Abstract
Campylobacter jejuni causes food- and waterborne gastroenteritis, and as such it must survive passage through the stomach in order to reach the gastrointestinal tract. While little is known about how C. jejuni survives transit through the stomach, its low infectious dose suggests it is well equipped to sense and respond to acid shock. In this study, the transcriptional profile of C. jejuni NCTC 11168 was obtained after the organism was exposed to in vitro and in vivo (piglet stomach) acid shock. The observed down-regulation of genes encoding ribosomal proteins likely reflects the need to reshuffle energy toward the expression of components required for survival. Acid shock also caused C. jejuni to up-regulate genes involved in stress responses. These included heat shock genes as well as genes involved in the response to oxidative and nitrosative stress. A role for the chaperone clpB in acid resistance was confirmed in vitro. Some genes showed expression patterns that were markedly different in vivo and in vitro, which likely reflects the complexity of the in vivo environment. For instance, transit through the stomach was characterized by up-regulation of genes that encode products that are involved in the use of nitrite as a terminal electron acceptor and down-regulation of genes that are involved in capsular polysaccharide expression. In conclusion, this study has enabled us to understand how C. jejuni modulates gene expression in response to acid shock in vitro and to correlate this with gene expression profiles of C. jejuni as it transits through the host stomach.
Campylobacter jejuni is the leading bacterial cause of food- and waterborne gastroenteritis worldwide. This bacterium can exist in a commensal relationship with poultry, which constitutes a major reservoir of C. jejuni. The consumption of contaminated poultry leads to acute diarrheal disease in humans. While most infections are self limiting, C. jejuni has been associated with the development of Guillain-Barré and Miller Fisher syndromes, autoimmune disorders that affect the peripheral nervous system and lead to temporary paralysis (reviewed in reference 112).
In order to cause disease in humans, C. jejuni must survive passage through the stomach, where it is exposed to low pH as well as reactive oxygen and nitrogen species. The acidity of the human stomach is dependent on physiological variables that include previous food intake. In the absence of food, the median luminal pH is around 2.0 and ranges from 1.5 to 5.5 (37, 43). Under a traditional Western diet, meal ingestion will increase the median gastric pH to about 6 (63). The ability to handle acid stress directly affects the infective dose (ID) of enteric pathogens. Indeed, enteric pathogens that have evolved efficient survival strategies to cope with acidic environments have a low ID (e.g., Shigella flexneri can survive extreme acid conditions of pH 2.5 for hours in vitro and has an ID of 100 organisms) (20). Thus, the low ID of 500 to 800 organisms for C. jejuni (19, 89) suggests that this bacterium is well equipped to sense and respond to a sudden drop in pH. Despite its importance for pathogenesis, the acid stress response in C. jejuni has been understudied. Nevertheless, a protein component secreted by C. jejuni strain CI120 has been shown to provide protection for this bacterium against acid stress (78). In addition, an adaptive tolerance response (ATR) to acid and/or aerobic conditions has been identified in this particular C. jejuni strain (77, 79). In fact, early-stationary-phase cells that were adapted to a mildly acidic pH (pH 5.5) for 5 h were found to exhibit increased survival against lethal pH (pH 4.5) (79). While this ATR was found to be dependent on protein synthesis, its mechanism awaits further elucidation. Furthermore, the induction of a similar ATR by other strains of C. jejuni still remains to be demonstrated.
Typically, bacteria respond to a drop in pH by activating systems that prevent H+ entry, extrude H+ from the cell, consume H+, and repair affected cellular material. In some bacteria, exposure to acid leads to the up-regulation of the cfa gene (26, 51), the product of which generates cyclopropane-containing phospholipids. Exposure to acid leads to increased levels of these modified phospholipids (23, 42), which are known to be important for acid stress resistance in Escherichia coli and Salmonella (30, 60). Acid also can cause bacteria to up-regulate the F1F0 ATPase (9, 17, 32, 46), which can pump protons out of the cell at the expense of ATP. In enteric bacteria, amino acid-dependent systems are activated at low pH. Amino acids (Glu, Arg, and Lys) are transported into the cell, protons are consumed during their decarboxylation, and the product and substrate are exchanged in an antiport reaction (reviewed in references 28 and 43). Exposure to acid also leads to the up-regulation of genes involved in the protection and repair of cellular components such as DNA and proteins. Genes encoding DNA binding proteins, components of DNA repair systems, and chaperones have been shown to be up-regulated under acid stress conditions (4, 51, 56, 71, 74, 101, 102, 108).
Acid shock, as studied in this work, involves exposing bacteria to a sudden drop in pH (generally below the threshold for growth) for a short duration. This reflects the situation naturally encountered by the bacterium as it goes from a food or water source into the host gastrointestinal (GI) tract. Acid shock studies have been undertaken with various enteric bacteria, but they often involve the exposure of the cells to a moderate acid stress prior to acid shock, which induces the expression of acid tolerance proteins that may not otherwise be expressed (reviewed in reference 44). These studies revealed the acid-induced up-regulation of genes involved in a number of cellular processes, such as metabolism, regulation, transport, and the biosynthesis of macromolecular structures. They also have confirmed the importance of known components of the acid tolerance and/or resistance systems in these bacteria. A limited number of studies, particularly of Helicobacter pylori, have focused on the bacterial acid shock response in the absence of a previous adaptation to a mildly acidic pH. In H. pylori, acid shock resistance is mediated in large part by the uptake of urea, its breakdown in the cytoplasm by urease, and the buffering activity of the resulting ammonia (reviewed in reference 104). A microarray analysis of the acid shock response of this bacterium also has revealed that a number of genes for flagellar biosynthesis were up-regulated at low pH, and this correlated with a larger proportion of motile cells swimming at faster speeds (74). Note that it is difficult to compare results from gene expression studies such as those described above, even for a single organism, given the variability in experimental protocols and methods used for data analysis.
Changes in gene expression in response to acid stress are mediated by a number of transcriptional regulators. In E. coli and other enteric bacteria, the alternative sigma factor RpoS is involved in the acid tolerance response (reviewed in references 43 and 88). RpoS is a global regulator associated with stationary-phase physiology that also induces the expression of a number of acid shock proteins in log-phase cells exposed to acid stress (43, 67). In Salmonella enterica serovar Typhimurium and H. pylori, the ferric uptake regulator (Fur) protein is involved in the acid stress response. Under iron-rich conditions, Fur binds iron and represses the expression of many iron acquisition genes. Mutations in fur render cells acid sensitive (9, 11, 12, 16, 45, 50) and cause the aberrant expression of several acid-regulated genes and proteins (45, 48). In S. enterica, a fur mutant unable to bind iron still could induce the expression of acid shock proteins, indicating that Fur regulation of acid shock gene expression was independent of iron, at least in this bacterium (50). In H. pylori, fur expression is regulated by NikR in response to acid and nickel concentrations (reviewed in reference 104). NikR also regulates genes involved in nickel uptake and metabolism as well as urease expression. Some two-component transcriptional regulators, such as ArsRS in H. pylori and PhoPQ in S. enterica serovar Typhimurium, also alter gene expression in response to acid stress (11, 82).
The C. jejuni genome lacks many of the elements of the acid stress responses of other bacteria. Notably, C. jejuni does not encode an RpoS homologue, amino acid-dependent acid tolerance systems, or a urease enzyme. C. jejuni does have a fur gene, which plays a role in iron-dependent gene regulation (53, 81). However, to date there are no data linking Fur to the acid stress response of C. jejuni. Given the absence of many typical acid stress response genes in the C. jejuni genome, it is likely that this bacterium uses novel means to survive exposure to acid shock.
The objective of this study was to identify C. jejuni genes for which expression was up- or down-regulated in response to a sudden drop in pH, such as that encountered by the bacterium as it transits through the stomach. To this end, the transcriptional profile of C. jejuni was determined after subjecting the cells to an in vitro acid shock, and this was correlated with gene expression levels seen in vivo after oral inoculation into a neonatal piglet. To the best of our knowledge, this is one of the only instances in which an in vivo model has been used to study the transcriptional profile of a bacterium in response to acid shock.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. jejuni NCTC 11168 was acquired from the National Collection of Type Cultures. The construction of the deletion mutants of hrcA and hspR used in this study was described previously (94). Bacteria were maintained at 37°C on Mueller-Hinton (MH) agar plates under microaerophilic conditions (83% N2, 4% H2, 8% O2, and 5% CO2) in a MACS-VA500 microaerophilic workstation (Don Whitley, West Yorkshire, England). For in vitro and in vivo pH studies, bacteria were grown to mid-logarithmic phase in biphasic MH medium (an equal volume of MH broth layered on top of MH agar in a tissue culture flask) (90) at 37°C in the MACS-VA500 workstation. In biphasic cultures under these growth conditions, mid-logarithmic phase coincides with optical densities at 600 nm (OD600s) of approximately 0.9 to 1.5 (86a).
In vitro acid shock study.
In order to identify genes for which expression was up- or down-regulated in response to acid shock in vitro, C. jejuni gene expression was monitored over a 20-min period following acid shock. Briefly, bacteria were grown under microaerophilic conditions in biphasic MH-MES medium containing 25 ml of MH broth buffered at pH 7.0 using 100 mM 2-(N-morpholino)ethanesulfonic acid (MES). The pH of the spent culture medium was within 0.1 of the starting pH. At mid-log phase (OD600 = 0.9), 5 ml of the bacterial culture was transferred to a biphasic medium containing 20 ml of MH-MES broth buffered to pH 4.5 (the addition of culture did not alter the pH of the medium). Total RNA was extracted from samples taken 2, 4, 12, 16, and 20 min after the acid challenge. For the control samples, 5 ml of the bacterial culture was transferred to a biphasic medium containing 20 ml of MH-MES broth buffered to pH 7.0, and samples were collected after 2, 4, 12, 16, and 20 min. Immediately after the collection of samples for RNA extraction, 0.1 volume of cold RNA degradation stop solution (10% [vol/vol] buffer-saturated phenol in ethanol) was added to prevent RNA turnover (14). Cells were collected by centrifugation (8,000 × g, 10 min, 4°C), and the cell pellet was resuspended in TE buffer (50 mM Tris-Cl, pH 8.0, 1 mM EDTA) containing 0.5 mg ml−1 (final concentration) lysozyme. Total RNA was extracted using a hot phenol-chloroform method (100), precipitated with ethanol, and resuspended in RNase-free H2O. The RNA preparation was treated with DNase I to remove any contaminating genomic DNA, further purified using a Qiagen RNeasy mini kit (Qiagen, Valencia, CA), and quantitated using either the RiboGreen RNA quantitation kit (Molecular Probes, Eugene, OR) or A260s. RNA integrity was assessed by agarose gel electrophoresis, and PCR amplification was used to confirm that the preparation was free of genomic DNA. RNA samples were stored at −80°C.
In vivo acid shock study.
The in vivo acid shock experiment was performed with colostrum-deprived piglets. This animal model was previously used in our laboratory to study the role of ferrous ion acquisition in gut colonization and Campylobacter pathogenesis (80). Piglets were fed a milk replacer (multipurpose milk replacer, Grade A Ultra24; Sav-A-Caf Products) upon arrival four times daily (between 60 to 120 ml per piglet per feeding). C. jejuni NCTC 11168 was grown in biphasic MH cultures under microaerophilic conditions. Cells were collected at mid-log phase (OD600 = 0.9) by centrifugation and gently resuspended in MH broth. Prior to inoculation, the piglets were starved for 8 h. Two 3-day-old piglets were orally inoculated with 10 ml of a bacterial suspension containing approximately 1013 viable bacteria. A third piglet was inoculated with 10 ml of sterile MH broth and served as a control animal. After 15 min, the piglets were euthanized, and the intact stomachs were excised from the abdominal cavity by ligating the lower end of the esophagus (next to the cardiac orifice) and the upper end of the duodenum (next to the pyloric orifice). Approximately 5 min following the euthanasia, the stomachs were linearly opened, and the stomach contents (∼20 ml per stomach) were recovered in 20 ml of a cold RNA degradation stop solution (consisting of 4 ml of 10% buffer saturated phenol in ethanol and 16 ml of phosphate-buffered saline [PBS] buffer). Large particles and epithelial cells were removed from the stomach suspensions by low-speed centrifugation (1,000 × g, 15 min, 4°C). Thereafter, Campylobacter cells were collected by centrifugation (8,000 × g, 10 min, 4°C). The same manipulations were carried out on both the control and infected animals. Total RNA was extracted from each sample using the hot phenol-chloroform protocol described above. Notably, no RNA was extracted from the sample originating from the control animal, while approximately 30 μg of RNA was extracted from each of the other two samples. Finally, the total RNA extracted from both Campylobacter-inoculated piglets were combined prior to their use in microarray experiments.
Probe labeling and slide hybridization.
RNA samples (16 μg) from each control and test condition were converted to cDNA using 10 μg random hexamers (Amersham Biosciences) and Superscript II reverse transcriptase (Invitrogen). Aminoallyl-dUTP was included in the reverse transcription reaction to permit the labeling of the cDNA with the monoreactive fluors indocarbocyanine (Cy3; used to label control samples) and indodicarbocyanine (Cy5; used to label test samples) (Amersham Biosciences). The Cy3 and Cy5 labeling of the probes was described previously (81). For each microarray experiment, the cDNA probes from one acid shock condition (e.g., 2 min after acid shock) were individually cohybridized with cDNA probes from the relevant control sample (cells exposed to MH-MES, pH 7.0, for the in vitro experiments or cells grown in vitro in biphasic MH cultures for the in vivo experiment). The C. jejuni NCTC 11168 microarray used in this study was described previously (93). This array was constructed using PCR-amplified fragments representing approximately 98% of the open reading frames (ORFs) identified in the NCTC 11168 genome.
Data collection and analysis.
Microarray slides were scanned at 532-nm (Cy3) and 635-nm (Cy5) wavelengths using a laser-activated confocal scanner (ScanArray Gx; Perkin Elmer) at a 10-μm resolution. Spot registration was optimized manually, and the fluorescence intensities of each spot were collected using ScanArray Express software (Perkin Elmer). Spots were excluded from the analysis if they were present in areas of slide abnormalities (these corresponded to spots flagged by the software as bad or not found) and if the spot intensity after background subtraction was below three times the standard deviation of the background in both channels. The fluorescence intensity of all remaining spots was normalized using locally weighted linear regression using the MIDAS software (available from The Institute for Genomic Research; http://www.tigr.org/software). The ratio of the mean Cy3:Cy5 values was log2 transformed, and the data were statistically analyzed using the empirical Bayes method as previously described (10). The in vitro acid stress data comprise three technical replicates for each of two biological replicates, while the in vivo data comprise six technical replicates from a single pool of RNAs obtained from two piglets. Genes were considered differentially expressed if their P value was below 10−4 and their change (n-fold) in relative transcript abundance was above 2. Differentially expressed genes were grouped by hierarchical clustering analysis using the Genesis software (available from Graz University of Technology, Graz, Austria; http://genome.tugraz.at).
Real-time qRT-PCR validation of microarray data.
The relative expression levels of 11 genes (cft, clpB, grpE, hrcA, katA, sdhA, Cj0264c, Cj0265c, Cj0358, Cj0414, and Cj0448c) deemed up-regulated in the piglet stomach by the microarray analysis and 3 genes (metC, dapB, and glyA) for which expression levels were essentially unchanged in vivo were analyzed by real-time quantitative reverse transcription PCR (qRT-PCR) using a 7300 real-time PCR system (Applied Biosystems) and the QuantiTect Sybr green RT-PCR kit (Qiagen), as described previously (93, 94) and according to the manufacturer's recommendations. The relative level of expression of each gene was normalized to that of lpxC, a gene for which the expression levels by microarray analysis remained unchanged in the piglet stomach compared to that from in vitro-grown C. jejuni. The qRT-PCRs were carried out at least in triplicate, and the specificity of the PCR amplification was confirmed by a melting curve analysis of the product according to the manufacturer's recommendations. The extent of induction of gene expression was obtained using the comparative threshold cycle (ΔΔCT) method. Primers were designed according to the manufacturer's recommendations using Primer3 software and are listed in Table 1.
TABLE 1.
Primers used in this study
| Primer (sense) | Sequence (5′ to 3′) |
|---|---|
| ak233 (+) | GCAAGAGTTTTGCTTATGTTAGCAG |
| ak234 (+) | GAAATGGGCAGAGTGTATTCTCCG |
| ak235 (+) | GTGCGGATAATGTTGTTTCTG |
| AR42 (+) | CGGGATCCCTCGGCGGTGTTCCTTTCCAAG |
| AR43 (−) | CGGGATCCCGCCCTTTAGTTCCTAAAGGG |
| AR48 (+) | TCCCCCGGGGATAAACCCAGCGAAC |
| AR49 (−) | TCCCCCGGGAAGCTTTCTAGACATCTAAATC |
| AR51 (−) | GCTCTAGATTAAGCTTTTTTGATGATTATTTC |
| AR56 (−) | CATCCTCTTCGTCTTGGTAGC |
| AR66 (+) | GCTCTAGAGCCGAAGTTAACAATACTG |
| cftF (+) | GAGTTCTTGGTGCTATGAAAACAG |
| cftR (−) | TCTTGAAGCTCTACATGAGAATCTG |
| Cj0264cF (+) | TCTTAAATGCTCTGGGTACAG |
| Cj0264cR (−) | GTAAGTAAGTGGAAAGGTGCTTC |
| Cj0265cF (+) | GCCTATACCGATGAAGTTGTATC |
| Cj0265cR (−) | CACAGAAGGTGCTTTAGGATTTAC |
| Cj0358F (+) | GAAAGAACTTTGATCACTCCATCTC |
| Cj0358R (−) | CCACCTAAATTCACACCATTATGAC |
| Cj0414F (+) | CTGTTTTAAAGGCAGCAGAACTTAC |
| Cj0414R (−) | CTTCTCCTTGCTCATCTTTAGG |
| Cj0448cF (+) | GGAACATTGCATAGAAGTGTAGATG |
| Cj0448cR (−) | CTAGTTTTCTTACTTCATCGGCAAC |
| clpBA (+) | GGAAGATCTCAAACCTTGAAAGTGCAGCAT |
| clpBB (−) | GGAAGATCTAGCACCAAAATCAGGGTCAA |
| clpBC (−) | CGCGGATCCTTTTCCTACGCCTGGTTCAC |
| clpBD (+) | CGCGGATCCAGGGCTTAATGCGGACAATA |
| clpBF (+) | AAGCCGTACGAAGAAAACCTTATAG |
| clpBR (−) | AATCCACTGTTACACCTTTGCTATC |
| dapBF (+) | TAAGTGGAAGAGATGGCATTATAGG |
| dapBR (−) | CCTTGAAGTAGCGGTATGATTAAG |
| glyAF (+) | CTAGTGCTTATGCAAGAGTGATTG |
| glyAR (−) | AGCTTACTACATGAGCGTATGG |
| grpEF (+) | GCTTTAGAAGCAGCTGTTAATG |
| grpER (−) | CATCTTTGATAAGAGCCACTCC |
| hrcAF (+) | GCTATAGAAGCAATGAAGAAAGAGC |
| hrcAR (−) | ACCTTAAGCCCCATAAAACCTTC |
| katAF (+) | CTTTAGTCCAAGCAATATCGTTCC |
| katAR (−) | CAGCGACATTGTAAGTATTCACTTC |
| lpxCF (+) | CTCCTGTTATGGATGGATCAAG |
| lpxCR (−) | GGGGTTCTTTAGTAGGAGTTAAACG |
| metCF (+) | CTAAACTTATTCATTGTGGCAGAGG |
| metCR (−) | CTCTGTATTTTTCCAAGTTGCGTG |
| sdhAF (+) | GTGGGGCTCATACTAGAGAAG |
| sdhAR (−) | TACTCTCCTTCCATCAAGTGAAAAC |
Construction and complementation of a clpB deletion mutant.
A 2,400-bp fragment of the clpB ORF was amplified from chromosomal DNA of C. jejuni NCTC 11168 using primers clpBA and clpBB (Table 1). These primers were designed to introduce a BglII site for cloning into BamHI-digested pUC19 (these enzymes generate compatible cohesive ends). A 1,136-bp deletion in the clpB ORF was created by inverse PCR with primers clpBC and clpBD (Table 1), which were designed to introduce BamHI restriction sites for the ligation of a Cm resistance cassette (cat) into the clpB gene. The Cm cassette with flanking BamHI sites was amplified from pRY111 (111) using primers AR42 and AR43 (Table 1). The resulting plasmid was introduced into C. jejuni NCTC 11168 by natural transformation (107), and mutants were selected by being plated onto MH plates containing 20 μg ml−1 chloramphenicol. The mutation was confirmed by PCR amplification of the chromosomal region flanking the clpB gene. As clpB does not appear to be in an operon (3, 83), the likelihood of polar effects due to this mutation is small.
For the complementation of the clpB deletion, we first constructed a derivative of the pRR plasmid (58) in which we inserted a kanamycin resistance cassette to allow for the selection of recombinants in our Cm-resistant deletion mutants. Plasmid pRR was digested with XbaI and blunt ended with T4 DNA polymerase. A kanamycin resistance cassette (aphA3) was amplified from pILL600 (65) using primers AR48 and AR49 (Table 1), digested with SmaI, and ligated into pRR to yield pRR-Km. The AR49 primer was designed to introduce an XbaI site just downstream of the aphA3 stop codon to permit the cloning of DNA of interest into the vector. The orientation of the aphA3 gene relative to the rRNA locus was determined by restriction enzyme digestion, and a plasmid with the aphA3 gene in the same orientation as the rRNA genes was chosen for further use.
The clpB gene and 243 bp of upstream sequence were amplified from chromosomal DNA of C. jejuni NCTC 11168 using primers AR66 and AR51 (Table 1) and the proofreading polymerase Pwo (Roche). Primers were designed to incorporate flanking XbaI sites for the ligation of the PCR product into pRR-Km. The resulting plasmid DNA was sequenced to confirm that the insert was in the same orientation as the aphA3 cassette and that the insert was free of PCR-induced errors. The plasmid was introduced into C. jejuni NCTC 11168 ΔclpB by natural transformation, and cells harboring the clpB gene and upstream DNA were selected by being plated on MH agar plates containing 20 μg ml−1 chloramphenicol and 10 μg ml−1 kanamycin. PCR amplification using primer ak233, ak234, or ak235 (58) and primer AR56 (binds within aphA3) confirmed that the clone used in our experiments had the clpB gene and upstream DNA integrated on the chromosome at the rRNA locus downstream of the Cj0029 gene.
Acid survival assays.
Cells were grown to logarithmic phase (OD600 = 0.8 to 1.5) in biphasic MH cultures. An aliquot of 2.5 ml of culture was added to 10 ml of MH medium adjusted to pH 2.6 using concentrated HCl (the final pH of the assay mix was 3). Samples were withdrawn at 0, 2, 4, 6, 8, 12, and 16 min after exposure to acid and diluted 1/50 into PBS, pH 7.4 (the pH of the resulting solution was ∼7.4). Samples were serially diluted, and 10 μl of each dilution was spotted in triplicate onto MH agar plates, which were incubated at 37°C under microaerophilic conditions for 72 h. Colonies were counted, and results were expressed as the percent survival (with time zero representing 100%) as a function of the duration of exposure to acid. For each strain and time point, percent survival values from a minimum of three independent experiments were pooled and compared to values for NCTC 11168 using a two-sample t test, assuming unequal variances (Microsoft Excel X). P values below 0.05 were considered significant.
Survival in SGF.
Bacterial survival was monitored in a synthetic gastric fluid (SGF) composed of proteose-peptone (8.3 g/liter), d-Glc (3.5 g/liter), NaCl (2.05 g/liter), KH2PO4 (0.6 g/liter), CaCl2 (0.11 g/liter), KCl (0.37 g/liter), bile (bovine; 0.05 g/liter), lysozyme (0.1 g/liter), and pepsin (13.3 mg/liter) (15). All of the components except the enzymes were dissolved in distilled water, the pH of the solution was adjusted to 4.0 with 1 M HCl to reflect the conditions observed in the piglet stomachs, and the solution was filter sterilized and stored at 4°C. Just prior to its use, lysozyme and pepsin were added from fresh stock solutions. Cells were grown to logarithmic phase (OD600 = 0.7 to 1.7) in biphasic MH cultures, and 1 ml of cells was collected by centrifugation and resuspended in 0.2 ml sterile distilled water. A volume of 4.8 ml of SGF was added to the cell suspension and mixed. Samples (20 μl) were withdrawn after 0, 4, 8, 12, and 16 min of exposure and added to 980 μl sterile PBS, pH 7.4. Dilutions, platings, and data analyses were done as described above for the acid survival assays.
Electron microscopy.
An aliquot of each sample used in the microarray analysis (before acid shock and 2, 4, 12, 16, and 20 min after acid shock) was analyzed by electron microscopy. C. jejuni cells were collected from each sample by centrifugation (3,000 × g, 20 min), and the supernatants were discarded. The pellets were resuspended in 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2). Cells were collected by centrifugation and washed three times in 0.1 M sodium cacodylate (pH 7.0). The samples were resuspended in 0.1 M sodium cacodylate (pH 7.0) and placed onto poly-l-lysine-coated coverslips. A secondary fixation using 2% glutaraldehyde in 0.2 M sodium cacodylate (pH 7.0) was carried out for 20 min. Excess glutaraldehyde was removed by washing the samples with 0.1 M phosphate buffer (pH 7.2). The samples were dehydrated according to the following series of ethanol concentrations (vol/vol), each for 20 min: 50, 70, 90, 95, and 100%. The samples then were subjected to critical point drying. The coverslips were mounted onto support stubs using silver glue, and the samples were coated with gold/palladium in a Balzer's Med 010 sputter coater. Finally, samples were observed in a JEOL JXM 6400 scanning electron microscope.
Microarray data accession numbers.
All microarray data have been deposited in the NCBI Gene Expression Omnibus database (accession numbers GSE9937 and GSE9938 and superSeries accession number GSE9942).
RESULTS
Microbes colonizing the GI tract must survive passage through the gastric acid barrier. While this life-threatening stress is encountered by many bacteria, very few studies have directly investigated the mechanism of resistance that allows bacteria to transit through the stomach in vivo. Most studies have relied on the investigation of the acid stress response in vitro by biochemical, physiological, or transcriptomic approaches to identify the components required for bacterial survival. However, both in vitro and in vivo approaches undoubtedly are required to fully characterize the bacterial acid shock response. Certainly, in vitro experimental conditions could never fully represent the gastric environment encountered by microorganisms. Furthermore, the gastric environment is defined by more than the low pH used in most in vitro experimental conditions. Consequently, a combined transcriptomic approach consisting of in vitro and in vivo experiments was developed to identify genes that are relevant to C. jejuni's ability to survive transit through the gastric acid barrier.
C. jejuni remains viable and helically shaped after 20 min of exposure to pH 4.5.
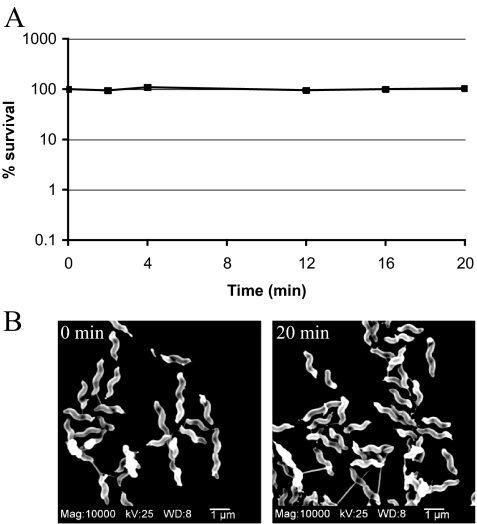
To investigate the C. jejuni gene expression profile in response to a sudden acid shock, the pH of a C. jejuni culture was shifted from 7.0 to 4.5, and transcript levels were measured for 20 min after the pH downshift. To determine whether a 20-min exposure at pH 4.5 affected cell viability, C. jejuni was grown to exponential phase in biphasic MES-buffered MH cultures at pH 7.0 and transferred to a medium at pH 4.5 (as described in Materials and Methods for the in vitro acid shock experiments). Viable counts were determined after 2, 4, 12, 16, and 20 min of exposure. As shown in Fig. 1A, C. jejuni cell numbers remained constant throughout this experiment, indicating that observed changes in gene expression were not caused by decreased cell viability at low pH. Of note, a pH of 4.5 does not support the growth of C. jejuni NCTC 11168 (36 and our unpublished observations).
FIG. 1.
C. jejuni remains viable and helically shaped after a 20-min exposure to MH-MES medium at pH 4.5. (A) C. jejuni NCTC 11168 was grown to exponential phase in biphasic MH medium and exposed to MH-MES medium at pH 4.5. Samples were withdrawn after 0, 2, 4, 12, 16, and 20 min for viable count determination. Values from seven independent experiments are represented as the percent survival (with 100% being the number of viable cells present at time zero). Error bars denoting the standard errors of the means are present but are too small to be seen. (B) Scanning electron micrographs of C. jejuni prior to (0 min) and after (20 min) exposure to MH-MES medium at pH 4.5. Size bars, 1 μm. Cells from samples taken 2, 4, 12, and 16 min after acid shock were indistinguishable from those shown. mag, magnification.
Unfavorable growth conditions can lead to morphological changes in C. jejuni, causing a shift from a spiral to a coccoid shape (22, 62, 86, 99). Microscopic analysis of the samples used for microarray analysis did not reveal the formation of coccoids or any other morphological variants under our experimental conditions (Fig. 1B).
Experimental design and global transcriptomic analysis.
In order to determine the transcriptional profile elicited in C. jejuni during transit through the host stomach and to understand the contribution of the acid stress response to this process, two sets of transcriptomic experiments were performed.
In the first set of experiments, the transcriptome profile of C. jejuni in response to acid shock in vitro was determined by exposing mid-logarithmic-phase C. jejuni to MH-MES medium at pH 4.5 for up to 20 min. Total RNA was extracted from cells 2, 4, 12, 16, and 20 min following the pH drop. The relative abundance of each transcript was monitored by competitive hybridization to our C. jejuni microarray of cDNA obtained from C. jejuni exposed to MH-MES medium at pH 7.0 and cDNA from C. jejuni exposed to MH-MES medium at pH 4.5 for each of the time points.
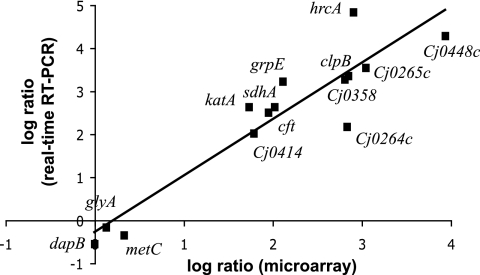
In the second set of experiments, the in vivo transcriptome profile of C. jejuni was obtained as the bacterium transited through the piglet stomach. Because of the similarities between the GI tracts of pigs and humans (70) and because C. jejuni infection in piglets mimics human campylobacteriosis (8, 80), the gene expression profile of C. jejuni recovered from a piglet stomach should closely resemble that of the transcriptome of C. jejuni during transit through the human stomach. Despite the apparent tolerance of NCTC 11168 to pH 4.5, we cannot discount the possibility that some of the C. jejuni cells used to inoculate the piglets were killed in the stomach. The piglets were fed ∼1013 viable bacteria, and we recovered between 109 to 1010 viable bacteria from the stomach contents. The loss of sample is inevitable during the feeding process and during the subsequent recovery of bacteria from the stomach contents. Likewise, we cannot discount the possibility that some of the inoculum already had moved beyond the stomach and into the intestine at the time of euthanasia. Thus, a true measure of gastric survival was not possible. Total RNA was extracted from the stomach contents of two piglets 20 min following oral inoculation with C. jejuni. Pilot studies with four piglets revealed that the pH of the stomach contents was between 3.8 and 4.2 after our experimental protocol (data not shown) and therefore was comparable to that of the in vitro acid shock conditions used. The extracted RNA was reverse transcribed, labeled, and subjected to microarray analysis by competitive hybridization with cDNA derived from C. jejuni grown to mid-log phase in vitro in MH medium (pH 7.0). The validity of the in vivo microarray data was confirmed by real-time qRT-PCR for a subset of genes, and the sets of data correlate well with each other (R2 = 0.86) (Fig. 2).
FIG. 2.
Comparison of in vivo gene expression levels measured by microarray and real-time qRT-PCR. The log2 ratio values of the microarray experiment were plotted against the log2 relative quantity values obtained from real-time qRT-PCR.
Transcriptome of C. jejuni during stomach transit.
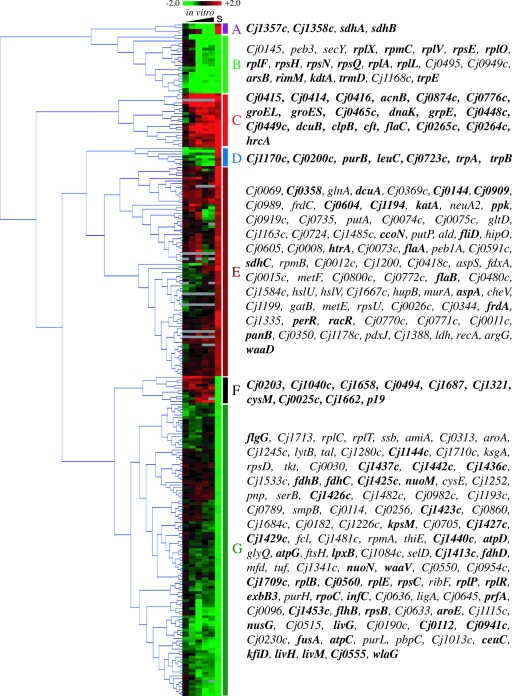
The stomach transcriptional profile was analyzed by merging the data from the in vivo and in vitro experiments. This analysis allowed the identification of genes that had their expression similarly affected in both experiments (these may represent the core genes involved in C. jejuni's acid shock response), those for which expression was affected solely in the stomach, and those showing opposite expression patterns in the two experiments. For this analysis, only the genes that were differentially expressed in vivo were selected and subjected to hierarchical clustering analysis. Genes were considered differentially expressed if the changes of their relative expression levels were ≥2-fold, with a Bayesian P value <10−4. The 250 genes thus identified grouped into seven major clusters, designated A to G (Fig. 3). Genes in cluster C were highly up-regulated in response to the in vitro acid shock condition and the gastric environment. These include a number of genes associated with bacterial heat shock responses, namely the transcriptional regulator hrcA and several chaperones and cochaperones (groEL, groES, dnaK, grpE, and clpB). This cluster also contains a number of genes involved in metabolism and energy generation, such as genes encoding products that are involved in trimethylamine N-oxide/dimethyl sulfoxide (TMAO/DMSO) respiration (Cj0264c and Cj0265c), C4-dicarboxylate transport (dcuB), the tricarboxylic acid cycle (acnB), and electron transport (Cj0874c). Genes involved in chemotaxis (Cj0448c), the nitrosative stress response (Cj0465c), the oxidative stress response (cft), and adhesion (flaC) also were up-regulated in response to acid shock, as were several genes of unknown function (Cj0414, Cj0415, Cj0416, Cj0776c, and Cj0449c).
FIG. 3.
Hierarchical clustering analysis of genes differentially expressed in the gastric environment. Genes differentially expressed (≥2-fold differential expression; P < 10−4) in the piglet stomach were subjected to hierarchical clustering using Genesis (Euclidian distance, average linkage). The genes of interest grouped into seven clusters, labeled A to G. Data from the in vitro shock experiments are shown in the first five columns (left to right: 2, 4, 12, 16, and 20 min of exposure), while in vivo gene expression ratios are shown in the last column (labeled S). A threshold log2 value of 2 (equivalent to fourfold differential gene expression) was used in this figure. Red boxes represent up-regulated genes, green boxes represent down-regulated genes, and gray boxes denote missing data. Genes in boldface are further discussed in the text.
Cluster B and the lower portion of cluster G contain genes that had their expression down-regulated in response to both the in vivo gastric environment and in vitro acid shock. A large number of the genes in these clusters encode ribosomal proteins (rplABEFLOPRVX, rpsBCEHNQ, and rpmC), and other genes are involved in transcription (rpoC and nusG), translation (infC, prfA, fusA, trmD, and Cj1453c), and ribosome modification (Cj1709c and rimM). Others are involved in uptake (exbB3, ceuC, Cj0941c, and Cj0555) and efflux (arsB, Cj0560, and Cj0112 [tolB]) processes as well as amino acid biosynthesis (aroE and trpE) and transport (livGHM).
Genes showing opposite in vivo and in vitro expression patterns are found in clusters A, D, and F (Fig. 3). Cluster A and D genes were up-regulated in vivo and down-regulated in vitro and include genes encoding succinate dehydrogenase subunits (sdhA and sdhB); proteins involved in nitrite respiration (Cj1357c and Cj1358c); enzymes for tryptophan (trpA and trpB), leucine (leuC), and purine biosynthesis (purB); the Omp50 porin (Cj1170c); a putative zinc metalloprotease (Cj0723c); and a putative periplasmic protein (Cj0200c). Nutrient availability likely accounts for at least some of these observed differences. Genes in cluster F were up-regulated in vitro and down-regulated in vivo. These include genes encoding putative transporters of iron (Cj1658 and p19) and citrate (Cj0203), putative major facilitator superfamily transporters (Cj1040c and Cj1687), a putative transferase involved in O-linked glycosylation (Cj1321), a putative exporting protein (Cj0494), a putative Na+/dicarboxylate symporter (Cj0025c), an enzyme involved in Cys biosynthesis (cysM), and a putative integral membrane protein (Cj1662).
The overall expression levels of cluster E genes were unchanged in vitro and were up-regulated in vivo. However, within this cluster are some genes that had their expression up-regulated under both conditions, albeit to a lesser degree than those of genes within cluster C. These include genes encoding the transcriptional regulators PerR (for peroxide stress regulator) and RacR (for reduced ability to colonize). Moderate increases in the levels of transcriptional regulators conceivably could cause much greater changes in expression levels of genes under their control. Other up-regulated genes in this cluster include those involved in phosphate uptake (Cj1194) and storage (Cj0604 and ppk) and peroxide detoxification (katA).
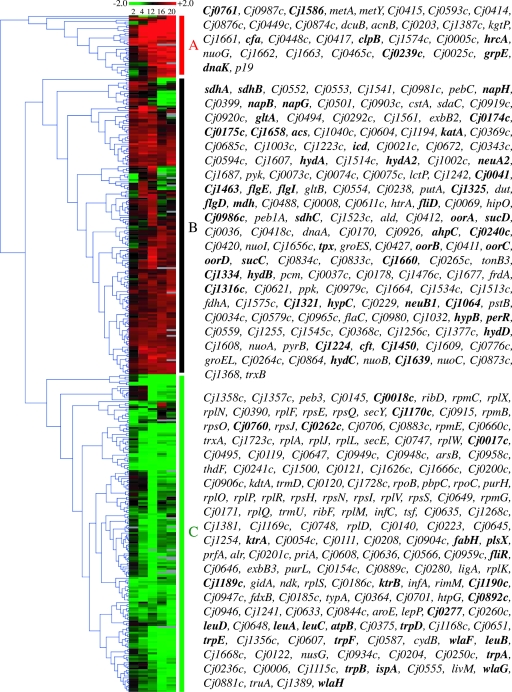
Transcriptome of C. jejuni in response to in vitro acid shock.
The transcriptional profile of C. jejuni in response to in vivo gastric conditions likely is a coordinated response to a number of factors, such as changes in nutrient availability, exposure to oxidative and nitrosative stress, and exposure to organic and inorganic acid shock. As such, it is possible that components of the acid shock response of this bacterium are not differentially expressed in vivo and/or are required only under particular environmental conditions. In addition, the selection of genes based on their differential expression at one time point in vivo would not permit the identification of transiently regulated genes. Consequently, in order to gain a more comprehensive view of the acid stress response in C. jejuni, the transcriptional profile of the cells exposed to in vitro acid shock was specifically analyzed by selecting the genes differentially expressed (≥2-fold change; P < 10−4) at one or more of the time points following the pH drop. The 360 genes that met these criteria grouped into three major clusters, designated A, B, and C (Fig. 4). Many of these genes already have been mentioned in the context of Fig. 3, and as such they will not be listed again. The genes of greatest interest in this analysis are those differentially expressed only in response to in vitro acid shock. Interestingly, 177 genes were found to be solely differentially expressed upon in vitro acid shock and not significantly affected during stomach transit (using a threshold of 1.5-fold differential expression for the in vivo data). These genes might be only transiently differentially expressed, or they might encode components of the acid shock response that are environment dependent. The genes in cluster A were highly up-regulated in vitro, and their expression levels increased with the length of exposure to acid. Of note among these genes are those encoding a cyclopropane fatty acid synthase (cfa) known to be involved in acid stress resistance in other bacteria, a single-domain hemoglobin (cgb; Cj1586) involved in oxidative stress defense, and a scaffold for FeS cluster assembly (nifU; Cj0239c). This cluster also contains a number of genes encoding transporters, most of which were down-regulated in the pig stomach, perhaps due to differences in nutrient availability, as suggested above.
FIG. 4.
Hierarchical clustering analysis of genes differentially expressed in response to in vitro acid shock. Genes differentially expressed (≥2-fold differential expression; P < 10−4) in response to in vitro acid shock were subjected to hierarchical clustering using Genesis (Euclidian distance, average linkage). The three main clusters are designated A, B, and C. Each column represents gene expression after a given exposure time (e.g., 2 indicates 2 min after acid shock). A threshold log2 value of 2 (equivalent to fourfold differential gene expression) was used in this figure. Red boxes denote up-regulated genes, green boxes designated down-regulated genes, and gray boxes represent missing data. Genes in boldface are discussed in the text.
Cluster B genes were transiently or consistently up-regulated in response to in vitro acid shock. Many of these genes were not differentially expressed in vivo. This cluster contains genes involved in flagellum biosynthesis (flgDEI and fliD) and glycosylation (Cj1316c, Cj1321, Cj1325, Cj1334, and neuA2), oxidative stress defense (ahpC, tpx, Cj1064, katA, perR, and cft), iron uptake (Cj0174c-175c, Cj1658, Cj1660, and Cj1224), and FeS cluster biogenesis (Cj0240c and Cj1639). In addition, genes encoding hydrogenases and hydrogenase assembly proteins were up-regulated in vitro (hydAA2BCD and hypBC), as were genes that encode products involved in the tricarboxylic acid (TCA) cycle (icd, mdh, gltA, sdhABC, and sucCD), nitrate respiration (napBGH), and subunits of an oxoglutarate:acceptor oxidoreductase (oorABCD). Of note, genes encoding TCA cycle enzymes (sucAB, icd, gltA, and mqo1) also were up-regulated in Staphylococcus aureus in response to acid shock (21).
Genes in cluster C were down-regulated upon exposure to acid shock in vitro. Unique to the in vitro situation was the down-regulation of genes involved in disulfide bond formation (Cj0017c-18c), cell shape determination (mreC and Cj0277), fatty acid biosynthesis (plsX, fabH, and ispA), potassium uptake (ktrAB), and leucine biosynthesis (leuABCD), among others. This cluster also contains a number of genes that encode products involved in chemotaxis (cetAB, Cj1190c-89c, and Cj0262c), protein glycosylation (pglABC [wlaGFH]), and tryptophan biosynthesis (trpABDEF). Of note, one gene involved in protein glycosylation also was down-regulated in vivo (pglA [wlaG]), as was one gene involved in tryptophan biosynthesis (trpE). While the expression of many genes encoding transporters, metabolic proteins, and transcription/translation proteins was repressed under both acid shock conditions, the specific genes affected differed somewhat between the two experiments.
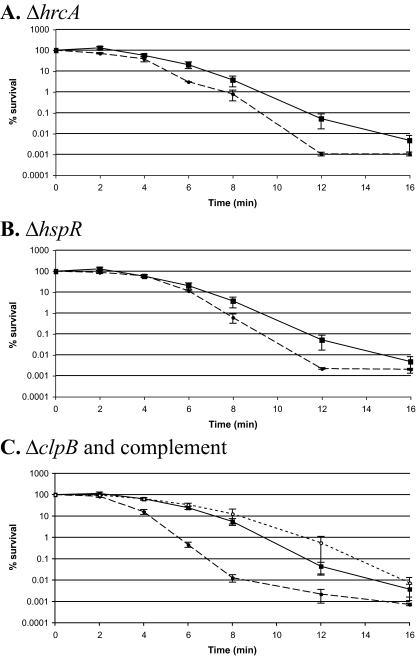
Genes involved in the heat shock response provide protection against acid killing.
The in vitro and in vivo transcriptome analyses revealed a link between the acid stress and heat shock responses with the up-regulation of most of the heat shock genes (clpB, groEL, groES, grpE, and dnaK) as well as the heat shock regulator gene hrcA. However, the up-regulation of a gene under a particular stress condition does not necessarily indicate that this gene is required for survival under that stress condition (18). In light of this, we examined the acid sensitivity of deletion mutants of the transcriptional repressors of the heat shock response, hrcA and hspR, as well as a deletion mutant of the chaperone clpB. The hrcA and clpB genes were highly up-regulated under both in vitro and in vivo acid shock conditions (Fig. 3). While the hspR gene was not selected by our statistical threshold as differentially expressed, it was found to be up-regulated 1.7-fold in response to in vitro acid shock (P < 10−5). In addition, HspR is a known repressor of the hrcA-grpE-dnaK operon and the clpB gene in C. jejuni (3), and as such it merited inclusion in our experiment. For this assay, we exposed log-phase bacteria to a lethal pH (MH medium at pH 3.0) for up to 16 min. As C. jejuni NCTC 11168 was killed after a 16-min exposure to pH 3.0 (Fig. 5), this assay allows the visualization of either improved or impaired resistance to acid shock in the deletion mutants. Deletion of hrcA or hspR caused a slight but statistically significant increase in the sensitivity of C. jejuni to acid killing (Fig. 5), with a P value below 0.05 after 2, 6, and 8 min of exposure for ΔhrcA and after 6 and 8 min of exposure for ΔhspR. However, deletion of clpB had a much greater effect on acid shock resistance in this bacterium (Fig. 5). A statistically significant increase in sensitivity was apparent after 4, 6, and 8 min of exposure to pH 3.0 (P ≤ 0.02). Of note, the exposure of the bacteria to unbuffered MH broth (pH ∼7) for the same duration did not lead to a change in cell numbers (data not shown), confirming that the assay manipulations themselves were not having adverse effects on cell viability. Complementation of the clpB mutant by recombination of the clpB gene and 243 bp of upstream sequence into the ribosomal locus restored wild-type levels of acid resistance (Fig. 5), confirming that the loss of clpB is responsible for the observed increase in acid sensitivity. Interestingly, attempts to complement the clpB mutant with only the clpB coding sequence did not improve acid resistance (data not shown), suggesting that the regulation of clpB gene expression is important for mediating acid resistance.
FIG. 5.
ClpB confers protection against acid shock. Wild-type C. jejuni NCTC 11168 and deletion mutants of hrcA, hspR, and clpB were grown to exponential phase in biphasic MH cultures and exposed to MH-HCl medium at pH 3.0. Samples were withdrawn after 0, 2, 4, 6, 8, 12, 14, and 16 min for viable count determination. Data from a minimum of three independent experiments are shown as the percent survival (with 100% being the viable counts at time zero for each strain) ± standard errors of the means. (A) NCTC 11168 (▪, solid line) and ΔhrcA (•, dashed line); (B) NCTC 11168 (▪, solid line) and ΔhspR (•, dashed line); (C) NCTC 11168 (▪, solid line), ΔclpB (•, dashed line), and the complemented ΔclpB mutant (○, dotted line).
In order to correlate in vivo and in vitro acid shock responses, we sought to determine if the clpB deletion mutant was more sensitive to killing in synthetic gastric fluid (SGF) (15). SGF has been used by others to study acid tolerance in E. coli (5) and the survival of Listeria monocytogenes (33) and S. enterica serovar Typhimurium (12) in the gastric environment. We chose to include 10 mM lactic acid in the SGF, as lactic acid values in the piglet stomach range from 1 to 163 mM (see reference 12 and references therein), and the addition of lactic acid to SGF yields data that are more similar to those from studies that used ex vivo gastric content (12). The pH of the SGF was adjusted to 4.0 to correspond to the measured pH of the piglet stomach contents (pH 3.8 to 4.2). C. jejuni NCTC 11168 was not killed by a 16-min exposure to the SGF (Fig. 6), while the clpB mutant showed a statistically significant increase in sensitivity to the SGF after 8, 12, and 16 min of exposure (P ≤ 0.01). As seen for the acid survival assays using MH medium, the complementation of the clpB mutant with clpB and its putative promoter region restored wild-type levels of resistance to the SGF (Fig. 6).
FIG. 6.
Increased sensitivity of a clpB deletion mutant in an SGF. C. jejuni NCTC 11168, ΔclpB, and the complemented ΔclpB mutant were grown to exponential phase in biphasic MH cultures and exposed to an SGF containing 10 mM lactic acid at pH 4.0. Samples were withdrawn after 0, 4, 8, 12, and 16 min for viable count determination. Data from a minimum of three independent experiments are shown as the percent survival (with 100% being the viable counts at time zero for each strain) ± standard errors of the means. Shown are NCTC 11168 (▪, solid line), the ΔclpB mutant (○, dashed line), and the complemented ΔclpB mutant (▴, dotted line).
DISCUSSION
Genome-wide expression studies of a number of bacteria have begun to reveal changes in gene expression induced by acid stress. However, many of these studies are difficult to compare to each other, given the different experimental protocols used (different strains, acid stress conditions, growth conditions, growth phases, end point versus time course experiments, etc.), and studies on the same organism sometimes show little or no overlap (74). In addition, these studies typically have been performed in vitro in order to identify genes of interest for subsequent genetic and pathogenesis studies. However, only in vivo analysis can reveal the role of the acid stress response in the ability of microbes to survive transit through the stomach and thus colonize the GI tract. In this work, we have correlated the in vitro and in vivo acid shock responses of C. jejuni. This approach is likely to identify genes with expression that is relevant to survival in the environment and the host, and it is apt to paint a more complete picture of the acid shock response of this pathogen.
Acid shock leads to extensive down-regulation of genes encoding ribosomal proteins.
The down-regulation of genes encoding ribosomal proteins under in vitro and in vivo acid shock conditions may reflect a cessation of protein synthesis, which would allow cells to redirect resources toward the expression of genes that encode products that are necessary for survival. Alternatively, increased expression of the many genes up-regulated under acid shock conditions could be occurring at the expense of ribosomal gene expression, as described for E. coli grown under conditions of carbon foraging (68). This decreased ribosomal gene expression also is seen when H. pylori (108), Staphylococcus aureus (21), and Shewanella oneidensis (66) are subjected to acid shock in vitro, when E. coli is grown under anaerobic and acidic conditions (51), and when E. coli is faced with nitrogen and sulfur starvation (49).
Acid shock activates a number of stress responses.
It is increasingly evident that there is overlap between stress responses, such as those mounted in response to acid, oxidative stress, and heat shock. A number of heat shock genes were highly up-regulated in response to acid shock. These include genes encoding the transcriptional regulator HrcA, chaperones DnaK, GroES, and GroEL, and cochaperones GrpE and ClpB. In Bacillus subtilis, HrcA represses gene expression by binding to CIRCE DNA elements in the promoter region of regulated genes. This binding requires the chaperones GroES and GroEL (75, 87). HrcA binding to the promoter regions of the groES-groEL and hrcA-grpE-dnaK operons of H. pylori has been demonstrated (92), and putative HrcA binding sites have been found upstream of dnaK and groEL in C. jejuni (3), suggesting that HrcA is involved in the heat shock response in C. jejuni. DnaK is a chaperone that mediates protein folding in conjunction with GrpE, a cochaperone that catalyzes nucleotide exchange in DnaK. A dnaK mutant of S. enterica serovar Typhimurium showed decreased survival when challenged with ex vivo swine stomach contents and in vitro acid shock (12). ClpB is a cochaperone that acts with the DnaK-GrpE-DnaJ chaperone system to dissolve protein aggregates in the cytoplasm and is known to improve the ability of E. coli to survive thermal stress (31). Heat shock proteins are part of a general stress response, and their expression is activated in response to unfolded or misfolded proteins. As exposure to a low-pH environment is likely to cause acidification of the cytoplasm and protein misfolding, such a response was not unexpected.
The transcriptional regulator HspR also is a part of the bacterial heat shock response. The transcriptional profile of a C. jejuni ΔhspR mutant has been reported previously (3). In that study, the authors identified a total of 17 genes directly or indirectly HspR activated and 13 genes directly or indirectly HspR repressed. Given that hspR was up-regulated 1.7-fold under in vitro acid shock conditions and that many heat shock genes were similarly affected, we sought to determine whether other known hspR-regulated genes were differentially expressed in our in vitro study. The up-regulation of hspR under acid shock conditions might be expected to increase the expression level of the HspR-activated genes. Indeed, the expression of 10 of these genes (flaD, flgE, flgE2, flgH, flgI, thiC, Cj0044c, Cj0986c, Cj1450, and acs) was transiently up-regulated in response to acid shock (using a cutoff of a 1.5-fold change in expression). Conversely, the up-regulation of hspR might be expected to decrease the expression levels of the 13 HspR-repressed genes. Transient repression of five such genes (def, dgkA, Cj0515, Cj0760, and Cj0892c) was apparent in our study. Notably, heat shock and other stress conditions are known to lift this repression for at least some of the genes, possibly by sequestering the chaperones required for HspR-DNA binding. The observed up-regulation in our in vitro study of six genes normally repressed by HspR (clpB, hrcA, dnaK, hspR, grpE, and Cj0761) supports this idea. Clearly, while the increased levels of hspR appear to be at least partly responsible for some of the gene expression changes brought about by in vitro acid shock, the complete picture likely is the result of a complex interplay of many transcriptional regulators.
We further examined the role of heat shock proteins in the acid shock response of C. jejuni by monitoring the survival of hrcA, hspR, and clpB deletion mutants under in vitro acid shock conditions. The increased sensitivity of the clpB mutant to acid killing may reflect a need for the cell to deal with protein aggregates accumulating in the cytoplasm upon its acidification and is consistent with the increased sensitivity of a Brucella suis clpB mutant to organic acid stress (pH 4) (38). The significance of the increased sensitivity of the hrcA and hspR deletion strains is less clear. Deletion of hspR in C. jejuni leads to increased levels of chaperones (3), which might be expected to improve acid resistance. The observed increase in acid sensitivity therefore may reflect roles for these proteins beyond the heat shock response and is consistent with the finding that an hspR deletion mutant in C. jejuni is more sensitive to growth at high temperatures (3). Consistent with a role for ClpB in acid shock tolerance, a clpB deletion mutant displayed impaired survival in an SGF containing 10 mM lactic acid.
Contrary to our findings for C. jejuni, the exposure of H. pylori to acid shock at pH 5 led to the up-regulation of dnaJ and the down-regulation of groEL, dnaK, and grpE (74). Down-regulation of clpB, dnaK, dnaJ, grpE, groES, and groEL also was apparent when S. oneidensis was exposed to acid shock at pH 4 (66). However, in S. aureus, clpB is highly up-regulated upon exposure to acid shock (21). In E. coli, the periplasmic chaperones hdeA and hdeB are up-regulated and activated specifically in response to acid stress (47, 51, 54, 59, 71, 102), suggesting a need for enhanced chaperone activity in the face of acid stress. While generalizations about the role of heat shock genes in bacterial acid stress responses cannot be made, it is clear that heat shock genes are up-regulated in C. jejuni in response to acid shock and that the activity of at least some of these components enhances resistance to this stress.
Acid shock also led to the up-regulation of the peroxide stress regulator gene (perR), the product of which plays an important role in the defense against oxidative stress in B. subtilis (25), S. aureus (55), and C. jejuni (103). Under acidic conditions, more iron is present in a soluble (ferrous) form, which may increase oxidative stress via the generation of reactive oxygen species by the Fenton reaction (85). PerR is a homologue of Fur, a transcriptional regulator important for resistance to acid stress in S. enterica serovar Typhimurium and H. pylori (9, 11, 12, 16, 45, 50). Fur plays a major role in iron acquisition, repressing iron uptake genes under iron-replete conditions. Fur also regulates genes involved in the oxidative stress response (katA and ahpC). This overlap between the Fur and PerR regulons has been noted for C. jejuni (103, 105). Our data raise the possibility that PerR is involved in the acid shock response in C. jejuni. Consistent with this, in vivo acid shock led to the up-regulation of katA (catalase) and cft (ferritin), the products of which are involved in protection from iron-induced oxidative stress, while in vitro acid shock led to the up-regulation of katA, cft, and additional oxidative stress defense genes (ahpC, tpx, and Cj1064). Our data are consistent with the up-regulation of cytochrome C551 peroxidase and catalase genes in H. pylori (108) and the up-regulation of katA, ahpC, and trxB in S. aureus (21) in response to acid shock.
In addition to the above-described overlaps with heat shock and oxidative stress responses, our data also suggest a link between acid and nitrosative stress regulons. The Cj0465c gene, encoding a truncated hemoglobin, was up-regulated in response to acid shock (in vitro and in vivo), while Cj1586, which encodes a single-domain hemoglobin, was up-regulated upon exposure to in vitro acid shock only. Cj0465c is involved in moderating O2 flux within C. jejuni cells (106), while Cj1586 is directly involved in the detoxification of nitric acid and related compounds (40). Both genes are members of the NssR regulon (39), which responds to nitrosative stress.
The up-regulation of ppk, a polyphosphate kinase gene, also was observed in response to acid shock. Polyphosphate can act as an energy storage molecule and can chelate cations such as Mg2+ and Fe3+, and polyphosphate accumulation may serve as a signal for changing environmental and/or growth conditions. Polyphosphate accumulation has been noted under conditions of osmotic shock and nutritional stress (nitrogen, amino acid, or phosphate limitation) (6, 61, 84). The mutation of ppk in bacterial pathogens leads to defects in general and stringent stress responses, biofilm formation, quorum sensing, and motility (reviewed in reference 24). H. pylori ppk mutants show impaired mouse colonization (7, 95), suggesting that Ppk function is important in vivo. Perhaps most relevant is the observed increase in phosphate uptake and polyphosphate accumulation when Burkholderia cepacia is grown at an acidic pH (76). Polyphosphate accumulation therefore may play a role in the acid stress response. Consistent with this, H. pylori gppA was up-regulated in response to acid shock (74, 108). GppA converts pppGpp into ppGpp, which alters gene expression as part of the stringent response.
The role of cell surface components in the acid shock response.
Components at the bacterial cell surface might be expected to play a protective role under many different stress conditions. Upon exposure to in vitro acid shock, H. pylori down-regulated 12 putative outer membrane proteins (OMPs), which likely resulted in a change in the permeability and antigenicity of the outer membrane (74). In Vibrio cholerae, the OmpT porin is repressed by acid, and the OmpU porin is required for acid resistance (73). From the data obtained in our experiments, it does not appear that C. jejuni drastically remodels its OMP composition in response to acid shock. However, we cannot discount a role for one or more OMPs in the acid shock response of this bacterium.
Acid shock led to the differential expression of some lipooligosaccharide (LOS) biosynthesis genes in C. jejuni. The gene encoding HldD (waaD), which is required for the synthesis of the heptose residue added to 3-deoxy-d-manno-octulosonic acid (Kdo)-lipid A by WaaC, was up-regulated in the gastric environment. However, the kdtA gene, which is required to transfer the Kdo residue onto lipid A, was down-regulated under both acid shock conditions, as it was in H. pylori (108). The gene required for LOS sialic acid biosynthesis (neuB1) was up-regulated in vitro, while a lipid A biosynthesis gene (lpxB), a glucosyltransferase gene (waaV), and a gene encoding a hypothetical protein of the LOS locus (Cj1144c) were down-regulated in vivo. Given these data, it is impossible to determine what role, if any, LOS plays in the resistance of C. jejuni to acid shock.
The link between flagella and the bacterial acid stress response remains unclear. Gene expression studies of H. pylori under conditions of acid shock reveal three conflicting scenarios. In one, a single flagellar gene (fliS) was down-regulated in response to acid shock, and none were up-regulated (2). In another, some flagellar genes (flaABG, flgBH, and fliDE) were up-regulated, while others (fliFS, flaA1, and flhF) were down-regulated (108). In the last, σ54-dependent flagellar genes (flaB and flgBCEK) were up-regulated, as was the anti-σ28 factor (flgM), which would block the expression of σ28-dependent genes (such as flaA) (74). These last data were supported by an observed increase in the number of motile cells and in their speed (74). A number of motility and chemotaxis genes also were up-regulated in H. pylori 10 days after the colonization of gerbil stomachs (91), further highlighting the importance of motility for H. pylori stomach colonization. Our data fail to provide a definitive answer regarding the role of flagella, if any, in the acid shock response of C. jejuni. The regulation of flagellar gene expression does occur in this bacterium in response to acid shock. The flagellin genes (flaAB) and the fliD gene (hook-associated protein) were up-regulated in the pig gastric environment, while flhB (biosynthetic protein) and flgG (basal body rod protein) expression was down-regulated. In vitro acid shock led to the transient up-regulation of flgD (putative hook assembly protein), flgE (hook protein), flgI (P-ring protein), and fliD and the transient down-regulation of fliR (biosynthesis protein), fliK (Cj0041; hook length control protein), and flgJ (Cj1463; hypothetical protein). Exposure to in vitro acid shock also led to the up-regulation of five genes involved in flagellar glycosylation (Cj1334, pseA [Cj1316c], Cj1321, Cj1325, and neuA2). It is possible that the up-regulation of flagellar genes is a general response to inhospitable environments, as suggested by Liu and coworkers (68). It also is possible that a drop in pH signals entry into a host environment, causing C. jejuni to express a more suitable gene complement. This may include increased levels of flagellar genes, as motility is thought to be required for rapid passage through the stomach and/or for localization to the protective mucus layer.
The expression of the methyl-accepting chemotaxis protein-type signal transduction protein, Cj0448c, was dramatically influenced by acid shock in C. jejuni. Both Cj0448c and Cj0449c, which appear to be in an operonic structure in NCTC 11168, were highly up-regulated under in vivo and in vitro acid shock conditions. Cj0448c is an atypical chemotaxis signal transduction protein, as it lacks a periplasmic sensing domain, transmembrane domains, and possible methylation sites (reviewed in reference 64). It is possible that Cj0448c senses a cytoplasmic signal in C. jejuni, which may be the acidification of the cytoplasm or a consequence of this acidification. Cj0448c also was up-regulated in the chick cecum (110), which raises the possibility that this protein is important for survival and/or growth in the host environment. The expression levels of other chemotaxis signal transduction genes also were affected by acid shock. The Cj0144 gene was up-regulated in vivo only, suggesting that it may sense and respond to signals within the host environment, while Cj0262c and cetAB (Cj1190c/Cj1189c) were down-regulated in vitro.
A number of bacteria modify inner membrane phospholipids in response to acid stress. The product of the cfa gene introduces cyclopropane groups on unsaturated fatty acyl chains (reviewed in reference 34). The effect of this modification on membrane properties is not well characterized, but increases in cyclopropane-containing phospholipids and cfa expression have been documented in bacteria exposed to stressful conditions such as acid shock (23, 26, 42), and cfa mutants display increased sensitivity to acid stress (30, 60). The cfa gene was up-regulated more than threefold after only 4 min of exposure to acid shock in vitro, with expression increasing to ∼5-fold after 20 min. Thus, it is likely that the product of the cfa gene is important for resistance to acid shock in C. jejuni, as it is in H. pylori, E. coli, and other bacteria. It is not clear why cfa expression is unchanged in the piglet stomach, but this may reflect the influence of additional factors in the more complex in vivo environment.
Acid shock alters the expression of genes involved in metabolism and energy generation.
A number of genes encoding proteins involved in energy generation were differentially expressed in response to acid shock. Acid shock induced the expression of genes encoding products that are involved in TMAO/DMSO respiration (Cj0264c and Cj0265c); a C4-dicarboxylate antiporter (dcuB), which is involved in fumarate respiration; and two putative oxidoreductase subunits (Cj0414c and Cj0415c) of unknown function. Cj0414 and Cj0415 share homology with gluconate dehydrogenase enzymes, but it is not clear how this activity would contribute to C. jejuni's acid shock response.
The aconitase gene acnB was highly up-regulated in response to acid shock. This could serve to increase acid consumption by the TCA cycle, or this may reflect a posttranscriptional regulatory role for AcnB. In E. coli and Bacillus, aconitases can act as iron and oxidative stress-responsive regulators, binding mRNA and altering transcript stability (1, 13, 52, 96, 98). In S. enterica serovar Typhimurium, AcnB prevents the expression of the FtsH protease by binding its mRNA, and thus it indirectly increases flagellin expression (97). Such a regulatory role for AcnB has not been shown to date for Campylobacter, but it certainly merits further study.
In response to in vitro acid shock, C. jejuni up-regulated the expression of hydrogenase (hydABCD and hydA2) and hydrogenase assembly (hypBC) genes. In H. pylori, hydrogenase assembly genes (hypEC) were up-regulated in response to acid shock (108), while hydrogenase genes were up-regulated in response to growth at low pH (4). Similarly, both hydrogenase and hydrogenase assembly genes were up-regulated in E. coli grown under anaerobic and acidic conditions (51). Finally, an E. coli strain deficient in hydrogenase activity (hypF mutant) showed impaired acid resistance (51). The authors suggested that hydrogenase expression may be important at low pH due to the ability of hydrogenases to extrude H+ from the cytoplasm.
Proton extrusion as a means to combat and/or prevent cytoplasm acidification.
An expected response to a sudden decrease in environment pH might include the induction of genes encoding products that consume or extrude protons. Thus, it may be expected that components of the ATP synthase are up-regulated, as this pump can extrude H+ from the cell at the expense of ATP. However, this response was not detected in C. jejuni, as these genes either were unaffected or were down-regulated (atpB in vitro, atpCDG in vivo) in response to acid shock. Down-regulation of ATPase subunit genes also was seen when H. pylori (108) and S. aureus (21) were subjected to in vitro acid shock and when H. pylori was grown in an acidic medium (27). In E. coli, an increase in growth medium pH led to the up-regulation of ATP synthase, presumably to increase H+ import and prevent the alkalinization of the cytoplasm (71). These findings are in agreement with the observed down-regulation of some of these components under acidic conditions in our studies.
The gene encoding a C4-dicarboxylate transporter (dcuB) was highly up-regulated under both acid shock conditions. In contrast, dcuA up-regulation was detected only in the pig stomach. Dicarboxylate carriers such as DcuA, DcuB, and DcuC are capable of the import, efflux, and exchange of C4-dicarboxylates such as succinate and fumarate (41, 113). The uptake and efflux reactions catalyzed by these transporters are electrogenic, resulting in the symport of succinate or fumarate and H+ (reviewed in reference 57). It is tempting to speculate that increased DcuA and/or DcuB expression helps C. jejuni resist cytoplasm acidification via the extrusion of protons in a symport reaction with a C4-dicarboxylate.
Survival in the host stomach: beyond acid stress.
Genes showing opposite expression patterns under in vivo and in vitro acid shock conditions can begin to reveal unique conditions encountered by the bacterium in the host environment, such as nutrient availability and exposure to additional stresses. In fact, Listeria monocytogenes is more sensitive to ex vivo porcine gastric fluid than to SGF at the same pH (33), suggesting that inorganic acid is not the only stress encountered in the stomach. In addition, S. enterica serovar Typhimurium mutants that were unable to survive exposure to an ex vivo swine gastric environment were not necessarily more sensitive to inorganic acid shock (12). The authors determined that the high levels of organic acids (up to 126 mM lactic acid) in the swine stomach contents were a major contributing factor to S. enterica killing.
In the pig stomach, genes required for the use of nitrite as a terminal electron acceptor (Cj1357c and Cj1358c) were highly up-regulated. In the mouth, oral bacteria reduce salivary nitrate to nitrite, which is converted into nitric oxide by the acidity of the stomach (72). The reduction of nitrite may provide a way for C. jejuni to combat this source of nitrogen stress.
The gene encoding the smaller Omp50 porin (Cj1170c) also was up-regulated exclusively in the pig stomach. pH regulation of porin expression has been reported previously, as the larger porin (MOMP) in C. jejuni is up-regulated under alkaline conditions (35). However, given that Cj1170c expression was down-regulated under conditions of in vitro acid shock, it is likely that Omp50 expression is modulated in response to the host environment rather than acid shock per se. In fact, the MOMP is an important antigen in C. jejuni (29, 69, 109), and increasing the ratio of Omp50:MOMP on the cell surface may be a means of evading the host immune response. Cj1170c expression was down-regulated in the chick cecum (110) and up-regulated in the rabbit ileal loop (94), supporting the idea that the expression of this OMP is tightly regulated in the host environment.
The genes encoding succinate dehydrogenase subunits (sdhAB) also were up-regulated in the pig stomach and down-regulated under in vitro acid shock conditions. In contrast, subunits of the NADH dehydrogenase (nuoMN) and formate dehydrogenase (fdhBCD) complexes were down-regulated only upon exposure to the gastric environment. Overall, it appears that in the pig stomach, electrons are fed into the electron transport chain primarily via the succinate dehydrogenase complex, while the NADH and formate dehydrogenase complexes are less important.
A number of genes were down-regulated in vivo and up-regulated in vitro. These gene products may not be needed for survival in the pig stomach, or their expression may need to be turned down in this environment. Possible differences in nutrient availability are illustrated by the opposite in vivo and in vitro expression patterns of a number of transporters (putative citrate transporter [Cj0203], putative iron transporter [p19], putative Na+:dicarboxylate family symporter [Cj0025c], and putative export proteins belonging to the major facilitator superfamily [Cj1687 and Cj1040c]).
Finally, cell surface polysaccharides (such as capsular polysaccharides [CPS] and exopolysaccharides) are known to provide protection against acid, heat, and desiccation as well as to confer resistance to the host immune response. The expression of genes involved in C. jejuni CPS assembly was not affected by exposure to acid shock. However, a number of CPS genes were down-regulated in the pig stomach (Cj1413c, Cj1423c, Cj1425c-27c, Cj1429c, Cj1436c-37c, Cj1440c, Cj1442c, kpsM, and kfiD), suggesting that C. jejuni needs to modulate the amount of CPS produced in the host, perhaps to expose adhesins or other factors required for colonization.
From this study, it is clear that the acid shock response of C. jejuni involves the down-regulation of genes involved in protein synthesis and the up-regulation of genes typically associated with numerous stress responses, such as the heat shock response, the stringent response, and the nitrosative and oxidative stress responses. Phospholipid modification and hydrogenase activity also may be important for acid resistance in vitro. Our data also have identified transcriptional responses specific to the pig stomach environment, which suggest that C. jejuni modulates the expression of surface components, metabolic enzymes, and transporters upon entry into a host.
Acknowledgments
This work was supported by grant number RO1-AI055612 from the National Institutes of Health. A. N. Reid acknowledges receipt of a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada.
We acknowledge Lisa Whitworth for the electron microscopy work reported in the manuscript.
Footnotes
Published ahead of print on 11 January 2008.
REFERENCES
- 1.Alén, C., and A. L. Sonenshein. 1999. Bacillus subtilis aconitase is an RNA-binding protein. Proc. Natl. Acad. Sci. USA 96:10412-10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan, E., C. L. Clayton, A. McLaren, D. M. Wallace, and B. W. Wren. 2001. Characterization of the low-pH responses of Helicobacter pylori using genomic DNA arrays. Microbiology 147:2285-2292. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, M. T., L. Brondsted, B. M. Pearson, F. Mulholland, M. Parker, C. Pin, J. M. Wells, and H. Ingmer. 2005. Diverse roles for HspR in Campylobacter jejuni revealed by the proteome, transcriptome and phenotypic characterization of an hspR mutant. Microbiology 151:905-915. [DOI] [PubMed] [Google Scholar]
- 4.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold, K. W., and C. W. Kaspar. 1995. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:2037-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ault-Riche, D., C. D. Fraley, C. M. Tzeng, and A. Kornberg. 1998. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180:1841-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayraud, S., B. Janvier, A. Labigne, C. Ecobichon, C. Burucoa, and J. L. Fauchere. 2005. Polyphosphate kinase: a new colonization factor of Helicobacter pylori. FEMS Microbiol. Lett. 243:45-50. [DOI] [PubMed] [Google Scholar]
- 8.Babakhani, F. K., G. A. Bradley, and L. A. Joens. 1993. Newborn piglet model for campylobacteriosis. Infect. Immun. 61:3466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baik, H. S., S. Bearson, S. Dunbar, and J. W. Foster. 1996. The acid tolerance response of Salmonella typhimurium provides protection against organic acids. Microbiology 142:3195-3200. [DOI] [PubMed] [Google Scholar]
- 10.Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509-519. [DOI] [PubMed] [Google Scholar]
- 11.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bearson, S. M., B. L. Bearson, and M. A. Rasmussen. 2006. Identification of Salmonella enterica serovar Typhimurium genes important for survival in the swine gastric environment. Appl. Environ. Microbiol. 72:2829-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beinert, H., M. C. Kennedy, and C. D. Stout. 1996. Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 96:2335-2374. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beumer, R. R., J. de Vries, and F. M. Rombouts. 1992. Campylobacter jejuni non-culturable coccoid cells. Int. J. Food Microbiol. 15:153-163. [DOI] [PubMed] [Google Scholar]
- 16.Bijlsma, J. J., A. L. M. Lie, I. C. Nootenboom, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J. Infect. Dis. 182:1566-1569. [DOI] [PubMed] [Google Scholar]
- 17.Bijlsma, J. J., B. Waidner, A. H. Vliet, N. J. Hughes, S. Hag, S. Bereswill, D. J. Kelly, C. M. Vandenbroucke-Grauls, M. Kist, and J. G. Kusters. 2002. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect. Immun. 70:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birrell, G. W., J. A. Brown, H. I. Wu, G. Giaever, A. M. Chu, R. W. Davis, and J. M. Brown. 2002. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl. Acad. Sci. USA 99:8778-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 20.Blaser, M. J., and L. S. Newman. 1982. A review of human salmonellosis: I. Infective dose. Rev. Infect. Dis. 4:1096-1106. [DOI] [PubMed] [Google Scholar]
- 21.Bore, E., S. Langsrud, O. Langsrud, T. M. Rode, and A. Holck. 2007. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153:2289-2303. [DOI] [PubMed] [Google Scholar]
- 22.Boucher, S. N., E. R. Slater, A. H. Chamberlain, and M. R. Adams. 1994. Production and viability of coccoid forms of Campylobacter jejuni. J. Appl. Bacteriol. 77:303-307. [DOI] [PubMed] [Google Scholar]
- 23.Brown, J. L., T. Ross, T. A. McMeekin, and P. D. Nichols. 1997. Acid habituation of Escherichia coli and the potential role of cyclopropane fatty acids in low pH tolerance. Int. J. Food Microbiol. 37:163-173. [DOI] [PubMed] [Google Scholar]
- 24.Brown, M. R., and A. Kornberg. 2004. Inorganic polyphosphate in the origin and survival of species. Proc. Natl. Acad. Sci. USA 101:16085-16087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 26.Budin-Verneuil, A., E. Maguin, Y. Auffray, S. D. Ehrlich, and V. Pichereau. 2005. Transcriptional analysis of the cyclopropane fatty acid synthase gene of Lactococcus lactis MG1363 at low pH. FEMS Microbiol. Lett. 250:189-194. [DOI] [PubMed] [Google Scholar]
- 27.Bury-Moné, S., J. M. Thiberge, M. Contreras, A. Maitournam, A. Labigne, and H. De Reuse. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:623-638. [DOI] [PubMed] [Google Scholar]
- 28.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cawthraw, S. A., R. A. Feldman, A. R. Sayers, and D. G. Newell. 2002. Long-term antibody responses following human infection with Campylobacter jejuni. Clin. Exp. Immunol. 130:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang, Y. Y., and J. E. Cronan, Jr. 1999. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol. Microbiol. 33:249-259. [DOI] [PubMed] [Google Scholar]
- 31.Chow, I. T., and F. Baneyx. 2005. Coordinated synthesis of the two ClpB isoforms improves the ability of Escherichia coli to survive thermal stress. FEBS Lett. 579:4235-4241. [DOI] [PubMed] [Google Scholar]
- 32.Cotter, P. D., C. G. Gahan, and C. Hill. 2000. Analysis of the role of the Listeria monocytogenes F0F1-ATPase operon in the acid tolerance response. Int. J. Food Microbiol. 60:137-146. [DOI] [PubMed] [Google Scholar]
- 33.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 34.Cronan, J. E., Jr. 2002. Phospholipid modifications in bacteria. Curr. Opin. Microbiol. 5:202-205. [DOI] [PubMed] [Google Scholar]
- 35.Dedieu, L., J. M. Pages, and J. M. Bolla. 2002. Environmental regulation of Campylobacter jejuni major outer membrane protein porin expression in Escherichia coli monitored by using green fluorescent protein. Appl. Environ. Microbiol. 68:4209-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doyle, M. P., and D. J. Roman. 1981. Growth and survival of Campylobacter fetus subsp. jejuni as a function of temperature and pH. J. Food Prot. 44:596-601. [DOI] [PubMed] [Google Scholar]
- 37.Dressman, J. B., R. R. Berardi, L. C. Dermentzoglou, T. L. Russell, S. P. Schmaltz, J. L. Barnett, and K. M. Jarvenpaa. 1990. Upper gastrointestinal (GI) pH in young, healthy men and women. Pharm. Res. 7:756-761. [DOI] [PubMed] [Google Scholar]
- 38.Ekaza, E., J. Teyssier, S. Ouahrani-Bettache, J. P. Liautard, and S. Kohler. 2001. Characterization of Brucella suis clpB and clpAB mutants and participation of the genes in stress responses. J. Bacteriol. 183:2677-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elvers, K. T., S. M. Turner, L. M. Wainwright, G. Marsden, J. Hinds, J. A. Cole, R. K. Poole, C. W. Penn, and S. F. Park. 2005. NssR, a member of the Crp-Fnr superfamily from Campylobacter jejuni, regulates a nitrosative stress-responsive regulon that includes both a single-domain and a truncated haemoglobin. Mol. Microbiol. 57:735-750. [DOI] [PubMed] [Google Scholar]
- 40.Elvers, K. T., G. Wu, N. J. Gilberthorpe, R. K. Poole, and S. F. Park. 2004. Role of an inducible single-domain hemoglobin in mediating resistance to nitric oxide and nitrosative stress in Campylobacter jejuni and Campylobacter coli. J. Bacteriol. 186:5332-5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engel, P., R. Kramer, and G. Unden. 1994. Transport of C4-dicarboxylates by anaerobically grown Escherichia coli. Energetics and mechanism of exchange, uptake and efflux. Eur. J. Biochem. 222:605-614. [DOI] [PubMed] [Google Scholar]
- 42.Flahaut, S., Y. Tierny, D. Watier, J. P. Hornez, and J. Jeanfils. 2000. Impact of thermal variations on biochemical and physiological traits in Pectinatus sp. Int. J. Food Microbiol. 55:53-61. [DOI] [PubMed] [Google Scholar]
- 43.Foster, J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2:898-907. [DOI] [PubMed] [Google Scholar]
- 44.Foster, J. W. 1999. When protons attack: microbial strategies of acid adaptation. Curr. Opin. Microbiol. 2:170-174. [DOI] [PubMed] [Google Scholar]
- 45.Foster, J. W., and H. K. Hall. 1992. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 174:4317-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foster, J. W., and H. K. Hall. 1991. Inducible pH homeostasis and the acid tolerance response of Salmonella typhimurium. J. Bacteriol. 173:5129-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gajiwala, K. S., and S. K. Burley. 2000. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295:605-612. [DOI] [PubMed] [Google Scholar]
- 48.Gancz, H., S. Censini, and D. S. Merrell. 2006. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect. Immun. 74:602-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gyaneshwar, P., O. Paliy, J. McAuliffe, D. L. Popham, M. I. Jordan, and S. Kustu. 2005. Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. J. Bacteriol. 187:1074-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall, H. K., and J. W. Foster. 1996. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J. Bacteriol. 178:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hayes, E. T., J. C. Wilks, P. Sanfilippo, E. Yohannes, D. P. Tate, B. D. Jones, M. D. Radmacher, S. S. BonDurant, and J. L. Slonczewski. 2006. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol. 6:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hentze, M. W., and L. C. Kuhn. 1996. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. USA 93:8175-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holmes, K., F. Mulholland, B. M. Pearson, C. Pin, J. McNicholl-Kennedy, J. M. Ketley, and J. M. Wells. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151:243-257. [DOI] [PubMed] [Google Scholar]
- 54.Hong, W., W. Jiao, J. Hu, J. Zhang, C. Liu, X. Fu, D. Shen, B. Xia, and Z. Chang. 2005. Periplasmic protein HdeA exhibits chaperone-like activity exclusively within stomach pH range by transforming into disordered conformation. J. Biol. Chem. 280:27029-27034. [DOI] [PubMed] [Google Scholar]
- 55.Horsburgh, M. J., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huesca, M., A. Goodwin, A. Bhagwansingh, P. Hoffman, and C. A. Lingwood. 1998. Characterization of an acidic-pH-inducible stress protein (hsp70), a putative sulfatide binding adhesin, from Helicobacter pylori. Infect. Immun. 66:4061-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Janausch, I. G., E. Zientz, Q. H. Tran, A. Kroger, and G. Unden. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39-56. [DOI] [PubMed] [Google Scholar]
- 58.Karlyshev, A. V., and B. W. Wren. 2005. Development and application of an insertional system for gene delivery and expression in Campylobacter jejuni. Appl. Environ. Microbiol. 71:4004-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kern, R., A. Malki, J. Abdallah, J. Tagourti, and G. Richarme. 2007. Escherichia coli HdeB is an acid stress chaperone. J. Bacteriol. 189:603-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim, B. H., S. Kim, H. G. Kim, J. Lee, I. S. Lee, and Y. K. Park. 2005. The formation of cyclopropane fatty acids in Salmonella enterica serovar Typhimurium. Microbiology 151:209-218. [DOI] [PubMed] [Google Scholar]
- 61.Kim, H. Y., D. Schlictman, S. Shankar, Z. Xie, A. M. Chakrabarty, and A. Kornberg. 1998. Alginate, inorganic polyphosphate, GTP and ppGpp synthesis coregulated in Pseudomonas aeruginosa: implications for stationary phase survival and synthesis of RNA/DNA precursors. Mol. Microbiol. 27:717-725. [DOI] [PubMed] [Google Scholar]
- 62.Klancnik, A., N. Botteldoorn, L. Herman, and S. S. Mozina. 2006. Survival and stress induced expression of groEL and rpoD of Campylobacter jejuni from different growth phases. Int. J. Food Microbiol. 112:200-207. [DOI] [PubMed] [Google Scholar]
- 63.Konturek, J. W., P. Thor, M. Maczka, R. Stoll, W. Domschke, and S. J. Konturek. 1994. Role of cholecystokinin in the control of gastric emptying and secretory response to a fatty meal in normal subjects and duodenal ulcer patients. Scand. J. Gastroenterol. 29:583-590. [DOI] [PubMed] [Google Scholar]
- 64.Korolik, V., and J. M. Ketley. 2005. Campylobacter chemotaxis. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter: molecular and cellular biology. Horizon Bioscience, London, United Kingdom.
- 65.Labigne-Roussel, A., J. Harel, and L. Tompkins. 1987. Gene transfer from Escherichia coli to Campylobacter species: development of shuttle vectors for genetic analysis of Campylobacter jejuni. J. Bacteriol. 169:5320-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leaphart, A. B., D. K. Thompson, K. Huang, E. Alm, X. F. Wan, A. Arkin, S. D. Brown, L. Wu, T. Yan, X. Liu, G. S. Wickham, and J. Zhou. 2006. Transcriptome profiling of Shewanella oneidensis gene expression following exposure to acidic and alkaline pH. J. Bacteriol. 188:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee, I. S., J. Lin, H. K. Hall, B. Bearson, and J. W. Foster. 1995. The stationary-phase sigma factor sigma S (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol. Microbiol. 17:155-167. [DOI] [PubMed] [Google Scholar]
- 68.Liu, M., T. Durfee, J. E. Cabrera, K. Zhao, D. J. Jin, and F. R. Blattner. 2005. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J. Biol. Chem. 280:15921-15927. [DOI] [PubMed] [Google Scholar]
- 69.Logan, S. M., and T. J. Trust. 1983. Molecular identification of surface protein antigens of Campylobacter jejuni. Infect. Immun. 42:675-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lunney, J. K. 2007. Advances in swine biomedical model genomics. Int. J. Biol. Sci. 3:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maurer, L. M., E. Yohannes, S. S. Bondurant, M. Radmacher, and J. L. Slonczewski. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187:304-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McKnight, G. M., C. W. Duncan, C. Leifert, and M. H. Golden. 1999. Dietary nitrate in man: friend or foe? Br. J. Nutr. 81:349-358. [DOI] [PubMed] [Google Scholar]
- 73.Merrell, D. S., C. Bailey, J. B. Kaper, and A. Camilli. 2001. The ToxR-mediated organic acid tolerance response of Vibrio cholerae requires OmpU. J. Bacteriol. 183:2746-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Merrell, D. S., M. L. Goodrich, G. Otto, L. S. Tompkins, and S. Falkow. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71:3529-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mogk, A., A. Volker, S. Engelmann, M. Hecker, W. Schumann, and U. Volker. 1998. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J. Bacteriol. 180:2895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mullan, A., J. P. Quinn, and J. W. McGrath. 2002. Enhanced phosphate uptake and polyphosphate accumulation in Burkholderia cepacia grown under low pH conditions. Microb. Ecol. 44:69-77. [DOI] [PubMed] [Google Scholar]
- 77.Murphy, C., C. Carroll, and K. N. Jordan. 2005. The effect of different media on the survival and induction of stress responses by Campylobacter jejuni. J. Microbiol. Methods 62:161-166. [DOI] [PubMed] [Google Scholar]
- 78.Murphy, C., C. Carroll, and K. N. Jordan. 2003. Identification of a novel stress resistance mechanism in Campylobacter jejuni. J. Appl. Microbiol. 95:704-708. [DOI] [PubMed] [Google Scholar]
- 79.Murphy, C., C. Carroll, and K. N. Jordan. 2003. Induction of an adaptive tolerance response in the foodborne pathogen Campylobacter jejuni. FEMS Microbiol. Lett. 223:89-93. [DOI] [PubMed] [Google Scholar]
- 80.Naikare, H., K. Palyada, R. Panciera, D. Marlow, and A. Stintzi. 2006. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74:5433-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pflock, M., N. Finsterer, B. Joseph, H. Mollenkopf, T. F. Meyer, and D. Beier. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 188:3449-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Price, M. N., K. H. Huang, E. J. Alm, and A. P. Arkin. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rao, N. N., S. Liu, and A. Kornberg. 1998. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J. Bacteriol. 180:2186-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 86.Reezal, A., B. McNeil, and J. G. Anderson. 1998. Effect of low-osmolality nutrient media on growth and culturability of Campylobacter species. Appl. Environ. Microbiol. 64:4643-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86a.Reid, A. N., R. Pandey, K. Palyada, L. Whitworth, E. Doukhanine, and A. Stintzi. 2008. Identification of Campylobacter jejuni genes contributing to acid adaptation by transcriptional profiling and genome-wide mutagenesis. Appl. Environ. Microbiol. 74:1598-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reischl, S., T. Wiegert, and W. Schumann. 2002. Isolation and analysis of mutant alleles of the Bacillus subtilis HrcA repressor with reduced dependency on GroE function. J. Biol. Chem. 277:32659-32667. [DOI] [PubMed] [Google Scholar]
- 88.Richard, H. T., and J. W. Foster. 2003. Acid resistance in Escherichia coli. Adv. Appl. Microbiol. 52:167-186. [DOI] [PubMed] [Google Scholar]
- 89.Robinson, D. A. 1981. Campylobacter infection. R. Soc. Health J. 101:138-140. [DOI] [PubMed] [Google Scholar]
- 90.Rollins, D. M., J. C. Coolbaugh, R. I. Walker, and E. Weiss. 1983. Biphasic culture system for rapid Campylobacter cultivation. Appl. Environ. Microbiol. 45:284-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scott, D. R., E. A. Marcus, Y. Wen, J. Oh, and G. Sachs. 2007. Gene expression in vivo shows that Helicobacter pylori colonizes an acidic niche on the gastric surface. Proc. Natl. Acad. Sci. USA 104:7235-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Spohn, G., A. Danielli, D. Roncarati, I. Delany, R. Rappuoli, and V. Scarlato. 2004. Dual control of Helicobacter pylori heat shock gene transcription by HspR and HrcA. J. Bacteriol. 186:2956-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stintzi, A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stintzi, A., D. Marlow, K. Palyada, H. Naikare, R. Panciera, L. Whitworth, and C. Clarke. 2005. Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect. Immun. 73:1797-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tan, S., C. D. Fraley, M. Zhang, D. Dailidiene, A. Kornberg, and D. E. Berg. 2005. Diverse phenotypes resulting from polyphosphate kinase gene (ppk1) inactivation in different strains of Helicobacter pylori. J. Bacteriol. 187:7687-7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tang, Y., and J. R. Guest. 1999. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology 145:3069-3079. [DOI] [PubMed] [Google Scholar]
- 97.Tang, Y., J. R. Guest, P. J. Artymiuk, R. C. Read, and J. Green. 2004. Post-transcriptional regulation of bacterial motility by aconitase proteins. Mol. Microbiol. 51:1817-1826. [DOI] [PubMed] [Google Scholar]
- 98.Tang, Y., M. A. Quail, P. J. Artymiuk, J. R. Guest, and J. Green. 2002. Escherichia coli aconitases and oxidative stress: post-transcriptional regulation of sodA expression. Microbiology 148:1027-1037. [DOI] [PubMed] [Google Scholar]
- 99.Tangwatcharin, P., S. Chanthachum, P. Khopaibool, and M. W. Griffiths. 2006. Morphological and physiological responses of Campylobacter jejuni to stress. J. Food Prot. 69:2747-2753. [DOI] [PubMed] [Google Scholar]
- 100.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thompson, S. A., and M. J. Blaser. 1995. Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resistance to low pH. Infect. Immun. 63:2185-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tucker, D. L., N. Tucker, and T. Conway. 2002. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 184:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van Vliet, A. H., M. L. Baillon, C. W. Penn, and J. M. Ketley. 1999. Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181:6371-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van Vliet, A. H., F. D. Ernst, and J. G. Kusters. 2004. NikR-mediated regulation of Helicobacter pylori acid adaptation. Trends Microbiol. 12:489-494. [DOI] [PubMed] [Google Scholar]
- 105.van Vliet, A. H., J. M. Ketley, S. F. Park, and C. W. Penn. 2002. The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense. FEMS Microbiol. Rev. 26:173-186. [DOI] [PubMed] [Google Scholar]
- 106.Wainwright, L. M., K. T. Elvers, S. F. Park, and R. K. Poole. 2005. A truncated haemoglobin implicated in oxygen metabolism by the microaerophilic food-borne pathogen Campylobacter jejuni. Microbiology 151:4079-4091. [DOI] [PubMed] [Google Scholar]
- 107.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wen, Y., E. A. Marcus, U. Matrubutham, M. A. Gleeson, D. R. Scott, and G. Sachs. 2003. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 71:5921-5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wenman, W. M., J. Chai, T. J. Louie, C. Goudreau, H. Lior, D. G. Newell, A. D. Pearson, and D. E. Taylor. 1985. Antigenic analysis of Campylobacter flagellar protein and other proteins. J. Clin. Microbiol. 21:108-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Woodall, C. A., M. A. Jones, P. A. Barrow, J. Hinds, G. L. Marsden, D. J. Kelly, N. Dorrell, B. W. Wren, and D. J. Maskell. 2005. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect. Immun. 73:5278-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 112.Young, V. B., and L. S. Mansfield. 2005. Campylobacter infection—clinical context. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter: molecular and cellular biology. Horizon Bioscience, London, United Kingdom.
- 113.Zientz, E., I. G. Janausch, S. Six, and G. Unden. 1999. Functioning of DcuC as the C4-dicarboxylate carrier during glucose fermentation by Escherichia coli. J. Bacteriol. 181:3716-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]