Abstract
In order to cause disease, the food- and waterborne pathogen Campylobacter jejuni must face the extreme acidity of the host stomach as well as cope with pH fluctuations in the intestine. In the present study, C. jejuni NCTC 11168 was grown under mildly acidic conditions mimicking those encountered in the intestine. The resulting transcriptional profiles revealed how this bacterium fine-tunes gene expression in response to acid stress. This adaptation involves the differential expression of respiratory pathways, the induction of genes for phosphate transport, and the repression of energy generation and intermediary metabolism genes. We also generated and screened a transposon-based mutant library to identify genes required for wild-type levels of growth under mildly acidic conditions. This screen highlighted the important role played by cell surface components (flagella, the outer membrane, capsular polysaccharides, and lipooligosaccharides) in the acid stress response of C. jejuni. Our data also revealed that a limited correlation exists between genes required for growth under acidic conditions and genes differentially expressed in response to acid. To gain a comprehensive picture of the acid stress response of C. jejuni, we merged transcriptional profiles obtained from acid-adapted cells and cells subjected to acid shock. Genes encoding the transcriptional regulator PerR and putative oxidoreductase subunits Cj0414 and Cj0415 were among the few up-regulated under both acid stress conditions. As a Cj0415 mutant was acid sensitive, it is likely that these genes are crucial to the acid stress response of C. jejuni and consequently are important for host colonization.
To cause disease, bacteria must survive passage through the gastrointestinal (GI) tract, where acid encountered in the stomach represents an important barrier to infection. Clearly, the ability to overcome this acid shock directly correlates with bacterial infectious doses (ID) and the likelihood of intestinal colonization. The oral ID of enteric pathogens typically ranges from 102 to 109, with Shigella flexneri being one of the most infectious and Salmonella species other than Salmonella typhi being among the least infectious (13). Following transit through the stomach, bacteria encounter a variety of environments within the intestine, ranging from mildly acidic (pH 5.5) to moderately alkaline (pH 7.4) (Table 1). Although the lumen of the intestine is less acidic than that of the stomach, successful enteric pathogens still must be able to withstand exposure to this intestinal acidity.
TABLE 1.
pH values associated with the human GI tract
| Site | pHa |
|---|---|
| Stomach | 1.5 |
| Duodenum | 6.0 |
| Ileum | 7.4 |
| Cecum | 5.7 |
| Right colon | 5.5 |
| Mid-colon | 5.5-6.5 |
| Left colon | 6.5 |
| Rectum | 6.7 |
The low ID (500 to 800 cells) of the enteric pathogen Campylobacter jejuni suggests that this bacterium can sense and respond to decreasing environmental pH (12, 73). While C. jejuni is among the leading causative agents of food- and waterborne gastroenteritis worldwide, few studies have sought to address how C. jejuni deals with acid stress. One strain of C. jejuni (CI120) has been shown to possess an adaptive tolerance response (ATR) that is induced by acid and/or oxygen and that increases the ability of Campylobacter to survive exposure to low pH (60, 62). The induction of the Campylobacter ATR is dependent on protein synthesis, which suggests the presence of pH-inducible proteins. C. jejuni CI120 also secretes a protein that provides some protection against acid stress (60, 61). However, this protein has not been identified or further characterized. Notably, C. jejuni is capable of colonizing both the large and small intestine (3, 63). This implies that C. jejuni can not only tolerate exposure to acid but also can adapt to grow under mildly acidic conditions. This is supported by the growth of C. jejuni in vitro at pHs as low as 5.0 (23). While adaptation to changing environmental pH clearly is important for pathogenesis, little is known about this process in C. jejuni.
Bacteria use many different strategies to combat acid stress. Some of these involve alterations of membranes and cell surfaces, perhaps as a means of preventing the influx of H+ into the cell. Examples of this include the modification of inner membrane phospholipids by cyclopropane fatty acyl synthase (21) as well as changes in the expression of outer membrane proteins (56). Other systems function to extrude protons, thus preventing and/or reversing the acidification of the cytoplasm. For instance, lactic acid bacteria use their ATPase to pump out H+ at the expense of ATP (76). Some of the best-characterized systems involve amino acid decarboxylases, which consume protons during amino acid decarboxylation, and antiporters that exchange product for substrate (29). Finally, DNA repair systems (10, 81, 82) and chaperones (8, 31, 41, 46, 50) appear to be important for the reparation of cellular material damaged by exposure to acid.
The regulation of bacterial acid stress responses is complex and varied. In enteric bacteria such as Escherichia coli, the global regulator RpoS is an important component of the acid stress response (29, 72). However, other regulators, such as the PhoPQ system and the ferric uptake regulator (Fur) in Salmonella enterica serovar Typhimurium, also play a role (4, 6, 8, 10, 30, 36). In Helicobacter pylori, the most important regulator of the acid stress response is the ArsSR two-component system, but Ni2+-dependent (NikR) and Fe2+-dependent (Fur) regulators also are involved (67). Adding to the complexity of these systems is the fact that many of these regulatory pathways overlap to some extent. Interestingly, the C. jejuni NCTC 11168 genome encodes relatively few transcriptional regulators and two-component systems (66), and the involvement of any of these in the acid stress response of C. jejuni has yet to be determined.
A number of studies have sought to examine the transcriptional profile of bacteria grown under acidic and/or basic conditions (2, 16, 18, 37, 53, 84). The small degree of overlap between these studies affirms the importance of using multiple approaches to characterize these responses as thoroughly as possible. These studies revealed that low pH changes the expression of genes involved in energy generation and metabolism, reflecting a switch to metabolic processes that minimize acid production (18, 37, 53). In addition, pH affects the expression of genes involved in motility and chemotaxis (2, 16, 37, 53, 84) as well as genes that encode products that take up and/or export H+ (e.g., ATPase and electron transport chain components) (2, 16, 18, 37, 53). These studies also have shown that bacterial acid stress responses show some overlap with oxidative stress (16, 53, 67) and heat shock (2, 18, 37, 67) responses. However, it is important that the individual genes affected and the nature of the effect (up- or down-regulation) are not often consistent between studies and might be bacterium and/or culture condition dependent.
In a complementary study, we identified genes for which expression was affected in response to acid shock (71). Here, we strive to understand how C. jejuni modulates gene expression to allow adaptation to growth under mildly acidic pH conditions. These are two distinct aspects of acid resistance. In H. pylori, survival under conditions of acid shock is largely mediated by urea uptake and its breakdown by urease (52, 55). However, growth of H. pylori at low pH does not depend on urease activity (9, 10) and requires genes such as lepA, uvrA, atpF, the flavin reductase gene, czcA, and aldo-keto reductase (10), some of which were not previously known to play a role in acid resistance. The objective of this study was to examine the gene expression profile of C. jejuni NCTC 11168 grown to exponential phase under mildly acidic conditions (pH 6.5, 6.0, and 5.5). As transcriptional profiles do not provide a complete picture of a bacterium's response to a given environmental condition, we also generated and screened a transposon insertion library of NCTC 11168 to identify mutants impaired for growth at pH 6.0 and/or 5.5.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. jejuni NCTC 11168 was acquired from the National Collection of Type Cultures. Bacteria were maintained and grown at 37°C on Mueller-Hinton (MH) agar plates or in biphasic MH cultures under microaerophilic conditions (83% N2, 4% H2, 8% O2, and 5% CO2) in a MACS-VA500 microaerophilic workstation (Don Whitley, West Yorkshire, England). The C. jejuni NCTC 11168 fur deletion mutant (65) and the kpsM insertion mutant (77) used in this study were described previously.
To study the effect of pH on C. jejuni growth and on its transcriptional profile, bacteria were grown to stationary phase in biphasic MH medium, and an aliquot of the culture equivalent to an optical density at 600 nm (OD600) of 0.05 was used to inoculate biphasic MH-2-(N-morpholino) ethanesulfonic acid (MES) medium at pH 7.0, 6.5, 6.0, and 5.5. This medium was prepared by layering MES-buffered MH broth (MH-MES; pH 7.0, 6.5, 6.0, or 5.5) over a layer of solid MH-MES agar at the same pH. MES was used at a concentration of 100 mM, and the medium pH was adjusted with NaOH. Bacterial growth was monitored spectrophotometrically at 600 nm by removing 100-μl samples at various time points for 40 h. At the stationary phase, the pH of the broth supernatant was measured using a pH meter (Accumet AR60).
Extraction of total RNA.
RNA was extracted from cells grown to early mid-logarithmic phase (OD600 = 0.9) at 37°C under microaerophilic conditions as described previously (65). Briefly, RNA turnover was prevented by mixing each sample (25 ml) with a cold RNA stabilization solution (2.5 ml of 10% [vol/vol] buffer-saturated phenol in ethanol) on ice. The cells next were collected by centrifugation (10 min, 6,000 × g, 4°C), cell pellets were resuspended in TE buffer (50 mM Tris-Cl, pH 8.0, 1 mM EDTA) containing 0.5 mg ml−1 (final concentration) lysozyme, and total RNA was extracted using a hot phenol-chloroform method (80). RNA was precipitated, resuspended in RNase-free H2O, and treated with DNase I (Epicenter Biotechnologies, Madison, WI) to remove contaminating DNA. Each RNA sample was further purified using a Qiagen RNeasy mini kit (Qiagen, Valencia, CA), and the RNA concentration was determined using the RiboGreen RNA quantification reagents (Molecular Probes, Eugene, OR). The samples were screened for contaminating DNA by PCR amplification and for degradation by agarose gel electrophoresis prior to conversion into cDNA.
Probe labeling and slide hybridization.
RNA samples (16 μg) from each control and test condition were converted to cDNA using 10 μg random hexamers (Amersham Biosciences) and Superscript II reverse transcriptase (Invitrogen). Aminoallyl-dUTP was included in the reverse transcription reaction to permit the labeling of the cDNA with the monoreactive fluors indocarbocyanine (Cy3 dye; used to label pH 7.0 control samples) and indodicarbocyanine (Cy5 dye; used to label test samples) (Amersham Biosciences). Fluorescent labeling of the probes was described previously (65). The labeled cDNA probes from each acid stress condition (e.g., pH 6.5) were individually cohybridized with labeled-cDNA probes from cells grown at pH 7.0 on microarray slides. The C. jejuni NCTC 11168 microarray used in this study was described previously (78) and was constructed using PCR-amplified fragments representing approximately 98% of the open reading frames identified in the NCTC 11168 genome.
Data collection and analysis.
Data were collected and analyzed as described previously (65). Briefly, microarray slides were scanned at 532-nm (Cy3) and 635-nm (Cy5) wavelengths using a laser-activated confocal scanner (ScanArray Gx; Perkin Elmer) at a 10-μm resolution. Spot registration was optimized manually, and the fluorescence intensities of each spot were collected using ScanArray Express software (Perkin Elmer). Spots were excluded from analysis if they were present in areas of slide abnormalities or if their fluorescent mean intensities were below three times the standard deviations from the background in both channels. The fluorescence intensities of all remaining spots were normalized using locally weighted linear regression, the ratio of the mean Cy5:Cy3 values was log2 transformed, and the data were statistically analyzed using the empirical Bayes method (5). The data represent three technical replicates for each of two biological replicates. A gene was considered differentially expressed if its P value was below 10−4 and its change (n-fold) in relative transcript abundance was ≥2. Differentially expressed genes were grouped by hierarchical clustering analysis using Genesis (Graz University of Technology, Graz, Austria).
Generation and screening of a transposon library of C. jejuni NCTC 11168.
A transposon library of C. jejuni NCTC 11168 was constructed using the EZ-Tn5 transposase and the EZ-Tn5 pMOD-3<R6Kγori/MCS> transposon construction vector (Epicenter Biotechnologies). The cat cassette was amplified from pRY111 (93) using primers RAA17 (5′ ATTATTAGGATCCCGGGTACCTGCAGAATTCAGCTGCTCGGCGGTGTTCCTTTCCAAG 3′) and RAA18 (5′-ATTATTAGGATCCCGGGTACCTGCAGAATTC AGCTGCGCCCTTTAGTTCCTAAAGGGT 3′) and cloned into BamHI-digested EZ-Tn5 pMOD-3<R6Kγori/MCS> to yield the EZ-Tn5-Cm transposon. Transposition reactions were carried out in vitro according to the manufacturer's instructions. The EZ-Tn5-Cm transposon was PCR amplified using primers PCRFP (5′ ATTCAGGCTGCGCAACTGT 3′) and PCRRP (5′ GTCAGTGAGCGAGGAAGCGGAAG 3′). Reaction mixtures contained 2 μg of C. jejuni NCTC 11168 chromosomal DNA (extracted from cultures grown to stationary phase), 1 μg of PCR-amplified EZ-Tn5-Cm transposon, 1× reaction buffer, and 1 U of EZ-Tn5 transposase in a 10-μl reaction volume. After a 2-h incubation at 37°C, the reactions were stopped by adding 1 μl of EZ-Tn5 10× stop solution and incubating them at 70°C for 10 min. The reactions then were extracted with phenol-chloroform-isoamyl alcohol (25:24:1), pH 6.7, and ethanol precipitated. The transposed DNA next was repaired by treating the DNA with 2.5 U of T4 DNA polymerase in the presence of 1 mM deoxynucleoside triphosphates (dNTPs) for 20 min at 11°C, followed by heat inactivation of the enzyme at 75°C for 15 min. Finally, the DNA was treated with 600 U of T4 DNA ligase (New England Biolabs) for 18 h at 16°C. The repaired transposed DNA was extracted with phenol-chloroform-isoamyl alcohol (25:24:1), pH 6.7, and ethanol precipitated prior to incubation with recipient C. jejuni NCTC 11168 cells and DNA uptake by natural transformation (89). Cells containing EZ-Tn5-Cm were selected by being plated on MH agar plates containing 20 μg ml−1 Cm. Colonies were individually patched to fresh MH-Cm plates to confirm resistance, inoculated into MH broth in 96-well plates, grown overnight under microaerophilic conditions, and stored at −80°C in 7% (vol/vol) dimethyl sulfoxide.
The resulting library was screened to identify mutants impaired for growth at low pH. Each mutant was grown from frozen stock on MH agar plates, patched to MH-Cm plates, and then transferred to MH-MES agar plates at pH 7.0, 6.0, and 5.5. Plates were incubated for 48 h at 37°C under microaerophilic conditions, and the growth of each mutant was scored. The growth of each strain at pH 6.0 and 5.5 was compared to growth at pH 7.0 and to growth of NCTC 11168 at pH 6.0 and 5.5. All of the acid-sensitive mutants reported here grew well on MH-MES plates at pH 7.0 but were either unable to grow at pH 6.0 and/or 5.5 or grew poorly on these acidic plates compared to the growth of NCTC 11168. The screen was repeated for mutants that showed impaired growth at pH 6.0 and/or 5.5, for a total of two to three independent screens per affected mutant. Only those mutants impaired for growth at pH 6.0 and/or 5.5 in all screens were considered affected for growth at low pH.
Mapping of EZ-Tn5-Cm insertion site in affected mutants.
Chromosomal DNA was extracted from the Tn mutants impaired for growth at low pH using standard protocols. This DNA was used as the template for either a single-primer PCR amplification (24, 45) or a semidegenerate PCR amplification (19, 74). PCR amplification using a single primer (also called RATE, for random amplification of transposon ends) was performed using 0.4 mM dNTPs, 0.8 μM of either primer Ori1 (5′ CCATGAGGGTTTAGTTCGTTAAA 3′) or SqFP (5′ GCCAACGACTACGCACTAGCCAAC 3′), which binds within the transposon, 4.4 mM MgCl2, and 5 U Taq DNA polymerase (hot start). The amplification reaction consists of an initial high-stringency phase that generates single-stranded products corresponding to the transposon end and flanking chromosomal DNA, a low-stringency second phase that allows the nonspecific amplification of the products generated in the first phase, and a final high-stringency phase to amplify all products generated in the second phase. The cycling conditions used were the following: 21 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 3 min; 31 cycles at 94°C for 30 s, 30°C for 30 s, and 72°C for 2 min; and 31 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min. Semidegenerate PCR amplification required two separate amplification steps. In the first step, primer Ori1 or SqFP (which binds within the transposon) and primer CEKG2C (5′ GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT 3′) (a semidegenerate primer with a 5′ overhang) (74) were used to amplify a portion of the transposon and flanking chromosomal DNA. The product of the first amplification reaction was diluted 10-fold, and 1 μl was used as the template for the second amplification reaction, which was carried out using a nested primer that binds within the transposon (SqFP or AR54 [5′ AGAGCTTAGTACGTTAAACATGA 3′]) and a primer (CEKG4; 5′ GGCCACGCGTCGACTAGTAC 3′) (74) complementary to the overhang sequence in CEKG2C. This second step is designed to further amplify the DNA of interest. The reaction mixture for both amplification reactions contained 0.2 mM dNTPs, 0.4 μM each primer, 4.4 mM MgCl2, and 2.5 U Taq polymerase, and the cycling conditions were those used by Salama and coworkers (74). The PCR product amplified using either strategy was sequenced using a nested transposon-specific primer (SqFP or AR54), and the transposon insertion site was identified by BLAST analysis of the DNA sequence immediately flanking the mosaic end of the transposon.
When the sequencing reaction failed to provide adequate data to map the exact insertion site or when the sequence data was of poor quality, the location of insertion sites was confirmed by PCR amplification using primers flanking the suspected Tn insertion site.
Creation of transposon mutations in a fresh NCTC 11168 background.
Because of the possibility that the acid-sensitive phenotype of some transposon mutants is not due to the insertion of the transposon itself but to a secondary mutation elsewhere on the chromosome, selected transposon insertions were transferred into a fresh background, and the acid sensitivity phenotype of the new mutants was determined. Chromosomal DNA was extracted from strains bearing transposon mutations in hisD, fliD, oorC, Cj1135, Cj1388, Cj1442c, and Cj1662, and 0.2 μg of DNA was used to transform C. jejuni NCTC 11168 (natural transformation). Mutants were selected by being plated on MH plates containing 20 μg ml−1 Cm, and the presence of the transposon at the expected chromosomal location was confirmed by PCR amplification using primers flanking the transposon insertion site. These new mutants were assayed for growth at pH 7, 6, and 5.5, as described above.
Phenotypic analysis of acid-sensitive transposon mutants.
The mutants harboring transposons in genes known or suspected to be involved in motility were screened for motility on soft agar plates. Briefly, cells were grown overnight in biphasic MH medium and standardized to an OD600 of 1.0, and 10 μl of this inoculum was spotted onto the center of MH plates containing 0.4% agar. Plates were incubated at 37°C in a MACS-VA500 microaerophilic workstation for 24 h, and the diameters of the zones of motility were measured. A minimum of three biological replicates was assayed per strain, each in technical duplicate. Data were converted to percent motility relative to the motility of NCTC 11168 (considered to be 100%). A two-sample t test assuming unequal variances (Microsoft Excel X) was performed to determine if the motility of the mutants differed from that of the wild type.
The carbohydrate profiles of strains with transposon insertions in genes involved in lipooligosaccharide (LOS) and capsular polysaccharide (CPS) expression were examined by deoxycholate-polyacrylamide gel electrophoresis (DOC-PAGE) of proteinase K-digested whole-cell lysates. Cell lysates were prepared using a modification of the method of Hitchcock and Brown (39). Briefly, cells were grown overnight in biphasic MH medium, and cells from 1 ml of culture at an OD600 of 1.0 were collected by centrifugation (8,000 × g, 5 min). Cells were washed in phosphate-buffered saline (pH 7.4), resuspended in 100 μl of lysis buffer (62.5 mM Tris-HCl, pH 6.8, 2% [wt/vol] sodium dodecyl sulfate, 10% [vol/vol] glycerol, 5% [vol/vol] β-mercaptoethanol, bromophenol blue), and heated at 95°C for 10 min. Proteinase K was added to a final concentration of 0.5 mg ml−1, and samples were incubated overnight at 55°C.
Digested cell lysates were separated on DOC-PAGE gels (16.5% resolving gel) and silver stained, as described by St. Michael and coworkers (77), except that the gels were developed using a solution of 0.025% (wt/vol) citric acid and 0.02% (vol/vol) formaldehyde.
Microarray data accession numbers.
All microarray data have been deposited in the NCBI Gene Expression Omnibus database (accession number GSE9920 and superSeries accession number GSE9942).
RESULTS
Growth of C. jejuni under mildly acidic conditions.
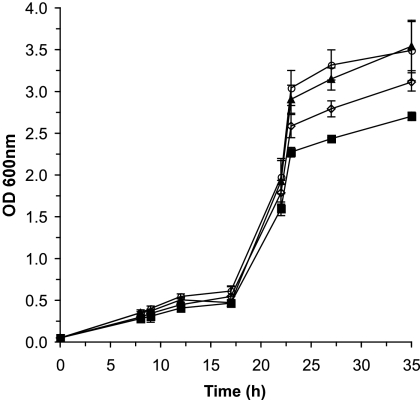
Microorganisms that inhabit the GI tract will encounter widely fluctuating pH environments. Indeed, the pH varies along the GI tract from highly acidic in the stomach to mildly acidic and neutral in the small and large intestines (Table 1). To determine the effect of pH on Campylobacter physiology, the growth of C. jejuni NCTC 11168 was investigated at various pHs (7.0, 6.5, 6.0, and 5.5), mimicking the mildly acidic conditions of the intestine (Fig. 1). While the growth rates of C. jejuni were similar at all four pHs, significant differences in the maximal cell yield were observed between bacteria grown in MH medium adjusted to pH 7.0 and bacteria grown in MH medium adjusted to pH 6.0 and 5.5. Of note, the pH of the MH broth medium did not vary by more than 0.2 pH units throughout the experiment. Consequently, mildly acidic conditions cause a substantial decrease in C. jejuni cell yield but do not affect the rate of cell growth.
FIG. 1.
Effect of pH on C. jejuni growth. C. jejuni was grown at 37°C in biphasic MH-MES medium at pH 7.0 (black triangles), 6.5 (white circles), 6.0 (white diamonds), and 5.5 (black squares) under microaerophilic conditions. The growth of C. jejuni was monitored spectrophotometrically. Error bars indicate standard deviations and in some cases are too small to be seen.
Global transcriptional profile of C. jejuni grown at low pH.
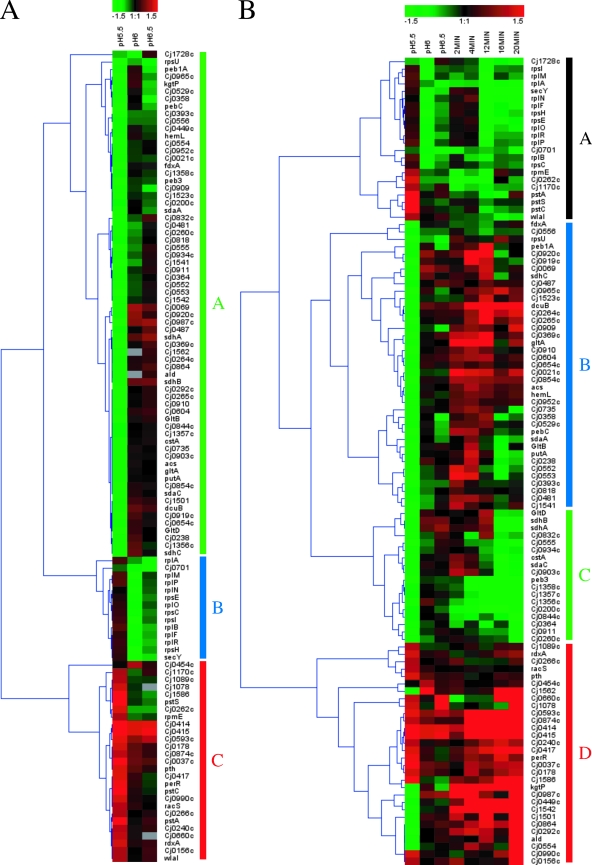
To study the effect of pH on gene expression, C. jejuni was grown to mid-log phase at pH 7.0, 6.5, 6.0, and 5.5 in biphasic MH-MES medium, and the resulting transcriptional profiles were analyzed by microarray technologies. Given a similar bacterial growth rate at the various pHs (Fig. 1), the transcriptional changes essentially should reflect the effect of adaptation to mildly acidic conditions. Genes were considered differentially expressed if their change in relative expression level was ≥2-fold, with a Bayesian P value of <10−4. Overall, the transcript abundance of 109 genes was altered in at least one of the three growth conditions. Not surprisingly, the magnitude of changes in gene expression was greatest when C. jejuni was grown at pH 5.5. To visually investigate the effect of mildly acidic pH on the transcriptome of C. jejuni, the 109 differentially expressed genes were subjected to hierarchical clustering analysis (Genesis; Euclidian distance, average linkage) and were found to group into three main clusters, designated A, B, and C (Fig. 2A).
FIG. 2.
Hierarchical clustering analysis of C. jejuni genes differentially expressed in response to growth under acidic conditions. (A) Genes differentially expressed (≥2-fold change; P < 10−4) under at least one growth condition (pH 6.5, 6.0, and/or 5.5) were subjected to hierarchical clustering analysis. (B) Transcriptional profiles of C. jejuni grown under acidic conditions (columns labeled pH 6.5, pH 6, and pH 5.5) were merged with those obtained after exposure of C. jejuni to acid shock (pH 4.5) in vitro for different lengths of time (columns labeled 2MIN to 20MIN). Only the genes differentially expressed (≥2-fold change; P < 10−4) in response to growth at low pH were included in our analysis. For both panels, up-regulated genes are shown in red, down-regulated genes are in green, and gray denotes missing data. The maximum color output for this figure was set at a log2 value of ±1.5 (equivalent to a threefold change in gene expression).
The 68 genes in cluster A were down-regulated at pH 5.5 and for the most part were unaffected at higher pHs. This cluster includes genes involved in energy generation and general intermediary metabolism, such as genes that encode products that are involved in electron transport (fdxA and Cj0369c) and trimethylamine N-oxide/dimethyl sulfoxide (TMAO/DMSO) (Cj0264c and Cj0265c) and nitrite (Cj1357c and Cj1358c) respiration and genes encoding succinate dehydrogenase subunits (sdhABC), a malate:quinone oxidoreductase (Cj0393c), citrate synthase (gltA), a putative periplasmic protein with a cytochrome c signature motif (Cj0854c), and an aldehyde dehydrogenase enzyme (ald). A number of genes involved in amino acid metabolism and transport also are in this cluster, including genes required for serine (sdaA) and proline (putA) catabolism, glutamate biosynthesis (gltBD), and serine (sdaC) and aspartate/glutamate (Cj0919c-Cj0920c-peb1A-pebC) transport, a gene involved in the conversion of aspartate to lysine (Cj0481), and genes encoding putative amino acid transport proteins (Cj0903c and Cj0934c). The gene encoding the α-ketoglutarate permease (kgtP), which imports a tricarboxylic acid cycle intermediate that is required for the biosynthesis of glutamate, proline, arginine, and glutamine, also is down-regulated at pH 5.5. Cluster A also includes genes encoding a number of known and putative transporters, including an anaerobic C4-dicarboxylate transporter (dcuB), a putative dicarboxylate carrier (Cj0555), a putative Na+/H+ antiporter family protein (Cj0832c), a peptide transporter (cstA), and a putative mechanosensitive ion channel (Cj0238). Genes that encode products that are required for cofactor biosynthesis (hemL, acs, and Cj0529c) also are present in this cluster, as are genes potentially involved in the defense against oxidative stress (Cj0358, a putative cytochrome C551 peroxidase) and the stringent response (Cj0604, a putative polyphosphate kinase). Consistent with the large number of genes in C. jejuni NCTC 11168 for which there is no ascribed function, cluster A comprises 21 genes of unknown function. The significance of their differential expression is impossible to interpret until these genes and their products have been further characterized.
Cluster B comprises 14 genes for which expression was significantly down-regulated at pH 6.5 and/or 6.0 and largely unchanged at pH 5.5. These genes encode ribosomal proteins (rplABFMNOPR and rpsCEHI), a component of the Sec translocon for protein secretion (secY), and a putative protease (Cj0701).
The expression of the 27 genes within cluster C was up-regulated at pH 5.5 and for the most part was not significantly altered at pH 6.5 and 6.0. Among these are genes encoding the peroxide stress regulator, PerR, as well as the sensor (RacS) of a two-component system involved in temperature-dependent signaling (14). Also in this cluster are genes encoding products that are involved in phosphate transport (pstSCA), Fe/S cluster biogenesis (Cj0240c), electron transport (Cj0037c and Cj0874c), protein synthesis (rimM, rpmE, truB, and pth), defense against nitrosative stress (Cj1586), N-linked protein glycosylation (Cj1123c and pglD), and chemotaxis (Cj0262c). This group also includes genes encoding transporters (Cj0178c, Cj1170c, and Cj0266c), a putative periplasmic protein with a glycosyl hydrolase motif (Cj1078), a putative membrane protein (Cj0454c) required for wild-type motility and chick colonization (38), and a nitroreductase (rdxA), as well as a number of genes of unknown function (Cj0417, Cj0593c, Cj0660c, Cj0990c, Cj1056c, and Cj1089c). Notably, within cluster C are two genes (Cj0414 and Cj0415) for which expression was highly up-regulated when C. jejuni was grown at pH 5.5, 6.0, and 6.5. These genes encode putative glucose-methanol-choline oxidoreductase subunits of unknown function.
Construction of a mutant library by transposon mutagenesis.
While transcriptional profiles provide a wealth of information about how a bacterium modulates gene expression in response to a given stress condition, transcriptome analysis suffers from two major weaknesses: mRNA abundance may not always correlate with the expression of functional proteins, and protein expression may not always correlate with its functional requirement. In particular, the up-regulation of a gene under a given stress condition does not necessarily indicate it is required for survival under that condition (11). Furthermore, the expression of a gene can be down-regulated yet still be required for survival under a given stress condition (58). Thus, it is important to supplement gene expression studies with experiments designed to assess the contribution of genes to stress resistance. Therefore, in order to directly identify the genes that are required for acidic pH survival, we first constructed a library of C. jejuni mutants by transposon mutagenesis and then screened individual mutants for their abilities to grow at low pH.
In order to efficiently generate a collection of mutants in C. jejuni NCTC 11168, a Tn5-based transposon containing a chloramphenicol resistance cassette was constructed. This transposon was used in in vitro transposition reactions with purified chromosomal DNA from C. jejuni NCTC 11168. The transposed chromosomal DNA was naturally transformed into C. jejuni, and transposon mutants were recovered on selective MH agar plates under microaerophilic conditions at 37°C. A total of 3,072 mutants were individually picked and grown in 96-well microtiter dishes and immediately frozen at −80°C. Of note, of these 3,072 mutants, only 2,577 remained viable (colonies were obtained on selective MH agar from the frozen stock). From this collection, 24 mutants were randomly chosen and shown by Southern blot analysis to contain a single transposon (data not shown). The transposon insertion site of 48 mutants (arbitrarily chosen) was further mapped using a single-primer PCR amplification method. This mapping confirmed the random insertion of the transposon (data not shown) and indicated that the transposons in the 2,577 mutants used in this study should be randomly distributed throughout the genome. Therefore, a library of this size is expected to contain one insertion at approximately every 640 bp.
Transposon mutants showing impaired growth at low pH.
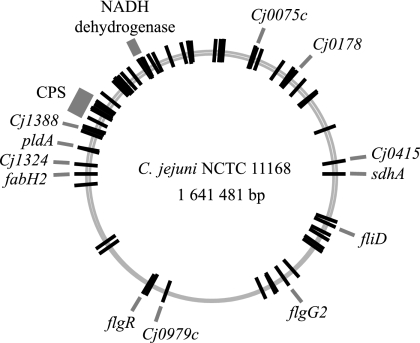
Each mutant from our collection of 2,577 clones was screened for growth on MES-MH plates at pH 7.0, 6.0, and 5.5. Mutants that showed impaired growth at one or both of the acidic pHs were selected, and the insertion site of the transposon was mapped using a PCR-based approach coupled with DNA sequencing. Of the 2,577 mutants screened, 86 were affected, and these mapped to 59 genes and five intergenic regions (Table 2). Mutations leading to a pH-sensitive phenotype were distributed throughout the chromosome (Fig. 3). Of note, we were unable to locate the transposon insertion site in six of the affected mutants.
TABLE 2.
Characterization of transposon mutants impaired for growth at low pH
| Gene and functional group or intergenic region | Description | Tn location (ORF size, in bp), orientationa | Operon (score)b | Expression in steady-state exptc,d | Expression in acid shock exptc,e |
|---|---|---|---|---|---|
| Chemotaxis and motility | |||||
| Cj0019c | Putative methyl-accepting chemotaxis signal transduction protein | 461 (1,779), − | Y (0.852) | ||
| Cj0144 | Probable methyl-accepting chemotaxis signal transduction protein | 224 (1,980), + | N | UP (4) | |
| Cj0262c | Putative methyl-accepting chemotaxis signal transduction protein | 915 (1,998), + | N | UP (5.5) | DOWN (2, 4, 12) |
| Cj0548 (fliD) | Flagellar hook-associated protein | 976 (1,929), + | Y (0.996) | UP (4, 12) | |
| Cj0697 (flgG2) | Flagellar basal body rod protein | 730 (813), + | Y (0.991) | ||
| Cj1024c (flgR) | σ54-Associated transcriptional activator, response regulator of FlgSR two-component system | 132 (1,302), + | Y (0.978) | DOWN (2, 12) | |
| Cj1324 | Hypothetical protein, O-linked glycosylation | 693 (1,122), + | Y (0.581) | ||
| LOS and CPS biosynthesis and expression | |||||
| Cj1135 | Putative two-domain glucosyltransferase, LOS locus | 1,111 (1,548), + | Y (0.943) | UP (5.5) | |
| Cj1150c (hldE) | d-β-d-Heptose 7-phosphate kinase/d-β-d-heptose 1-phosphate adenylyltransferase, LOS locus | 851 (1,386), − | Y (0.992) | NDh | ND |
| Cj1413c (kpsS) | Possible polysaccharide modification protein | 560 (1,185), + | Y (0.998) | ||
| Cj1431c (hddC) | Heptosyltransferase, CPS locus | 1,404 (1,749), − | Y (0.829) | ||
| Cj1432c | Putative sugar transferase, CPS locus | 1,842 (3,096), + | Y (0.829) | ||
| Cj1437c | Aminotransferase, CPS locus | 790 (1,104), − | Y (0.734) | ||
| Cj1442c | Putative sugar transferase, CPS locus | 49 (1,635), + | Y (0.892) | ||
| Lipoproteins and outer membrane proteins | |||||
| Cj0599 | OmpA family protein | 516 (954), − | Y (0.979) | UP (4) | |
| Cj1279c | Putative fibronectin domain-containing lipoprotein | 965 (1,236), + | Y (0.984) | DOWN (4) | |
| Cj1351 (pldA) | Phospholipase A | 777 (990), + | Y (0.930) | DOWN (5.5) | |
| Cj1677 | Possible lipoprotein | 2,953 and 2,779 (3,363), − | N | UP (12, 20) | |
| DNA restriction/modification and repair | |||||
| Cj0208 | DNA modification methylase (adenine specific) | 115 and 750 (1,092), − | N | DOWN (4, 12) | |
| Cj1731c (ruvC) | Crossover junction endodeoxyribonuclease, DNA repair | 144 (483), + | N | UP (5.5) | |
| Oxidative stress | |||||
| Cj0020c | Cytochrome C551 peroxidase | 463 (915), + | Y (0.852) | ||
| Metabolism and bioenergetics | |||||
| Cj0075cf | Putative oxidoreductase iron-sulfur subunit | 153 (741), + | Y (0.992) | DOWN (5.5) | UP (12) DOWN (16) |
| Cj0081 (cydA) | Cytochrome bd oxidase, subunit I | 369 (1,563), + | Y (0.995) | DOWN (12, 16) | |
| Cj0437 (sdhA) | Succinate dehydrogenase, flavoprotein subunit | 1,132 (1,836), + | Y (0.999) | UP (6.5) DOWN (5.5) | UP (12) DOWN (16, 20) |
| Cj0538 (oorC) | Subunit of 2-oxoglutarate:acceptor oxidoreductase | 149 (558), − | Y (0.999) | UP (12, 16) | |
| Cj1509c (fdhC) | Formate dehydrogenase, cytochrome subunit | 228 (933), + | Y (0.972) | ||
| Cj1566c (nuoN) | NADH dehydrogenase I, chain N | 509 (1,389), + | Y (0.999) | ||
| Cj1570c (nuoJ) | NADH dehydrogenase I, chain J | 476 (519), + | Y (0.999) | UP (12, 16) | |
| Cj1571c (nuoI) | NADH dehydrogenase I, chain I | 428 (642), − | Y (0.998) | UP (12, 16) | |
| Cj1573c (nuoG) | NADH dehydrogenase I, chain G | 362 (2,463), + | Y (0.998) | UP (12, 16, 20) | |
| Cj1682c (gltA) | Citrate synthase | 352 (1,269), +; 823 (1,269), − | N | DOWN (5.5) | UP (2, 4, 12, 20) |
| Amino acid biosynthesis and transport | |||||
| Cj0007 (gltB) | Glutamate synthase, large subunit | 2,447 (4,491), + | Y (0.61) | DOWN (5.5) | UP (4) DOWN (16) |
| Cj0013 (ilvD) | Dihydroxyacid dehydratase, branched chain amino acid biosynthesis | 444 (1,677), − | N | ||
| Cj1015c (livG) | Putative branched-chain amino acid ABC transport system, ATP binding protein | 58 (771), − | Y (0.992) | DOWN (12, 16) | |
| Cj1393 (metC′) | Cystathionine β-lyase | 210 (1,047), +; 161 (1,047), − | N | ND | ND |
| Cj1598 (hisD) | Histidinol dehydrogenase | 557 (1,287), − | Y (0.998) | ||
| Transporters | |||||
| Cj0175c (cfbpA) | Putative iron uptake ABC transport system, periplasmic iron-binding protein | 767 (1,005), − | Y (0.998) | UP (12, 16) | |
| Cj0178 | Putative TonB-dependent outer membrane receptor | 1,550 (2,268), + | Y (0.982) | UP (5.5) | UP (4, 20) |
| Cj0607 | ABC-type transmembrane transporter | 623 (1,926), − | Y (0.995) | DOWN (all) | |
| Cj0679 (kdpD) | Truncated KdpD protein, contains osmosensitive K+ channel His kinase sensor domain | 1,767 (1,821), + | N | ND | |
| Cj0727 | Putative periplasmic solute-binding protein | 259 (1,047), + | Y (0.584) | DOWN (4, 16) | |
| Cj0732 | ABC transport system ATP-binding protein | 811 (990), + | Y (0.987) | ND | ND |
| Cj1352 (ceuB) | Enterobactin uptake permease | 215 (969), − | Y (0.994) | ND | ND |
| Cj1539c | Putative anion-uptake ABC transport permease protein | 233 (720), + | Y (0.964) | ||
| Cj1587c | Putative ABC transporter | 1,432 (1,632), − | N | ||
| Cj1630 (tonB2) | Putative TonB transporter | 620 (684), + | Y (0.976) | ||
| Cj1662 | Putative integral membrane protein, ABC transporter cluster | 958 (1,119), − | Y (0.99) | UP (12, 16) | |
| Purine biosynthesis and nucleoside salvage | |||||
| Cj0340 | Putative nucleoside hydrolase | 8 (1,008), − | Y (0.848) | UP (4, 12) | |
| Cj1498c (purA) | Adenylosuccinate synthase | 798 (1,251), − | Y (0.984) | ||
| Degradation of DNA, RNA and proteins | |||||
| Cj0979c | Putative secreted nuclease | 391 (528), + | Y (0.874) | UP (5.5) | UP (12, 16, 20) |
| Cj1388 | Putative endoribonuclease L-PSP | 198 (363), + | Y (0.742) | UP (6.0) | UP (16) |
| Synthesis and modification of aminoacyl-tRNA synthetases | |||||
| Cj1378 (selA) | l-Seryl-tRNA selenium transferase | 231 (1,323), + | Y (0.999) | ||
| Cj1504c (selD) | Selenophosphate synthetase | 681 (927), − | Y (0.588) | ||
| Biosynthesis of cofactors, prosthetic groups and carriers | |||||
| Cj1518 (moaE) | Putative molybdopterin converting factor, subunit 2 | 140 (447), − | Y (0.995) | ||
| Fatty acid biosynthesis | |||||
| Cj1303 (fabH2) | Putative 3-oxoacyl-[acyl-carrier-protein] synthase | 330 (1,062), + | Y (0.968) | ||
| Miscellaneous and unknown functions | |||||
| Cj0184c | Putative Ser/Thr protein phosphatase | 129 (1,155), + | Y (0.852) | ||
| Cj0256 | Putative sulfatase family protein | ∼1,328 (1,539), − | Y (0.967) | UP (6.0) | |
| Cj0415 | Putative glucose-methanol-choline oxidoreductase subunit | 1,507 (1,722), + | Y (0.983) | UP (all) | UP (all) |
| Cj0569 | Hypothetical protein | 309 (870), − | Y (0.764) | ||
| Intergenic regionsg | |||||
| Cj0091-Cj0092 | 101,369 | N | |||
| Cj0363c (hemN)-Cj0364 | 330,736 | N | |||
| Cj0755 (cfrA)-Cj0757 (hrcA) | 707,606 | N | |||
| Cj1463 (flgJ)-Cj1464 (flgM) | 1,399,902 | N | |||
| Cj1508c (fdhD)-Cj1509c (fdhC) | 1,444,609 | N |
The number indicates the nucleotide before which the Tn is inserted; the Cm cassette within the Tn is either in the same orientation as the gene (+) or is in the opposite orientation (−). ORF, open reading frame.
The operon score indicates whether a gene is predicted to be part of an operon; the strength of the prediction is listed in parentheses (values near 1 are confident predictions that the gene is part of an operon, while values near 0.5 are low-confidence predictions) (68); the highest score is shown for genes predicted to be in an operonic structure with both flanking genes.
UP and DOWN indicate up-regulated and down-regulated gene expression, respectively (>1.5-fold difference in relative gene expression; P < 10−4).
For gene expression from steady-state experiments (this study), the number in parentheses indicates the pH value at which the gene was differentially expressed.
For gene expression from in vitro acid shock experiments (71), the value in parentheses indicates the time point after acid shock (2, 4, 12, 16, and 20 min) at which the gene was differentially expressed.
Boldface highlights genes for which the change in expression was similar in response to that for in vitro acid shock and steady-state growth at low pH.
The location of intergenic transposons is given as the base pair number within the C. jejuni NCTC 11168 genome before which the Tn was inserted.
ND, not determined. The data either were excluded from microarray analysis or the gene was not represented on the array.
FIG. 3.
Distribution of transposon insertion sites leading to an acid-sensitive phenotype. The 59 genes and five intergenic regions described in Table 2 are represented by bars on a chromosomal map of C. jejuni NCTC 11168. Only some of these genes are labeled.
The genes required for growth at low pH are involved in a variety of cellular processes, including motility and chemotaxis, CPS and LOS biosynthesis, metabolism and bioenergetics, DNA repair, stress responses, amino acid biosynthesis, transport, macromolecule degradation, and cofactor biosynthesis. While gene expression data were not available for 4 of the 59 genes identified in our library screen, the expression of 42 of the remaining 55 genes was not significantly affected in our microarray analysis (Table 2), revealing a limited correlation between gene requirement and gene expression. As described above, this is not an unusual finding. However, in our case, part of this discrepancy may stem from the use of a biphasic growth medium for gene expression analysis and a solid medium for the screening of the library. In addition, it should be noted that the mutants are likely to encounter an initial pH stress when they are transferred from MH agar plates at pH 7.0 to MH agar plates at pH 5.5. Genes required for coping with acid shock might be different than those required to grow under mildly acidic pH conditions. In agreement with this hypothesis, 27 of the 55 genes mentioned above were found to be differentially expressed upon acid shock exposure (Table 2, last column). Interestingly, among the 13 differentially expressed genes required for growth at low pH, 4 of them were down-regulated, suggesting that gene expression homeostasis is essential for cell survival. The up-regulated genes required for growth under mildly acidic conditions are Cj0262c (chemotaxis), Cj1135 (LOS biosynthesis), ruvC (DNA repair), Cj0178 (outer membrane receptor), Cj0979c (secreted nuclease), Cj1388 (endoribonuclease), Cj0256 (sulfatase), and Cj0415 (putative oxidoreductase). The down-regulated genes required for growth at low pH are pldA (phospholipase A), Cj0075c (oxidoreductase), gltA (citrate synthase), and gltB (Glu synthase). The sdhA gene (succinate dehydrogenase subunit) was up-regulated at pH 6.5 and down-regulated at pH 5.5.
It is possible that the acid-sensitive phenotypes of some of the mutants are not due to the interruption of the gene in question but rather to polar effects on downstream genes, effects on transcript stability, or spontaneous mutations elsewhere on the chromosome. The cat cassette used in the transposon is not known to cause polar effects (87), but we cannot discount the possibility that the transposon itself affects the expression of downstream genes. In Table 2, we have indicated which genes are predicted to be in operonic structures (68). These represent ∼80% of the genes identified in our screen. Many of these are clustered with other genes involved in the same biological process (e.g., flagellum biosynthesis). So while the interruption of the gene in question may not in itself be the reason for the observed acid sensitivity, the biological process in which it is involved likely is implicated. Likewise, the identification of multiple mutants with transposon insertions in various genes involved in a common function (e.g., CPS expression) strengthens the case for that function being important for acid adaptation. Spontaneous mutations elsewhere on the chromosome also are not likely explanations for cases in which we identified more than one mutant (separate insertion sites) for a given gene (Cj1677, Cj0208, and gltA). Nevertheless, to address the possible contribution of spontaneous mutations to the acid sensitivity of the mutants, we recreated seven mutants (representing ∼12% of the genes identified in our screen: hisD, fliD, oorC, Cj1135, Cj1388, Cj1442c, and Cj1662) in a fresh NCTC 11168 background. Like their parents, these mutants displayed impaired growth at pH 5.5, suggesting that the interruption of the gene and/or cluster in question likely is responsible for the observed phenotype.
Phenotypic analysis of selected acid-sensitive transposon mutants.
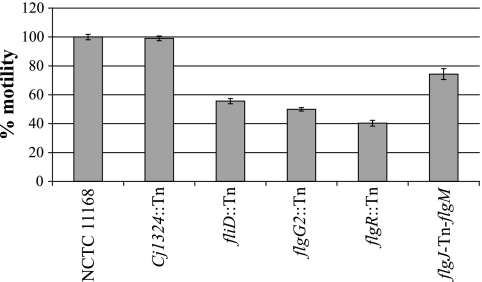
Four acid-sensitive transposon mutants harbored insertions in genes known or predicted to be involved in motility (fliD, flgG2, flgR, and Cj1324), which suggests a possible role for flagellum biosynthesis and/or modification in C. jejuni's ability to grow under mildly acidic conditions. In addition, a transposon inserted in the intergenic region between flgJ and flgM led to an acid-sensitive phenotype. Given these data, the motilities of these five strains on soft agar plates at 37°C were compared to that of NCTC 11168 (Fig. 4). Transposon mutants in fliD, flgG2, and flgR and in the intergenic region between flgJ and flgM were significantly less motile than the wild type (P = 10−4 to 10−12), while the Cj1324:Tn mutant was unaffected (P = 0.82).
FIG. 4.
Motility of selected transposon mutants on 0.4% agar plates. The motility of selected transposon mutants was assayed on MH plates containing 0.4% agar. The diameters of the zones of motility were recorded after 24 h of incubation at 37°C in a microaerophilic workstation and are expressed as the percent motility relative to the motility of NCTC 11168 (considered to be 100%). Values reported are percent motility ± standard errors of the means and represent a minimum of three independent experiments, each performed in technical duplicate.
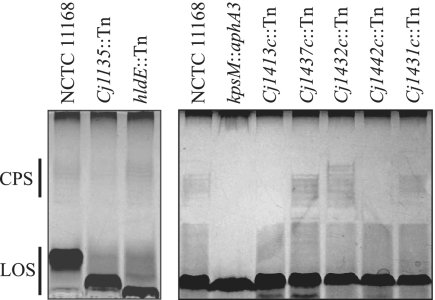
Given that the insertion sites of several transposons mapped to genes located within LOS and CPS loci, we wanted to determine whether these surface structures play a role in C. jejuni adaptation to growth at low pH. Total carbohydrate samples were prepared from cell lysates, separated on sodium DOC polyacrylamide gels, and detected by silver staining (Fig. 5). In these gels, LOS appears as a dark, fast-migrating band. As is apparent from Fig. 5, both LOS transposon mutants (Cj1135:Tn and hldE:Tn) have a severely truncated LOS structure compared to that of the wild type, suggesting that a more complete LOS structure is needed for acid survival and/or adaptation. The CPS can be seen in the wild type as a collection of bands migrating about midway into the gel. A kpsM insertional mutant is known to be acapsular (44, 77) and was included in the gel as a negative control. While strains bearing transposon insertions in Cj1413c (kpsS) and Cj1442c appear to be acapsular, strains with insertions in Cj1437c, Cj1431c, and Cj1432c still express CPS, although a subtle change in CPS migration is apparent in the Cj1432c mutant. Note that minor differences in the amount of polysaccharide produced and/or the extent of modification would not be detected in this experiment.
FIG. 5.
LOS and CPS phenotypes of selected transposon mutants. Proteinase K-digested cell lysates were run on 16.5% DOC polyacrylamide gels and silver stained. The range of migration of CPS and LOS is indicated by bars at the left.
Merged transcriptional profiles of C. jejuni's response to growth at low pH and exposure to acid shock.
In order to gain a more comprehensive picture of the acid stress response of C. jejuni, we merged the transcriptional profiles obtained after the growth of C. jejuni under acidic conditions with those obtained after exposure to acid shock (pH 4.5) in vitro (71). Genes differentially expressed in response to growth at pH 6.5, 6.0, and/or 5.5 were included in our analysis. The data were subjected to hierarchical clustering analysis (Genesis; Euclidian distance, average linkage) and were found to group into four clusters (Fig. 2B). Many of these genes showed opposite expression patterns between response to growth at acidic pH and that after acid shock (Fig. 2B, cluster B and a subset of clusters A and D). The genes with similar expression profiles under both experimental conditions are of particular interest, because they might encode key players of the acid stress survival mechanism. For instance, C. jejuni down-regulated genes encoding ribosomal proteins at pH 6 and 6.5 and upon exposure to acid shock (Fig. 2, cluster A). In addition, growth at pH 5.5 and exposure to acid shock led to the down-regulation of succinate dehydrogenase subunits (sdhAB), a gene encoding a Na+/H+ antiporter (Cj0832c), genes for nitrite respiration (Cj1357c and Cj1358c), and a number of genes of unknown function (Fig. 2B, cluster C). The genes up-regulated in response to both acid stress conditions included a single-domain globin (Cj1586) involved in nitric oxide stress defense, the peroxide regulator gene (perR), a putative ferric-siderophore transporter (Cj0178), genes encoding putative oxidoreductase subunits (Cj0414 and Cj0415), and genes of unknown function (Fig. 2B, cluster D).
DISCUSSION
It is well established that the ability of bacteria to grow and survive in acidic environments is dependent on the regulation and/or maintenance of their internal pH close to neutrality. While this process has been extensively studied in E. coli and other food-borne pathogens, there is little information on how C. jejuni survives exposure to low pH. To initiate the study of this process, we compared the gene expression profile of C. jejuni NCTC 11168 grown to mid-log phase in neutral (pH 7.0) medium to that of the same strain grown in mildly acidic media (pH 5.5, 6.0, and 6.5). Such gene expression profiles reveal how the bacterium fine-tunes its transcriptome to adapt to a given environment and thus provide information about which genes and/or systems might be involved in acidic pH adaptation. However, it is clear that despite the wealth of information generated by gene expression profiling, this technology suffers from two major limitations. First, microarray technology is not sensitive enough to capture small changes in gene expression that can have a significant physiological effect. Second, transcript abundances may not necessarily correlate with the expression of functional proteins. Moreover, transcriptome studies cannot identify proteins that are regulated at the posttranslational level. In fact, the pH-dependent regulation of protein function is an important feature of antiporters that participate in pH homeostasis (64). Consequently, to gain a more comprehensive look at the steady-state acid stress response of C. jejuni, we supplemented transcriptional profiles with the screening of a collection of transposon-based mutants in which genes essential for wild-type levels of growth at mildly acidic pH were identified. This integrated approach revealed the importance of a number of cellular processes in the mechanism of C. jejuni survival and/or adaptation to low pH, ranging from general metabolism and energy generation to the synthesis of surface structure components.
Effect of acid stress on metabolism and energy generation.
The growth of C. jejuni at low pH led to the differential expression of genes that encode products that are involved in the tricarboxylic acid cycle as well as in various electron transport pathways. Succinate dehydrogenase (sdhABC) and malate:quinone oxidoreductase (mqo; Cj0393c) genes were down-regulated at pH 5.5, as were genes encoding products that are involved in the use of TMAO/DMSO (Cj0264c and Cj0265c) and nitrite (Cj1357c and Cj1358c) as terminal electron acceptors. While one cytochrome c gene (Cj0854c) was down-regulated at pH 5.5, two other cytochrome c genes (Cj0037c and Cj0874c) were up-regulated. Down-regulation of sdhAB and Cj1357-58c and up-regulation of Cj0037c also were seen when C. jejuni was exposed to acid shock (Fig. 2B). While it may be tempting to speculate that growth at low pH does not rely on succinate dehydrogenase feeding electrons into the electron transport chain (ETC), our transposon library screen identified an sdhA mutant as being impaired for growth at pH 5.5. This phenotype in turn suggests that the ETC contributes to the ability of C. jejuni to grow under acidic conditions and highlights the importance of an integrated experimental approach combining transcriptomics and genome-wide mutagenesis. The mutation of a number of subunits of the NADH dehydrogenase complex (nuoGIJN) led to impaired growth at low pH. Given that succinate dehydrogenase subunits are down-regulated at pH 5.5, a functional NADH dehydrogenase complex may be required for energy generation. Alternatively, the requirement for NADH dehydrogenase expression may reflect the fact that this complex pumps four H+/electron pairs into the periplasm, which may help prevent and/or reverse cytoplasm acidification. Similarly, the inability of strains harboring mutations in the cyanide-insensitive oxidase (cydA), the formate dehydrogenase (fdhC), and the cytochrome c peroxidase (Cj0020c) genes to grow at low pH further highlights the importance of electron transport pathways for low-pH tolerance. Taking our results together, it is tempting to speculate that the translocation of protons across the cytoplasmic membrane during electron transfer contributes significantly to the ability of C. jejuni to survive in mildly acidic environments. In contrast, the expression of succinate dehydrogenase genes in S. flexneri is induced by either acidic or basic growth conditions (18), while in E. coli, succinate and NADH dehydrogenase genes show opposite pH responses under aerobic (induced by acid) and anaerobic (repressed by acid) growth conditions (37, 53). Thus, it is possible that the pH-dependent regulation of energy generation systems is dependent on the organism and/or the growth conditions used.
The growth of E. coli and Shigella under acidic or basic conditions leads to the preferential expression of metabolic systems that are compatible with the bacterium's environment, i.e., that minimize acid production at low pH and maximize it at high pH (17, 18, 37, 53). For instance, growth under acidic conditions leads to the up-regulation of genes encoding products that are involved in amino acid catabolism, which generates amines (decarboxylases) or ammonia (dehydratases) that can buffer the cellular environment and prevent and/or reverse the acidification of the cytoplasm. It therefore is surprising that C. jejuni down-regulates a gene (sdaA) for the catabolism of serine, which generates ammonia, when grown on medium at pH 5.5. On the other hand, the biosynthesis of methionine, histidine, and glutamate and the biosynthesis and transport of branched-chain amino acids appear to be required for growth at pH 5.5, as metC, hisD, gltB, ilvD, and livM mutants were acid sensitive. The metC gene encodes a putative cystathionine beta-lyase, a key enzyme in methionine biosynthesis, the activity of which generates homocysteine, pyruvate, and ammonia. In agreement with our observations, methionine biosynthesis appears to play a role in bacterial acid stress responses. In H. pylori, metB expression is activated at low pH by the ArsSR two-component system (67), and a metB-Tn mutant of Mycobacterium smegmatis was recovered in a screen designed to identify mutants unable to grow at low pH in the presence of a proton-motive force uncoupler (83). Finally, the main difference between an acid-resistant mutant of Bifidobacterium longum and its more sensitive parent is the overproduction of MetE, CysD, and MetB in the mutant (75).
Adaptation to growth in a medium at low pH might be expected to involve the down-regulation of genes encoding products that take up H+ and the up-regulation of genes encoding proteins capable of H+ extrusion. Consistent with this, growth of C. jejuni at pH 5.5 caused the down-regulation of a putative Na+/H+ antiporter (Cj0832c) and a pseudogene encoding a H+/oligopeptide symporter (Cj0654c). Cj0832c also was down-regulated in response to acid shock, suggesting that decreased expression of this gene is an important component of C. jejuni's response to acid stress. In H. pylori, growth at acidic pH also led to the down-regulation of a Na+/H+ antiporter (16), while in E. coli the NhaA Na+/H+ antiporter is essential for adaptation to high salinity and alkaline pH, extruding Na+ in exchange for H+ (reviewed in reference 64). The down-regulation at pH 5.5 of dcuB, which can extrude H+ in an antiport reaction with C4-dicarboxylates, was surprising, as dcuB was highly up-regulated in response to acid shock. However, our data are consistent with those of Cheng and coworkers (18), who found that both dcuA and dcuB were down-regulated in S. flexneri grown at low pH. While the F1F0 ATPase also can extrude H+ at the expense of ATP, thus protecting lactic acid bacteria against acid shock (76), the genes encoding this enzyme were not differentially expressed in our experiments.
The growth of C. jejuni under moderately acidic conditions (pH 6.5 and 6.0) led to the repression of 12 genes encoding ribosomal proteins. This is reminiscent of the widespread repression of these genes in C. jejuni in response to acid shock (71) and heat shock (78). With the exception of rpsU (30S ribosomal protein S21), which was down-regulated at pH 6.5 and 5.5, and rpmE (50S ribosomal protein L31), which was up-regulated at pH 5.5 only, the genes for ribosomal proteins were not differentially expressed at pH 5.5. One possible explanation for the failure of growth at pH 5.5 to cause the repression of genes encoding ribosomal proteins is that growth at this lower pH is damaging to ribosomes, and survival and/or adaptation relies on the continued synthesis of ribosomal proteins. In fact, the loss of ribosomes has been reported for E. coli exposed to medium at pH 3 (25).
Role of cell surface components in the acid stress response.
Components at the cell surface are in direct contact with the bacterium's environment, and as such they might be expected to play a role in protecting the cell against external assaults. In E. coli, the exopolysaccharide colanic acid is known to confer protection against acid and a number of other stresses (49, 51). Cells harboring mutations in a number of CPS genes (Cj1413c, Cj1431-32c, Cj1437c, and Cj1442c) were impaired for growth at pH 5.5, suggesting that the capsule plays a protective role, possibly by decreasing the influx of H+ into the cell. These genes are predicted to be involved in diverse stages of CPS expression, from biosynthesis (Cj1442c, Cj1432c, and Cj1431c) of the polysaccharide to transport (Cj1413c). Loss of CPS expression was apparent in the acid-sensitive mutants with insertions in Cj1413c (kpsS) and Cj1442c (Fig. 5). Our data are consistent with the acapsular phenotype of a kpsS insertion mutant in C. jejuni (44) and with the loss of polysaccharide transport in E. coli and Neisseria meningitidis kpsS mutants (20, 85). In contrast, transposon insertions in Cj1431c, Cj1432c, and Cj1437c did not result in the loss of CPS (Fig. 5). The mutation of Cj1431c (hddD) in C. jejuni leads to the loss of the O-methyl heptose side branch of the CPS repeat unit (43). Our data suggest that the heptose side branch is required for the capsule's protective effect. Given that roles for Cj1432c and Cj1437c in CPS expression have yet to be ascribed, it is difficult to interpret the observed phenotypes of these mutants.
The C. jejuni LOS also may play a similar protective role. Mutants with transposon insertions in two genes involved in LOS biosynthesis, Cj1135 and Cj1150c (hldE), displayed acid-sensitive phenotypes. The hldE gene is believed to be involved in the biosynthesis of nucleotide-activated heptose, which then is transferred to lipid A-3-deoxy-d-manno-octulosonic acid to form the LOS inner core (34, 43, 77). Cj1135 encodes a putative two-domain glucosyltransferase thought to transfer a glucose residue onto the first inner core heptose (34, 43). The products of these genes act early in LOS biosynthesis, suggesting that a complete LOS structure is required for wild-type levels of acid resistance. Silver-stained DOC-PAGE analysis of carbohydrate samples from these mutants revealed the presence of severely truncated LOS structures (Fig. 5), which strengthens the notion that a minimum LOS structure is required for acid resistance and/or adaptation. In H. pylori, acid shock caused the up-regulation of a lipopolysaccharide gene (wbcJ) encoding a product that is required for O antigen synthesis and Lewis X and/or Y expression (54). In the absence of external urea, a wbcJ mutant was more sensitive to acid killing (54), supporting the idea that lipopolysaccharide is protective against acid.
The role of flagella in bacterial acid stress responses remains unclear, and gene expression studies so far have remained inconclusive. While some studies report the up-regulation of flagellar genes under conditions of acid stress (37, 53, 57, 90), others report the down-regulation of these genes (18, 84, 90). Furthermore, these data remain difficult to interpret, as only a subset of the flagellar genes is ever found to be differentially expressed. In our study, the growth of C. jejuni at low pH was not associated with differential expression of flagellar genes. The only gene related to motility that was differentially expressed (up-regulated at pH 6.0) is Cj0454c. This gene encodes a putative membrane protein of unknown function that previously was shown to be required for full motility (38). Interestingly, despite the absence of differential expression of the flagellum biogenesis genes, the screening of our mutant library indicates a role for the flagellum in the ability of C. jejuni to grow under acidic conditions. Indeed, mutants harboring insertions in genes encoding components of the flagellar apparatus (fliD and flgG2) as well as the response regulator (flgR) of the two-component system that activates the expression of some flagellar genes (92) were unable to grow at acidic pH. An intergenic transposon located between flgJ, which encodes a hypothetical protein, and flgM, which encodes a putative anti-σ28 factor, also rendered cells acid sensitive. All of these mutants displayed motility defects on soft agar plates (Fig. 4), which is consistent with the reported loss of flagella in C. jejuni fliD (35) and flgR mutants (42). These data suggest that motility or, minimally, flagellum expression is required for survival and/or adaptation to acidic conditions. It is not clear at this time why motility would be required for acid tolerance, particularly on solid media as we have used here. Interestingly, a mutant bearing an insertion in Cj1324, which encodes a hypothetical protein in the O-linked protein glycosylation locus, was unable to grow at pH 5.5. This mutant displayed wild-type levels of motility on soft agar plates (Fig. 4), suggesting that flagellar glycosylation plays a role in enabling growth under acidic conditions. Finally, chemotaxis also appears to play a role in acid adaptation, as a mutation in any one of three genes encoding methyl-accepting chemotaxis protein-type signal transduction proteins (Cj0019c, Cj0144, and Cj0262c) led to an acid-sensitive phenotype. Altogether, these data indicate a key role for motility in acidic pH tolerance, warranting a more in-depth study of the mechanism of flagellum-mediated pH resistance.
The modulation of membrane composition also might be expected to alter cell wall permeability to H+. Our mutant library screen identified a pldA mutant as being impaired for growth at pH 5.5. The pldA gene encodes phospholipase A, an enzyme that can cleave membrane phospholipids to yield lysophospholipids (22). In H. pylori, the expression of active phospholipase A leads to the increased production of lysophospholipids and the improved acid stress survival of the strain (15, 79). It therefore appears that pldA plays a role in the acid stress response of both H. pylori and C. jejuni. Finally, fatty acid biosynthesis appears to be important for C. jejuni's adaptation to growth at low pH. The gene encoding 3-oxoacyl-[acyl-carrier-protein] synthase was up-regulated 1.7-fold at pH 5.5 (fabH), and a fabH2 mutant was acid sensitive. In agreement with this observation, fatty acid metabolism was found to be an important component of the adaptation of Mycobacterium to growth at low pH (28, 83).
Phosphate acquisition and the acid stress response.
Genes encoding products that are involved in phosphate uptake (pstSCA) were up-regulated at pH 5.5. In C. jejuni and other bacteria, the transcription of pstSCAB is activated by phosphate limitation (1, 91, 94), and in C. jejuni, pstS and pstC are up-regulated in a mutant (ΔspoT) unable to mount a stringent response (33). Most relevant to this study is the observed increase in phosphate uptake and polyphosphate accumulation when Burkholderia cepacia was grown at an acidic pH (59). Phosphate uptake and polyphosphate accumulation may therefore play a role in the acid stress response of some bacteria. While the data from this study support a role for phosphate uptake in the acid stress response of C. jejuni, the down-regulation of a polyphosphate kinase homologue (Cj0604) at pH 5.5 appears to speak against a role for polyphosphate accumulation in this response. The C. jejuni genome encodes an additional ppk homologue, the expression of which was not affected under steady-state acid stress conditions. In fact, the ppk gene (Cj1359) was up-regulated in response to both in vitro and in vivo acid shock in C. jejuni (71), supporting a role for polyphosphate accumulation in the acid stress response of this bacterium. Phosphate uptake genes also were up-regulated under acid shock conditions (pH 4) in Shewanella oneidensis (47).
In C. jejuni, the two-component system encoded by phoSR activates the transcription of the phosphate regulon, which includes pstSCA as well as a number of other genes (Cj0145, pstB, and the Cj0727-Cj0733 operon) (91), that were not differentially expressed in response to growth at pH 5.5. The Cj0727-Cj0733 operon encodes a putative ABC transporter system for phosphate uptake. The up-regulation of one uptake system (pstSCA) and not the other may be a response to the specific growth conditions used in this experiment and may reflect an as-yet unknown aspect of the regulation of phosphate uptake.
Overlap between acid stress and other stress responses.
Acid stress in bacteria is known to induce cross-resistance to other stresses (e.g., heat shock and oxidative and osmotic stress) (7), often by the induction of genes required to cope with these stresses (e.g., chaperones typical of heat shock responses) (18, 37, 53).
In this study, the gene encoding the peroxide stress regulator PerR was up-regulated at pH 5.5. In C. jejuni, as in B. subtilis, PerR represses the expression of genes such as ahpC (alkyl hydroperoxide reductase) and katA (catalase), the products of which detoxify peroxides (86). Oxidative stress leads to the irreversible oxidation of His residues in PerR that inactivate this repressor, leading to the expression of these genes (48). The up-regulation of perR under mildly acidic conditions might be expected to decrease the levels of PerR-regulated genes. However, the genes encoding the key components of the peroxide stress regulon were not differentially expressed. This observation suggests that the expression of the PerR protein is posttranscriptionally controlled and/or that the PerR protein is deactivated under acidic conditions and thus is unable to repress the PerR-regulated genes. Interestingly, the perR gene also was highly up-regulated in response to both in vitro and in vivo acid shock (71), further suggesting that the modulation of PerR levels is a general response to acid stress in C. jejuni. While there are no previous reports implicating PerR in bacterial acid stress responses, its homologue, the ferric uptake regulator Fur, is required for the induction of acid shock genes and proteins in H. pylori and S. enterica (30, 32, 36), and the mutation of fur in these bacteria renders cells acid sensitive (4, 6, 8, 10, 30, 36). Studies of S. enterica suggest that the role of Fur in the acid stress response is independent of iron (36) and consequently is distinct from its role in mediating iron homeostasis. The C. jejuni fur gene was not differentially expressed under either steady-state conditions of acid stress or in response to acid shock, and our mutant library screen failed to identify a fur mutant. Given that this mutant might be absent from our library, the role of Fur in the acid stress response of C. jejuni was determined by assessing the ability of a defined fur deletion mutant to grow at pH 5.5 (data not shown). Interestingly, the fur mutant was impaired in its ability to grow at pH 5.5, indicating that Fur plays a role in acid resistance in C. jejuni.
The experiments reported in this paper provide limited evidence of overlap between acid and nitrosative stress responses. The cgb gene, encoding a single-domain hemoglobin, was up-regulated at pH 5.5 and in response to acid shock. Cgb is an important player in the nitrosative stress response, scavenging and detoxifying nitric oxide (27). The expression of cgb and other members of the nitrosative stress regulon is controlled by NssR (26). Under conditions of acid shock, the gene encoding a truncated hemoglobin (Cj0465c) was highly up-regulated in C. jejuni (71). While this gene was implicated in mediating O2 flux in Campylobacter (88) rather than being directly involved in detoxification, it is nonetheless a member of the NssR regulon (26). Other members of the nitrosative stress regulon in C. jejuni showed no significant changes in gene expression.
While the heat shock response is an important component of the adaptation of C. jejuni to acid shock (71), no heat shock genes were up-regulated under steady-state acidic growth conditions. In addition, although a clpB mutant showed increased susceptibility to killing by acid shock (71), this same mutant was not impaired for growth at pH 5.5 (data not shown), supporting the idea that different mechanisms are required to protect cells against a lethal acid shock than are required for growth under mildly acidic conditions.
Shared acid stress response elements.
Strikingly, only two genes, Cj0414 and Cj0415, were up-regulated at all three acidic pHs (pH 6.5, 5.5, and 5.0) and upon exposure to acid shock (Fig. 2B, cluster D). Cj0414 and Cj0415 are annotated as oxidoreductases and bear some similarity to gluconate dehydrogenase subunits of other bacteria. Gluconate dehydrogenase activity was detected in C. jejuni 81 - 176 but not in an insertion mutant of Cj0415 (70), supporting the assignment of this gene as a gluconate dehydrogenase. Interestingly, a transposon insertion in Cj0415 affected the growth of C. jejuni NCTC 11168 at low pH (Table 2). Consequently, these genes likely play an important role in the survival and adaptation to acidic conditions and might be essential for efficient host colonization. Understanding the role of these components in the acid stress response of C. jejuni will necessitate further characterization of these genes and their products.
Conclusions.
While environmental stresses usually cause the induction of a large set of genes involved in coping with the particular stress and/or repairing ensuing damages, the adaptation of C. jejuni to acidic conditions is characterized, most notably, by the down-regulation of genes. In fact, only 26 genes were significantly induced at pH 5.5 (Fig. 2A, cluster C), whereas 68 genes were repressed (Fig. 2A, cluster A). The transcriptional profile at mildly acidic pH is characterized by the differential expression of respiratory pathways, by the induction of genes involved in phosphate transport, and by the repression of genes involved in energy generation and intermediary metabolism. In addition, our data support a role for Cj0415 in survival and adaptation to acidic conditions.
In summary, our work highlights the power of an integrated experimental approach combining a gene expression study with genome-wide mutagenesis to investigate bacterial responses to environmental conditions. While the transcriptome profile revealed the preferential gene expression levels in C. jejuni after adaptation to mildly acidic pH conditions, the genome-wide mutagenesis approach identified genes that are absolutely required for growth at low pH. Importantly, both approaches are complementary, as genome-wide mutagenesis does not identify genes involved in bacterial fitness under a particular growth condition, whereas gene expression studies reveal the best-fit transcriptome. On the other hand, genome-wide mutagenesis identifies genes that are required for growth at low pH even if those genes are not differentially expressed, thus revealing the genes involved in the intrinsic resistance of Campylobacter to low pH.
Acknowledgments
This work was supported by grant number RO1-AI055612 from the National Institutes of Health. A. N. Reid acknowledges receipt of a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada.
We thank Christine Szymanski for providing the C. jejuni kpsM mutant.
Footnotes
Published ahead of print on 11 January 2008.
REFERENCES
- 1.Aguena, M., E. Yagil, and B. Spira. 2002. Transcriptional analysis of the pst operon of Escherichia coli. Mol. Genet. Genomics 268:518-524. [DOI] [PubMed] [Google Scholar]
- 2.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babakhani, F. K., G. A. Bradley, and L. A. Joens. 1993. Newborn piglet model for campylobacteriosis. Infect. Immun. 61:3466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baik, H. S., S. Bearson, S. Dunbar, and J. W. Foster. 1996. The acid tolerance response of Salmonella typhimurium provides protection against organic acids. Microbiology 142:3195-3200. [DOI] [PubMed] [Google Scholar]
- 5.Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509-519. [DOI] [PubMed] [Google Scholar]
- 6.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. [DOI] [PubMed] [Google Scholar]
- 8.Bearson, S. M., B. L. Bearson, and M. A. Rasmussen. 2006. Identification of Salmonella enterica serovar Typhimurium genes important for survival in the swine gastric environment. Appl. Environ. Microbiol. 72:2829-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bijlsma, J. J., M. M. Gerrits, R. Imamdi, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 1998. Urease-positive, acid-sensitive mutants of Helicobacter pylori: urease-independent acid resistance involved in growth at low pH. FEMS Microbiol. Lett. 167:309-313. [DOI] [PubMed] [Google Scholar]
- 10.Bijlsma, J. J., A. L. M. Lie, I. C. Nootenboom, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J. Infect. Dis. 182:1566-1569. [DOI] [PubMed] [Google Scholar]
- 11.Birrell, G. W., J. A. Brown, H. I. Wu, G. Giaever, A. M. Chu, R. W. Davis, and J. M. Brown. 2002. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl. Acad. Sci. USA 99:8778-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 13.Blaser, M. J., and L. S. Newman. 1982. A review of human salmonellosis. I. Infective dose. Rev. Infect. Dis. 4:1096-1106. [DOI] [PubMed] [Google Scholar]
- 14.Brás, A. M., S. Chatterjee, B. W. Wren, D. G. Newell, and J. M. Ketley. 1999. A novel Campylobacter jejuni two-component regulatory system important for temperature-dependent growth and colonization. J. Bacteriol. 181:3298-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bukholm, G., T. Tannaes, P. Nedenskov, Y. Esbensen, H. J. Grav, T. Hovig, S. Ariansen, and I. Guldvog. 1997. Colony variation of Helicobacter pylori: pathogenic potential is correlated to cell wall lipid composition. Scand. J. Gastroenterol. 32:445-454. [DOI] [PubMed] [Google Scholar]
- 16.Bury-Moné, S., J. M. Thiberge, M. Contreras, A. Maitournam, A. Labigne, and H. De Reuse. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:623-638. [DOI] [PubMed] [Google Scholar]
- 17.Chagneau, C., M. Heyde, S. Alonso, R. Portalier, and P. Laloi. 2001. External-pH-dependent expression of the maltose regulon and ompF gene in Escherichia coli is affected by the level of glycerol kinase, encoded by glpK. J. Bacteriol. 183:5675-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng, F., J. Wang, J. Peng, J. Yang, H. Fu, X. Zhang, Y. Xue, W. Li, Y. Chu, and Q. Jin. 2007. Gene expression profiling of the pH response in Shigella flexneri 2a. FEMS Microbiol. Lett. 270:12-20. [DOI] [PubMed] [Google Scholar]
- 19.Chun, K. T., H. J. Edenberg, M. R. Kelley, and M. G. Goebl. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13:233-240. [DOI] [PubMed] [Google Scholar]
- 20.Cieslewicz, M., and E. Vimr. 1996. Thermoregulation of kpsF, the first region 1 gene in the kps locus for polysialic acid biosynthesis in Escherichia coli K1. J. Bacteriol. 178:3212-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cronan, J. E., Jr. 2002. Phospholipid modifications in bacteria. Curr. Opin. Microbiol. 5:202-205. [DOI] [PubMed] [Google Scholar]
- 22.Dekker, N. 2000. Outer-membrane phospholipase A: known structure, unknown biological function. Mol. Microbiol. 35:711-717. [DOI] [PubMed] [Google Scholar]
- 23.Doyle, M. P., and D. J. Roman. 1981. Growth and survival of Campylobacter fetus subsp. jejuni as a function of temperature and pH. J. Food Prot. 44:596-601. [DOI] [PubMed] [Google Scholar]
- 24.Ducey, T. F., and D. W. Dyer. 2002. Rapid identification of EZ:Tn transposon insertion sites in the genome of Neisseria gonorrhoeae. Epicentre Forum 9:6-7. [Google Scholar]
- 25.El-Sharoud, W. M., and G. W. Niven. 2007. The influence of ribosome modulation factor on the survival of stationary-phase Escherichia coli during acid stress. Microbiology 153:247-253. [DOI] [PubMed] [Google Scholar]
- 26.Elvers, K. T., S. M. Turner, L. M. Wainwright, G. Marsden, J. Hinds, J. A. Cole, R. K. Poole, C. W. Penn, and S. F. Park. 2005. NssR, a member of the Crp-Fnr superfamily from Campylobacter jejuni, regulates a nitrosative stress-responsive regulon that includes both a single-domain and a truncated haemoglobin. Mol. Microbiol. 57:735-750. [DOI] [PubMed] [Google Scholar]
- 27.Elvers, K. T., G. Wu, N. J. Gilberthorpe, R. K. Poole, and S. F. Park. 2004. Role of an inducible single-domain hemoglobin in mediating resistance to nitric oxide and nitrosative stress in Campylobacter jejuni and Campylobacter coli. J. Bacteriol. 186:5332-5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foster, J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2:898-907. [DOI] [PubMed] [Google Scholar]
- 30.Foster, J. W., and H. K. Hall. 1992. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 174:4317-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gajiwala, K. S., and S. K. Burley. 2000. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295:605-612. [DOI] [PubMed] [Google Scholar]
- 32.Gancz, H., S. Censini, and D. S. Merrell. 2006. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect. Immun. 74:602-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaynor, E. C., D. H. Wells, J. K. MacKichan, and S. Falkow. 2005. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol. Microbiol. 56:8-27. [DOI] [PubMed] [Google Scholar]
- 34.Gilbert, M., P. C. R. Godschalk, C. T. Parker, H. P. Endtz, and W. W. Wakarchuk. 2005. Genetic basis for the variation in the lipooligosaccharide outer core of Campylobacter jejuni and possible association of glycosyltransferase genes with post-infectious neuropathies. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter: molecular and cellular biology. Horizon Bioscience, London, United Kingdom.
- 35.Golden, N. J., and D. W. Acheson. 2002. Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect. Immun. 70:1761-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall, H. K., and J. W. Foster. 1996. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J. Bacteriol. 178:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayes, E. T., J. C. Wilks, P. Sanfilippo, E. Yohannes, D. P. Tate, B. D. Jones, M. D. Radmacher, S. S. BonDurant, and J. L. Slonczewski. 2006. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol. 6:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 39.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holzer, P. 2007. Taste receptors in the gastrointestinal tract. V. Acid sensing in the gastrointestinal tract. Am. J. Physiol. Gastrointest. Liver Physiol. 292:G699-G705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong, W., W. Jiao, J. Hu, J. Zhang, C. Liu, X. Fu, D. Shen, B. Xia, and Z. Chang. 2005. Periplasmic protein HdeA exhibits chaperone-like activity exclusively within stomach pH range by transforming into disordered conformation. J. Biol. Chem. 280:27029-27034. [DOI] [PubMed] [Google Scholar]
- 42.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 183:2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karlyshev, A. V., J. M. Ketley, and B. W. Wren. 2005. The Campylobacter jejuni glycome. FEMS Microbiol. Rev. 29:377-390. [DOI] [PubMed] [Google Scholar]
- 44.Karlyshev, A. V., D. Linton, N. A. Gregson, A. J. Lastovica, and B. W. Wren. 2000. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35:529-541. [DOI] [PubMed] [Google Scholar]
- 45.Karlyshev, A. V., M. J. Pallen, and B. W. Wren. 2000. Single-primer PCR procedure for rapid identification of transposon insertion sites. Bio Techniques 28:1078-1082. [DOI] [PubMed] [Google Scholar]
- 46.Kern, R., A. Malki, J. Abdallah, J. Tagourti, and G. Richarme. 2007. Escherichia coli HdeB is an acid stress chaperone. J. Bacteriol. 189:603-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leaphart, A. B., D. K. Thompson, K. Huang, E. Alm, X. F. Wan, A. Arkin, S. D. Brown, L. Wu, T. Yan, X. Liu, G. S. Wickham, and J. Zhou. 2006. Transcriptome profiling of Shewanella oneidensis gene expression following exposure to acidic and alkaline pH. J. Bacteriol. 188:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee, J. W., and J. D. Helmann. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440:363-367. [DOI] [PubMed] [Google Scholar]
- 49.Lee, S. M., and J. Chen. 2004. Survival of Escherichia coli O157:H7 in set yogurt as influenced by the production of an exopolysaccharide, colanic acid. J. Food Prot. 67:252-255. [DOI] [PubMed] [Google Scholar]
- 50.Lemos, J. A., Y. Y. Chen, and R. A. Burne. 2001. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183:6074-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mao, Y., M. P. Doyle, and J. Chen. 2001. Insertion mutagenesis of wca reduces acid and heat tolerance of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:3811-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marshall, B. J., L. J. Barrett, C. Prakash, R. W. McCallum, and R. L. Guerrant. 1990. Urea protects Helicobacter (Campylobacter) pylori from the bactericidal effect of acid. Gastroenterology 99:697-702. [DOI] [PubMed] [Google Scholar]
- 53.Maurer, L. M., E. Yohannes, S. S. Bondurant, M. Radmacher, and J. L. Slonczewski. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187:304-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGowan, C. C., A. Necheva, S. A. Thompson, T. L. Cover, and M. J. Blaser. 1998. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol. Microbiol. 30:19-31. [DOI] [PubMed] [Google Scholar]
- 55.McGowan, C. C., T. L. Cover, and M. J. Blaser. 1996. Helicobacter pylori and gastric acid: biological and therapeutic implications. Gastroenterology 110:926-938. [DOI] [PubMed] [Google Scholar]
- 56.Merrell, D. S., C. Bailey, J. B. Kaper, and A. Camilli. 2001. The ToxR-mediated organic acid tolerance response of Vibrio cholerae requires OmpU. J. Bacteriol. 183:2746-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merrell, D. S., M. L. Goodrich, G. Otto, L. S. Tompkins, and S. Falkow. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71:3529-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merrell, D. S., D. L. Hava, and A. Camilli. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 43:1471-1491. [DOI] [PubMed] [Google Scholar]
- 59.Mullan, A., J. P. Quinn, and J. W. McGrath. 2002. Enhanced phosphate uptake and polyphosphate accumulation in Burkholderia cepacia grown under low pH conditions. Microb. Ecol. 44:69-77. [DOI] [PubMed] [Google Scholar]
- 60.Murphy, C., C. Carroll, and K. N. Jordan. 2005. The effect of different media on the survival and induction of stress responses by Campylobacter jejuni. J. Microbiol. Methods 62:161-166. [DOI] [PubMed] [Google Scholar]
- 61.Murphy, C., C. Carroll, and K. N. Jordan. 2003. Identification of a novel stress resistance mechanism in Campylobacter jejuni. J. Appl. Microbiol. 95:704-708. [DOI] [PubMed] [Google Scholar]
- 62.Murphy, C., C. Carroll, and K. N. Jordan. 2003. Induction of an adaptive tolerance response in the foodborne pathogen, Campylobacter jejuni. FEMS Microbiol. Lett. 223:89-93. [DOI] [PubMed] [Google Scholar]
- 63.Naikare, H., K. Palyada, R. Panciera, D. Marlow, and A. Stintzi. 2006. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74:5433-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Padan, E., T. Tzubery, K. Herz, L. Kozachkov, A. Rimon, and L. Galili. 2004. NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+ antiporter. Biochim. Biophys. Acta 1658:2-13. [DOI] [PubMed] [Google Scholar]
- 65.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 67.Pflock, M., N. Finsterer, B. Joseph, H. Mollenkopf, T. F. Meyer, and D. Beier. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 188:3449-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Price, M. N., K. H. Huang, E. J. Alm, and A. P. Arkin. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rao, K. A., E. Yazaki, D. F. Evans, and R. Carbon. 2004. Objective evaluation of small bowel and colonic transit time using pH telemetry in athletes with gastrointestinal symptoms. Br. J. Sports Med. 38:482-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rathbun, K. M., M. Pajaniappan, S. A. Cawthraw, D. G. Newell, J. E. Hall, J. F. Nechtman, E. C. Gaynor, C. M. Burns, and S. A. Thompson. 2005. 13th Int. Wkshp. Campylobacter Helicobacter Relat. Organ., abstr. B31. Gold Coast, Queensland, Australia.
- 71.Reid, A. N., R. Pandey, K. Palyada, H. Naikare, and A. Stinzi. 2008. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl. Environ. Microbiol. 74:1583-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richard, H. T., and J. W. Foster. 2003. Acid resistance in Escherichia coli. Adv. Appl. Microbiol. 52:167-186. [DOI] [PubMed] [Google Scholar]
- 73.Robinson, D. A. 1981. Campylobacter infection. R. Soc. Health J. 101:138-140. [DOI] [PubMed] [Google Scholar]
- 74.Salama, N. R., B. Shepherd, and S. Falkow. 2004. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 186:7926-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sánchez, B., M. C. Champomier-Verges, C. Collado Mdel, P. Anglade, F. Baraige, Y. Sanz, C. G. de Los Reyes-Gavilan, A. Margolles, and M. Zagorec. 2007. Low-pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum. Appl. Environ. Microbiol. 73:6450-6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sheng, J., and R. E. Marquis. 2006. Enhanced acid resistance of oral streptococci at lethal pH values associated with acid-tolerant catabolism and with ATP synthase activity. FEMS Microbiol. Lett. 262:93-98. [DOI] [PubMed] [Google Scholar]
- 77.St. Michael, F., C. M. Szymanski, J. Li, K. H. Chan, N. H. Khieu, S. Larocque, W. W. Wakarchuk, J. R. Brisson, and M. A. Monteiro. 2002. The structures of the lipooligosaccharide and capsule polysaccharide of Campylobacter jejuni genome sequenced strain NCTC 11168. Eur. J. Biochem. 269:5119-5136. [DOI] [PubMed] [Google Scholar]
- 78.Stintzi, A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tannaes, T., N. Dekker, G. Bukholm, J. J. Bijlsma, and B. J. Appelmelk. 2001. Phase variation in the Helicobacter pylori phospholipase A gene and its role in acid adaptation. Infect. Immun. 69:7334-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thompson, S. A., and M. J. Blaser. 1995. Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resistance to low pH. Infect. Immun. 63:2185-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thompson, S. A., R. L. Latch, and J. M. Blaser. 1998. Molecular characterization of the Helicobacter pylori uvrB gene. Gene 209:113-122. [DOI] [PubMed] [Google Scholar]
- 83.Tran, S. L., M. Rao, C. Simmers, S. Gebhard, K. Olsson, and G. M. Cook. 2005. Mutants of Mycobacterium smegmatis unable to grow at acidic pH in the presence of the protonophore carbonyl cyanide m-chlorophenylhydrazone. Microbiology 151:665-672. [DOI] [PubMed] [Google Scholar]
- 84.Tucker, D. L., N. Tucker, and T. Conway. 2002. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 184:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tzeng, Y. L., A. K. Datta, C. A. Strole, M. A. Lobritz, R. W. Carlson, and D. S. Stephens. 2005. Translocation and surface expression of lipidated serogroup B capsular polysaccharide in Neisseria meningitidis. Infect. Immun. 73:1491-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Vliet, A. H., M. L. Baillon, C. W. Penn, and J. M. Ketley. 1999. Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181:6371-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Vliet, A. H., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 180:5291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wainwright, L. M., K. T. Elvers, S. F. Park, and R. K. Poole. 2005. A truncated haemoglobin implicated in oxygen metabolism by the microaerophilic food-borne pathogen Campylobacter jejuni. Microbiology 151:4079-4091. [DOI] [PubMed] [Google Scholar]
- 89.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wen, Y., E. A. Marcus, U. Matrubutham, M. A. Gleeson, D. R. Scott, and G. Sachs. 2003. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 71:5921-5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wösten, M. M., C. T. Parker, A. van Mourik, M. R. Guilhabert, L. van Dijk, and J. P. van Putten. 2006. The Campylobacter jejuni PhosS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Mol. Microbiol. 62:278-291. [DOI] [PubMed] [Google Scholar]
- 92.Wösten, M. M., J. A. Wagenaar, and J. P. van Putten. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 279:16214-16222. [DOI] [PubMed] [Google Scholar]
- 93.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 94.Yuan, Z. C., R. Zaheer, and T. M. Finan. 2006. Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium meliloti. J. Bacteriol. 188:1089-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]