Abstract
Incomplete and/or sluggish maltotriose fermentation causes both quality and economic problems in the ale-brewing industry. Although it has been proposed previously that the sugar uptake must be responsible for these undesirable phenotypes, there have been conflicting reports on whether all the known α-glucoside transporters in Saccharomyces cerevisiae (MALx1, AGT1, and MPH2 and MPH3 transporters) allow efficient maltotriose utilization by yeast cells. We characterized the kinetics of yeast cell growth, sugar consumption, and ethanol production during maltose or maltotriose utilization by several S. cerevisiae yeast strains (both MAL constitutive and MAL inducible) and by their isogenic counterparts with specific deletions of the AGT1 gene. Our results clearly showed that yeast strains carrying functional permeases encoded by the MAL21, MAL31, and/or MAL41 gene in their plasma membranes were unable to utilize maltotriose. While both high- and low-affinity transport activities were responsible for maltose uptake from the medium, in the case of maltotriose, the only low-affinity (Km, 36 ± 2 mM) transport activity was mediated by the AGT1 permease. In conclusion, the AGT1 transporter is required for efficient maltotriose fermentation by S. cerevisiae yeasts, highlighting the importance of this permease for breeding and/or selection programs aimed at improving sluggish maltotriose fermentations.
Several important industrial applications of the yeast Saccharomyces cerevisiae, such as brewing, baking, and the production of distilled beverages, rely on the efficient fermentation of starch hydrolysates rich in glucose and in the α-glucosides maltose and maltotriose. In brewer's wort, for example, the most abundant fermentable sugar is maltose (50 to 60%), followed by maltotriose (15 to 20%) and glucose (10 to 15%). Of these sugars, glucose is preferentially and rapidly utilized by the yeast cells, but process efficiency also requires the complete fermentation of both maltose and maltotriose. After glucose exhaustion, maltose is easily fermented by the majority of S. cerevisiae strains, and not only is maltotriose the least preferred sugar for uptake by these yeast cells, but many yeasts may not use it at all (23, 37, 44). The difficulty that some strains have in consuming maltotriose leads to one of the problems experienced by many breweries, namely, a high content of residual sugar in the finished beer, low ethanol yields, and atypical beer flavor profiles. Therefore, the rate of uptake and fermentation of maltotriose is one of the major determinants of fermentation efficiency and product quality. Due to its relevance for beer fermentation, maltotriose utilization has been studied mostly with the two classes of yeast strains used for brewing, S. cerevisiae ale strains and S. pastorianus lager strains. These studies have revealed that maltotriose utilization by ale strains is significantly slower than that by lager yeasts and that, consequently, residual maltotriose is more common at the end of ale fermentations (22, 23, 44).
Previous genetic and biochemical studies focusing mainly on the maltose fermentation system revealed a series of five unlinked telomere-associated MAL loci: MAL1 through MAL4 and MAL6. Each locus contains at least one copy of three different genes (MALx1, where x stands for one of the five loci) encoding a maltose permease responsible for the active uptake of maltose, a gene (MALx2) for an intracellular maltase or α-glucosidase that hydrolyzes the sugar into glucose molecules, and a gene (MALx3) encoding a positive regulatory protein that induces the transcription of the two previous genes in the presence of maltose (25). The genes in these MAL loci show high degrees of sequence and functional homology, but there may also be extensive variability since these genes are telomeric and several different alleles that determine distinct phenotypes (i.e., MAL-inducible and MAL-constitutive phenotypes) have been described previously. The MAL1 locus is considered to be the progenitor locus from which all other MAL loci were derived, as all S. cerevisiae strains contain MAL1 sequences at chromosome VII. This pattern holds true even for many non-maltose-fermenting yeast strains, which may harbor partially functional mal1p (MAL13 mal11 mal12), mal1g (mal13 MAL11 MAL12), or mal10 (mal13 mal11 MAL12) loci containing just a functional regulator, just a maltose permease gene and a maltase gene, or just a maltase gene, respectively (16, 24).
Several studies on sugar utilization by yeast cells have also revealed that maltose and maltotriose are transported by different permeases, while the intracellular maltases are capable of hydrolyzing both sugars (37, 42). All yeast α-glucoside transport systems so far characterized are H+ symporters that use the electrochemical proton gradient to actively transport these sugars into the cell, even for downhill transport (7, 30, 36). Maltose transport into the cell is required for the full induction of MAL genes, and several previous studies have also shown that maltose uptake is the rate-limiting step for fermentation (21, 27, 41). Maltose transport in both laboratory and industrial yeast strains has thus been extensively studied, revealing complex kinetics that indicates the presence of high- and low-affinity transporters. The analysis of yeast strains carrying the MAL11, MAL21, or MAL61 maltose transporter revealed that these permeases have a high affinity (Km, ∼2 to 4 mM) for maltose, allowing the efficient fermentation of maltose and turanose, but not maltotriose or other α-glucosides, including α-methylglucoside, palatinose, isomaltose, and melezitose (5-7, 16, 21, 27, 32). Significantly less well characterized than maltose transport, maltotriose uptake by yeast cells also shows complex kinetics indicating the presence of permeases with different affinities for maltotriose (32, 42, 45). The molecular identity of maltotriose transporters in S. cerevisiae remained largely unknown until some years ago when a new permease gene, designated AGT1, in a partially functional mal1g locus was characterized (16). The AGT1 H+ symporter transports a series of α-glucosides (35) and has high affinity (Km, ∼8 mM) for trehalose and sucrose and significantly lower affinity (Km, ∼20 to 35 mM) for maltose, maltotriose, and α-methylglucoside (1, 4, 32, 33). In accordance with the importance of maltotriose transport for the industrial application of yeasts, practically all brewing strains have the AGT1 permease gene in their genomes, and transcriptome studies have also shown high levels of expression of this gene during wort fermentation (19, 20, 23, 39).
A further level of complexity in maltotriose utilization by S. cerevisiae yeast cells is shown by the fact that several industrial strains can utilize maltotriose aerobically, i.e., growth on this carbon source occurs in the absence of ethanol production, while other strains ferment maltotriose efficiently (22, 28, 42, 43). Since sugar influx in glycolysis seems to be the rate-limiting step for the fermentation of the sugar, very likely, the relative fraction of fermented or respired maltotriose varies with the number of functional maltotriose transporters present in each strain. However, there have been controversial reports regarding the molecular mechanism responsible for the observed patterns of maltotriose utilization by yeast cells. Some years ago, Day and coworkers presented data indicating that all known α-glucoside transporters present in S. cerevisiae, including the MAL31 and MAL61 maltose permeases and two new members of this family of permeases discovered after the sequencing of the yeast genome (encoded by MPH2 and MPH3), allow the growth of the yeast cells on both maltose and maltotriose (9, 10). Furthermore, their kinetic analysis of maltose and maltotriose uptake by the cells indicated that all these permeases can transport both sugars with practically the same affinities (Km, ∼3 to 7 mM) and capacities (maximum rate, 43 ± 6 nmol min−1 mg of yeast−1). Although they made no attempt to explain why maltose is preferably transported by the cells, they proposed that nutritional and environmental stresses occurring at the later stages of fermentation, and not necessarily differences in the transporters responsible for the uptake of maltose and maltotriose, would be responsible for the inability of the yeast cells to utilize maltotriose (9, 10). In the present study, we have reevaluated the contribution of the maltose permeases, encoded by the MALx1 genes, to the active transport and fermentation of maltotriose by S. cerevisiae cells. Our results indicate that these permeases do not allow efficient maltotriose uptake or utilization and that the AGT1 permease is required for the efficient fermentation of this sugar by S. cerevisiae.
MATERIALS AND METHODS
Media and culture conditions.
Cells were routinely grown on rich YP medium (1% yeast extract and 2% peptone) supplemented with 2% of the indicated carbon source. The pH of each medium was adjusted to 5.0 with HCl. When required, 2% agar and 200 mg of Geneticin (G-418) sulfate/liter were added to the medium. Cells were grown aerobically at 28°C with shaking (160 rpm) in cotton-plugged Erlenmeyer flasks filled to one-fifth of the volume with medium. Cellular growth was monitored by turbidity measurements at 570 nm, culture samples were harvested regularly and centrifuged (5,000 × g for 1 min), and their supernatants were used for the evaluation of sugars and ethanol. For batch fermentations using malt extract, yeasts were pregrown on YP-2% maltose until the exponential phase (∼1 mg [dry weight] of yeast/ml), centrifuged (3,500 × g for 3 min), washed twice with cold water, and inoculated at a high cell density (10 mg [dry weight] of yeast/ml) into synthetic medium (0.4% yeast nitrogen base without amino acids and with 1% ammonium sulfate) containing 5% malt extract (grade X; Sigma). Samples were collected regularly and processed as described above.
Yeast strains.
The S. cerevisiae strains and oligonucleotides used in the present study are described in Table 1. The three wild-type strains 1403-7A, CEN.PK2-1C, and MC966A had unrelated genetic backgrounds. Standard methods for yeast transformation, DNA manipulation, and analysis were employed (2). The AGT1 gene was deleted according to a previously described PCR-based gene replacement procedure (4). Briefly, the kanMX6 cassette from plasmid pFA6a-kanMX6 (26) was amplified with primers AGT1-pFA6-F1 and AGT1-pFA6-R1, and the resulting PCR product of 1,579 bp was used to transform competent yeast cells. After 2 h of cultivation on YP-2% glucose, the transformed cells were plated onto the same medium containing G-418 and incubated at 28°C. G-418-resistant isolates were tested for proper genomic integration of the kanMX6 cassette at the AGT1 locus by Southern analysis (see below) or analytical colony PCR using three primers (V-AGT1-F, V-AGT1-R, and V-kanr-R) (Table 1). This set of three primers amplified a 1,938-bp fragment from a normal AGT1 locus or yielded an 813-bp fragment if the kanMX6 cassette was correctly integrated at this locus and replaced the AGT1 gene.
TABLE 1.
S. cerevisiae strains and oligonucleotides used in this study
| Yeast strain or primer | Relevant genotype or description | Source or reference(s) |
|---|---|---|
| Yeast strainsa | ||
| 1403-7A | MATaMAL4cMGL3 suc2 gal3 gal4 trp1 ura3 | 24 |
| CEN.PK2-1C | MATaMAL2-8cura3-52 his3Δ1 leu2-3,112 trp1-289 | 12 |
| MC966A | MATaMAL2 ura3-52 his3-11,15 leu2-3,112 SUC2 | 4 |
| LCM001 | agt1Δ::kanMX6 derivative of 1403-7A | This study |
| LCM003 | agt1Δ::kanMX6 derivative of CEN.PK2-1C | This study |
| BSY09 | agt1Δ::kanMX6 derivative of MC966A | 4 |
| S288C | MATα mal gal2 mel flo1 flo8-1 hap1 SUC2 | 14, 40 |
| Primers | ||
| AGT1-pFA6-F1 | AAGCAAGAAGAAGGCTGCCTCAAAAAATGAGGATAAAAACATTTCTGAGCGGATCCCCGGGTTAATTAA | Invitrogen |
| AGT1-pFA6-R1 | AAAGGGATTCCTTATTTCTTCCAAAAAAAAAAAAACAACCCTTTTACTTAGAATTCGAGCTCGTTTAAAC | Invitrogen |
| AGT1-F | AGGAGCTCATGAAAAATATCATTTCATTGG | GIBCO BRL |
| AGT1-R | TTGGATCCACATTTATCAGCTGC | GIBCO BRL |
| MALx1-F | CCATACTTGTTGTGAGTGG | GIBCO BRL |
| MALx1-R | TCATTTGTTCACAACAGATG | Invitrogen |
| V-AGT1-F | GAATTTTCGGGTTGGTG | GIBCO BRL |
| V-AGT1-R | TTGGATCCACATTTATCAGCTGC | GIBCO BRL |
| V-kanr-R | GGAATCGAATGCAACCGG | GIBCO BRL |
The wild-type strains 1403-7A, CEN.PK2-1C, and MC966A have unrelated genetic backgrounds.
PFGE, chromosome blotting, and hybridization.
Yeast chromosomes were prepared as previously described (15) from 1 ml of yeast cells pregrown in YP medium containing 2% glucose and collected at the stationary phase of growth. Cells were washed with 10 mM Tris-HCl, pH 7.5, containing 50 mM EDTA and resuspended in 0.4 ml of 4 mM Tris-HCl, pH 7.5, containing 95 mM EDTA, 130 μg of Zymolyase 20T/ml, and 0.7% molten (42°C) low-melting-point agarose. After solidification in a mold (Pharmacia Biotech), the agarose blocks were immersed in 10 mM Tris-HCl, pH 7.5, containing 0.5 M EDTA and incubated at 37°C for 8 h. Following a subsequent incubation in 10 mM Tris-HCl, pH 9.5, containing 0.5 M EDTA, 1% N-lauroylsarcosine, and 2 mg of proteinase K/ml at 50°C overnight, the blocks were washed in 10 mM Tris-HCl, pH 7.5, containing 50 mM EDTA and stored at 4°C in the same buffer. Each low-melting-point agarose block was transferred onto a 1% agarose gel in 50 mM Tris-HCl, pH 8.3, containing 50 mM boric acid and 1 mM EDTA. Pulsed-field gel electrophoresis (PFGE) was performed at 10°C using a Gene Navigator pulsed-field system (Pharmacia Biotech) for a total of 27 h at 200 V. After 15 h, the pulse time of 70 s was stepped up to 120 s for 12 h. Following electrophoresis, the gel was stained with ethidium bromide and photographed.
The chromosomes separated by PFGE were transferred onto a nylon filter (Biodyne A; GIBCO BRL) by capillary blotting (2), and the labeling of DNA probes (see below), including prehybridization, hybridization, stringency washing, and chemiluminescent signal generation and detection, was performed by using an AlkPhos kit as recommended by the manufacturer (GE Healthcare/Amersham Biosciences). After hybridization, an autoradiography film (Hiperfilm ECL; Kodak) was exposed to the membrane for 2 to 3 h before the film was developed. Images were obtained by scanning with an ImageScanner (Amersham Biosciences) and annotated with Microsoft PowerPoint. Probes corresponding to nucleotides +1 through +1848 relative to the translation start site on the AGT1 open reading frame or −73 through +1845 relative to the translation start site of the MAL31 gene were generated by PCR using primer pairs AGT1-F and AGT1-R and MAL31-F and MAL31-R (Table 1), respectively, and genomic DNA from strain CEN.PK2-1C as a template.
Analytical methods.
Glucose was assessed by the glucose oxidase and peroxidase method using a commercial kit (BioDiagnostica-Laborclin). Maltose and maltotriose were assessed spectrophotometrically at 540 nm with methylamine in 0.25 M NaOH as described previously (1). Sugars were also analyzed by thin-layer chromatography (TLC) on silica gel 60 TLC sheets using 5:2:1 (vol/vol/vol) n-butanol-ethanol-water as the solvent. The positions of different sugars (10 μg each of glucose, maltose, and maltotriose) in each sample and the positions of standard compounds on the TLC plates were determined with sulfuric acid as described elsewhere (34). Ethanol was assessed with alcohol oxidase and peroxidase as described previously (1, 3).
The α-glucosidase activity in cells collected by centrifugation (2,500 × g for 3 min) at the exponential phase of growth was determined in situ with permeabilized yeast cells as described previously (31) by using 100 mM MOPS (morpholinepropanesulfonic acid)-NaOH (pH 6.8) buffer and 2 mM p-nitrophenyl-α-d-glucopyranoside (pNPαG) or 100 mM maltose or maltotriose as substrates. The activity of the AGT1 permease was determined with a specific colorimetric assay (17) using 5 mM pNPαG in 100 mM succinate-Tris, pH 5.0. All assays were done in triplicate, and controls with previously boiled yeast cells were used. The rates and kinetics of active H+-trehalose, H+-maltose, or H+-maltotriose symport were determined as previously described (32) using a PHM84 research pH meter attached to a TT1 Servograph (Radiometer, Copenhagen). Initial rates of sugar-induced proton uptake were calculated from the slope of the initial (<10-s) part of the curve obtained in the recorder by subtracting the basal rate of proton uptake observed before the addition of 0.1 to 100 mM sugar. All determinations were done at least in duplicate, and assays were monitored so that no more than 5% of the substrate was depleted. All activities were expressed as nanomoles of the substrate transported (or hydrolyzed) per milligram (dry weight) of cells per minute.
RESULTS
MALx1 and AGT1 genotypes of yeast strains.
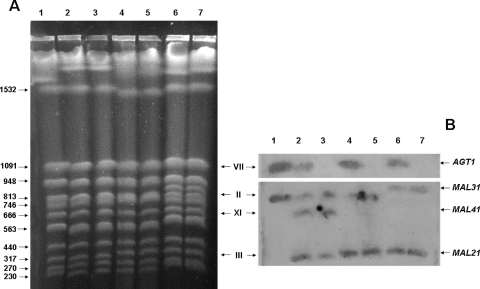
In the present study, we analyzed two MAL-constitutive S. cerevisiae strains (strain CEN.PK2-1C carries MAL2-8c, and strain 1403-7A carries MAL4c) and a MAL-inducible S. cerevisiae strain (MC966-A carries MAL2) (Table 1). Although a single MAL locus is sufficient for maltose fermentation, many yeast strains contain more than one of these loci in their genomes (16, 20, 23, 24, 39). Thus, we first confirmed the presence of the AGT1 gene in chromosomes VII of all these three strains (Fig. 1) and also identified which MALx1 genes were present in their chromosomes. Strain 1403-7A contains three MALx1 genes, the permease gene from the MAL4c locus (MAL41, chromosome XI), and also MAL21 (chromosome III) and MAL31 (chromosome II). Both genetic and molecular studies (16, 24) have shown that in strain 1403-7A, the MAL41, AGT1, and MAL31 permease genes are functional, the two latter genes belonging to the partially functional mal1g (AGT1 MAL12 mal13) and mal3g (MAL31 MAL32 mal33) loci lacking a MALx3 regulator. On the other hand, strains CEN.PK2-1C (MAL2-8c) and MC966A (MAL2) have, besides the MAL21 and AGT1 permease genes, the MAL31 gene (Fig. 1).
FIG. 1.
Detection of MALx1 and AGT1 transporter genes in yeast strains. (A) Separation of chromosomes of yeast strains by PFGE and staining with ethidium bromide. (B) Detection of MALx1 and AGT1 transporter genes after the chromosomes shown in panel A were blotted onto a nylon membrane and hybridized with a probe for AGT1 (upper panel) or MALx1 (lower panel). Lanes 1, the reference strain S288C that contains the AGT1 and MAL31 genes in its genome (14, 40); lanes 2, 1403-7A; lanes 3, LCM001; lanes 4, CEN.PK2-1C; lanes 5, LCM003; lanes 6, MCY996A; lanes 7, BSY09. The values on the right in panel A are the sizes (in kilobase pairs) of selected S288C chromosomes. The Roman numerals between panels A and B are chromosome numbers. The positions of chromosomes carrying AGT1, MAL21, MAL31, and MAL41 are indicated on the right in panel B. Besides those represented by the bands shown in the upper part of panel B, no chromosome hybridized with the AGT1 probe. Conversely, besides those represented by the bands shown in the lower part of panel B, no chromosome hybridized with the MALx1 probe.
In order to verify the importance of the permease encoded by the AGT1 gene for maltotriose fermentation, we deleted this gene from the genomes of strains 1403-7A, CEN.PK2-1C, and MCY966A. Our PFGE and blotting results for the isogenic counterparts of these strains with the AGT1 gene deleted (strains LCM001, LCM003, and BSY09, respectively) (Table 1) confirmed that this gene was specifically removed from their genomes (Fig. 1). Indeed, the deletion of the AGT1 gene was also confirmed by assaying the capacity of maltose-grown cells to transport trehalose or pNPαG, both sugars transported with high affinity by the AGT1 permease (17, 32). Table 2 shows the data obtained with strain CEN.PK2-1C and its agt1Δ counterpart, strain LCM003, which showed no trehalose or pNPαG transport activity. Similar results were obtained with the other unrelated strain pairs 1403-7A and LCM001 and MC966A and BSY09.
TABLE 2.
Active sugar transport and hydrolysis by yeast strains
| Strain and growth substrate | Rate of transporta (nmol mg [dry wt] of cells−1 min−1) of:
|
Rate of hydrolysisb (nmol mg [dry wt] of cells−1 min−1) of:
|
|||||
|---|---|---|---|---|---|---|---|
| Maltose | Maltotriose | Trehalose | pNPαG | Maltose | Maltotriose | pNPαG | |
| CEN.PK2-1C (wild type) | |||||||
| 2% Maltose | 62 | 7 | 21 | 5 | 1,083 | 495 | 1,107 |
| 2% Maltotriose | 104 | 10 | 39 | 10 | 1,027 | 463 | 1,192 |
| LCM003 (agt1Δ) | |||||||
| 2% Maltose | 74 | <1 | <1 | 0 | 997 | 412 | 1,171 |
| 2% Maltotriosec | 11 | <1 | <1 | 0 | 2 | 0 | 15 |
Estimated by the rate of H+ cotransport (for maltose, maltotriose, and trehalose) or p-nitrophenol production (for pNPαG) by the yeast cells using a 5 mM final sugar concentration. Assays were carried out in triplicate, with standard errors of less than 15%.
Determined with permeabilized yeast cells using a 100 mM (maltose and maltotriose) or 2 mM (pNPαG) final sugar concentration. Assays were carried out in triplicate, with standard errors of less than 10%.
Cells were collected after 75 h of incubation in rich YP-2% maltotriose medium, at an optical density of ∼3.5 (i.e., about 1 mg [dry weight] of cells/ml).
Maltose and maltotriose fermentation.
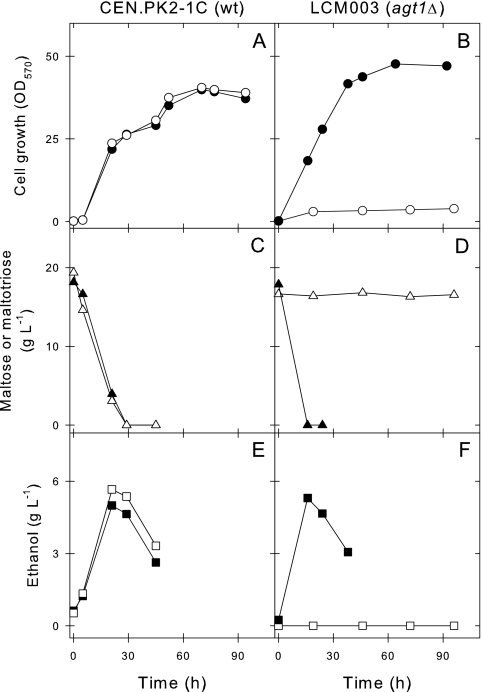
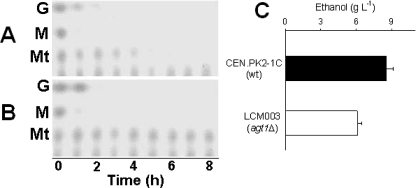
To avoid any repressive effect on the utilization of maltotriose triggered by glucose (22), all yeast strains used in the present study were pregrown in rich YP medium containing 2% maltose as the carbon source. When the maltose-pregrown cells of any of the three wild-type strains 1403-7A, CEN.PK2-1C, and MCY966A were inoculated into rich medium containing maltose or maltotriose, the maltose or maltotriose was fermented rapidly by the yeast cells. Data for strain CEN.PK2-1C are shown in Fig. 2, and the results for the two other wild-type strains 1403-7A and MC966A were similar to those shown for strain CEN.PK2-1C. Thus, our results indicated that maltose-adapted yeast cells can efficiently ferment maltotriose, with kinetics of growth and ethanol yields very similar to those obtained during maltose fermentation (Fig. 2). Nevertheless, when these yeast cells pregrown on maltose were used to ferment a sugar mixture resembling brewer's wort (prepared with 5% malt extract and containing approximately 10 g each of glucose, maltose, and maltotriose/liter), different patterns of sugar utilization were observed (Fig. 3A). As previously described for other industrial yeast strains (13), maltose-adapted cells rapidly consumed the maltose present in the mixture, even more quickly than they consumed glucose, but still consumed maltotriose very slowly, taking roughly twice the time required for the consumption of maltose and glucose to remove maltotriose from the medium (Fig. 3).
FIG. 2.
Maltose and maltotriose fermentation by yeast strains. The rates of cell growth (A and B), sugar consumption (C and D), and ethanol production (E and F) in rich YP medium containing 2% maltose (black symbols) or maltotriose (white symbols) inoculated with maltose-pregrown cells of the wild-type (wt) strain CEN.PK2-1C (left panels A, C, and E) or its agt1Δ counterpart strain LCM003 (right panels B, D, and F) were determined. OD570, optical density at 570 nm.
FIG. 3.
Fermentation of sugars from malt extract by yeast strains. (A and B) TLC analysis of sugar consumption during batch fermentation of a 5% malt extract by 10-g/liter concentrations of yeast cells of the wild-type (wt) CEN.PK2-1C strain (A) or its agt1Δ counterpart strain LCM003 (B), pregrown in YP-2% maltose. The positions of glucose (G), maltose (M), and maltotriose (Mt) are indicated on the left. (C) Final ethanol concentration after malt extract fermentation by each strain.
A very different phenotype was observed when the AGT1 permease gene was deleted from the genomes of these S. cerevisiae strains. The agt1Δ yeast strains continued to grow on and ferment maltose efficiently, but growth on and fermentation of maltotriose by these cells were totally impaired (data for strain LCM003 are shown in Fig. 2). This lack of maltotriose consumption was also observed during the fermentation of malt extract (Fig. 3B), and consequently, the agt1Δ LCM003 strain produced less ethanol than the parental wild-type CEN.PK2-1C yeast strain (Fig. 3C). Strains LCM001 or BSY09 were also incapable of maltotriose utilization, while maltose fermentation by these agt1Δ yeast strains was unaffected. Although the data shown in Fig. 2 are restricted to just ∼100 h of cultivation, strain LCM003 was unable to utilize maltotriose even after an extensive (>200-h) incubation in rich YP medium containing this sugar. It should be stressed that strain LCM003, pregrown on maltose, had normal α-glucosidase activity (even using maltotriose as the substrate) and active maltose uptake but appeared to lack H+-maltotriose symport activity (Table 2).
Kinetics of active maltose and maltotriose transport.
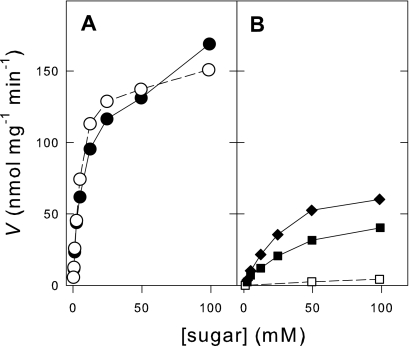
The results shown above prompted us to determine the kinetics of active sugar uptake by these strains. Figure 4 shows the kinetics of the H+-maltose symport activities of strain CEN.PK2-1C and strain LCM003 pregrown on maltose. As already extensively reported in the literature (7, 27, 32, 42), the kinetics of maltose transport by the wild-type strain CEN.PK2-1C indicated the presence of both high-affinity (Km, ∼5 mM) and low-affinity (Km, ∼30 mM) transport activities. When the AGT1 gene was deleted from the genome, only the low-affinity H+-maltose symport activity was affected (Fig. 4), the cells retaining the ability to take up maltose from the medium through the high-affinity (and high-capacity) component. In the case of the active H+-maltotriose symport activity, quite a different pattern was observed, as the wild-type strain presented only a single low-affinity (Km, 36 ± 2 mM) component, which was completely lost when the AGT1 gene was deleted from the genome (see the data for strain LCM003 in Fig. 4). This low-affinity H+-maltotriose symport activity was also the only transport activity present in the CEN.PK2-1C cells grown in maltotriose (Fig. 4), although these cells had greater maltotriose uptake capacity than cells pregrown in maltose, in agreement with the data presented in Table 2. The same results were obtained when the kinetics of active sugar uptake were determined for the two other wild-type strains, 1403-7A and MC966A (the presence of high- and low-affinity maltose transport and just low-affinity maltotriose uptake), as well as the corresponding agt1Δ yeast strains LCM001 and BSY09 (the loss of the low-affinity maltose uptake and the absence of active maltotriose transport).
FIG. 4.
Kinetics of active H+-maltose and H+-maltotriose symport activity in yeast strains. The initial rates of active H+ cotransport with maltose (A) or maltotriose (B) in yeast cells of the wild-type CEN.PK2-1C strain (• and ▪) or its agt1Δ counterpart strain LCM003 (○ and □) pregrown in YP-2% maltose were determined. The kinetics of active H+-maltotriose symport in the wild-type CEN.PK2-1C strain pregrown in YP-2% maltotriose (⧫) was also determined. V, transport rate.
DISCUSSION
The reasons leading to the incomplete or delayed consumption of maltotriose by S. cerevisiae strains in starch-based industrial processes, such as brewing, remain controversial. Since many industrial yeast strains unable to utilize maltotriose nevertheless have intracellular α-glucosidases capable of hydrolyzing this sugar, it was proposed that the uptake of the sugar must be responsible for these undesirable phenotypes (23, 37, 42, 43). However, this point of view was recently challenged (9, 10) by results that indicated that all known α-glucoside transporters (i.e., MALx1, AGT1, and MPH2 and MPH3 transporters), when expressed constitutively in yeast cells, allow growth on maltotriose. The authors of these studies also claimed that all these permeases transport both maltose and maltotriose with practically the same affinities and capacities.
In the present study, we analyzed the patterns of active transport and fermentation of maltose and maltotriose by several unrelated S. cerevisiae yeast strains, both MAL constitutive and MAL inducible, and their isogenic counterparts lacking the AGT1 gene. Although they were haploid laboratory strains, the wild-type S. cerevisiae strains analyzed in this study showed all the characteristics of ale-brewing strains. These include the presence of multiple MAL loci in their genomes (Fig. 1) and efficient maltose and maltotriose fermentation (Fig. 2); these strains also displayed the typical slow kinetics of maltotriose utilization from mixtures of glucose, maltose, and maltotriose resembling brewer's wort (Fig. 3). Our results clearly show that the presence of MALx1 (MAL21, MAL31, or MAL41) transporters in the plasma membranes did not allow efficient maltotriose consumption or fermentation by S. cerevisiae cells. These results are in full agreement with the results from the early genetic work done by Stewart and coworkers that showed that both haploid and diploid MAL1 yeast strains cannot consume maltotriose from wort (37). The maltose transporter encoded by the MAL1 locus, the MAL11 permease, shows over 99% homology to other (e.g., MAL31- and MAL61-encoded) maltose transporters from S. cerevisiae. Obviously, yeast strains harboring the normal MAL11 gene at chromosome VII cannot have the mal1g allele encoding the AGT1 permease at this locus and, thus, are incapable of maltotriose utilization. Indeed, the inability of the MAL11 permease to allow maltotriose consumption and fermentation from wort has recently been confirmed (reference 18 and our unpublished observations) and is also in accordance with previous data showing that the highly homologous MAL21 (32), MAL31 (28), and MAL61 (5, 16) permeases are unable to transport maltotriose.
Although we have not specifically addressed the involvement of the MPH2 and MPH3 genes in maltotriose utilization, the MAL-constitutive CEN.PK strains have been reported to have these two genes in their genomes (8). We have also confirmed the presence of these two genes in another MAL-constitutive strain, 1403-7A, by comparative genotyping using microarrays (B. U. Stambuk et al., unpublished data). Since in both cases the deletion of the AGT1 permease completely abolished maltotriose fermentation by these MAL-constitutive strains, we can also conclude that the MPH2 and MPH3 genes do not allow efficient maltotriose uptake by the yeast cells. It should be stressed that when the MAL-inducible strain MC996A or its agt1Δ counterpart, BSY09, was transformed with a plasmid containing a constitutive MAL63c gene, the patterns of maltotriose utilization described above were not affected, while transformation with a centromeric plasmid containing the AGT1 gene restored completely the ability of strain BSY09 to ferment maltotriose (data not shown).
Our results also show significant differences in the kinetics of active maltose and maltotriose transport by the yeast strains analyzed (Fig. 4). All the wild-type strains had two H+-maltose transport components, one with high affinity (Km, ∼5 mM) and another with low affinity (Km, ∼30 mM) for the sugar. Our results also show that the deletion of the AGT1 gene did not affect the high-affinity component and that, thus, the agt1Δ strains were fully competent for the efficient uptake of maltose from the medium (Fig. 2 and 3). Regarding active H+-maltotriose uptake, a single low-affinity (Km, 36 ± 2 mM) component could be observed in the wild-type strains grown in either maltose or maltotriose. This low-affinity H+-maltotriose transporter disappeared when the AGT1 gene was deleted from the genomes of the yeast strains (Fig. 4), indicating that the AGT1 transporter is the only fully competent maltotriose permease in S. cerevisiae. The low affinity of the AGT1 permease for maltose and maltotriose in the wild-type strains analyzed is in accordance with data obtained with other unrelated strains (1, 32) and would explain the lower rates of maltotriose consumption from malt extract (Fig. 3) than of maltose uptake by the same cells.
It should be noted that yeast cells grown on maltotriose had higher levels of maltotriose transport activity than cells pregrown on maltose (Fig. 4 and Table 2). This result suggests that maltotriose is probably a better inducer of the AGT1 permease, as recently described for another industrial yeast strain (28) as well as for a strain containing this permease as its unique α-glucoside transporter (1). Although there has been significant progress in understanding how the MALx3 regulators activate the transcription of MAL genes through the MAL upstream activation sequence, we still do not know how this regulatory protein senses maltose and/or maltotriose. Like maltose (41), it seems that maltotriose requires internalization by the yeast cells in order to induce the MAL genes (consider the data on the very low levels of maltose permease and α-glucosidase expression in the agt1Δ strain LCM003 incubated in rich medium containing maltotriose, presented in Table 2). Indeed, our results are in accordance with very recent data showing that both maltose and maltotriose are potent inducers of filamentation in S. cerevisiae (38). The authors of the previous study showed that pseudohyphal signaling and differentiation on maltotriose, but not on maltose or other sugars, was abolished in an agt1Δ/agt1Δ mutant.
We believe that most of the conflicting data reported by Day and coworkers results from the transport assays they employed, which used commercial [14C]maltotriose (9, 10). Not only did they use extensive (150-s) incubation times to measure maltose and maltotriose uptake, which can lead to serious errors in the estimation of the amount of the radiolabeled substrate inside the cells due to the rapid metabolism of the sugar (29), but also it has been recently shown that commercial [14C]maltotriose (from American Radiolabeled Chemicals Inc.) is heavily contaminated (at levels corresponding to up to 16% of the total radioactivity) with both [14C]maltose and [14C]glucose, causing gross overestimations of maltotriose uptake by the yeast cells (11). In zero-trans uptake assays, only a very small proportion (usually less than 1%) of the substrate should enter the cell, and thus, the uptake of any impurity would cause severe overestimation of the ability of a particular permease (e.g., the MALx1, MPH2, or MPH3 permease) to transport maltotriose (9, 10). In our experiments, the kinetic parameters were determined by the initial rates of H+ influx (<10 s) triggered by the cotransport of the sugar, and therefore, these experimental artifacts were avoided. The importance of the AGT1 permease for maltotriose active transport and fermentation is also evident from recent data showing that constitutive and enhanced expression of this gene is required for efficient maltotriose fermentation by S. cerevisiae (1, 36). The recent discovery of new AGT1 alleles in industrial yeast strains (39), some of which seem to transport maltotriose more efficiently (30), opens new opportunities to increase yeast fitness for the fermentation of starch hydrolysates, preventing incomplete or sluggish maltotriose fermentations. These findings may be of particular importance for the brewing industry, as several studies indicate that many industrial ale-brewing strains have dominant AGT1-type permeases in their plasma membranes, independent of their MAL genotypes and/or the presence of several other MALx1 genes (23, 27, 39, 42).
Acknowledgments
This work was supported in part by a grant from the Brazilian agencies CNPq and FAPESP (no. 04/10067-6). S.L.A., R.A.H., and C.H. were recipients of scholarships from CAPES-Brazil, and D.T., L.C.M., and B.U.S. were recipients of research fellowships from CNPq.
We thank R. Needleman (Wayne State University), M. Longtine (University of North Carolina), J. François (Centre de Bioingénierie Gilbert Durand), and A. Kruckeberg (University of Amsterdam) for providing plasmids and yeast strains.
Footnotes
Published ahead of print on 18 January 2008.
REFERENCES
- 1.Alves, S. L., Jr., R. A. Herberts, C. Hollatz, L. C. Miletti, and B. U. Stambuk. 2007. Maltose and maltotriose active transport and fermentation by Saccharomyces cerevisiae. J. Am. Soc. Brew. Chem. 65:99-104. [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, New York, NY.
- 3.Badotti, F., A. S. Batista, and B. U. Stambuk. 2006. Sucrose active transport and fermentation by Saccharomyces cerevisiae. Braz. Arch. Biol. Technol. 49:s115-s123. [Google Scholar]
- 4.Batista, A. S., L. C. Miletti, and B. U. Stambuk. 2004. Sucrose fermentation by Saccharomyces cerevisiae lacking hexose transport. J. Mol. Microbiol. Biotechnol. 8:26-33. [DOI] [PubMed] [Google Scholar]
- 5.Chang, Y. S., R. A. Dubin, E. Perkins, C. A. Michels, and R. B. Needleman. 1989. Identification and characterization of the maltose permease in a genetically defined Saccharomyces strain. J. Bacteriol. 171:6148-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, Q., and C. A. Michels. 1991. MAL11 and MAL61 encode the inducible high-affinity maltose transporter of Saccharomyces cerevisiae. J. Bacteriol. 173:1817-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crumplen, R. M., J. C. Slaughter, and G. G. Stewart. 1996. Characteristics of maltose transporter activity in an ale and lager strain of the yeast Saccharomyces cerevisiae. Lett. Appl. Microbiol. 23:448-452. [DOI] [PubMed] [Google Scholar]
- 8.Daran-Lapujade, P., J.-M. Daran, P. Kotter, T. Petit, M. D. W. Piper, and J. T. Pronk. 2003. Comparative genotyping of the Saccharomyces cerevisiae laboratory strains S288C and CEN.PK113-7D using oligonucleotide microarrays. FEMS Yeast Res. 4:259-269. [DOI] [PubMed] [Google Scholar]
- 9.Day, R. E., P. J. Rogers, I. W. Dawes, and V. J. Higgins. 2002. Molecular analysis of maltotriose transport and utilization by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 68:5326-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day, R. E., V. J. Higgins, P. J. Rogers, and I. W. Dawes. 2002. Characterization of the putative maltose transporters encoded by YDL247w and YJR160c. Yeast 19:1015-1027. [DOI] [PubMed] [Google Scholar]
- 11.Dietvorst, J., J. Londesborough, and H. Y. Steensma. 2005. Maltotriose utilization in lager yeast strains: MTT1 encodes a maltotriose transporter. Yeast 22:775-788. [DOI] [PubMed] [Google Scholar]
- 12.Entian, K.-D., and P. Kotter. 1998. Yeast mutant and plasmid collections. Methods Microbiol. 26:431-449. [Google Scholar]
- 13.Ernandes, J. R., J. W. Williams, I. Russell, and G. G. Stewart. 1993. Effect of yeast adaptation to maltose utilization on sugar uptake during the fermentation of brewer's wort. J. Inst. Brew. 99:67-71. [Google Scholar]
- 14.Feuermann, M., L. Charbonnel, J. de Montigny, J. C. Bloch, S. Potier, and J. L. Souciet. 1995. Sequence of a 9.8 kb segment of yeast chromosome II including the three genes of the MAL3 locus and three unidentified open reading frames. Yeast 11:667-672. [DOI] [PubMed] [Google Scholar]
- 15.Guerring, S. L., C. Connelly, and P. Hieter. 1991. Positional mapping of genes by chromosome blotting and chromosome fragmentation. Methods Enzymol. 194:57-77. [DOI] [PubMed] [Google Scholar]
- 16.Han, E. K., F. Cotty, C. Sottas, H. Jiang, and C. A. Michels. 1995. Characterization of AGT1 encoding a general α-glucoside transporter from Saccharomyces. Mol. Microbiol. 17:1093-1107. [DOI] [PubMed] [Google Scholar]
- 17.Hollatz, H., and B. U. Stambuk. 2001. Colorimetric determination of active α-glucoside transport in Saccharomyces cerevisiae. J. Microbiol. Methods 46:253-259. [DOI] [PubMed] [Google Scholar]
- 18.Houghton-Larsen, J., and A. Brandt. 2006. Fermentation of high concentrations of maltose by Saccharomyces cerevisiae is limited by the COMPASS methylation complex. Appl. Environ. Microbiol. 72:7176-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James, T. C., S. Campbell, D. Donnelly, and U. Bond. 2003. Transcription profile of brewery yeast under fermentation conditions. J. Appl. Microbiol. 94:432-448. [DOI] [PubMed] [Google Scholar]
- 20.Jespersen, L., L. B. Cesar, P. G. Meaden, and M. Jakobsen. 1999. Multiple α-glucoside transporter genes in brewer's yeast. Appl. Environ. Microbiol. 65:450-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kodama, Y., N. Fukui, T. Ashikari, Y. Shibano, K. Morioka-Fujimoto, Y. Hiraki, and K. Nakatani. 1995. Improvement of maltose fermentation efficiency: constitutive expression of MAL genes in brewing yeasts. J. Am. Soc. Brew. Chem. 53:24-29. [Google Scholar]
- 22.Londesborough, J. 2001. Fermentation of maltotriose by brewer's and baker's yeast. Biotechnol. Lett. 23:1995-2000. [Google Scholar]
- 23.Meneses, F. J., and V. Jiranek. 2002. Expression patterns of genes and enzymes involved in sugar catabolism in industrial Saccharomyces cerevisiae strains displaying novel fermentation characteristics. J. Inst. Brew. 108:322-335. [Google Scholar]
- 24.Michels, C. A., and R. B. Needleman. 1984. The dispersed, repeated family of MAL loci in Saccharomyces spp. J. Bacteriol. 157:949-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novak, S., V. Zechner-Krpan, and V. Marie. 2004. Regulation of maltose transport and metabolism in Saccharomyces cerevisiae. Food Technol. Biotechnol. 42:213-218. [Google Scholar]
- 26.Petracek, M. E., and M. S. Longtine. 2002. PCR-based engineering of yeast genome. Methods Enzymol. 350:445-469. [DOI] [PubMed] [Google Scholar]
- 27.Rautio, J., and J. Londesborough. 2003. Maltose transport by brewer's yeast in brewer's wort. J. Inst. Brew. 109:251-261. [Google Scholar]
- 28.Salema-Oom, M., V. V. Pinto, P. Goncalves, and I. Spencer-Martins. 2005. Maltotriose utilization by industrial Saccharomyces strains: characterization of a new member of the α-glucoside transporter family. Appl. Environ. Microbiol. 71:5044-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serrano, R. 1997. Energy requirements for maltose transport in yeasts. Eur. J. Biochem. 80:97-102. [DOI] [PubMed] [Google Scholar]
- 30.Smit, A., R. R. Cordero-Otero, and I. S. Pretorius. 2007. Differences among AGT1-encoded α-glucoside transporters and their ability to transport maltotriose in Saccharomyces yeasts. Ann. Microbiol. 57:77-84. [Google Scholar]
- 31.Stambuk, B. U. 1999. A simple experiment illustrating metabolic regulation, induction versus repression of yeast α-glucosidase. Biochem. Educ. 27:177-180. [Google Scholar]
- 32.Stambuk, B. U., and P. S. de Araujo. 2001. Kinetics of active α-glucoside transport by Saccharomyces cerevisiae. FEMS Yeast Res. 1:73-78. [DOI] [PubMed] [Google Scholar]
- 33.Stambuk, B. U., A. S. Batista, and P. S. de Araujo. 2000. Kinetics of active sucrose transport in Saccharomyces cerevisiae. J. Biosci. Bioeng. 89:212-214. [DOI] [PubMed] [Google Scholar]
- 34.Stambuk, B. U., J. H. Crowe, L. M. Crowe, A. D. Panek, and P. S. de Araujo. 1993. A dependable method for the synthesis of [14C]trehalose. Anal. Biochem. 212:150-153. [DOI] [PubMed] [Google Scholar]
- 35.Stambuk, B. U., M. A. da Silva, A. D. Panek, and P. S. de Araujo. 1999. Active α-glucoside transport in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 170:105-110. [DOI] [PubMed] [Google Scholar]
- 36.Stambuk, B. U., S. L. Alves, Jr., C. Hollatz, and C. R. Zastrow. 2006. Improvement of maltotriose fermentation by Saccharomyces cerevisiae. Lett. Appl. Microbiol. 43:370-376. [DOI] [PubMed] [Google Scholar]
- 37.Stewart, G. G., J. Erratt, I. Garrison, T. Goring, and I. Hancock. 1979. Studies on the utilization of wort carbohydrates by brewer's yeast strains. Tech. Q. Master Brew. Assoc. Am. 16:1-7. [Google Scholar]
- 38.Van de Velde, S., and J. M. Thevelein. 21 September 2007. Cyclic AMP-protein kinase A and Snf1 signaling mechanisms underlie the superior potency of sucrose for induction of filamentation in Saccharomyces cerevisiae. Eukaryot. Cell. doi: 10.1128/EC.00276-07. [DOI] [PMC free article] [PubMed]
- 39.Vidgren, V., L. Ruohonen, and J. Londesborough. 2005. Characterization and functional analysis of the MAL and MPH loci for maltose utilization in some ale and lager yeast strains. Appl. Environ. Microbiol. 71:7846-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volckaert, G., M. Voet, and J. Robben. 1997. Sequence analysis of a near-subtelomeric 35.4 kb DNA segment on the right arm of chromosome VII from Saccharomyces cerevisiae carrying the MAL1 locus reveals 15 complete open reading frames, including ZUO1, BGL2 and BIO2 genes and an ABC transporter gene. Yeast 13:251-259. [DOI] [PubMed] [Google Scholar]
- 41.Wang, X., M. Bali, I. Medintz, and C. A. Michels. 2002. Intracellular maltose is sufficient to induce MAL gene expression in Saccharomyces cerevisiae. Eukaryot. Cell 1:696-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zastrow, C. R., C. Hollatz, P. S. de Araujo, and B. U. Stambuk. 2001. Maltotriose fermentation by Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 27:34-38. [DOI] [PubMed] [Google Scholar]
- 43.Zastrow, C. R., M. A. Mattos, C. Hollatz, and B. U. Stambuk. 2000. Maltotriose metabolism by Saccharomyces cerevisiae. Biotechnol. Lett. 22:455-459. [Google Scholar]
- 44.Zheng, X., T. D'Amore, I. Russell, and G. G. Stewart. 1994. Factors influencing maltotriose utilization during brewery wort fermentations. J. Am. Soc. Brew. Chem. 52:41-47. [Google Scholar]
- 45.Zheng, X., T. D'Amore, I. Russell, and G. G. Stewart. 1994. Transport kinetics of maltotriose in strains of Saccharomyces. J. Ind. Microbiol. 13:159-166. [Google Scholar]