a major portion of interference matrices. By use of the one-step cell capture and modified RNA extraction method combined with the RT-PCR electronic DNA microarray detection, E. coli O157:H7 bacteria (7 to 70 CFU/liter) in the river water samples were successfully detected, as demonstrated by the S/B ratio results reported in Table 1. When the S/B ratio was larger than 5, a positive identification was assigned for the sample. Use of the ethanol precipitation of polysaccharides and of the phenol-chloroform extraction procedures described above was required for detection of as few as 7 CFU/liter of E. coli O157:H7 bacteria in highly turbid river water.
This detection limit for E. coli O157:H7 bacteria in river water samples represents an improvement of several orders of magnitude over those of previous methods for water analysis (7, 8, 12). Direct PCR analysis of DNA was able to detect 2 × 105 CFU/liter of E. coli O157:H7 bacteria in ground water (8). As few as 6,000 CFU/liter in surface water could be detected using 21 h of enrichment and real-time PCR (with DNA as a target) (7). A sandwich enzyme-linked immunosorbent assay was able to detect 7.5 × 105 CFU/liter in lake water (12).
Detection of VBNC E. coli O157:H7 bacteria in river water.
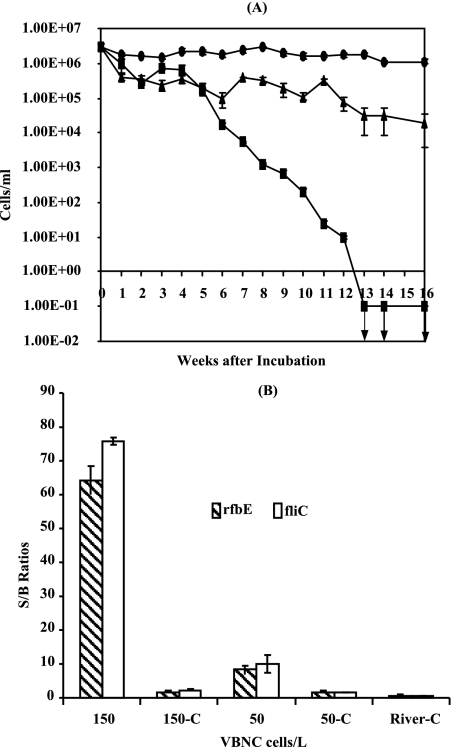
The capability of our method for detection of VBNC cells was further evaluated because conventional methods cannot detect VBNC cells (30). The VBNC cells of E. coli O157:H7 were generated through starvation in deionized water by following the procedures described above. The VBNC state of E. coli O157:H7 was confirmed by both culture plating and viability staining experiments in parallel. The results of determinations of the numbers of the total counts of the bacterial cells, culturable cells, and viable but nonculturable cells are shown in Fig. 3A. After 13 weeks of incubation under starvation conditions, the E. coli O157:H7 cells were not detected on the culture plates (<0.1 CFU/ml), and viable cell numbers were determined by the staining method at 3 × 104 cells. The numbers of VBNC cells were determined to account for 1% of the initial cell volume (3 × 106), and the rest of the cells were dead.
FIG. 3.
(A) Induction of VBNC E. coli O157:H7 cells in deionized water at room temperature. Culturability was assessed by agar plate counting (in CFU per milliliter) (▪); the numbers of total cells (•) and of viable cells (▴) were determined by the Live/Dead method. Error bars represent SD. Downward-pointing arrows indicate culturability results below the level of detection. (B) Electronic microarray analysis of E. coli O157:H7 cells in a VBNC state spiked in 1 liter river water for the rfbE and fliC genes. The columns represent the fluorescence S/B ratios obtained with the electronic microarray chip. The S/B ratios were obtained from the E. coli O157:H7 rfbE and fliC targets amplified from the reverse transcription products obtained with 150 and 50 VBNC cells, and the ratios were positive; the S/B ratios obtained with the DNase I controls (150-C and 50-C) for each sample as described above (and with the river water control [River-C]) were negative. Error bars represent one SD of the S/B ratios from triplicate samples.
These VBNC cells were spiked into 1 liter of the river water. A set of spiked water samples containing 300 VBNC cells and 100 VBNC cells were prepared through serial dilution in triplicate. The numbers of the VBNC cells were determined by staining and microscopic counting in parallel. The cells were concentrated onto a low-protein-binding membrane, and their RNA was extracted using the new procedures for the river water analysis. When the rfbE and fliC genes were used as viability and confirmation markers, as few as 50 VBNC cells of E. coli O157:H7 in 1 liter of river water were detected using the RT-PCR microelectronic array technique; the results are shown in Fig. 3B. Such detection sensitivity for VBNC cells in river water containing complex matrices has never been demonstrated before. Triplicate river water control samples were analyzed using the same preparation procedures and detection method. No viable cells, including VBNC cells, were detected in the control samples.
A 105-fold improvement in the sensitivity of detection of VBNC E. coli O157:H7 was demonstrated using our method compared to that previously reported (41). This is the first report demonstrating the detection of as few as 50 VBNC E. coli O157:H7 cells in 1 liter of river water. In addition, our results also confirmed the existence of the rfbE and fliC genes in the VBNC cells, in agreement with previous findings (24, 41). The electronic DNA microarray detection used here allows for concentration of RT-PCR products on the chip for high sensitivity and parallel analysis of up to 100 samples per chip. In conclusion, a set of new procedures have been demonstrated for rapid and sensitive capture and detection of a few culturable or VBNC cells of E. coli O157:H7 in 1 liter of tap water or river water, and our method could be suitable for monitoring of viable E. coli O157:H7 cells, including VBNC cells, in environmental water. This technique contributes to the development of microarray technology for applications in microbial ecology and environmental research (11).
Acknowledgments
This study was supported financially by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) and Alberta Health and Wellness, an Alberta Heritage Foundation for Medical Research (AHFMR) summer studentship (A.G.), and an NSERC University Faculty Award (X.-F.L.).
We thank Carol Goertz of the Agri-Food Laboratories Branch, Alberta Agriculture and Food, Canada, for supplying the bacterial strain used in this study. The help of Jessica Boyd, Katerina Carastathis, Christina Fung, and Elizabeth Hrudey in the preparation of this paper is also acknowledged.
Footnotes
Published ahead of print on 18 January 2008.
REFERENCES
- 1.Armstrong, G. L., J. Hollingsworth, and J. G. Morris. 1996. Emerging foodborne pathogens: E. coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 18:29-51. [DOI] [PubMed] [Google Scholar]
- 2.Asakura, H., S. Igimi, K. Kawamoto, S. Yamamoto, and S. Makino. 2005. Role of in vivo passage on the environmental adaption of enterohemorrhagic Escherichia coli O157:H7: cross-induction of the viable but nonculturable state by osmotic and oxidative stresses. FEMS Microbiol. Lett. 253:243-249. [DOI] [PubMed] [Google Scholar]
- 3.Awais, R., H. Fukudomi, K. Miyanaga, H. Unno, and Y. Tanji. 2006. A recombinant bacteriophage-based assay for the discriminative detection of culturable and viable but nonculturable Escherichia coli O157:H7. Biotechnol. Prog. 22:853-859. [DOI] [PubMed] [Google Scholar]
- 4.Barak, J. D., K. Sananikone, and M. J. Delwiche. 2005. Comparison of primers for the detection of pathogenic Escherichia coli using real-time PCR. Lett. Appl. Microbiol. 41:112-118. [DOI] [PubMed] [Google Scholar]
- 5.Bekal, S., R. Brousseau, L. Masson, G. Profontaine, J. Fairbrother, and J. Harel. 2003. Rapid identification of Escherichia coli pathotypes by virulence gene detection with DNA microarrays. J. Clin. Microbiol. 41:2113-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulos, L., M. Prevost, B. Barbeau, J. Coallier, and R. Desjardins. 1999. LIVE/DEAD BacLight: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J. Microbiol. Methods 37:77-86. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, G. R., J. I. Prosser, L. A. Glover, and K. Killham. 2001. Detection of Escherichia coli O157:H7 in soil and water using multiplex PCR. J. Appl. Microbiol. 91:1-7. [DOI] [PubMed] [Google Scholar]
- 8.Fode-Vaughan, K. A., J. S. Maki, J. A. Benson, and M. L. P. Collins. 2003. Direct PCR detection of Escherichia coli O157:H7. Lett. Appl. Microbiol. 37:239-243. [DOI] [PubMed] [Google Scholar]
- 9.Gagliardi, J. V., and J. S. Karns. 2000. Leaching of Escherichia coli O157:H7 in diverse soils under various agricultural management practices. Appl. Environ. Microbiol. 66:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, H., Z. K. Yang, T. J. Gentry, L. Wu, C. W. Schadt, and J. Zhou. 2007. Microarray-based analysis of microbial community RNAs by whole-community RNA amplification. Appl. Environ. Microbiol. 73:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentry, T. J., G. S. Wickham, C. W. Schadt, Z. He, and J. Zhou. 2006. Microarray applications in microbial ecology research. Microb. Ecol. 52:159-175. [DOI] [PubMed] [Google Scholar]
- 12.Guttikonda, S., X. L. Tang, B. M. Yang, G. D. Armstrong, and M. R. Suresh. 2007. Monospecific and bispecific antibodies against E. coli O157 for diagnostics. J. Immunol. Methods 327:1-9. [DOI] [PubMed] [Google Scholar]
- 13.He, Z., T. J. Gentry, C. W. Schadt, L. Wu, J. Liebich, S. C. Chong, Z. Huang, W. Wu, B. Gu, P. Jardine, C. Criddle, and J. Zhou. 2007. Geochip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1:67-77. [DOI] [PubMed] [Google Scholar]
- 14.Hrudey, S. E., and E. J. Hrudey. 2004. Safe drinking water: lessons from recent outbreaks in affluent nations. IWA Publishing, London, United Kingdom.
- 15.Huang, Y., J. Shirajian, A. Schroder, Z. Yao, T. Summers, D. Hodko, and R. Sosnowski. 2004. Multiple sample amplification and genotyping integrated on a single electronic microarray. Electrophoresis 25:3106-3116. [DOI] [PubMed] [Google Scholar]
- 16.Iijima, Y., M. Matsumoto, K. Higuchi, T. Furuta, and T. Honda. 1996. Resistance to dryness of Escherichia coli O157:H7 strains from outbreak in Saki City, Japan. Emerg. Infect. Dis. 4:340-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juhna, T., D. Birzniece, and J. Rubulis. 2007. Effect of phosphorus on survival of Escherichia coli in drinking water biofilms. Appl. Environ. Microbiol. 73:3755-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolling, G. L., and K. R. Matthews. 2001. Examination of recovery in vitro and in vivo of nonculturable Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:3928-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kushner, S. R. 1996. mRNA decay, p. 849-860. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 20.Lee, S. H., D. A. Levy, G. F. Craun, M. J. Beach, and R. L. Calderon. 2002. Surveillance for waterborne-disease outbreaks—United States, 1999-2000. MMWR Surveill. Summ. 51:1-47. [PubMed] [Google Scholar]
- 21.Lewinsohn, E., C. L. Steele, and R. Croteau. 1994. Simple isolation of functional RNA from woody stems of gymnosperms. Plant Mol. Biol. Rep. 12:20-25. [Google Scholar]
- 22.Li, J., and C. J. Hovde. 2007. Expression profiles of bovine genes in the rectoanal junction mucosa during colonization with Escherichia coli O157:H7. Appl. Environ. Microbiol. 73:2380-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, Y., Z. Gong, N. Morin, O. Pui, M. Cheung, H. Zhang, and X. F. Li. 2006. Electronic deoxyribonucleic acid (DNA) microarray detection of viable pathogenic Escherichia coli, Vibrio cholerae, and Salmonella Typhi. Anal. Chim. Acta 578:75-81. [DOI] [PubMed] [Google Scholar]
- 24.Mcdougald, D., S. A. Rice, D. Weichart, and S. Kjelleberg. 1998. Nonculturablity: adaptation or debilitation? FEMS Microbiol. Ecol. 25:1-9. [Google Scholar]
- 25.McKay, A. M. 1992. Viable but non-culturable forms of potentially pathogenic bacteria in water. Lett. Appl. Microbiol. 14:29-135. [Google Scholar]
- 26.Mizunoe, Y., S. N. Wai, A. Takade, and S. Yoshida. 1999. Restoration of culturability of starvation-stressed and low-temperature-stressed Escherichia coli O157 cells by using H2O2-degrading compounds. Arch. Microbiol. 172:63-67. [DOI] [PubMed] [Google Scholar]
- 27.Morin, N., Z. Gong, and X. F. Li. 2004. Reverse transcription-multiplex PCR assay for simultaneous detection of Escherichia coli O157:H7, Vibrio cholerae O1, and Salmonella Typhi. Clin. Chem. 50:2037-2044. [DOI] [PubMed] [Google Scholar]
- 28.Nilsson, L., J. D. Oliver, and S. Kjelleberg. 1991. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. J. Bacteriol. 173:5054-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliver, J. D. 2000. Public health significance of viable but nonculturable bacteria, p. 277-300. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, DC.
- 30.Oliver, J. D. 2005. The viable but non-culturable state in bacteria. J. Microbiol. 43:93-100. [PubMed] [Google Scholar]
- 31.Oyofo, B. A., and D. M. Rollins. 1993. Efficacy of filter types for detecting Campylobacter jejuni and Campylobacter coli in environmental water samples by polymerase chain reaction. Appl. Environ. Microbiol. 59:4090-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips, C. A. 1999. The epidemiology, detection and control of Escherichia coli O157. J. Sci. Food Agric. 79:1367-1381. [Google Scholar]
- 33.Reissbrodt, R., I. Rienaecker, J. M. Romanova, P. P. E. Freestone, R. D. Haigh, M. Lyte, H. Tschape, and P. H. Williams. 2002. Resuscitation of Salmonella enterica serovar Typhimurium and enterohemorrhagic Escherichia coli from the viable but nonculturable state by heat-stable enterobacterial autoinducer. Appl. Environ. Microbiol. 68:4788-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee, S.-K., X. Liu, L. Wu, S. C. Chong, X. Wan, and J. Zhou. 2004. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., p. A8.9. Cold Spring Harbor Laboratory Press, New York, NY.
- 36.Stevens, K. A., and L. A. Jaykus. 2004. Bacterial separation and concentration from complex sample matrices: a review. Crit. Rev. Microbiol. 30:7-24. [DOI] [PubMed] [Google Scholar]
- 37.Wang, G., and M. P. Doyle. 1998. Survival of enterohemorrhagic Escherichia coli O157:H7 in water. J. Food Prot. 61:662-667. [DOI] [PubMed] [Google Scholar]
- 38.Wu, C. F., J. J. Valdes, W. E. Bentley, and J. W. Sekowski. 2003. DNA microarray for discrimination between pathogenic O157:H7 EDL933 and non-pathogenic Escherichia coli strains. Biosens. Bioelectron. 19:1-8. [DOI] [PubMed] [Google Scholar]
- 39.Wu, L., D. K. Thompson, X. Liu, M. W. Fields, C. E. Bagwell, J. M. Tiedje, and J. Zhou. 2004. Development and evaluation of microarray-based whole genome hybridization for detection of microorganisms within the context of environmental applications. Environ. Sci. Technol. 38:6775-6782. [DOI] [PubMed] [Google Scholar]
- 40.Wu, L., X. Liu, C. W. Schadt, and J. Zhou. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl. Environ. Microbiol. 72:4931-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yaron, S., and K. R. Matthews. 2002. A reverse transcriptase-polymerase chain reaction assay for detection of viable Escherichia coli O157:H7: investigation of specific target genes. J. Appl. Microbiol. 92:633-640. [DOI] [PubMed] [Google Scholar]
- 42.Yergeau, E., S. Kang, Z. He, J. Zhou, and G. A. Kowalchuk. 2007. Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect. ISME J. 1:63-179. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, X., L. R. Hilliard, S. J. Mechery, Y. Wang, R. P. Bagwe, and S. Jin. 2004. A rapid bioassay for single bacterial cell quantitation using bioconjugated nanoparticles. Proc. Natl. Acad. Sci. USA 101:15027-15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, J., and D. K. Thompson. 2004. Microarray technology and applications in environmental microbiology. Adv. Agron. 82:183-270. [Google Scholar]