Abstract
Spatial heterogeneity in physical, chemical, and biological properties of soils allows for the proliferation of diverse microbial communities. Factors influencing the structuring of microbial communities, including availability of nutrients and water, pH, and soil texture, can vary considerably with soil depth and within soil aggregates. Here we investigated changes in the microbial and functional communities within soil aggregates obtained along a soil profile spanning the surface, vadose zone, and saturated soil environments. The composition and diversity of microbial communities and specific functional groups involved in key pathways in the geochemical cycling of nitrogen, Fe, and sulfur were characterized using a coupled approach involving cultivation-independent analysis of both 16S rRNA (bacterial and archaeal) and functional genes (amoA and dsrAB) as well as cultivation-based analysis of Fe(III)-reducing organisms. Here we found that the microbial communities and putative ammonia-oxidizing and Fe(III)-reducing communities varied greatly along the soil profile, likely reflecting differences in carbon availability, water content, and pH. In particular, the Crenarchaeota 16S rRNA sequences are largely unique to each horizon, sharing a distribution and diversity similar to those of the putative (amoA-based) ammonia-oxidizing archaeal community. Anaerobic microenvironments within soil aggregates also appear to allow for both anaerobic- and aerobic-based metabolisms, further highlighting the complexity and spatial heterogeneity impacting microbial community structure and metabolic potential within soils.
Owing to their immense physical, chemical, and biological heterogeneity, soils are considered the most microbially diverse environments on earth (15). For instance, 1 gram of soil can contain up to 109 microbial cells, representing more than 10,000 genomes (82). While the importance of Bacteria in soil ecosystem function has long been recognized, it has only recently become evident that Archaea, and Crenarchaeota in particular, are also ubiquitous and abundant organisms within soils (10, 36, 39, 58). Although little is known regarding the metabolic potential of Crenarchaeota, they have been directly linked to chemoautotrophic nitrification (43) and speculated to be involved in carbon metabolism (39) and amino acid uptake/assimilation (59, 81).
The abundance, composition, and diversity of microbial communities within soils are strongly depth dependent. For instance, the bacterial biomass (23), concentration of bacterial 16S rRNA genes (39, 92), numbers of terminal restriction fragment length polymorphism peaks (44) and denaturing gradient gel electrophoresis bands (1) (representative of richness), and proportion of gram-negative to gram-positive bacteria (23) are lower in subsurface than in surface soils. Similarly, declines in archaeal 16S rRNA genes (39) and pronounced archaeal community shifts, including a decrease in the proportion of euryarchaeal to crenarchaeal ribosomal DNA sequences, have been observed along depth gradients within forest soils (61). Interestingly, while numbers of Bacteria were consistently higher than those of Archaea, the ratio of archaeal 16S rRNA genes to the total of both Bacteria and Archaea 16S rRNA genes increased with soil depth from 12 to 38% (39).
Changes in microbial community structure with soil depth are attributed to the response of microbes to the contrasting physical and chemical conditions associated with surface, vadose zone, and saturated soils (31). Environmental factors that influence microbial community composition and diversity include (but are not limited to) pH (19), particle size (74), organic carbon content (93), nutrient availability (22), water content (84), and oxygen concentration (53). The magnitude and variation of these parameters vary between surface and subsurface soils. For instance, the availability of water, plant-derived resources (carbon, nitrogen, and other nutrients), and mineralizable carbon and nitrogen and the oxygen concentration decline steeply with depth (31). Thus, the physiology and metabolic potential of microbial communities will vary greatly with location along a soil profile.
Soils are structured media where the mineral and organic matter components are organized into aggregates that vary in size, porosity, pore size and continuity, and composition. Microaggregates (2 to 20 μm) are considered the most favorable habitat for bacteria in most types of soil (65), with a higher abundance of bacteria located in micropores (∼2 μm) of the inner aggregate fraction (27, 64). While little is known regarding the impact of soil structure on bacterial diversity (64), microbial community composition appears to vary as a function of location within (35, 66) and size of (37, 74, 85) soil aggregates. Soil aggregates may support a different microbial community than the bulk soil by imposing size exclusion on select biota from restricted pore domains (4, 12), minimizing predation (90), and/or decreasing competition as a consequence of lower water tensions (84). Furthermore, anaerobic microsites have been observed within both natural (75) and artificial (46, 68) soil aggregates, resulting as a consequence of O2 consumption rates exceeding diffusive flux rates (14, 25). Anaerobiosis within aggregates may result in the colonization of microbes able to utilize alternative electron acceptors, such as nitrate (63), sulfate, and Fe(III), present as (hydr)oxides comprising the mineral fraction of the aggregate. In fact, aggregate-induced anaerobiosis has been attributed to an enhanced denitrification potential of microaggregate fractions relative to nonfractionated soil (49). Anaerobic microsites within aggregates in aerobic soils may therefore allow for a diverse range of aerobic- and anaerobic-based metabolisms over micrometer scales that contribute to the bulk chemical fluxes of nutrients and metabolites within soils.
Here we investigated the composition and diversity of microbial communities and functional groups within soil aggregates obtained along a continuous profile spanning the surface, vadose zone, and saturated soil environments. The presence and diversity of microbes involved in key pathways in the geochemical cycling of nitrogen (nitrification/denitrification), Fe (ferric iron reduction), and sulfur (sulfate reduction) were investigated. In particular, aggregate-associated microbial communities were characterized using a coupled approach involving cultivation-independent analysis of both 16S rRNA (bacterial and archaeal) and functional (amoA, nirK/nirS, dsrAB) genes, as well as cultivation-based analysis of Fe(III)-reducing organisms. Together, these analyses revealed that microbial and functional communities varied greatly along a geochemically variable soil profile, further highlighting the complexity and spatial heterogeneity of the soil environment.
METHODS AND MATERIALS
Site description and sampling.
Soil samples were obtained from a continuous soil profile within the Melton Branch Watershed, Oak Ridge, TN. The Melton Branch Watershed is underlain by Maryville limestone, which is a limy shale formation. The weathered parent material of interbedded shale and limestone is an Inceptisol and is near the ground surface, ranging from 0.5 to 3 m in depth. The Melton Branch shale saprolite is a loamy, skeletal, mixed thermic, shallow typic Dystrocept (33). The soil profile contains distinct horizonation into A, B, and C horizons. The C horizon is split by the water table, which fluctuates in response to the water level in an adjacent creek, resulting in a permanently unsaturated upper C horizon and a saturated lower C horizon. Bulk soil samples were obtained from the four horizons by vertically coring into the exposed soil profile, excluding exposed soil. Samples were stored immediately on either ice (geochemistry/enrichments) or dry ice (DNA extraction) and shipped to Stanford, CA, for analysis. Soil aggregates (ranging from 5 to 20 mm) were separated from the bulk soil from four horizons, the A, B, unsaturated C, and saturated C horizons. Using sterile techniques under anaerobic conditions, the exterior 1 mm of soil was removed from the surface of ∼10-mm aggregates. The aggregates (ca. 5 to 20) from each horizon were then pooled, ground, and homogenized for DNA extraction and bacterial enrichments.
Soil physiochemical analysis.
Soil aggregates were dried, ground, homogenized, and analyzed for pH, particle size, elemental composition, water content, and bulk Fe mineralogy. Soil pH was obtained by measuring the equilibrium pH of soil pastes containing 1 g soil homogenized in 2 ml of CaCl2 (5 mM) solution. Particle size separation was obtained by centrifugation and the hydrometer method with samples pretreated to remove organic matter (30% H2O2 extraction) (34). Organic carbon content was determined using a Leco total carbon analyzer before and after pretreatment of the sample with 25% HCl to remove inorganic C. Nitrogen was measured using a total nitrogen analyzer and the ammonia ion activity was measured directly using an ion selective electrode. The total elemental composition was obtained by inductively coupled plasma-optical emission spectroscopy of soil extracts obtained by successive cycles of acid (hydrochloric, nitric, perchloric, and hydrofluoric) addition resulting in the decomposition of metal salts, carbonates, sulfides, silicates, sulfates, and oxides. The bulk Fe mineralogy of the soil aggregates was determined using extended X-ray absorption fine-structure spectroscopy as described in detail previously (26).
Fe(III)-reducing enrichments.
Enrichments were conducted within gas-tight 24-ml culture tubes containing 9 ml of artificial freshwater medium (NaCl, 1 g/liter; MgCl2·6H2O, 0.4 g/liter; CaCl2·2H2O, 0.1 g/liter; NH4Cl, 0.25 g/liter; KH2PO4, 0.2 g/liter; KCl, 0.5 g/liter; and mineral and vitamin solutions, 1 ml/liter each [3]) buffered with 30 mM NaHCO3 and amended with 50 mM ferrihydrite and either lactate, acetate, or ethanol (10 mM). Two-line ferrihydrite [Fe(OH)3] was synthesized according to the procedures of Schwertmann and Cornell (73). The medium pH was adjusted to values ranging from 4.5 to 7, corresponding to the pH of the host soil horizon. Soil samples (1 g) were added to sterile medium tubes inside a glove bag with a 95% N2-5% H2 atmosphere, and the headspace was replaced with pure N2. Initial inoculates were serially diluted (10−2 to 10−8) aseptically and anaerobically via N2 gas-flushed syringes. Enrichments were incubated at room temperature (25°C) in the dark and successively transferred (10% inoculum) every 2 months for 1 year to select for Fe(III)-reducing communities.
DNA extraction and PCR amplification.
DNA was extracted from soil aggregates (0.5 g) and Fe(III)-reducing enrichments (9 ml) using either the FastDNA spin kit for soil (Qbiogene, Inc.) or the UltraClean soil DNA kit (Mo Bio, Inc.), respectively, according to the manufacturer's instructions. Cell lysis was achieved using the FastPrep instrument according to the manufacturer's instructions (Qbiogene, Inc.) for optimization of lysis of microbial cells within environmental samples, including gram-positive bacteria. Partial 16S rRNA genes for Bacteria and Archaea and various functional gene fragments were amplified from DNA extracts using the primers listed and protocols referenced in Table 1 (9, 16, 24, 45, 69, 70, 78, 88). The following reaction chemistry was used: 25 μl 2× PCR premix E or F (Epicentre), a 0.5 μM concentration of each primer, and 1.25 U Taq polymerase (dsrAB, amoA, nirK, nirS, and archaeal 16S rRNA) or 5 μl 10× PCR buffer, 2 mM MgCl2, a 0.5 μM concentration of each primer, a 200 μM concentration of each deoxynucleoside triphosphate, and 1.25 U Taq polymerase (bacterial 16S rRNA). Triplicate PCR products were pooled, gel purified, and cloned into the vector pCR2.1 by using a TOPO-TA cloning kit (Invitrogen). White transformants were transferred to 96-well plates containing LB broth (with 50 μg/ml kanamycin), grown overnight at 37°C, and PCR screened directly for the presence of inserts using T7 and M13R vector primers (0.1 μg/reaction).
TABLE 1.
Primers used for PCR amplification of target groups in the A, B, unsaturated C (C-u), and saturated C (C-s) horizons
| Target domain or metabolic group | Target gene | Primer | Sequence | Reference(s) for protocol | PCR amplificationa
|
|||
|---|---|---|---|---|---|---|---|---|
| A | B | C-u | C-s | |||||
| Bacteria | 16S rRNA | 27F | 5′-AGAGTTTGATCCTGGCTCAG-3′ | 45 | + | + | + | + |
| 1492R | 5′-GGTTACCTTGTTACGACTT-3′ | |||||||
| Archaea | 16S rRNA | A21F | 5′-TTCCGGTTGATCCYGCCGGA-3′ | 16 | − | + | + | + |
| A958R | 5′-YCCGGCGTTGAMTCCAATT-3′ | |||||||
| Denitrifying bacteria | nirK | 583Fdg | 5′-TCATGGTGCTGCCGCGYGANGG-3′ | 9, 70 | − | − | − | − |
| nirK5R | 5′-GCCTCGATCAGRTTRTGG-3′ | |||||||
| Denitrifying bacteria | nirS | nirS1F | 5′-CCTAYTGGCCGCCRCART-3′ | 9 | − | − | − | − |
| nirS6R | 5′-CGTTGAACTTRCCGGT-3′ | |||||||
| Ammonia-oxidizing bacteria | amoA | AmoA-1F* | 5′-GGGGHTTYTACTGGTGGT-3′ | 69, 78 | − | − | − | − |
| AmoA-2R | 5′-CCCCTCKGSAAAGCCTTCTTC-3′ | |||||||
| Ammonia-oxidizing archaea | amoA | Arch-amoAF | 5′-STAATGGTCTGGCTTAGACG-3′ | 24 | − | + | + | + |
| Arch-amoAR | 5′-GCGGCCATCCATCTGTATGT-3′ | |||||||
| Sulfate-reducing bacteria | dsrAB | DSR1F | 5′-ACSCACTGGAAGCACG-3′ | 88 | − | − | − | + |
| DSR4R | 5′-GTGTAGCAGTTACCGCA-3′ | |||||||
Successful (+) or unsuccessful (−) PCR amplification for target groups within soil horizon aggregates.
Sequencing and phylogenetic analysis.
Sequencing of T7/M13 PCR products was performed using vector primers T7 and M13R on ABI 3730xl capillary sequencers (PE Applied Biosystems). A total of 653 clones were randomly sequenced, resulting in 384 bacterial (∼1,450-bp) and 127 archaeal (∼940-bp) 16S rRNA gene sequences and 91 archaeal amoA (∼635-bp), 26 dsrAB (∼1.9-kb), and 20 Fe(III)-reducing enrichment culture 16S rRNA (∼1,450-bp) sequences. Nucleotide sequences were assembled and edited using Sequencher v.4.2 (GeneCodes Corp.). The entire ∼2.1-kb dsrAB gene fragment (encoding the α and β subunits of DSR) were successfully amplified, cloned, and sequenced. The included phylogenetic tree is based on a ca. 500-bp region of the dsrA gene to allow for inclusion of the nearest neighbors in final alignments, which were based solely on the α subunit. However, tree topologies and clade designations were stable and consistently recovered by analysis of the DSR α subunit, β subunit, or partial α and β subunit genes. Initial alignment of amplified sequences and closely related sequences identified via BLAST (2, 8) was performed using McClade for the dsrAB and amoA genes or ARB (54) for bacterial and archaeal 16S rRNA (aligned against the small-subunit prokaryote database from the Ribosomal Database Project). Ambiguously and incorrectly aligned positions were aligned manually on the basis of conserved primary sequence and secondary structure. Phylogenetic trees were created based on distance analysis using the neighbor-joining algorithm of the software program PAUP, version 4.0b10, with a Jukes-Cantor correction (79). The robustness of inferred topologies was tested by bootstrap resampling using the same distance model (1,000 replicates).
Operational taxonomic units (OTUs) were defined as clones sharing 90% dsrAB (42), 95% amoA (7), or 97% 16S rRNA (20) nucleotide sequence similarity. OTUs, diversity statistics, and rarefaction curves were computed using the software program DOTUR (71). The similarity of bacterial 16S rRNA clone libraries was statistically evaluated using ∫-LIBSHUFF (72), with 10,000 randomizations and a distance interval (D) of 0.01 on ARB-generated Jukes-Cantor pairwise distance matrices.
Nucleotide sequence accession numbers.
Sequences obtained in this study have been deposited in the GenBank database under accession numbers EU335141 to EU335453 (bacterial 16S rRNA clones), EU306957 to EU307083 (archaeal 16S rRNA clones), EU307084 to EU307103 [16S rRNA Fe(III)-reducing enrichment clones], EU339372 to EU339462 (archaeal amoA clones), and EU301743 to EU301766 (dsrAB).
RESULTS
Soil aggregate geochemistry.
The water content, pH, soil texture, and concentrations of carbon and nutrients varied among the four horizons within the vertical soil profile (Table 2). The A horizon had higher concentrations of organic carbon and N than the other horizons. While the A, B, and unsaturated C horizons had low pH values (4.5 to 5.3), the saturated C horizon was circumneutral (pH 6.9). All four soil horizon aggregates contained high concentrations of Fe, ranging from 3.3 to 4.8%. The Fe solid phases present were Fe-rich clays (i.e., ferrosmectite), hornblende (Fe silicate), and the Fe(III) (hydr)oxides ferrihydrite [Fe(OH)3] and goethite (α-FeOOH).
TABLE 2.
Physical and geochemical characteristics of soil aggregates from the A, B, unsaturated (C-u), and saturated (C-s) C soil horizons
| Horizon | Particle size (%)
|
Water content (g g−1) | pH | %
|
mg kg−1
|
mol% Feb
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sand | Silt | Clay | Organic C | Inorganic C | Na | P | S | Fe | Mn | Ferrihydrite | Goethite | Ferrosmectite | Hornblende | |||
| A | 22 | 46 | 32 | 0.33 | 5.3 | 3.13 | 1.02 | 0.31 | 0.08 | 0.05 | 33,000 | 2,800 | 51 | 30 | 0 | 19 |
| B | 31 | 50 | 19 | 0.20 | 4.5 | 0.14 | 0.12 | 0.03 | 0.06 | 0.01 | 48,300 | 252 | 40 | 31 | 9 | 20 |
| C-u | 50 | 36 | 14 | 0.21 | 5.1 | 0.06 | 0.14 | 0.01 | 0.13 | NDc | 35,100 | 670 | 33 | 22 | 16 | 29 |
| C-s | 42 | 30 | 28 | 0.33 | 6.9 | 0.10 | 0.01 | 0.02 | 0.10 | 0.02 | 39,200 | 1,260 | 28 | 33 | 15 | 24 |
NH4 was below the detection limit (<0.01%).
Extended X-ray absorption fine structure = ±5%.
ND, not detected.
Soil aggregate microbial diversity. (i) Bacteria.
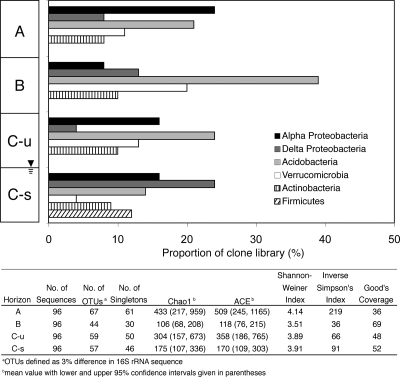
The four soil horizon aggregates contained a diverse bacterial community, which varied in composition and richness along the soil profile (Table 3; Fig. 1). Fourteen phyla were identified within the bacterial clone libraries, with the majority of the soil clones affiliated with nine major phylum-level groups, ranging from 89% (saturated C horizon) to 97% (A horizon) (Table 3). Similarly, Janssen (32) noted that although soil bacteria are affiliated with at least 32 phylum-level groups, an average of 92% of soil clone libraries are members of 9 dominant groups: Proteobacteria (39%), Acidobacteria (20%), Actinobacteria (13%), Verrucomicrobia (7%), Bacteroidetes (5%), Chloroflexi (3%), Planctomycetes (2%), Gemmatimonadetes (2%), and Firmicutes (2%). In particular, five major phyla constitute 73% of the total clone sequences from the soil horizons investigated here, which include the phyla Acidobacteria (25%), Alphaproteobacteria (16%), Deltaproteobacteria (12%), Verrucomicrobia (12%), and Actinobacteria (9%).
TABLE 3.
Frequency of bacterial phyla in 16S rRNA clone libraries derived from the A, B, unsaturated (C-u), and saturated (C-s) C horizon soil aggregates
| Phylum | % of clone sequencesa in horizon:
|
||||
|---|---|---|---|---|---|
| A | B | C-u | C-s | Totalb | |
| Alphaproteobacteria | 24 | 8 | 16 | 16 | 16 |
| Betaproteobacteria | 1 | 0 | 1 | 1 | 1 |
| Deltaproeteobacteria | 8 | 13 | 4 | 24 | 12 |
| Gammaproteobacteria | 7 | 3 | 3 | 3 | 4 |
| Firmicutes | 0 | 0 | 0 | 12 | 3 |
| Actinobacteria | 8 | 10 | 10 | 9 | 9 |
| Verrucomicrobia | 11 | 20 | 13 | 4 | 12 |
| Planctomycetes | 7 | 1 | 6 | 0 | 4 |
| Acidobacteria | 21 | 39 | 24 | 14 | 25 |
| Chloroflexi | 3 | 0 | 10 | 4 | 4 |
| Bacteroidetes | 7 | 1 | 6 | 0 | 4 |
| Gemmatimonadetes | 3 | 1 | 1 | 3 | 2 |
| OP10 | 0 | 3 | 1 | 1 | 1 |
| WS3 | 0 | 0 | 1 | 0 | 0 |
| Unknown | 3 | 3 | 4 | 9 | 4 |
A total of 384 clones were sequenced.
Values in bold represent the dominant phyla.
FIG. 1.
Proportions of major bacterial groups within clone libraries (384 random clones sequenced) obtained from soil aggregates from the A, B, unsaturated C (C-u), and saturated C (C-s) horizons. The table lists the diversity indices for the bacterial sequences recovered from the four horizons. Estimates of phylotype richness were calculated according to the abundance-based coverage estimate (ACE) and the bias-corrected Chao1 estimator. The Shannon-Weiner diversity index, which takes into account species richness and evenness, was also calculated. The evenness of the population is estimated by the inverse of the Simpson's index (1/D), which is sensitive to the level of phylotype dominance. The percentage of coverage was calculated using Good's coverage equation.
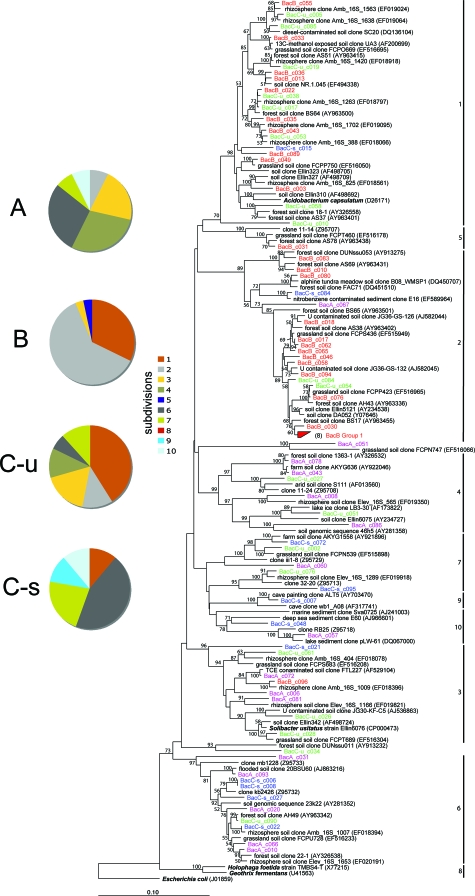
The bacterial community composition and proportions of the five major phyla within clone libraries varied along the soil profile (Fig. 1). While Actinobacteria (8 to 10%) were relatively evenly distributed, the relative contributions of Acidobacter (14 to 39%), Verrucomicrobia (4 to 20%), Alphaproteobacteria (8 to 24%), and Deltaproteobacteria (4 to 24%) varied more dramatically between the horizons. Interestingly, Firmicutes-like bacterial sequences were not observed in the A, B, and unsaturated C horizons, yet they comprised 12% of the saturated C horizon library. Similarly, an increase in the proportional abundance of gram-positive bacteria with depth in a soil profile has previously been observed (23). The proportions of Alpha- and Deltaproteobacteria are substantially higher in the A and saturated C horizons, respectively. Of particular interest is that nearly 40% of the B horizon soil library is affiliated with the phylum Acidobacteria. The phylum Acidobacteria has eight currently recognized subdivisions (designated 1 to 8) that have class-level rank (32). The subdivisions have been expanded to 11 and more recently to 26, based on new clones recovered from Paleolithic cave paintings (94) and uranium-contaminated subsurface sediments (5), respectively. Here, the soil aggregate Acidobacteria-like clones are affiliated with nine subdivisions (1, 2, 3, 4, 5, 6, 7, 9, and 10) (Fig. 2) but are primarily within subdivisions 1 (25%), 2 (31%), 3 (10%), 4 (8%), 6 (13%), and 7 (7%). The composition and evenness of the Acidobacteria varies along the soil profile (Fig. 2). The A horizon contains nearly equal proportions of Acidobacteria within subdivisions 3 (21%), 4 (29%) and 6 (29%), as well as 21% of the clones from subdivisions 2, 7, and 10. In contrast, 93% of the Acidobacteria within the B horizon are affiliated with subdivisions 1 (32%) and 2 (61%). The unsaturated and saturated C horizon clone libraries are dominated by subdivisions 1 (41%) and 6 (44%), respectively. A few Acidobacteria clones from subdivisions 5, 9, and 10 are also detected in the A, B, and saturated C horizons.
FIG. 2.
Neighbor-joining phylogenetic tree of Acidobacteria-affiliated 16S rRNA sequences for the A, B, unsaturated C (C-u), and saturated C (C-s) horizons. As identified by Zimmerman et al. (94), intersubdivision tree topologies differ depending on the tree-building methods (e.g., neighbor joining, maximum parsimony, or maximum likelihood) utilized, yet relationships within subdivisions are consistently stable regardless of the algorithm utilized. Here we utilize the distance-based neighbor-joining method, which results in a different ordering of the previously assigned subdivision numbers while maintaining a stable branching order within each subdivision. Bootstrap values (n = 1,000) of greater than 50% are indicated at nodes. Escherichia coli is the outgroup.
The Acidobacteria also are also the most widespread phyla along the soil profile, having the most overlapping phylotypes among the four soil horizons. Out of the 384 bacterial 16S rRNA clones sequenced in this study, 227 OTUs were defined based on a 97% nucleotide sequence identity threshold. Only 13 of these OTUs occurred in more than one horizon, 5 of which are Acidobacteria. The A, B, and unsaturated C horizons all shared OTUs, with members in the Alphaproteobacteria (2 OTUs), Acidobacteria (5 OTUs), and Verrucomicrobia (2 OTUs). The saturated C horizon shared OTUs with those from only the unsaturated C horizon, belonging to the Alpha- and Deltaproteobacteria. The lack of substantial sequence overlap between the soil clone libraries suggests that the bacterial communities are unique to each soil horizon. This observation was statistically supported using ∫-LIBSHUFF (72), whereby significant P values (<0.05) representative of distinct populations were obtained for all soil horizon comparisons (Table 4). Therefore, while five phyla encompass the majority of the bacterial communities within the four contrasting horizons, species-level differences are more pronounced. For example, the surface A horizon has the highest predicted bacterial richness (Chao1 = 433), while the B horizon is the least diverse (Chao1 = 106) (Fig. 1) at the 97% identity OTU cutoff. Furthermore, the bacterial 16S rRNA sequences are widely dispersed throughout the bacterial groups, rather than grouped into horizon-specific clusters, reflecting high inter- and intrahorizon phylotype richness.
TABLE 4.
P values estimating similarity among bacterial 16S rRNA clone libraries generated using ∫-LIBSHUFF
| Horizon used for homologous library (X) |
P value comparison of heterologous library (Y) with Xa
|
|||
|---|---|---|---|---|
| A | B | C-u | C-s | |
| A | 0.0093 | 0.5338 | 0.0445 | |
| B | 0.0000 | 0.0033 | 0.0000 | |
| C-u | 0.0001 | 0.0045 | 0.0000 | |
| C-s | 0.0000 | 0.0000 | 0.0000 | |
P values comparing either X to Y or Y to X indicate that the libraries are drawn from significantly (P < 0.05) different communities. C-u, unsaturated C; C-s, saturated C.
(ii) Archaea.
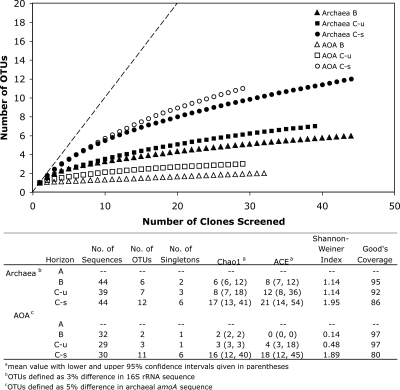
Amplification of archaeal 16S rRNA genes was successful for the B and two C horizons but not for the A horizon, despite multiple screenings (Table 1). A total of 127 sequences were obtained for the three horizons, revealing a total of 25 OTUs (Fig. 3). Based on rarefaction analysis and richness estimators (Chao1 and abundance-based coverage estimate), the observed OTUs appear to be a good representation of the predicted archaeal community present within the soil aggregates (Fig. 3). Compared to the bacterial clone libraries, archaeal 16S rRNA gene diversity/richness is quite low within the three soil horizons, with the B horizon having the lowest (Chao1 = 6) and the saturated C horizon having the highest (Chao1 = 17) richness. The level of archaeal diversity is comparable with previous diversity indices from lake sediments (47) and radioactive thermal springs (89) but lower than that observed for marine sediments (40).
FIG. 3.
Rarefaction curves indicating archaeal 16S rRNA and amoA richness within clone libraries derived from the B, unsaturated C (C-u), and saturated C (C-s) horizons. The dashed line represents 1:1, indicative of infinite diversity. The table lists the archaeal 16S rRNA and amoA sequence diversity indices as described for Fig. 1. OTUs were defined as groups of sequences sharing 97% (16S rRNA) and 95% (amoA) nucleotide sequence identity.
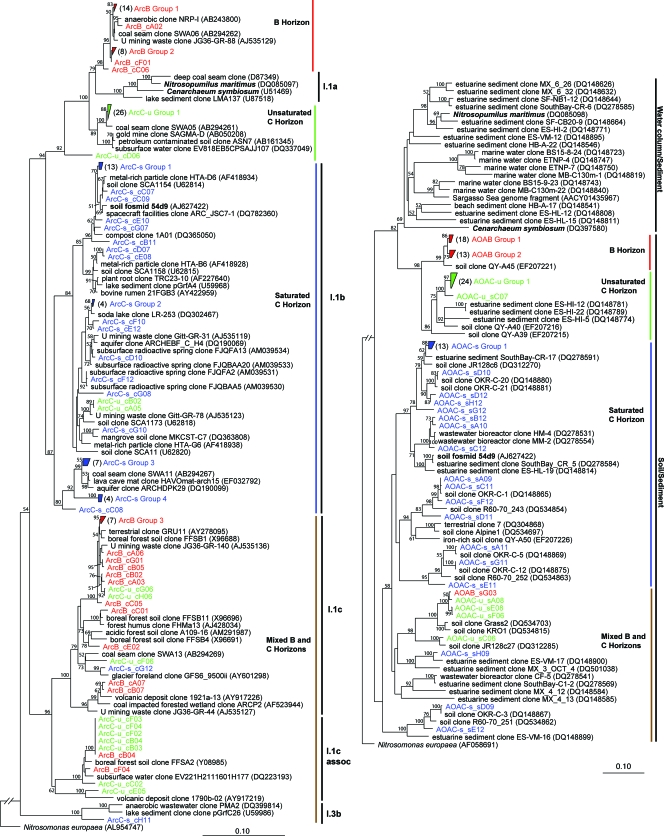
The archaeal community in this study appears to be composed solely of members of the Crenarchaeota. In contrast to the bacterial community (Fig. 1), the archaeal 16S rRNA gene sequences reveal distinct archaeal communities within the three horizons (Fig. 4). The crenarchaeal sequences fall into four major clusters; three of which are horizon specific, and one contains 16S rRNA gene sequences obtained from all three horizons. The majority (95%) of the saturated C horizon sequences (10 of 12 OTUs) are affiliated with the group I.1b Crenarchaeota, the most dominant crenarchaeal group in terrestrial environments (58), comprised primarily of soil, freshwater, and subsurface clones. The saturated C horizon archaeal sequences are closely related to other environmental clones obtained from a variety of habitats, including ferromanganous nodules from a freshwater reservoir (77), uranium-mining waste, and subsurface radioactive thermal springs (89). Furthermore, the dominant group of saturated C horizon 16S rRNA gene clones (C-s group 1 in Fig. 4) is closely related to that of a 43-kb group I.1b crenarchaeal metagenomic clone (54d9), which also contains functional genes (amoA and amoB) putatively encoding subunits of ammonia monooxygenase, derived from a calcareous grassland soil (83). Fifty-seven percent of the B horizon sequences and 69% of the unsaturated C horizon sequences are affiliated with two distinct clades that are most closely related to clones from a number of unusual environments, including coal seams, deep groundwater aquifers, petroleum-contaminated soil (38), and a deep South African gold mine (80). These sequences do not fall clearly into one of the previously defined groups within the Crenarchaeota but rather are most closely related, albeit distantly, to the subsurface/marine/freshwater-associated group I.1a-like sequences. One cluster contains sequences obtained from all three soil horizons, consisting primarily of B horizon and unsaturated C horizon clones. The cluster contains two dominant clades, affiliated with Crenarchaeota group I.1c and I.1c-associated sequences, comprised of forest soil, sediment, and freshwater clones (30, 36). The mixed cluster contains only two sequences (2 OTUs) from the saturated C horizon yet contains most of the 16S rRNA sequence diversity within the B and unsaturated C horizon libraries, accounting for 83% (five of six OTUs) and 57% (four of seven OTUs) of the OTUs for those horizons.
FIG. 4.
Neighbor-joining phylogenetic trees of archaeal 16S rRNA (left) and amoA (right) clone sequences for the B, unsaturated C (C-u), and saturated C (C-s) horizons. Bootstrap values (n = 1,000) of greater than 50% are indicated at nodes. The tree is based on 690 (16S rRNA) and 575 (amoA) masked nucleotide positions. Both the archaeal 16S rRNA and amoA gene trees are rooted with the amoA-containing ammonia-oxidizing bacterium Nitrosomonas europaea.
Presence and diversity of functional groups.
The presence and diversity of microbial communities involved in the cycling of N, Fe, and S within soil aggregates were investigated by identification of bacterial 16S rRNA sequences closely related to known functional/metabolic groups, amplification of functional genes, and selective culture-based enrichments.
(i) Denitrification/nitrification.
Two primary pathways in the cycling of N within soil environments are denitrification, i.e., the dissimilatory reduction of nitrate (NO3−) and nitrite (NO2−) to gaseous products (NO, N2O, and N2), and nitrification, i.e., the oxidation of ammonia (NH3) to nitrate via a nitrite intermediate. Denitrifying bacteria are phylogenetically diverse, spanning over 50 genera, which complicates the identification of denitrifiers based on 16S rRNA taxonomy. Nevertheless, a survey of the 16S rRNA clone library sequences obtained from the four soil horizon aggregates did not reveal the presence even of sequences related to well-known denitrifying bacterial species. Common ammonia-oxidizing (e.g., Nitrosospira and Nitrosomonas) and nitrite-oxidizing (e.g., Nitrobacter and Nitrospira) bacteria were also not detected within our 16S rRNA clone libraries. Correspondingly, amplification of functional genes encoding the dissimilatory nitrite reductases (nirK and nirS) and the α subunit of the betaproteobacterial ammonia monooxygenase (amoA) from the four soil horizons was not successful, despite repeated screening using various PCR primers and protocols (Table 1) commonly used in our laboratory (7, 70). The inability to amplify these functional genes certainly does not preclude the presence of ammonia-oxidizing or denitrifying bacteria in these soils and may simply be due to a low abundance of target organisms, PCR or DNA extraction biases, etc.
Recently, genes resembling bacterial amoA sequences were identified on archaeal metagenomic clones from seawater (87) and soil (83), suggesting the possibility of ammonia oxidation within the domain Archaea. Isolation of an ammonia-oxidizing member of the marine “group 1” Crenarchaeota (Nitrosopumilus maritimus) allowed these amoA-like genes to be definitely linked both to the Archaea and to the critical process of nitrification (43). Here, in contrast to the case for bacterial amoA, amplification of archaeal amoA was successful in three of the four soil horizons (Table 1). In agreement with the archaeal 16S rRNA analysis, archaeal amoA genes were not detected in the A horizon. A total of 90 clones were sequenced from the B, unsaturated C, and saturated C horizons, which corresponded to 16 OTUs (at a 5% cutoff [7]). The curvilinear rarefaction curves and Chao1 richness estimators indicate that nearly all of the predicted OTUs were sampled within our amoA clone libraries (Fig. 3). The richness of amoA sequences from putative ammonia-oxidizing archaea (AOA) in the B and C horizons parallels that of the archaeal 16S rRNA, where the B horizon is the least (Chao1 = 2) and the saturated C horizon is the most (Chao1 = 16) diverse. The archaeal amoA richness within the soil aggregate samples is comparable, albeit on the lower end, to previous observations of diversity in wastewater treatment plant bioreactors (60), marine waters and sediments (24), and nutrient-rich estuary sediments (7).
Like the archaeal 16S rRNA gene sequence distribution, the archaeal amoA-based AOA community composition appears to be distinct to each soil horizon (Fig. 4). Ninety-seven percent of the B, 86% of the unsaturated C, and 90% of the saturated C horizon amoA sequences fall within horizon-specific clusters. Also, similar to the archaeal 16S rRNA sequences, the saturated C horizon amoA sequences were the most diverse, containing 11 OTUs, 8 of which comprise 90% of the sequences. These sequences are most closely related to amoA sequences obtained from wastewater treatment bioreactors (60) and various soils (48), including previously reported clones obtained from the same saturated C horizon (OKR-C clones) (24). As observed for the archaeal 16S rRNA-based community distribution, the amoA sequence from the crenarchaeal fosmid clone 54d9 (83) is situated within the saturated C horizon cluster. The B horizon amoA sequences are defined by two OTUs, one of which contains 97% of the amoA sequences and is most closely related (95% identical) to a clone obtained from an iron-rich soil undergoing fertilization (28). The unsaturated C horizon library contains three OTUs, one of which comprises 86% of the sequences and is distantly related (91% identical) to a fertilized soil clone. One cluster contains sequences from all three soil horizons, accounting for 50% (1 of 2 OTUs), 67% (2 of 3 OTUs), and 27% (3 of 11 OTUs) of the OTUs defined for the B, unsaturated C, and saturated C horizons, respectively.
(ii) Iron reduction.
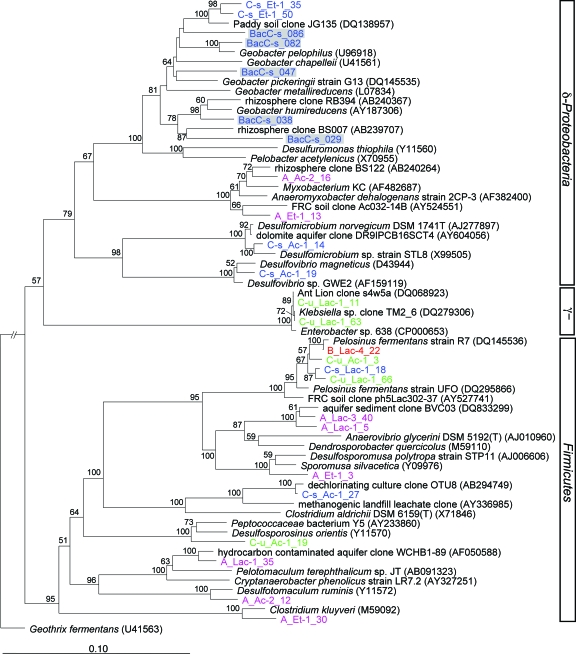
Bacterial sequences closely related to known Fe(III)-reducing bacteria were observed only in the saturated C horizon 16S rRNA clone libraries (Fig. 5). Interestingly, 28% of the Deltaproteobacteria and 7% of the saturated C horizon Bacteria clone sequences were closely related to the Fe(III)-reducing genus Geobacter. Incubation of soil aggregates in synthetic medium supplemented with organic carbon (lactate, acetate, or ethanol) and ferrihydrite resulted in growth and Fe(III) reduction for all four soil horizons at pH values ranging from 4.5 to 7. However, as frequently observed in cultivation-based studies, the Fe(III)-reducing communities enriched were not well represented in the bacterial 16S rRNA PCR clone libraries derived directly from the soil aggregates. In fact, the only Fe(III)-reducing enrichment clones related to bacteria represented in the soil aggregate bacterial clone libraries are members of the Geobacter clade (Fig. 5), but the sequences were not identical.
FIG. 5.
Neighbor-joining phylogenetic tree of 16S rRNA clone sequences obtained from the Fe(III)-reducing enrichments and of the closely affiliated saturated C horizon bacterial clones (BacC-s; gray shading). The A (pink), B (red), unsaturated C (C-u) (green), and saturated C (C-s) (blue) horizons were enriched with ferrihydrite as the electron acceptor and lactate (Lac), acetate (Ac), or ethanol (Et) as the electron donor. The tree is based on 1,060 masked nucleotide positions and Geothrix fermentans is the outgroup. Bootstrap values (n = 1,000) of greater than 50% are indicated at nodes.
Despite the high bacterial diversity present within the soil aggregates (Fig. 1), only three dominant groups were recovered within our Fe(III)-reducing enrichment cultures. Following enrichment, Firmicutes and Delta- and Gammaproteobacteria constituted 58, 32, and 7% of the enriched bacterial species, respectively (Fig. 6). In contrast, Firmicutes (0 to 12%), Deltaproteobacteria (4 to 24%), and Gammaproteobacteria (3 to 7%) clones accounted for a lesser fraction of the initial bacterial communities prior to stimulation/enrichment (Table 2). The enriched Firmicutes bacterial clones are comprised primarily of Clostridia-like species (84%), including 16S rRNA gene sequences most closely related to Dendrosporobacter quercicolus (A horizon) and Pelosinus fermentans (B and C horizons) (Fig. 6). Pelosinus fermentans sequences dominated (49 to 100%) the lactate-based enrichments of the B, unsaturated C, and saturated C soil horizons. The second-largest fraction (32%) of the enriched clones belongs to the Deltaproteobacteria, consisting of 16S rRNA sequences similar to Geobacter (42%), Myxobacteria (34%), and a number of sulfate-reducing genera (23%). Sequences within the Geobacter clade constituted 14% of the Fe(III)-reducing enrichment clones but, interestingly, were obtained only within ethanol-based cultures from the saturated C horizon (Fig. 5). The Myxobacteria clones are closely related to the Fe(III)-reducing bacterium Anaeromyxobacterium dehalogenans strain 2CP-3 (29) and an acetate-stimulated Fe(III)-reducing enrichment clone, Ac032-14B, from an adjacent, contaminated watershed within the U.S. Department of Energy Field Research Center at Oak Ridge (62). Although Gammaproteobacteria constitute only a minor (7%) fraction of the phylotypes within the Fe(III)-reducing enrichments (Fig. 6), two Klebsiella-like species within the Enterobacteriaceae family of Gammaproteobacteria account for nearly half (49%) of the lactate-stimulated organisms from the unsaturated C horizon (Fig. 5).
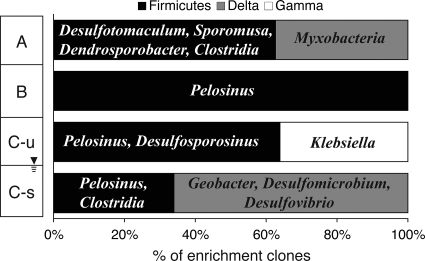
FIG. 6.
Proportion of bacterial phyla enriched under Fe(III)-reducing conditions as a function of soil horizon. Dominant species are indicated in the representative bar graph for each horizon.
The relative abundance of the three dominant bacterial phyla within the enrichment clone libraries varied among the four geochemically distinct soil horizons (Fig. 6). Bacteria within the Firmicutes division constituted a major fraction of the Fe(III)-reducing enrichment clones for all four soil horizons (Fig. 7). The C horizons supported the majority of the Fe(III)-reducing enrichment clones that are not related to Firmicutes- or Myxobacterium-like bacteria (Fig. 6). While most enriched species were unique to each soil horizon, three of the four horizons (B, unsaturated C, and saturated C) contained 16S rRNA sequences most closely related Pelosinus fermentans strain R7, a fermentative Fe(III)-reducing bacterium recently isolated from a subsurface kaolin lense (76) (Fig. 5).
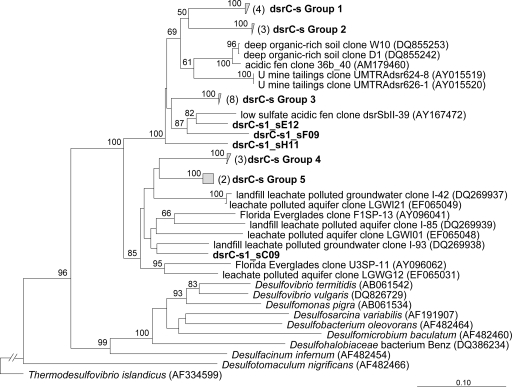
FIG. 7.
Phylogenetic tree reflecting the relationship of sulfate-reducing bacteria within the saturated C horizon (C-s) based on an ∼500-bp region of dsrAB gene fragment. The tree was constructed by neighbor joining based on Jukes-Cantor-corrected DNA distances. Bootstrap values (n = 1,000) of greater than 50% are indicated at nodes. Tree topologies and clade designations were stable and consistently recovered by analysis of the DSR α subunit, β subunit, or partial α and β subunit genes. Thermodesulfovibrio islandicus of the Nitrospira phylum is the outgroup.
(iii) Sulfate reduction.
16S rRNA sequences closely related to known sulfate-reducing bacteria were not observed in any of the four soil horizons. However, amplification of the ∼2-kb dissimilatory sulfite reductase gene fragment (dsrAB) was achieved for the saturated C horizon, yet amplification was unsuccessful for the A, B, and unsaturated C soil horizons (Table 1).
As expected from a temperate soil environment, dsrAB sequences obtained from the saturated C horizon clone libraries were affiliated only with the domain Bacteria and not with the Archaea (Fig. 7). Based on a 90% dsrAB sequence identity cutoff (42), nine bacterial OTUs were identified within the 26 sequenced clones, representing excellent coverage of the nine OTUs predicted by the Chao1 richness estimator. The dsrAB diversity observed for the saturated C horizon is comparable to that previously observed for a number of environments, including nutrient-impacted and pristine sediments from the Florida Everglades (11), hypersaline sediments from the Great Salt Lake (42), hypersaline anoxic basins in the Eastern Mediterranean deep sea (86), and marine sediments (67).
The nine dsrAB OTUs are located within five groups that contain between two and nine sequences and four unique phylotypes. The soil phylotypes are situated within two larger clades consisting of only environmental clones that are deeply divergent from common cultured sulfate-reducing bacteria within the Deltaproteobacteria (e.g., Desulfovibrio spp.) and the Firmicutes (e.g., Desulfotomaculum spp.). Furthermore, our soil dsrAB sequences share only 84 to 91% nucleotide sequence identity with currently available environmental clones within these two clades. Twenty-seven percent of the clones are present as two phylotypes (groups 1 and 2) and are most closely related (84 to 85% identical) to a clone obtained from a deep organic-rich soil. Phylotype group 3 along with three unique phylotypes constitute 42% of the dsrAB sequences and are most closely related (86 to 91% identical) to a clone obtained from a low-sulfate acidic fen (52). Groups 4 and 5, containing 19% of the sequences along with one unique phylotype, are most closely related (84 to 89% identical) to two environmental clones obtained from two different landfill leachate-polluted aquifers.
DISCUSSION
The bacterial communities associated with aggregates from four geochemically distinct soil horizons are composed of highly diverse organisms widely distributed within nine major phyla (Proteobacteria, Acidobacteria, Actinobacteria, Verrucomicrobia, Bacteroidetes, Chloroflexi, Planctomycetes, Gemmatimonadetes, and Firmicutes) (Fig. 1). The relative abundances of the phyla are similar to those recently reported in a survey of 32 Bacteria clone libraries obtained from various soils ranging from pristine forest and grassland to agricultural soils (32). Here, Acidobacteria, in particular, constitute a significant fraction (14 to 39%) of the bacterial 16S rRNA clone libraries from all four soil horizons (Table 3). Similarly, Acidobacteria have been reported to make up an average of 20% of the bacterial clones, with contributions of as high as 50% of cloned 16S rRNA sequences within various soils (17). Acidobacter 16S rRNA sequences have been retrieved from a wide variety of environments, ranging from neutral soils (18) to acid mine drainage (41) to the surfaces of paleolithic cave paintings (94). Furthermore, the phylogenetic depth of Acidobacteria is considered nearly as great as that in the phylum Proteobacteria (32). Acidobacteria are grouped into as many as 26 subdivisions, with subdivisions 1, 3, 4, and 6 being the most abundant within a diverse range of soils (6, 32). Here, subdivisions 1, 2, 3, 4, and 6 are most abundant along the soil profile, with 1 and 2 accounting for over half of the Acidobacteria clones (Fig. 2). To date, little information is known regarding the metabolic potential of Acidobacteria, and there are only four formally described genera within the phylum Acidobacteria, including subdivision 1 members Acidobacterium capsulatum (41) and Terriglobus roseus (19) and subdivision 8 members Geothrix fermentans (13) and Holophaga foetida (50). The variability in the subdivision distribution among the horizons may, however, lend insight into the conditions that favor the different Acidobacteria subdivisions. For instance, unsaturated, low-nutrient/carbon, acidic soils (horizons B and unsaturated C) appear to favor subdivisions 1 and 2, while higher-nutrient/carbon conditions (A horizon) favor subdivisions 3, 4, and 6. The saturated, circumneutral pH soils within the saturated C horizon favor subdivisions 6 and 7. Interestingly, Acidobacteria within subdivision 1 were not observed in the A horizon, despite their ubiquitous distribution in a wide range of soils (32) and their proliferation at low pH (19). Furthermore, Acidobacteria within subdivision 6 made up a substantial proportion of the Acidobacteria community within the contrasting A (29%) and saturated C (44%) horizons, suggesting a wide tolerance to pH, water content, and nutrient/carbon availability. The paucity of physiological information and wide ecological diversity of the Acidobacteria make metabolic/functional inferences problematic. Yet, based on their abundance and phylogenetic depth, they appear to play an important role in the ecology of soils.
The diversity and composition of the bacterial community varied along the soil profile (Fig. 1). The change in the relative proportions of the Alphaproteobacteria, Acidobacteria, Deltaproteobacteria, and Firmicutes along the soil profile is particularly apparent. Based on previous soil surveys, the concentration of carbon appears to be the primary control on the composition and richness of bacterial communities (21, 44, 93). The variable carbon content may therefore explain the high bacterial diversity within the A horizon and contrasting low diversity within the B horizon (Fig. 1). Furthermore, a recent survey of the relative abundances of bacterial phyla within 71 soils from a wide range of ecosystems revealed that the net carbon mineralization rate (an index of C availability) was the best predictor of the abundance of some bacterial phyla, including Acidobacteria (21). In fact, the strong and ubiquitous inverse relationship between carbon availability and abundance of Acidobacteria suggests that Acidobacteria may be specifically classified as oligotrophic (21). In contrast, the abundance of Alphaproteobacteria has been found to be higher in the rhizosphere than in comparatively carbon-poor bulk soil. The impact of carbon may therefore explain the higher abundance of Alphaproteobacteria and lower abundance of Acidobacteria clones in the carbon-rich A horizon relative to the carbon-poor B and C horizons. An inverse relationship between Acidobacteria diversity and abundance and soil pH has also been previously observed (19). Thus, the particularly low-carbon and -pH environment of the B horizon may account for the development of an Acidobacter-dominated (40% of clones) bacterial community.
In lower-carbon soils (such as the B and C horizons), the structure of bacterial communities is also shaped by competition, which is often a function of water content (84, 93). The higher moisture content associated with the saturated conditions of the C horizon, coupled with a higher pH and lower oxygen concentration, may account for a different bacterial community composition compared to that of the B and unsaturated C horizons, including a smaller proportion of Alphaproteobacteria and Verrucomicrobia clones (Fig. 1). Similarly, a survey of flooded paddy soils indicated a predominance of Alpha- and Betaproteobacteria in the surface oxic layers, while Clostridia-like Firmicutes dominated the anoxic zones (53). Here, the saturated C horizon contains 16S rRNA gene sequences from the phylum Firmicutes, which are not observed in the other horizons, as well as a substantially higher abundance of Deltaproteobacteria (Fig. 1). These soil clones are most closely related to obligate anaerobes involved in fermentation (e.g., Clostridia spp.) and Fe(III) respiration (e.g., Geobacter spp.). While Fe(III)-reducing enrichments reveal the presence of Firmicutes within the A, B, and unsaturated C horizons (Fig. 5), their absence in the soil 16S rRNA gene clone libraries suggests that they may represent a minor fraction of the bacterial community. In contrast, bacteria belonging to the Fe(III)-reducing genus Geobacter were recovered in the Fe(III)-reducing enrichments and as a substantial fraction of the Deltaproteobacteria in the bacterial 16S rRNA gene clone libraries from the saturated C horizon (Fig. 5). The presence of organisms considered obligate anaerobes (e.g., Clostridia spp.) within the Fe(III) enrichments and 16S rRNA clone libraries suggests that anaerobic microsites are present within the aerobic soil horizons.
The Fe(III)-reducing consortia enriched from the soil aggregates were dominated by Firmicutes and Deltaproteobacteria, with a variable distribution and composition along the soil profile (Fig. 6). The acidic-pH (A, B, and unsaturated C horizons), low-carbon and -nutrient (B and C horizons) conditions and the variable water contents (Table 2) associated with these soils may select for more resilient spore- and spore-like-forming Firmicutes and Anaeromyxobacter bacteria, respectively. As observed previously for acidic, contaminated sediments (62), fermentative organisms dominated the Fe(III)-reducing enrichment cultures. Furthermore, the predominance of the Firmicutes bacterium Pelosinus fermentans within enrichments from three geochemically contrasting soils (B and C horizons) suggests metabolic and environmental flexibility within this species. As observed for other Clostridia-like species, P. fermentans donates electrons to Fe(III) during fermentative growth rather than using Fe(III) as an electron acceptor for respiration (76). Thus, fermentation-based Fe(III) reduction could play an important role within anaerobic microsites in unsaturated soils, as well as contribute to Fe(III) reduction within saturated soils.
While acetate is considered the most important electron donor for Fe(III) reduction by Geobacter-like species (51), interestingly, acetate-based Fe(III)-reducing enrichments derived from soil aggregates from the saturated C horizon belonged to known sulfate-reducing bacteria within the genera Desulfomicrobium and Desulfovibrio (Fig. 5). The ability to conserve energy via Fe(III) reduction varies greatly among the sulfate-reducing bacterial species, and the mechanisms of Fe(III) reduction are not resolved (51). Screening for the dissimilatory sulfite reductase gene (dsrAB) recovered sequences only within the saturated C horizon, which were not closely related to known sulfate-reducing genera (Fig. 7). Instead, the dsrAB sequences belong to novel lineages of sulfate-reducing bacteria and may represent organisms suited to conditions found within soil aggregates, including low carbon, sulfate, and nutrient fluxes.
In contrast to the bacteria, the archaeal communities have a low diversity (Fig. 3), and the sequences (both archaeal 16S rRNA and amoA) cluster within horizon-specific clades (Fig. 4), a potential indicator of “environmental specificity” among the archaeal species. The phylogenetic groups I.1b and I.1c are considered the most dominant crenarchaeal groups within a wide range of soils (36, 39, 61). Here, group I.1b is comprised almost entirely of sequences recovered from the saturated C horizon, which, interestingly, also accounts for all but two of the saturated C horizon sequences (Fig. 4). Similarly, group I.1b was often the only lineage of Archaea found in a broad survey of soils varying in pH, nutrient status, and temperature, which led the authors to conclude that group I.1b may be the most capable crenarchaea to effectively compete with the high diversity of bacteria in soils (58). However, here, group I.1b crenarchaea are poorly represented (two phylotypes) or not represented in the unsaturated C and B horizon archaeal 16S rRNA clone libraries, respectively (Fig. 4). Instead, the archaeal communities within the B and unsaturated C horizons appear to be dominated by group I.1c and two unique lineages most closely related to group I.1a. The majority of the B and unsaturated C horizon sequences fall within the novel lineages, yet the sequence diversity is mostly affiliated with group I.1c and I.1c-associated creanarchaea. Recent surveys of glacier foreland soils indicated that the distribution of group I.1c sequences was restricted to mature (lower-pH, higher-nutrient) soils (56, 57). Changes in archaeal community composition with depth in acidic forest soils have been attributed to changes in organic carbon composition and availability (39, 61). Considering the similar carbon and nutrient conditions in the B and C horizons in this study, however, other geochemical variables, including water content, oxygen partial pressure, and pH, are more likely to be controlling the archaeal community diversity and distribution. In particular, the lower pH and more stable (developed) conditions associated with the B and unsaturated C horizons may favor the proliferation of group I.1c crenarchaea relative to the C horizon situated below the fluctuating water table.
To date, little is known regarding the metabolic potential of the mesophilic Crenarchaeota, with the ammonia-oxidizing Nitrosopumilus maritimus being the only isolate (43). Here, the distribution and diversity of archaeal amoA sequences within three of the four horizons along the soil profile suggest that the archaeal community may be capable of ammonia oxidation. Similar to the archaeal 16S rRNA clone libraries, the archaeal amoA sequences cluster within horizon-specific clades (Fig. 4). Also, as observed for the archaeal 16S rRNA sequences, one clade contains archaeal amoA sequences from all three horizons, possibly representing AOA more tolerant to the wide range of environmental conditions (e.g., pH and water content) found within the three soil horizons. The diversity, number, and clustering of the archaeal amoA phylotypes are remarkably similar to those of the crenarchaeal 16S rRNA gene sequences (Fig. 3 and 4), suggesting that many of the archaea present within the soil aggregates are putative AOA. Since the recent discovery of AOA (43), archaeal amoA sequences have been recovered from a wide range of environments, including soils (48), marine waters (24, 55), estuarine sediments (7, 24), wastewater treatment plants (60), and radioactive thermal springs (89), highlighting the broad ecological and phylogenetic diversity of AOA (24). Furthermore, the high relative abundance of archaeal compared to bacterial amoA genes (48, 55, 91) and confirmation of expressional activity of AOA in situ (48) within soils reveal that Crenarchaeota may play a dominant role in ammonia oxidation within the soil environment.
Here, changes in carbon availability, water content, and pH as a function of soil depth appear to correlate with differences in resident microbial and functional communities within surface and subsurface soils. Furthermore, the presence of both aerobic- and anaerobic-based metabolisms (inferred from 16S rRNA, functional gene, and cultivation-based analyses) suggests that anaerobic microsites within aggregates allow for the development of functionally diverse communities over small spatial scales. Thus, the physical and chemical heterogeneity in soils further amplifies the complexity and diversity of microbial communities within the soil environment. The findings reported here provide a glimpse into this diversity by interrogating homogenized aggregates from four distinct horizons along a soil profile and highlight the need for a more exhaustive sampling effort to fully reveal the diversity and composition of soils at the microscale.
Acknowledgments
This work was funded by the Office of Science Biological and Environmental Research ERSD Program, the U.S. Department of Energy (grant ER 63609-1021814), and the Stanford NSF Environmental Molecular Sciences Institute (grant NSF-CHE-0431425). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program.
We thank two anonymous reviewers for their detailed comments, which substantially improved the manuscript.
Footnotes
Published ahead of print on 11 January 2008.
REFERENCES
- 1.Agnelli, A., J. Ascher, G. Corti, M. T. Ceccherini, P. Nannipieri, and G. Pietramellara. 2004. Distribution of microbial communities in a forest soil profile investigated by microbial biomass, soil respiration and DGGE of total and extracellular DNA. Soil Biol. Biochem. 36:859-868. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bales, R. C., C. P. Gerba, G. H. Grondin, and S. L. Jensen. 1989. Bacteriophage transport in sandy soil and fractured tuff. Appl. Environ. Microbiol. 55:2061-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barns, S. M., E. C. Cain, L. Sommerville, and C. R. Kuske. 2007. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 73:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barns, S. M., S. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beman, J. M., and C. A. Francis. 2006. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl. Environ. Microbiol. 72:7767-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson, D. A., I. Karsh-Mizrachi, D. J. Lipman, J. Ostell, B. A. Rapp, and D. L. Wheeler. GenBank. Nucleic Acids Res. 28:15-18. [DOI] [PMC free article] [PubMed]
- 9.Braker, G., A. Fesefeldt, and K. P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckley, D. H., J. R. Graber, and T. M. Schmidt. 1998. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Appl. Environ. Microbiol. 64:4333-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castro, H., K. R. Reddy, and A. Ogram. 2002. Composition and function of sulfate-reducing prokaryotes in eutrophic and pristine areas of the Florida Everglades. Appl. Environ. Microbiol. 68:6129-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Champ, D. R., and J. Schroeter. 1988. Bacterial transport in fractured rock: a field-scale tracer test at the Chalk River Nuclear Laboratories. Water Sci. Technol. 20:81-87. [Google Scholar]
- 13.Coates, J. D., D. Ellis, C. W. Gaw, and D. R. Lovley. 1999. Geothrix fermentans gen. nov., sp. nov., a novel Fe(III)-reducing bacterium from a hydrocarbon-contaminated aquifer. Int. J. Syst. Bacteriol. 49:1615-1622. [DOI] [PubMed] [Google Scholar]
- 14.Curie, J. A. 1961. Gaseous diffusion in porous media. III. Wet granular materials. Br. J. Appl. Phys. 12:275-281. [Google Scholar]
- 15.Daniel, R. 2005. The metagenomics of soil. Nat. Rev. Microbiol. 3:470-478. [DOI] [PubMed] [Google Scholar]
- 16.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichorst, S. A., J. A. Breznak, and T. M. Schmidt. 2007. Isolation and characterization of soil bacteria that define Terriglobus gen. nov. in the phylum Acidobacteria. Appl. Environ. Microbiol. 73:2708-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fields, M. W., T. Yan, S.-K. Rhee, S. L. Carroll, P. M. Jardine, D. B. Watson, C. S. Criddle, and J. Zhou. 2005. Impacts on microbial communities and cultivable isolates from groundwater contaminated with high levels of nitric acid-uranium waste. FEMS Microbiol. Ecol. 53:417-428. [DOI] [PubMed] [Google Scholar]
- 21.Fierer, N., M. A. Bradford, and R. B. Jackson. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354-1364. [DOI] [PubMed] [Google Scholar]
- 22.Fierer, N., J. P. Schimel, and P. A. Holden. 2003. Controls on microbial CO2 production: a comparison of surface and subsurface soil horizons. Global Change Biol. 9:1322-1332. [Google Scholar]
- 23.Fierer, N., J. P. Schimel, and P. A. Holden. 2003. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 35:167-176. [Google Scholar]
- 24.Francis, C. A., K. J. Roberts, J. M. Beman, A. E. Santoro, and B. B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenwood, D. J., and D. Goodman. 1967. Direct measurement of the distribution of oxygen in soil aggregates and in columns of fine soil crumbs. J. Soil Sci. 18:182-196. [Google Scholar]
- 26.Hansel, C. M., S. G. Benner, J. Neiss, A. Dohnalkova, R. K. Kukkadapu, and S. Fendorf. 2003. Secondary mineralization pathways induced by dissimilatory iron reduction of ferrihydrite under advective flow. Geochim. Cosmochim. Acta 67:2977-2992. [Google Scholar]
- 27.Hattori, T. 1988. Soil aggregates as microhabitats of microorganisms. Rep. Inst. Agric. Res. Tohoku Univ. 37:23-26. [Google Scholar]
- 28.He, J.-Z., J.-P. Shen, L.-M. Zhang, Y.-G. Zhu, Y.-M. Zheng, M.-G. Xu, and H. Di. 2007. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ. Microbiol. 9:2364-2374. [DOI] [PubMed] [Google Scholar]
- 29.He, Q., and R. A. Sanford. 2003. Characterization of Fe(III) reduction by chlororespiring Anaeromyxobacter dehalogenans. Appl. Environ. Microbiol. 69:2712-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hershberger, K. L., S. M. Barns, A. L. Reysenbach, S. C. Dawson, and N. R. Pace. 1996. Wide diversity of Crenarchaeota. Nature 384:420. [DOI] [PubMed] [Google Scholar]
- 31.Holden, P. A., and N. Fierer. 2005. Microbial processes in the vadose zone. Vadose Zone J. 4:1-21. [Google Scholar]
- 32.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jardine, P. M., G. K. Jacobs, and G. V. Wilson. 1993. Unsaturated transport processes in undisturbed heterogeneous porous-media. 1. Inorganic contaminants. Soil Sci. Soc. Am. J. 57:945-953. [Google Scholar]
- 34.Jardine, P. M., N. L. Weber, and J. F. McCarthy. 1989. Mechanisms of dissolved organic carbon adsorption on soil. Soil Sci. Soc. Am. J. 53:1378-1385. [Google Scholar]
- 35.Jocteur-Monrozier, L., J. N. Ladd, A. W. Fitzpatrick, R. C. Foster, and M. Raupach. 1991. Components and microbial biomass content of size fractions in soils of contrasting aggregation. Geoderma 49:37-62. [Google Scholar]
- 36.Jurgens, G., K. Lindstrom, and A. Saano. 1997. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl. Environ. Microbiol. 63:803-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanazawa, S., and Z. Filip. 1986. Distribution of microorganisms, total biomass, and enzyme activities in different particles of brown soil. Microb. Ecol. 12:205-215. [DOI] [PubMed] [Google Scholar]
- 38.Kasai, Y., Y. Takahata, T. Hoaki, and K. Watanabe. 2005. Physiological and molecular characterization of a microbial community established in unsaturated, petroleum-contaminated soil. Environ. Microbiol. 7:806-818. [DOI] [PubMed] [Google Scholar]
- 39.Kemnitz, D., S. Kolb, and R. Conrad. 2007. High abundance of Crenarchaeota in a temperate acidic forest soil. FEMS Microbiol. Ecol. 60:442-448. [DOI] [PubMed] [Google Scholar]
- 40.Kendall, M. M., G. D. Wardlaw, C. F. Tang, A. S. Bonin, Y. Liu, and D. L. Valentine. 2007. Diversity of Archaea in marine sediments from Skan Bay, Alaska, including cultivated metanogens, and description of Methanogenium boonei sp. nov. Appl. Environ. Microbiol. 73:407-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kishimoto, N., Y. Kosako, and T. Tano. 1991. Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr. Opin. Microbiol. 22:1-7. [DOI] [PubMed] [Google Scholar]
- 42.Kjeldsen, K. U., A. Loy, T. F. Jakobsen, T. R. Thomsen, M. Wagner, and K. Ingvorsen. 2007. Diversity of sulfate-reducing bacteria from an extreme hypersaline sediment, Great Salt Lake (Utah). FEMS Microbiol. Ecol. 60:287-298. [DOI] [PubMed] [Google Scholar]
- 43.Konneke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 44.LaMontagne, M. G., J. P. Schimel, and P. A. Holden. 2003. Comparison of subsurface and surface soil bacterial communities in California grassland as assessed by terminal restriction fragment length polymorphisms of PCR-amplified 16S rRNA genes. Microb. Ecol. 46:216-227. [DOI] [PubMed] [Google Scholar]
- 45.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackbrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 46.Leffelaar, P. A. 1986. Dynamics of partial anaerobiosis, denitrification, and water in a soil aggregate: experimental. Soil Sci. 142:352-365. [Google Scholar]
- 47.Lehours, A.-C., P. Evans, C. Bardot, K. Joblin, and F. Gerard. 2007. Phylogenetic diversity of archaea and bacteria in the anoxic zone of a meromictic lake (Lake Pavin, France). Appl. Environ. Microbiol. 73:2016-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 49.Lensi, R., A. Clays-Josserand, and L. J. Monrozier. 1995. Denitrifiers and denitrifying activity in size fractions of a mollisol under permanent pasture and continuous cultivation. Soil Biol. Biochem. 27:61-69. [Google Scholar]
- 50.Liesack, W., U. Kreft, and E. Stackbrandt. 1994. Holophaga foetida gen. nov., spec. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch. Microbiol. 162:85-90. [DOI] [PubMed] [Google Scholar]
- 51.Lovley, D. R., D. E. Holmes, and K. P. Nevin. 2004. Dissimilatory Fe(III) and Mn(IV) Reduction. Adv. Microb. Physiol. 49:219-286. [DOI] [PubMed] [Google Scholar]
- 52.Loy, A., K. Kusel, A. Lehner, H. L. Drake, and M. Wagner. 2004. Microarray and functional gene analysis of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal cooccurrence of recognized genera and novel lineages. Appl. Environ. Microbiol. 70:6998-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ludemann, H., I. Arth, and W. Liesack. 2000. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl. Environ. Microbiol. 66:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ludwig, W., W. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Foster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mincer, T. J., M. J. Church, L. T. Taylor, C. Preston, D. M. Karl, and E. F. DeLong. 2007. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9:1162-1175. [DOI] [PubMed] [Google Scholar]
- 56.Nicol, G. W., D. Tscherko, L. Chang, U. Hammesfahr, and J. I. Prosser. 2006. Crenarchaeal community assembly and microdiversity in developing soils at two sites associated with deglaciation. Environ. Microbiol. 8:1382-1393. [DOI] [PubMed] [Google Scholar]
- 57.Nicol, G. W., D. Tscherko, T. M. Embley, and J. I. Prosser. 2005. Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ. Microbiol. 7:337-347. [DOI] [PubMed] [Google Scholar]
- 58.Ochsenreiter, T., D. Selezi, A. Quaiser, L. Bonch-Osmolovskaya, and C. Schleper. 2003. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 5:787-797. [DOI] [PubMed] [Google Scholar]
- 59.Ouverney, C. C., and J. A. Fuhrman. 2000. Marine planktonic archaea take up amino acids. Appl. Environ. Microbiol. 66:4829-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park, H.-D., G. F. Wells, H. Bae, C. S. Criddle, and C. A. Francis. 2006. Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl. Environ. Microbiol. 72:5643-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pesaro, M., and F. Widmer. 2002. Identification of novel Crenarchaeota and Euryarchaeota clusters associated with different depth layers of a forest soil. FEMS Microbiol. Ecol. 42:89-98. [DOI] [PubMed] [Google Scholar]
- 62.Petrie, L., N. N. North, S. L. Dollhopf, D. L. Balkwill, and J. E. Kostka. 2003. Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI). Appl. Environ. Microbiol. 69:7467-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Phillipot, L., P. Renault, J. Sierra, C. Henault, A. Clays-Josserand, C. Chenu, R. Chaussod, and R. Lensi. 1996. Dissimialtory nitrite-reductase provides a competitive advantage to Pseudomonas sp. RTC01 to colonise the centre of soil aggregates. FEMS Microbiol. Ecol. 21:175-185. [Google Scholar]
- 64.Ranjard, L., S. Nazaret, F. Gourbiere, J. Thioulouse, P. Linet, and A. Richaume. 2000. A soil microscale study to reveal the heterogeneity of Hg(II) impact on indigenous bacteria by quantification of adapated phenotypes and analysis of community DNA fingerprints. FEMS Microbiol. Ecol. 31:107-115. [DOI] [PubMed] [Google Scholar]
- 65.Ranjard, L., and A. Richaume. 2001. Quantitative and qualitative microscale distribution of bacteria in soil. Res. Microbiol. 152:707-716. [DOI] [PubMed] [Google Scholar]
- 66.Ranjard, L., A. Richaume, L. Jocteur-Monrozier, and S. Nazaret. 1997. Response of soil bacteria to Hg(II) in relation to characteristics and cell location. FEMS Microbiol. Ecol. 24:321-331. [Google Scholar]
- 67.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Renault, P., and P. Stengel. 1994. Modeling oxygen diffusion in aggregated soils. I. Anaerobiosis inside the aggregates. Soil Sci. Soc. Am. J. 58:1017-1023. [Google Scholar]
- 69.Rotthauwe, J.-H., K.-P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santoro, A. E., A. B. Boehm, and C. A. Francis. 2006. Denitrifier community composition along a nitrate and salinity gradient in a coastal aquifer. Appl. Environ. Microbiol. 72:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwertmann, U., and R. M. Cornell. 2000. Iron oxides in the laboratory: preparation and characterization. Wiley-VCH, New York, NY.
- 74.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sexstone, A. J., N. P. Revsbech, T. B. Parkin, and J. M. Tiedje. 1985. Direct measurement of oxygen profiles and denitfication rates in soil aggregates. Soil Sci. Soc. Am. J. 49:645-651. [Google Scholar]
- 76.Shelobolina, E. S., K. P. Nevin, J. D. Blakeney-Hayward, C. V. Johnsen, T. W. Plaia, P. Krader, T. Woodard, D. E. Holmes, C. G. VanPraagh, and D. R. Lovley. 2007. Geobacter pickeringii sp. nov., Geobacter argillaceus sp. nov. and Pelosinus fermentans gen. nov. sp. nov., isolated from subsurface kaolin lenses. Int. J. Syst. Evol. Microbiol. 57:126-135. [DOI] [PubMed] [Google Scholar]
- 77.Stein, L. Y., M. T. La Duc, T. J. Grundl, and K. H. Nealson. 2001. Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments. Environ. Microbiol. 3:10-18. [DOI] [PubMed] [Google Scholar]
- 78.Stephen, J. R., Y.-J. Chang, S. J. MacNaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil b-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Swofford, D. L. 1999. Paup*: phylogenetic analysis using parsimony (*and other methods), 4.0b10 ed. Sinauer Associates, Sunderland, MA.
- 80.Takai, K., D. P. Moser, M. DeFlaun, T. C. Onstott, and J. K. Fredrickson. 2001. Archaeal diversity in waters from deep south African gold mines. Appl. Environ. Microbiol. 67:5750-5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Teira, E., H. van Aken, C. Veth, and G. J. Herndl. 2006. Archaeal uptake of enantiomeric amino acids in the meso- and bathypelagic waters of the North Atlantic. Limnol. Oceanogr. 51:60-69. [Google Scholar]
- 82.Torsvik, V., and L. Ovreas. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240-245. [DOI] [PubMed] [Google Scholar]
- 83.Treusch, A. H., S. Leininger, A. Kletzin, S. C. Schuster, H.-P. Klenk, and C. Schleper. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985-1995. [DOI] [PubMed] [Google Scholar]
- 84.Treves, D. S., B. Xia, and J. M. Tiedje. 2003. A two-species test of the hypothesis that spatial isolation influences microbial diversity in soil. Microb. Ecol. 45:20-28. [DOI] [PubMed] [Google Scholar]
- 85.Vaisanen, R. K., M. S. Roberts, J. L. Garland, S. D. Frey, and L. A. Dawson. 2005. Physiological and molecular characterisation of microbial communities associated with different water-stable aggregate size classes. Soil Biol. Biochem. 37:2007-2016. [Google Scholar]
- 86.van der Wielen, P. W. J. J., and S. K. Heijs. 2007. Sulfate-reducing prokaryotic communities in two deep hypersaline anoxic basins in the Eastern Mediterranean deep sea. Environ. Microbiol. 9:1335-1340. [DOI] [PubMed] [Google Scholar]
- 87.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Y. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 88.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weidler, G. W., M. Dornmayr-Pfaffenhuemer, F. W. Gerbl, W. Heinen, and H. Stan-Lotter. 2007. Communities of Archaea and Bacteria in a subsurface radioactive thermal spring in the Austrian Central Alps and evidence of ammonia-oxidizing Crenarchaeota. Appl. Environ. Microbiol. 73:259-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wright, D. A., K. Killham, L. A. Glover, and J. I. Prosser. 1995. Role of pore size location in determining bacterial activity during predation by protozoa in soil. Appl. Environ. Microbiol. 61:3537-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wuchter, C., B. Abbas, M. J. L. Coolen, L. Herfort, J. van Bleijswijk, P. Timmers, M. Strous, E. Teira, G. J. Herndl, J. J. Middleburg, S. Schouten, and J. S. S. Damste. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. USA 103:12317-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou, J., B. Xia, H. Huang, A. V. Palumbo, and J. M. Tiedje. 2004. Microbial diversity and heterogeneity in sandy subsurface soils. Appl. Environ. Microbiol. 70:1723-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou, J., B. Xia, D. S. Treves, L.-Y. Wu, T. L. Marsh, R. V. O'Neill, A. V. Palumbo, and J. M. Tiedje. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zimmerman, J., J. M. Gonzalez, C. Saiz-Jimenez, and W. Ludwig. 2005. Detection and phylogenetic relationships of highly diverse uncultured acidobacterial communities in Altamira Cave using 23S rRNA sequence analysis. Geomicrob. J. 22:379-388. [Google Scholar]