Abstract
Dinophysis acuminata cells were isolated from Narragansett Bay water samples in June 2005 using flow cytometry. Dinoflagellate-specific PCR primers were used to isolate small-subunit rRNA (18S rRNA), mitochondrial cytochrome b (cob), and cytochrome c oxidase I (cox1) genes and the encoded cDNAs. Maximum-likelihood analysis of a concatenated data set of ribosomal DNA and cDNA sequences of cob and cox1 showed that D. acuminata was sister to Gonyaulacoids, but without strong bootstrap support. The approximately unbiased test could not reject alternative positions of D. acuminata. To gain better resolution, mRNA editing of cob and cox1 was inferred for D. acuminata and 13 other dinoflagellate species. The location and type of editing as well as the distribution pattern in D. acuminata were generally similar to those in other dinoflagellates except for two edited sites that are unique to this species. Bayesian analyses of a matrix that recorded the location and type of editing, and of a matrix that included the protein sequences of COB and COX1 with the editing data yielded tree topologies similar to the three-gene tree but again failed to resolve the phylogenetic position of D. acuminata. However, the density of edited sites in the D. acuminata mitochondrial genes, consistent with phylogenetic trees, indicated that Dinophysis is a derived dinoflagellate lineage, diverging after other lineages such as Oxyrrhis, Amphidinium, and Symbiodinium. We demonstrate that dinoflagellate-specific PCR coupled with flow cytometry can be a useful tool to analyze genes and their transcripts from a natural dinoflagellate population.
Many members of the dinoflagellate genus Dinophysis produce potent polyether toxins that can accumulate in filter-feeding bivalves, leading to a syndrome known as diarrhetic shellfish poisoning in humans consuming tainted shellfish. These harmful algal bloom species are important not only for their potential impact on public health but also from an ecological point of view because of their dual role as primary and secondary producers in complex microbial food webs. Unlike the majority of photosynthetic dinoflagellates, many Dinophysis species harbor plastids of cryptophyte origin (13), and mounting evidence points to a temporary acquisition of plastids (i.e., kleptoplastidy) in this mixotrophic life style (16, 25, 28). It is of interest to understand the evolutionary history of toxin production, its possible association with plastid acquisition and function (18, 47), and the multiple trophic roles and the phylogenetic affiliation of Dinophysis. Given the limited knowledge of this genus and the fact that until recently most Dinophysis species were unculturable under laboratory conditions (28), molecular analyses, particularly those that can be applied to sorted or unsorted natural populations, provide an important window into the evolution of this group. Thus far, only limited work has been done in this regard with Dinophysis. Existing work has generally focused on the ribosomal DNA (rDNA) locus (>200 entries in GenBank) because of its high copy number in the cell that allows ready isolation of the gene from a small number of cells or even from a single cell (24). Few protein-encoding genes (e.g., the photosystem II D1 protein [psbA] and Rubisco large-subunit genes) have been studied.
Given the challenge of obtaining pure Dinophysis samples from natural microbial assemblages for DNA and RNA extraction, isolation of these cells can be made more efficient with the use of flow cytometry (6). Furthermore, dinoflagellate-specific primers can significantly reduce the amplification of gene homologs in other organisms still present in the sorted cell samples. In this study, we combined these two methods to maximize the chance of obtaining gene sequences from the target species. We chose to analyze three genes: the small-subunit (SSU) rRNA (18S rDNA), mitochondrial cytochrome b (cob), and cytochrome c oxidase I (cox1). The 18S rDNA gene was included because a large data set is available for this conserved gene. Mitochondrial genes were selected because of their higher mutation rates and a more clock-like behavior relative to 18S rDNA (see references 10 and 45 and references therein). In addition, mRNA editing characteristics of mitochondrial genes can potentially act as species-specific molecular markers (20). Thus, taking advantage of dinoflagellate-specific gene primers developed in recent years (19, 42), we sequenced these three genes and cDNAs of cob and cox1 from Dinophysis acuminata collected in 2005 from a field population from Narragansett Bay, RI. We then characterized mRNA editing of cob and cox1 for this and other dinoflagellate species. Our main goal was to develop an approach that would lead to an efficient genetic analysis of environmental samples for ecological and phylogenetic purposes and allow for the study of gene expression in a natural dinoflagellate population.
MATERIALS AND METHODS
Sample collection.
A population of D. acuminata was collected between the surface and a depth of 0.5 m from Greenwich Cove, RI (41o39′30‴N, 71o27′00‴W) with a 10-μm mesh plankton net on 20 June 2005, 5 hours after sunrise. The water temperature at the 0.5-m depth was 20.5°C, and the salinity was 25.7 practical salinity units (PSU). To obtain an estimate of the original population density, a whole water sample was collected using a 2-m-long tube to integrate the entire water column and later quantitatively concentrated by reverse filtration and counted with a Sedgwick-Rafter chamber. The plankton sample was further enriched in D. acuminata by collecting the reddish “clouds” of swimming cells that formed a swarm near the surface of the sample. D. acuminata was identified as described by Steidinger and Tangen (37). Individual cells of this species were isolated the next day from the live enriched plankton sample using a BD FACSVantage flow cytometer cell sorter at the Northeast Fisheries Science Center Milford Laboratory, Connecticut, with adjustment for fluorescence, size, and scatter characteristics of the D. acuminata population. About 1,200 cells were sorted and preserved in 2% Lugol's solution for later DNA extraction (42), whereas 1,100 cells were sorted and kept alive for RNA extraction. Two additional sortings of ca. 1,000 cells each were inoculated in f/2-Si medium (11) to monitor potential growth of contaminant cells for 2 months. Within 2 hours, isolated cells were brought to the University of Connecticut Avery Point campus for molecular analyses. Cells for RNA extraction were harvested by centrifugation at 3,000 × g at 4°C for 20 min, resuspended in Trizol, and kept at −80°C until RNA extraction (21).
Dinoflagellate cultures and sample collection.
The dinoflagellate taxa used in this study are listed in Table 1. All photosynthetic species were grown in f/2-Si medium except D. acuminata collected from Narragansett Bay and the heterotrophic species grown with an algal prey (Rhodomonas sp. strain CCMP768) (46). Salinity was adjusted to 28 PSU for most species and to 15 PSU for Rhodomonas, Karlodinium veneficum, and heterotrophic taxa (46). Cultures were maintained at 20 ± 1°C under a 12 h:2 h light-dark cycle with a photon flux density of ca. 100 μmol photons·m−2·s−1. The growth rate was monitored by microscopic cell counts using a Sedgwick-Rafter chamber.
TABLE 1.
Dinoflagellate species included in the phylogeny and mitochondrial cob and cox1 editing analyses
| Taxon | Abbreviation used in figures | Strain and/or sourcea | Trophic modee | Accession no.i
|
||||
|---|---|---|---|---|---|---|---|---|
| SSU rDNA | cob cDNA | cob gDNA | cox1 cDNA | cox1 gDNA | ||||
| Adenoides eludens | CCMP1891 | P | AF274249 | EF036541 | EF036565 | |||
| Akashiwo sanguinea | Asa | LIS1b | P | AY456106 | EF036542 | AY456105 | EF036566 | EU126138g |
| Alexandrium affine | CCMP112 | P | AY831409 | EF036543 | EF377324 | |||
| Alexandrium tamarense | Ata | CB307; D. M. Anderson | P | AF022191f | DQ082987 | AY456116 | EF036567 | EU126139g |
| Amphidinium carterae | Aca | CCMP1314 | P | AF274251 | EF036544 | EU126130 | EF036568 | EU126140 |
| Amphidinium operculatum | Aop | CCMP123 | P | AY443011f | EF036545 | EU126131 | EF036569 | EU126141 |
| Ceratium longipes | CCMP1770 | P | DQ388462 | EF036546 | EF036570 | |||
| Ceratocorys horrida | CCMP157 | P | DQ388456 | EF036547 | EF036571 | |||
| Coolia monotis | CCMP304 | P | AJ415509f | EF036572 | ||||
| Crypthecodinium cohnii | Cco | WHd; M. Gray | H | M64245 | AF403221 | AF403220 | AF487783 | AF186994 |
| Dinophysis acuminata | Dac | Narragansett Bay sample | P | EU130569g | EU130567g | EU130568g | EU130565g | EU130566g |
| Gambierdiscus toxicus | CCMP401 | P | DQ388463 | EF036550 | EF036575 | |||
| Gonyaulax cochlea | CCMP1592 | P | DQ388465 | EF036551 | EF036576 | |||
| Gymnodinium catenatum | CCMP1937 | P | AF022193f | EF036552 | ||||
| Gymnodinium simplex | CCMP419 | P | DQ388466 | EF036553 | EF036577 | |||
| Heterocapsa triquetra | Htr | CCPM449 | P | AJ415514f | EF036554 | EU126132 | EF036578 | EU126142 |
| Heterocapsa rotundata (= Katodinium rotundatum) | Hro | CCMP1542 | P | DQ388464 | EF036556 | EU126133 | EF036582 | EU126143 |
| Karenia brevis | Kbr | CCPM2229 | P | AF274259f | EF036555 | AY456104h | EF036580 | EU126144g |
| Karenia mikimotoi | CCPM429 | P | AF009131f | EF036581 | ||||
| Karlodinium veneficum (formerly K. micrum) | Kve | CCMP1975 | M | EF036540 | DQ082989 | AY345908 | EF036579 | AF463416g |
| Noctiluca scintillans | Nsc | NS3; E. J. Buskey | H | DQ388461 | EF036583 | EU126145 | ||
| Oxyrrhis marina | Oma | CCMP1795 | H | AF482425f | EF036557 | EU126134 | EF036584 | EU126146 |
| Peridinium aciculiferum | PAER1 | P | AY970653 | DQ094825 | ||||
| Pfiesteria piscicida | Ppi | CCMP1831 | H | AF330620 | AF357518 | AF357519 | AF463413 | AF463412 |
| Pfiesteria-like | Ppi-like | CCMP1828 | H | AY590476 | EF036558 | AY456119 | EF036585 | EU126147g |
| Prorocentrum cassubicum (= Exuviaella cassubica) | Pca | LB1596c | P | DQ388460 | EF036548 | EU126135g | EF036573 | |
| Prorocentrum dentatum | Pde | CCMP1517 | P | DQ336057 | DQ336059 | DQ336058 | ||
| Prorocentrum donghaiense | Pdo | S. Lü | P | DQ336054 | DQ336056 | DQ336055 | EF036587 | |
| Prorocentrum lima (= Exuviaella lima) | CCMP1966 | P | EF377326 | EF036559 | EF377325 | |||
| Prorocentrum micans | Pmic | CCMP1589 | P | AY585526 | AY745238 | AY585525 | EF036588 | EU126148g |
| Prorocentrum minimum strain 696 | Pmin | CCMP696 | P | DQ336072 | AY030285 | AY030286 | AF463415 | AF463414 |
| Prorocentrum minimum strain EXUV | PmEXUV | CCMP1329 | P | DQ336060 | DQ336062 | DQ336061 | ||
| Prorocentrum minimum strain JA01 | Pm01 | JA01; P. Glibert | P | DQ336063 | DQ336065 | DQ336064 | ||
| Prorocentrum minimum strain PTPM | PmPTPM | PTPM; P. Tester | P | DQ336069 | DQ336071 | DQ336070 | ||
| Prorocentrum nanum (= Exuviaella pusilla) | Pna | LB1008c | P | DQ388459 | EF036549 | EU126136 | EF036574 | |
| Protoceratium reticulatum | Pre | CCMP1721 | P | AF274273f | EF036560 | EU126137g | EF036589 | EU126149g |
| Pseudopfiesteria shumwayae | Psh | T4; P. Tester, | H | AF218805f | DQ082988 | AF502593 | EF036586 | EU130570g |
| Pyrocystis lunula | J. W. Hastings | P | AF274274f | EF036561 | EF036590 | |||
| Pyrocystis noctiluca | CCMP732 | P | AF022156 | EF036562 | EF036591 | |||
| Scrippsiella sp. | Scrsp | LISb | P | AY743960 | AY743962 | AY743961 | EF036592 | EU130571g |
| Scrippsiella sweeneyae | CCCM280d | P | AF274276 | EF036563 | EF036593 | |||
| Symbiodinium goreaui | Sgo | CCMP2466 | P | EF036539 | EF036564 | EU130574g | ||
| Symbiodinium microadriaticum | Smi | CCMP830 | P | AY456111 | DQ082985 | AY456110 | EF036594 | EU130573g |
| Symbiodinium sp. | Symsp | CCMP832 | P | AY456113 | DQ082986 | AY456112 | EF036595 | EU130572g |
CCMP strains are from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton, West Boothbay Harbor, ME.
Isolated from Long Island Sound in April 2003, morphologically similar to Akashiwo sanguinea based on the description in reference 37 and SSU rDNA sequence (identical to AF276818). The LIS isolate of Scrippsiella sp. is most closely related to S. trochoidea based on morphology (37) and SSU rDNA identity (1,738 bp out of 1,754 bp identical to AY421792).
The Culture Collection of Algae (UTEX), The University of Texas at Austin, TX.
Canadian Center for the Culture of Microorganisms, University Boulevard, Vancouver, BC, Canada.
P, photoautotrophic; H, heterotrophic; M, mixotrophic.
Sequence from GenBank originally obtained from other strains of the same species as used in this study.
Sequences and editing analyzed in the present study.
Editing analyzed in the present study.
For autotrophic dinoflagellates, samples were collected when cultures were in the exponential growth phase; for heterotrophic species, samples were collected after feeding was discontinued for over 2 days, when very few (<2% of total cells) prey algae could be detected by microscopic examination. The cells were harvested and kept in Trizol as described above for field-collected samples.
Cloning and sequencing of 18S rDNA, cob, and cox1.
DNA and total RNA were extracted and cDNA synthesized as previously described (21). The resulting cDNAs and genomic DNA were subjected to PCR amplification with dinoflagellate specific primer sets 18ScomF1 (5′-GCTTGTCTCAAAGATTAAGCCATGC-3′)-Dino18SR1 (5′-GAGCCAGATRCDCACCCA-3′) and Dino18SF1 (5′-AAGGGTTGTGTTYATTAGNTACARAAC-3′)-18ScomR1 (5′-CACCTACGGAAACCTTGTTACGAC-3′) for 18S rDNA (17), Dinocob1F (5′-ATGAAATCTCATTTACAWWCATATCCTTGTCC-3′)-Dinocob1R (5′-TCTCTTGAGGKAATTGWKMACCTATCCA-3′) for cob, and Dinocox1F (5′-AAAAATTGTAATCATAAACGCTTAGG-3′)-Dinocox1R (5′-TGTTGAGCCACCTATAGTAAACATTA-3′) for cox1 (21, 44). PCRs comprised 35 cycles of 25 s at 94°C, 30 s at 52°C, and 40 s at 72°C, followed by 10 min at 72°C. PCR products were directly sequenced as reported by Zhang and Lin (43).
Inference of RNA editing and analysis of editing characteristics.
Genomic DNA and corresponding cDNA sequences of cob and cox1 were aligned using CLUSTAL W (1.8) (http://clustalw.ddbj.nig.ac.jp/top-e.html). Differences between the genomic and cDNA sequences were identified as an indication of mRNA editing. Editing density, or the percent nucleotides of the gene sequence that are edited, was determined for each species. Editing densities were compared between groups of taxa that were clustered on phylogenetic trees. Furthermore, types of nucleotide and amino acid changes as a result of editing were analyzed. The percent edited sites that were mapped to the same location (location of editing [LOE]) between two species and that underwent the same type of nucleotide substitution (type of editing [TOE]) between the two species were also calculated. In addition, the frequency of editing at each position of the codon was analyzed.
Phylogenetic analysis.
The DNA sequences encoding COB and COX1 were aligned using REVTRANS (http://www.cbs.dtu.dk/services/RevTrans/) with the default values. The cob and cox1 cDNA sequences were combined to produce a two-gene data set (1,963 nucleotides [nt]) and then with SSU rDNA (1,714 nt) to generate a cob-cox1-SSU rDNA three-gene (3,677-nt) data set. The apicomplexans Plasmodium yoelii and Plasmodium berghei were used to root the COB and COX1 protein tree, whereas the early diverging dinoflagellate Oxyrrhis marina was used to root the three-gene DNA tree (46). In the previous study (45), the congruence of COB, COX1, and rDNA data from dinoflagellates was tested (with the D. acuminata data not included) using the partition homogeneity test (ILD test in PAUP*; 1,000 replicates). An absence of significant incongruence between the COB and COX1 protein alignments (P = 0.136) and significant incongruence between the DNA data from these two partitions (P = 0.003) were noted. However, the single protein and DNA trees did not differ substantially from each other in these analyses, suggesting that gene concatenation would not mask a clear topological conflict (for details, see Fig. 1 and 3 in reference 45), but rather increased overall bootstrap support in the phylogenies. In this study we repeated the analysis for each gene pair in the three-gene alignment. These ILD test results again showed that P = 0.001, 0.001, and 0.003 for the rDNA-cob, rDNA-cox1, and cob-cox1 data, respectively (see also reference 45). Although these were significant P values, we chose to combine these data because of our previous work (45) and because there was considerable debate about the usefulness of the ILD test (4, 14). We also did a preliminary maximum-parsimony (MP) analysis with each data partition to determine whether the position of D. acuminata was resolved in any of the single-gene trees with an MP bootstrap support value of ≥70%. This analysis revealed that this taxon emerged as an independent lineage in all three gene trees, leading us to concatenate the data in the hope of achieving greater phylogenetic resolution.
For the three-gene DNA data set, the “best” tree was inferred using PAUP* and the site-specific GTR model (ssGTR) (31) with different evolutionary rates for each amino acid codon position and for the rDNA data. Bootstrap analyses were done using PHYML (200 replicates) with the GTR + I + Γ model over all nucleotide positions. Bayesian posterior probabilities for the ssGTR tree were calculated using MrBayes and the ssGTR +I + Γ model over the four data partitions. These analyses were run as described by Zhang et al. (45). Unweighted MP bootstrap analyses were also performed with the three-gene data set (2,000 replications) using heuristic searches and TBR branch-swapping to find the shortest trees (38). The number of random-addition replicates was set to 10 for each bootstrap tree search, and best-scoring trees were held at each step.
For the COB-COX1 data set, ProtTest V1.3 (1) was used to identify the best-fit model with “Fast” optimization and a BIONJ tree. The ProtTest parameter values were then used in maximum-likelihood analyses with the RAxML (VI-HPC, v2.2.1) computer program using the hill-climbing algorithm (36). The results of a PHYML V2.4.3 (12) bootstrap analysis (200 replicates) with tree optimization were used to assess the robustness of monophyletic clades in the RAxML tree. The protein data set was also analyzed using Bayesian inference (MrBayes V3.0b4) (15). The ProtTest best-fit evolutionary model was used in this analysis with Metropolis-coupled Markov chain Monte Carlo from a random starting tree. Four chains were run simultaneously, of which three were heated and one was cold, and the nrun = 2 command was used to monitor tree standard deviations. To increase the probability of chain convergence, trees were sampled after the standard deviations of the two runs were <0.05 to calculate the posterior probabilities (i.e., after 56,200 generations). The remaining phylogenies were discarded as burn-in. For the COB-COX1 data set, an unweighted MP bootstrap analysis was also performed as described above.
Testing the tree topology.
To assess the position of D. acuminata in the three-gene DNA tree, we generated a backbone phylogeny that was identical to the “best” ssGTR topology but excluded this species. Using this backbone tree, D. acuminata was then added individually using MacClade V4.05 to each branch in the tree to generate a set of topologies that addressed all possible positions for this taxon (23). The site-by-site likelihoods for the trees in this analysis were calculated using the three-gene data set and TREEPUZZLE (V5.2) (33) with the GTR + I + Γ evolutionary model (the alpha value for the gamma distribution was identified using RAxML) and the default settings. The approximately unbiased (AU) test was implemented using CONSEL V0.1i (35) to identify the pool of probable trees in this test and to assign their probabilities. The phylogenetic relationship of LOE and TOE was analyzed for 17 dinoflagellates. Each edited site was designated as a character with presence (1) or absence (0) of editing in each species as its state. For the TOE-based MP analysis, each of the six TOEs that was observed was treated as a character, each of which was assigned a state value of 1 or 0 for presence or absence of that editing type in each species. The six characters for each species were recorded and concatenated into a single alignment for unweighted MP bootstrap analysis as described above. In addition, Bayesian analysis was conducted for the LOE and TOE concatenated data set in an alignment that also included the COB and COX1 protein sequences. We used an “unlinked” analysis in which independent model parameter estimates were calculated for each data partition, i.e., independent gamma parameter estimate for the editing data and independent invariants-gamma model parameter estimates for the protein data (i.e., cpREV + I + Γ as chosen by ProtTest V 1.3 [1]). The Bayesian analysis was done as described above, and the trees were sampled after the standard deviations of the two runs were <0.01 to calculate the posterior probabilities (i.e., after 59,000 generations).
RESULTS
Dinophysis cells isolated by flow cytometry.
On the day of collection, all D. acuminata cells observed using epifluorescence microscopy displayed a typical phycoerythrin pigmentation localized to digitated chloroplasts. The original population density was 52,500 cells liter−1; we noticed a few cells (fewer than 0.5%) parasitized by the perkinsozoan Parvilucifera infectans. On 21 June, once the settings on the flow cytometer were deemed satisfactory, all samples were sorted into tubes and inspected for cell viability and purity by microscopy (phase contrast; magnification, ×200) within 2 hours. After 2 months, microscopic examination of the sorted additional samples showed that some D. acuminata cells were still alive; no contaminant phytoplankters were detected.
18S rDNA, cob, and cox1 sequences.
The use of two primer sets of the 18S rDNA yielded a longer gene fragment than possible with a single primer set, resulting in a 1,741-bp fragment. Genomic sequences of cob (926 bp) and cox1 (1,338 bp) and corresponding cDNA sequences (cob, 926 bp; cox1, 1,338 bp) were also obtained. The 18S rDNA was identical to that reported for D. acuminata (accession no. AJ506972) and the cox1 sequence obtained here was similar to those of dinoflagellates reported previously (21, 43). These sequences were colinear with homologs in other dinoflagellates, with no indels detected. When translated to amino acid sequences, the critical histidines conserved in COB (4 His; ligands for the heme b group in apocytochrome b) and COX1 (6 His; ligands for heme a, CuB, and heme a3) of other organisms were identified in the D. acuminata proteins.
mRNA editing characteristics.
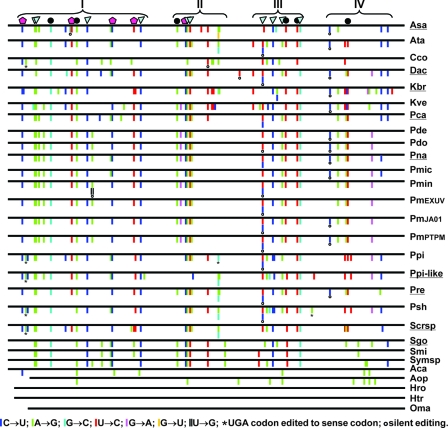
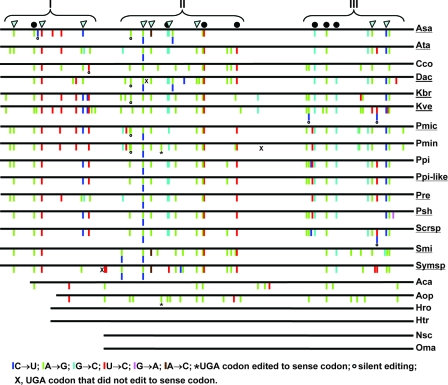
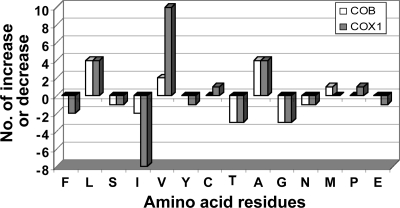
Comparison of the colinear genomic and corresponding cDNA sequences of D. acuminata revealed edited sites for cob and cox1. Five TOEs were detected for cob; A→G, U→C, and C→U were predominant, followed by G→C and G→A (Table 2). For cox1, six TOEs were observed, with additional A→C edits in comparison to cob. Similar to the case for cob, cox1 editing was dominated by A→G and U→C, followed by G→C and C→U. There were 31 editing events in cob and 30 in cox1, accounting for 3.35% and 2.24%, respectively, of the sequence length covered in the study. These edited sites were distributed in four (Fig. 1) and three (Fig. 2) discrete clusters for cob and cox1, respectively, as found in other dinoflagellates. Unique to D. acuminata was a silent U→C change that occurred in the gap between clusters II and III (Fig. 1). In cox1, there were two unique editing sites (site 3 and 17, both A→G) in D. acuminata; however, they resided in clusters I and II, respectively. Similar to the case for other dinoflagellates documented previously (21, 43), editing in both cob and cox1 of D. acuminata occurred predominantly in the first and second positions of the affected codons, resulting in changes in the encoded amino acid residues. These changes did not result in an increase of similarity in amino acid sequences between Dinophysis and other lineages (not shown). Rather, editing caused some changes in the proportions of several amino acid residues (Fig. 3). For COB in D. acuminata, leucine and alanine increased and threonine and glycine decreased most markedly. For COX1, the greatest increase occurred in valine, leucine, and alanine whereas, isoleucine, threonine, and glycine decreased. These changes led to only a small increase in the average hydrophobicity for both COB and COX1 (from 8.4 to 9.1 and from 9.0 to 9.3, respectively, as estimated using PepTool [BioTools, Inc., Alberta, Canada]).
TABLE 2.
Distribution of number and type of editing in cob and cox1 mRNAs in dinoflagellates
| Edit | No. (%) in mRNA from the indicated speciesa
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
cob
|
cox1d
|
||||||||||||||||
| Dac | Kbr | Pca | Pna | Ppi-like | Pre | Dac | Kbr | Kve | Pmic | Pmin | Ppi-like | Pre | Psh | Scrsp | Smi | Sysp | |
| Totalb | 31 (3.35) | 40 (4.30) | 32 (3.45) | 33 (3.54) | 32 (3.52) | 30 (3.22) | 30 (2.27) | 30 (2.27) | 39 (2.95) | 32 (2.42) | 34 (2.58) | 27 (2.05) | 27 (2.05) | 25 (1.89) | 28 (2.12) | 26 (1.97) | 28 (2.12) |
| A→G | 7 (22.6) | 11 (27.5) | 12 (37.5) | 13 (39.4) | 11 (34.4) | 11 (36.7) | 15 (50.0) | 16 (53.3) | 20 (51.3) | 15 (46.9) | 18 (52.9) | 13 (48.1) | 12 (44.4) | 12 (48.0) | 14 (50.0) | 18 (69.2) | 15 (53.9) |
| G→A | 1 (3.2) | 1 (2.5) | 0 (0) | 2 (6.1) | 1 (3.2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (4.0) | 0 (0) | 0 (0) | 0 (0) |
| C→U | 11 (35.5) | 11 (27.5) | 10 (31.3) | 9 (27.3) | 8 (25.0) | 8 (26.7) | 5 (16.7) | 3 (10.0) | 5 (12.8) | 3 (9.4) | 2 (5.9) | 3 (11.1) | 1 (3.7) | 2 (8.0) | 4 (14.3) | 2 (7.7) | 3 (10.7) |
| U→C | 9 (29.0) | 14 (35.0) | 8 (25.0) | 7 (21.2) | 8 (25.0) | 8 (26.7) | 5 (16.7) | 6 (20.0) | 9 (23.1) | 8 (25.0) | 8 (23.5) | 6 (22.2) | 7 (25.9) | 6 (24.0) | 6 (21.4) | 3 (11.5) | 9 (28.6) |
| U→G | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| G→U | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| U→Ac | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| A→C | — | — | — | — | — | — | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (3.8) | 1 (3.6) |
| G→C | 3 (9.7) | 3 (7.5) | 2 (6.3) | 2 (6.1) | 4 (12.5) | 3 (10.0) | 5 (16.7) | 5 (16.7) | 5 (12.8) | 6 (18.8) | 6 (17.6) | 5 (18.5) | 7 (25.9) | 4 (16.0) | 4 (14.3) | 2 (7.7) | 1 (3.6) |
| A, U→ G, C | 16 (51.6) | 25 (62.5) | 20 (60.6) | 19 (59.4) | 19 (63.3) | 20 (66.7) | 22 (73.3) | 29 (74.4) | 23 (71.9) | 26 (76.5) | 19 (70.4) | 19 (70.4) | 18 (72.0) | 20 (71.4) | 21 (80.8) | 24 (82.8) | |
Types of editing that are not detected in any species for an mRNA are denoted by dashes. Abbreviations of species names are as listed in Table 1.
Editing density.
One edit of this type has been documented in Ppi (21), but it is outside the region of sequence considered in this study (i.e., nt 88 to 1012 of Pfiesteria piscicida cob mRNA, GenBank accession no. AF357518 [for Amphidinium operculatum, the corresponding region is nt 126 to 1012]).
The region of sequence considered in this study is equivalent to Crypthecodinium cohnii cox1 genomic clone coding region nt 85 to 1404, GenBank accession no. AF186994 (for Amphidinium carterae and Amphidinium operculatum, the corresponding regions are nt 215 to 1404 and nt 274 to 1404, respectively).
FIG. 1.
Schematic diagram of the distribution of editing events in mitochondrial cytochrome b (cob) in dinoflagellates. The sites at which editing increases similarity between dinoflagellate and nondinoflagellate species are designated by either a closed circle (editing to an amino acid identical to the consensus) or a pentagon (editing to an amino acid chemically similar to the consensus). A triangle denotes dinoflagellate-specific editing. Brackets with Roman numerals indicate discrete clusters of editing events. Species abbreviations are as listed in Table 1, and those underlined are taxa whose gene sequences were analyzed in this study.
FIG. 2.
Schematic diagram of distribution of editing events in mitochondrial cytochrome c oxidase subunit I (cox1) in dinoflagellates. Symbols are as in Fig. 1. Brackets with Roman numerals indicate discrete clusters of editing events. Species abbreviations are as listed in Table 1, and those underlined are taxa whose gene sequences were analyzed in this study.
FIG. 3.
Editing-mediated increase (+) or decrease (−) in the content of the 20 amino acid substituents of COB and COX1. Amino acids are denoted by their standard one-letter abbreviations.
18S rDNA-cob-cox1 three-gene phylogeny.
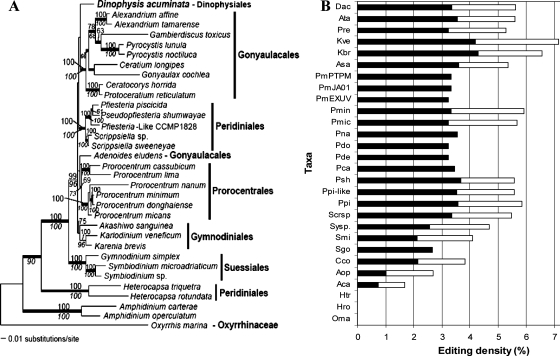
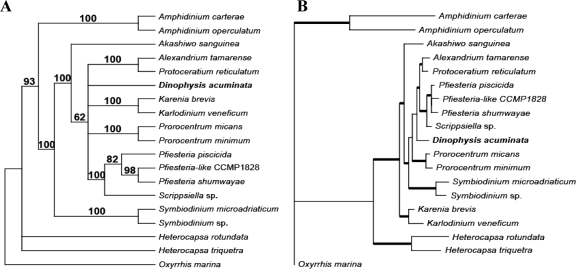
Maximum-likelihood analysis of the three-gene concatenated DNA alignment yielded the tree topology shown in Fig. 4A. Dinophysis appeared to be an independent lineage that is sister (<60% bootstrap support) to the cluster consisting of Gonyaulacales lineages. Assessment of alternate positions for D. acuminata in the three-gene tree using the AU test did not turn up strong evidence in favor of the “best” position shown in Fig. 4A (P = 0.783) over several other divergence points. For example, the position of D. acuminata on the branch uniting Adenoides eludens and the Prorocentrales was also supported (P = 0.325). Many other alternate positions were also not rejected by the AU test. These results left in question the phylogenetic placement of D. acuminata. Similar results were found with maximum-likelihood and MP analysis of the COB-COX1 protein data (data not shown).
FIG. 4.
Maximum-likelihood tree inferred from 18S rDNA-cob-cox1 three-gene concatenated data set (A) and mapping of editing density (B). (A) The three-gene maximum-likelihood tree was inferred using PAUP with an ssGTR model (four categories of nucleotide substitution rates). Bootstrap support was assessed using PHYML with a GTR + I + Γ model (numbers above branches) and MP analysis (numbers below branches). Posterior probabilities from Bayesian analysis when ≥0.95 are indicated as thick branches. The tree is rooted with Oxyrrhis, the lineage that is consistently placed at the base of all dinoflagellates when trees are rooted with the apicomplexan Plasmodium. (B) Editing density is plotted against taxa (abbreviations of species names are as in Table 1), with filled bars of cob stacked by open bars of cox1. Note that in the phylogenetic tree, there is a well-supported early-to-later branching order from Oxyrrhis, to Amphidinium, to Heterocapsa, to Symbiodinium, to the clade of Akashiwo/Karlodinium/Karenia. Accordingly, editing density increases in this order. For the remaining lineages, phylogenetic resolution is less clear, and editing densities among those lineages are similar.
Phylogenetic trend of editing density.
The mRNA editing of cob and cox1 was analyzed for 13 non-Dinophysis dinoflagellates (Table 1). Editing density, calculated for each species as the percentage of nucleotides in the gene that are edited, was mapped to the three-gene phylogenetic tree (Fig. 4B). This procedure revealed that editing did not occur in basal lineages (e.g., Oxyrrhis), and its density increased progressively in later-diverging lineages, reaching an equilibrium density of ca. 3% in the derived dinoflagellates in our study. Therefore, based on the editing density of Dinophysis and the general phylogenetic trend of increasing editing density over evolutionary time, Dinophysis most likely does not occupy a basal position among dinoflagellates and is more closely related to a derived lineage (i.e., as in Fig. 4A). In addition, if these editing data reflect true phylogenetic signals, then they clearly support the early divergence of Amphidinium spp. and Heterocapsa spp. (46), two long-branched taxa that have little or no editing of mitochondrial cob and cox1.
Editing-based phylogeny.
The Bayesian consensus tree based on the concatenated data (i.e., six categories) of editing information is generally congruent with the three-gene DNA tree although providing far less resolution of branching order among derived dinoflagellate clades due to the paucity of informative editing characteristics for these taxa (Fig. 5A versus Fig. 4A), However, again we found no significant Bayesian support for a specific affiliation of D. acuminata (Fig. 5A). These data showed that D. acuminata diverged somewhere in the region occupied by gonyaulacoid, gymnodinioid, prorocentroid, and peridinioid lineages that formed a multifurcation (Fig. 5A). Similarly, the Bayesian consensus tree (burn-in = 9,000 generations) inferred using the unlinked models for editing (+ Γ) and for each protein partition (cpREV + I + Γ) revealed the affiliation of D. acuminata in the neighborhood of the prorocentroid, gonyaulacoid, and peridinioid clusters (Fig. 5B).
FIG. 5.
(A) Bayesian consensus tree that was inferred from the concatenated editing data. The posterior probabilities that were calculated after the standard deviation between the two runs was <0.05 (i.e., 11,000 generations) are shown at the branches. (B) Bayesian consensus tree (burn-in = 9,000 generations) inferred using the unlinked models for editing (+ Γ) and for each protein partition (cpREV + I + Γ). The average standard deviation between two runs was <0.01. Posterior probabilities of ≥0.95 are indicated by thick lines.
DISCUSSION
This is the first report of cob/cox1 genomic and cDNA sequences from Dinophysis or any other dinoflagellate natural population. The effort was made possible with the use of flow cytometric analysis and dinoflagellate-specific PCR approaches. This approach can be used for other microbial organisms that are currently not amenable to single-species culture, such as heterotrophic and mixotrophic species and many unclassified picoplanktonic dinoflagellates that have recently been discovered (19, 22, 26, 32).
Amplifying multiple genes (nuclear and mitochondrial) and deciphering characteristics of editing density proved useful in characterizing dinoflagellates, albeit still not allowing us to elucidate the phylogenetic relationships of D. acuminata. Low taxon sampling within this genus may explain this result. It has already been demonstrated that a combination of cob and nuclear genes provides robust phylogenetic trees for alveolates and other organisms (30, 34) and that cob and cox1 can be used for inferring dinoflagellate phylogeny (44, 45). Our study indicates that the characteristics of editing density bolster the results of the multigene phylogeny approach even for a natural population.
In our analysis of the editing of cob and cox1, the putative basal lineages exhibited low (Amphidinium) or no (Oxyrrhis and Heterocapsa) editing, whereas more-derived lineages (including Dinophysis) consistently had higher levels of editing. This result extends the previous observation of cob mRNA editing density increasing from basal to derived lineages of dinoflagellates (43, 46). The editing alone and editing-plus-protein phylogenetic trees are generally similar to the gene sequence tree, in that Oxyrrhis, Heterocapsa, Noctiluca, and Amphidinium occupied basal positions in the tree. The clustering of Pfiesteria and related taxa with Scrippsiella is in full agreement with our current understanding that Pfiesteria and related lineages are peridinioid, and the affiliation of Karenia with Karlodinium in all of the trees is also consistent with our current understanding of their phylogenetic positions (see, e.g., references 9, 41, and 45). These data allowed us to draw two important conclusions. First, D. acuminata is not related to any of the basal lineages identified in this study based on the phylogenetic analyses and the frequency and distribution of edited sites in the cob and cox1 genes. In support of this conclusion are the intermediate level of editing density and the fact that D. acuminata was never placed near the root in any of the phylogenetic trees inferred using various combinations of data.
Second, D. acuminata is no more closely related to Prorocentrales than to Gonyaulacales or Gymnodiniales. This is despite the fact that Dinophysiales and Prorocentrales share major synapomorphies. Both lineages have a theca divided into lateral halves and have two apical pores, one large and the other small, with both flagella arising from the larger pore (39), and many Dinophysis and some Prorocentrum species (e.g., P. lima) produce polyether-type toxins (27). The apparently close relationship between the two dinoflagellate orders based on these features is not strongly supported by phylogenies based on multiple genes and editing characteristics. This weak correspondence stresses the need to include as many analytic approaches as possible and to use caution when inferring relationships between groups of organisms. Indeed, increasing evidence shows that algal lineages that produce the same type of metabolites can be phylogenetically distantly related. For instance, some cyanobacteria (e.g., Anabaena circinalis) and Alexandrium species produce saxitoxin (3, 29). Amphidinium and Karlodinium produce linear polyketide toxins (2, 8, 17). Our study provides another piece of evidence that similar toxin-producing attributes do not necessarily indicate a close phylogenetic affiliation, in our case between Prorocentrum and Dinophysis. The AU test rejects the positioning of D. acuminata at the base of the Prorocentrales (P = 0.007) or as sister to any of its members (P < 0.01).
In summary, our study presents a novel approach that could prove successful in characterizing environmental samples, especially when harmful algal bloom species are considered. Currently, our approach is time-consuming and requires highly skilled workers, and it thus can be used only for research purposes. Nevertheless, it can contribute to the development of molecular markers specific to certain target organisms, especially when genes involved in toxin production are incorporated in the approach.
Acknowledgments
We are very grateful to Gary H. Wikfors and Jennifer H. Alix (Northeast Fisheries Science Center Milford Laboratory) for help in using their flow cytometer. We also thank Donald Anderson, Pat Glibert, Woody Hastings, and Pat Tester for providing cultures. Two reviewers provided valuable comments that significantly improved the manuscript.
This research was supported by NSF grants DEB-0344186 (to S.L. and H.Z.), EF-0629624 (to S.L.), DEB-0107754 (to D.B.), and MCB-0236631 (to D.B.).
Footnotes
Published ahead of print on 28 December 2007.
REFERENCES
- 1.Abascal, F., R. Zardoya, and D. Posada. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104-2105. [DOI] [PubMed] [Google Scholar]
- 2.Adolf, J. E., T. R. Bachvaroff, D. N. Krupatkina, H. Nonogaki, P. J. P. Brown, A. J. Lewitus, H. R. Harvey, and A. R. Place. 2006. Species specificity and potential roles of Karlodinium micrum toxin. African J. Mar. Sci. 28:415-419. [Google Scholar]
- 3.Anderson, D. M. 1994. Red tides. Sci. Am. 271:62-68. [DOI] [PubMed] [Google Scholar]
- 4.Barker, F. K., and F. M. Lutzoni. 2002. The utility of the incongruence length difference test. Syst. Biol. 51:625-637. [DOI] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Carvalho, W. F., and E. Granéli. 2006. Acidotropic probes and flow cytometry: a powerful combination for detecting phagotrophy in mixotrophic and heterotrophic protists. Aquat. Microb. Ecol. 44:85-96. [Google Scholar]
- 7.Reference deleted.
- 8.Deeds, J. R., D. E. Terlizzi, J. E. Adolf, D. Stoecker, and A. R. Place. 2002. Hemolytic and ichthyotoxic activity from cultures of Karlodinium micrum (Dinophyceae) associated with fish mortalities in an estuarine aquaculture facility. Harmful Algae 1:169-189. [Google Scholar]
- 9.Gottschling, M., H. Keupp, J. Plötner, R. Knop, H. Willems, and M. Kirsch. 2005. Phylogeny of calcareous dinoflagellates as inferred from ITS and ribosomal sequence data. Mol. Phylogen. Evol. 36:444-455. [DOI] [PubMed] [Google Scholar]
- 10.Gray, M. W., G. Burger, and B. F. Lang. 1999. Mitochondrial evolution. Science 283:1476-1481. [DOI] [PubMed] [Google Scholar]
- 11.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 26-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, NY.
- 12.Guindon, S., and O. Gascuel. 2003. A Simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 13.Hackett, J. D., L. Maranda, H. S. Yoon, and D. Bhattacharya. 2003. Phylogenetic evidence for the cryptophyte origin of the plastid of Dinophysis (Dinophysiales, Dinophyceae). J. Phycol. 39:440-448. [Google Scholar]
- 14.Hipp, A. L., J. C. Hall, and K. J. Sytsma. 2004. Congruence versus phylogenetic accuracy: revisiting the incongruence length difference test. Syst. Biol. 53:81-89. [DOI] [PubMed] [Google Scholar]
- 15.Huelsenbeck, J. P., and F. Ronquist. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 16.Koike, K., H. Sekiguchi, A. Kobiyama, K. Takishita, M. Kawachi, K. Koike, and T. Ogata. 2005. A novel type of kleptoplastidy in Dinophysis (Dinophyceae): presence of haptophyte-type plastid in Dinophysis mitra. Protist 156:225-237. [DOI] [PubMed] [Google Scholar]
- 17.Kubota, T., Y. Iinuma, and J. Kobayashi. 2006. Cloning of polyketide synthase genes from amphidinolide-producing dinoflagellate Amphidinium sp. Biol. Pharm. Bull. 29:1314-1318. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence, J. E., and A. D. Cembella. 1999. An immunolabeling technique for the detection of diarrhetic shellfish toxins in individual dinoflagellate cells. Phycologia 38:60-65. [Google Scholar]
- 19.Lin, S., H. Zhang, Y. Hou, L. Miranda, and D. Bhattacharya. 2006. Development of a dinoflagellate-oriented PCR primer set leads to the detection of picoplanktonic dinoflagellates from Long Island Sound. Appl. Environ. Micribiol. 72:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, S., H. Zhang, and N. Jiao. 2006. Potential utility of mitochondrial cytochrome b and its mRNA editing in resolving closely related dinoflagellates: a case study of Prorocentrum (Dinophyceae). J. Phycol. 42:646-654. [Google Scholar]
- 21.Lin, S., H. Zhang, D. Spencer, J. Norman, and M. Gray. 2002. Widespread and extensive editing of mitochondrial mRNAs in dinoflagellates. J. Mol. Biol. 320:727-739. [DOI] [PubMed] [Google Scholar]
- 22.López-García, P., F. Rodriguez-Valera, C. Pedros-Allo, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 23.Maddison, D. R., and W. P. Maddison. 2002. MacClade V4.05. Sinauer, Sunderland, MA.
- 24.Marín, I., A. Aguilera, B. Reguera, and J. P. Abad. 2001. Preparation of DNA suitable for PCR amplification from fresh or fixed single dinoflagellate cells. BioTechniques 30:88-90, 92-93. [DOI] [PubMed] [Google Scholar]
- 25.Minnhagen, S., and S. Janson. 2006. Genetic analyses of Dinophysis spp. support kleptoplastidy. FEMS Microbiol. Ecol. 57:47-54. [DOI] [PubMed] [Google Scholar]
- 26.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 27.Nascimento, S. M., D. A. Purdie, and S. Morris. 2005. Morphology, toxin composition and pigment content of Prorocentrum lima strains isolated from a coastal lagoon in southern UK. Toxicon 45:633-649. (Erratum, 46:360-361, 2005.) [DOI] [PubMed] [Google Scholar]
- 28.Park, M. G., S. Kim, H. S. Kim, G. Myung, Y. G. Kang, and Y. Wonho. 2006. First successful culture of the marine dinoflagellate Dinophysis acuminata. Aquat. Microb. Ecol. 45:101-106. [Google Scholar]
- 29.Pomati, F., R. Kellmann, R. Cavalieri, B. P. Burns, and B. A. Neilan. 2006. Comparative gene expression of PSP-toxin producing and non-toxic Anabaena circinalis strains. Environ. Int. 32:743-748. [DOI] [PubMed] [Google Scholar]
- 30.Rathore, D., A. M. Wahl, M. Sullivan, and T. F. McCutchan. 2001. A phylogenetic comparison of gene trees constructed from plastid mitochondrial and genomic DNA of Plasmodium species. Mol. Biochem. Parasitol. 114:89-94. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez, F., J. L. Oliver, A. Marin, and J. R. Medina. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142:485-501. [DOI] [PubMed] [Google Scholar]
- 32.Romari, K., and D. Vaulot. 2004. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol. Oceanograph. 49:784-798. [Google Scholar]
- 33.Schmidt, H., A. K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 34.Serizawa, K., H. Suzuki, and K. Tsuchiya. 2000. A phylogenetic view on species radiation in Apedemus inferred from variation of nuclear and mitochondrial genes. Biochem. Genet. 38:27-40. [DOI] [PubMed] [Google Scholar]
- 35.Shimodaira, H., and M. Hasegawa. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246-1247. [DOI] [PubMed] [Google Scholar]
- 36.Stamatakis, A., T. Ludwig, and H. Meier. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456-463. [DOI] [PubMed] [Google Scholar]
- 37.Steidinger, K. A., and K. Tangen. 1997. Dinoflagellates, p. 387-584. In C. R. Tomas (ed.), Identifying marine phytoplankton. Academic Press, New York, NY.
- 38.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony and other methods, v. 4.0b10. Sinauer Associates, Sunderland, MA.
- 39.Taylor, F. J. R. 2004. Illumination or confusion? Dinoflagellate molecular phylogenetic data viewed from a primary morphological standpoint. Phycol. Res. 52:308-324. [Google Scholar]
- 40.Reference deleted.
- 41.Yoon, H. S., J. D. Hackett, G. Pinto, and D. Bhattacharya. 2002. A single, ancient origin of the plastid in the Chromista. Proc. Natl. Acad. Sci. USA 99:15507-15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, H., and S. Lin. 2002. Identification and quantification of Pfiesteria piscicida by using the mitochondrial cytochrome b gene. Appl. Environ. Microbiol. 68:989-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, H., and S. Lin. 2005. Mitochondrial cytochrome b mRNA editing in dinoflagellates: possible ecological and evolutionary associations? J. Eukaryot. Microbiol. 52:538-545. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, H., D. Bhattacharya, and S. Lin. 2005. Phylogeny of dinoflagellates based on mitochondrial cytochrome b and nuclear small subunit rDNA sequence comparisons. J. Phycol. 41:411-420. [Google Scholar]
- 45.Zhang, H., D. Bhattacharya, and S. Lin. 2007. A three-gene dinoflagellate phylogeny suggests monophyly of Prorocentrales and a basal position for Amphidinium and Heterocapsa. J. Mol. Evol. 65:463-474. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, H., and S. Lin. Status of mRNA editing and SL RNA trans-splicing groups Oxyrrhis, Noctiluca, Heterocapsa, and Amphidinium as basal lineages of dinoflagellates. J. Phycol., in press. [DOI] [PubMed]
- 47.Zhou, J., and L. Fritz. 1994. Okadaic acid antibody localizes to chloroplasts in the DSP-toxin-producing dinoflagellates Prorocentrum lima and Prorocentrum maculosum. Phycologia 33:455-461. [Google Scholar]