Abstract
The biocontrol activity of the root-colonizing Pseudomonas fluorescens strain CHA0 is largely determined by the production of antifungal metabolites, especially 2,4-diacetylphloroglucinol. The expression of these metabolites depends on abiotic and biotic environmental factors, in particular, elements present in the rhizosphere. In this study, we have developed a new method for the in situ analysis of antifungal gene expression using flow cytometry combined with green fluorescent protein (GFP)-based reporter fusions to the phlA and prnA genes essential for the production of the antifungal compounds 2,4-diacetylphloroglucinol and pyrrolnitrin, respectively, in strain CHA0. Expression of phlA-gfp and prnA-gfp in CHA0 cells harvested from the rhizosphere of a set of plant species as well as from the roots of healthy, leaf pathogen-attacked, and physically stressed plants were analyzed using a FACSCalibur. After subtraction of background fluorescence emitted by plant-derived particles and CHA0 cells not carrying the gfp reporters, the average gene expression per bacterial cell could be calculated. Levels of phlA and prnA expression varied significantly in the rhizospheres of different plant species. Physical stress and leaf pathogen infection lowered phlA expression levels in the rhizosphere of cucumber. Our results demonstrate that the newly developed approach is suitable to monitor differences in levels of antifungal gene expression in response to various plant-derived factors. An advantage of the method is that it allows quantification of bacterial gene expression in rhizosphere populations at a single-cell level. To our best knowledge, this is the first study using flow cytometry for the in situ analysis of biocontrol gene expression in a plant-beneficial bacterium in the rhizosphere.
In this study, we present a novel approach to monitor in the rhizosphere the expression of genes substantially contributing to the biocontrol ability of fluorescent pseudomonads. Root-colonizing fluorescent pseudomonads are well known for their ability to suppress plant diseases caused by soil-borne pathogenic fungi (19, 29, 36, 57, 73). The production of antifungal metabolites such as 2,4-diacetylphloroglucinol (DAPG), pyoluteorin (PLT), pyrrolnitrin (PRN), phenazines, and hydrogen cyanide has emerged as a key mechanism of disease suppression by these bacteria (29, 30, 36, 57). There is evidence that the expression of these metabolites is strongly influenced by biotic and abiotic factors which may vary under different environmental conditions. In order to improve the consistency of biocontrol pseudomonads, it is therefore important to gain information about how the production of these substances is regulated by environmental factors. Direct monitoring of antifungal compounds at the site of interest, the rhizosphere, remains a difficult task (13, 37, 49, 55). Indirect approaches have often been chosen by monitoring the expression of antifungal genes using reporter genes. Studies investigating gene expression in plant-associated beneficial bacteria have thus far been based on the use of different reporter genes such as lacZ encoding β-galactosidase (30, 55), inaZ encoding an ice nucleation protein (22, 42), and lux reporter genes encoding luciferase (18, 35). In strain Pseudomonas fluorescens CHA0, lacZ-based reporter fusions have been applied to determine the contribution of pathway-specific and global regulatory elements and of environmental signals in the regulation of biocontrol trait expression such as production of the antimicrobial compounds DAPG, PLT, and hydrogen cyanide (40, 48, 55, 56, 59, 69).
Autofluorescent proteins are another class of reporter proteins widely used for studying the biology of plant-associated bacteria and their interaction with the environment. The green fluorescent protein (GFP) (17) from the jellyfish Aequorea victoria has been used in many different ways, for example, as a tag to monitor the colonization patterns and ecological behavior of bacteria living on plant surfaces (11, 33, 39, 54) or associated with mycorrhizal fungi (2). Furthermore, GFP-based reporters combined with fluorescence microscopy have been applied to monitor in situ the expression of bacterial genes responding to N-acylhomoserine lactone signal molecules (28, 63) or to the availability of micro- and macronutrients (22, 34, 51) as well as of genes involved in the production of different metabolites such as the phytohormone indoleacetate in Erwinia herbicola (15) and the phytotoxin coronatine in Pseudomonas syringae (72) on plant surfaces. The fluorescent signal emitted by GFP can be monitored in a nondestructive manner in a living organism, which presents another major advantage in comparison to other reporter systems such as the one based on β-galactosidase encoded by the lacZ gene (21, 33). Moreover, there are several advantages of using GFP as a reporter or marker such as its minimal toxicity, its noninvasive detection, and its ability to generate green light without the addition of external cofactors, all together conferring an almost real-time monitoring of gene expression in living cells. A recent review by G. Bloemberg (10) summarizes new approaches using GFP and other autofluorescent proteins to study plant-bacteria interactions.
We have previously developed a set of reporter fusions based on stable GFP variants for monitoring antifungal gene expression in P. fluorescens CHA0 (5). Baehler et al. (5) demonstrated that expression curves of these reporter constructs adequately reflect antifungal compound production in liquid cultures and that the reporters are suitable tools for studying the regulation of DAPG, PLT, and PRN biosynthesis in vitro. Here, we developed a novel method using flow cytometry based on fluorescence-activated cell sorting (FACS) for measuring the expression of these GFP-based reporter fusions in the rhizosphere. FACS-based flow cytometry is a powerful method for the detection and quantification of fluorescence from individual cells within a population. FACS provides data on optical properties of thousands of single cells per second as they pass through a laser beam (52). With this method, antifungal gene expression in a Pseudomonas population harvested from the rhizosphere can be quantified at a single-cell level. Flow cytometry furthermore has the advantage that the size of a bacterial population can be directly quantified without using traditional culture-dependent methods such as plating serial dilutions on selective agar.
The aims of the present study were first to evaluate whether this FACS-based method is suitable to quantify the expression of antifungal genes in P. fluorescens CHA0 colonizing plant roots and, second, to determine whether this method can be used to detect changes in antifungal gene expression triggered by plant-derived signals. For this purpose, the expression of phlA-gfp, pltA-gfp, and prnA-gfp fusions in P. fluorescens CHA0 was monitored in the rhizosphere of plants. Expression patterns were studied (i) on different plant species and (ii) on both healthy plants and plants undergoing physiological stress following either infection with a leaf pathogen or mechanical injury.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are described in Table 1. For monitoring antifungal gene expression, derivatives of P. fluorescens strain CHA0 carrying rhizosphere-stable plasmids with transcriptional fusions of a stable variant of the gfp gene to the phlA, pltA, and prnA genes (5) required for the biosynthesis of the antifungal compounds DAPG, PLT, and PRN, respectively, were used. P. fluorescens strains were routinely cultivated on King's medium B agar (KMB) at 30°C, unless specified otherwise. When required, antibiotics were added to the growth medium at the following concentrations: chloramphenicol, 20 μg/ml; tetracycline hydrochloride, 125 μg/ml. For plant inoculations, bacterial strains were grown for 4 h in Luria-Bertani broth (58) on a rotary shaker (180 rpm) at 27°C. Bacterial cultures were centrifuged and washed twice with autoclaved distilled water. Bacterial suspensions were adjusted with autoclaved distilled water to a concentration of 108 cells/ml for the inoculation of seedlings in growth pouches or 107 cells/ml for the inoculation of cucumber plants in the gnotobiotic system.
TABLE 1.
Fungi, bacterial strains, and plasmids used in this study
| Fungal or bacterial strain or plasmid | Characteristics or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| Botrytis cinerea D110 | Causal agent of mold of various plant species | Agroscope Changins-Wädenswil, Switzerland |
| Glomerella lagenaria D323 | Causal agent of leaf spot of cucumber | J.-P. Métraux, University of Fribourg, Switzerland |
| Puccinia triticina MEL | Causal agent of leaf rust of wheat | F. Mascher, Agroscope Changins-Wädenswil, Switzerland |
| Pseudomonas fluorescens CHA0 | Wild type; Cmr | 71 |
| Plasmids | ||
| pME7100 | phlA-gfp transcriptional fusion; Tcr | 5 |
| pME7109 | pltA-gfp transcriptional fusion; Tcr | 5 |
| pME7116 | prnA-gfp transcriptional fusion; Tcr | 5 |
Cmr, resistance to chloramphenicol; Tcr, resistance to tetracycline.
Fungal strains, culture conditions, and preparation of conidia suspensions.
Glomerella lagenaria D323 and Botrytis cinerea D110 (Table 1) were grown and maintained on malt extract agar (15 g/liter Oxoid malt extract agar and 12 g/liter Oxoid technical agar, pH 7.0) at 24°C in the dark. Conidia formation by B. cinerea was induced by irradiation of cultures with near-UV light for 12 h, followed by a further incubation in the dark at 24°C. For the preparation of conidia suspensions, 3- to 4-week-old agar plate cultures were flooded with autoclaved double-distilled water, and both mycelium and conidia were scraped off with a spatula. The resulting suspension was filtered through glass wool, and the filtrate containing conidia was adjusted to 107 conidia/ml for B. cinerea and to 5 × 105 conidia/ml for G. lagenaria. Conidia suspensions of B. cinerea were amended with 10 mM ammonium phosphate (pH 6.5) and 1% (wt/vol) sucrose.
Puccinia triticina MEL (Table 1), the causal agent of leaf rust, is an obligate biotrophic pathogen of wheat. The leaf rust pathogen was cultivated and multiplied on 1- to 3-week-old wheat plants in a growth chamber with a relative humidity of 70% and 16 h of light (160 μE/m2/s) at 22°C, followed by an 8-h dark period at 18°C. For spore multiplication, 10-day-old wheat plants were inoculated with spores from freshly sporulating lesions on older plants using a brush. Plants were slightly sprayed with distilled water, transferred to a plastic tent containing 100% relative humidity, and kept in the dark for 48 h at 22°C. Thereafter, the light and temperature conditions described above were restored. For the infection regimen, 20 mg of fresh spores was harvested with a brush, resuspended in 1 ml of paraffin oil, and used to inoculate wheat plants in 12 growth pouches using an atomizer.
Growth pouch assay for monitoring antifungal gene expression on roots of different plant species.
For experiments in which the influence of plant species on antifungal gene expression on roots was evaluated, seeds of cucumber (Cucumis sativus cv. Chinesische Schlange), winter wheat (Triticum aestivum cv. Arina), hybrid maize (Zea mais cv. Swissgold), winter triticale (Triticosecale cv. Prader), winter barley (Hordeum vulgare cv. Baretta), tomato (Solanum lycopersicum cv. Marmande), garden cress (Lepidium sativum), and purple turnip (Brassica rapa cv. Rapa) from DSP, Delley, Switzerland (for cereals), and Samen Mauser, Winterthur, Switzerland (for dicotyledones), were surface-sterilized for 10 min in 4% NaClO (vol/vol) and subsequently rinsed with autoclaved distilled water. Seeds were germinated on soft agar (Oxoid technical agar at 8.5 g/liter) for 48 h at 24°C in the dark. Three sterile-grown seedlings were then placed into an autoclaved growth pouch (Mega International, West St. Paul, MN). Growth pouches contain a filter paper allowing root development on a support with uniform nutrient distribution. The artificial plant support system has the advantage of being free of soil particles which could interfere with flow cytometric analysis. Prior to autoclaving, growth pouches were amended with 15 ml of modified Knop plant nutrition solution (38). P. fluorescens suspensions containing 108 cells/ml were prepared as described above, and 1 ml was added to the seedlings in the growth pouch. This resulted in an initial bacterial concentration per g of root of 2.1 × 106 CFU for wheat, 1.8 × 106 CFU for triticale, 2.2 × 106 CFU for barley, 0.5 × 106 CFU for maize, 1.0 × 106 CFU for turnip, 1.4 × 106 CFU for cress, 1.1 × 106 CFU for tomato, and 0.7 × 106 CFU for cucumber as determined by colony plating on KMB agar. The pouches were wrapped into aluminum foil to protect roots from light and were incubated in a growth chamber with 70% relative humidity for 16 h with light (160 μE/m2/s) at 22°C, followed by an 8-h dark period at 18°C (standard conditions). The experiment consisted of the following treatments: control without added bacteria, inoculation with wild-type CHA0 (for setting the background for FACS analysis), inoculation with CHA0/pME7100 carrying a phlA-gfp fusion, inoculation with CHA0/pME7109 carrying a pltA-gfp fusion, and inoculation with CHA0/pME7116 carrying a prnA-gfp fusion. After a 10-day incubation period, plants were removed from the pouches. Roots from each pouch were pooled and transferred into sterile 50-ml Erlenmeyer flasks containing 7 ml of sterile distilled water. Flasks were vigorously shaken at 400 rpm for 20 min. From the resulting suspensions, samples of 2 ml were taken, transferred on ice, and immediately analyzed by FACS. Roots were assessed for fresh weight. Each treatment was performed in triplicate, and the entire experiment was repeated twice.
Growth pouch assay for monitoring DAPG gene expression on roots of leaf rust-infected wheat.
Wheat seedlings were inoculated with wild-type CHA0 or its derivative CHA0/pME7100 (phlA-gfp) and grown in growth pouches under the conditions described above. Six days later, the leaves of the young wheat plants were inoculated with a P. triticina spore-oil emulsion as described above. Control plants were sprayed with paraffin oil only. Plants were transferred to a plastic tent maintained at 100% relative humidity and kept in the dark for 2 days. Thereafter, the standard light and temperature conditions were resumed except that plants were maintained in a semipermeable plastic tent to keep a high humidity conducive to pathogen infection. Nine days after pathogen inoculation, roots were harvested, and suspensions were prepared for FACS analysis as described above. Roots were assessed for fresh weight, and the amount of culturable bacteria was determined by plating serial dilutions of root washes onto KMB agar containing tetracycline (125 μg/ml) and subsequent CFU counting. Each treatment consisted of 15 replicates with one growth pouch containing three plants per replicate. The entire experiment was repeated twice.
Gnotobiotic assay for monitoring DAPG gene expression on roots of healthy, diseased, or mechanically injured cucumber plants.
The effect of physiological stress triggered either by infection with leaf pathogens or by mechanical injury on the expression of phlA-gfp in CHA0 growing on the roots of cucumber was investigated. Cucumber seeds were surface sterilized and pregerminated as described above. One seedling was planted into an autoclaved 1,000-ml Erlenmeyer flask containing 200 ml of soft Knop agar (Oxoid technical agar, 1.6 g; Knop nutrient solution, 200 ml) and sealed with cotton wool stoppers. Flasks were incubated in a growth chamber under the same conditions as described above. Ten-day-old cucumber plants were inoculated with wild-type CHA0 or CHA0/pME7100 carrying a phlA-gfp fusion. Five aliquots of 400 μl of a bacterial suspension containing 107 cells/ml were injected into the Knop agar. One aliquot was injected directly beneath the hypocotyl; the other four were injected above the roots that had grown into the Knop agar. The same day, primary leaves were inoculated with either B. cinerea or G. lagenaria or were mechanically injured. For B. cinerea or G. lagenaria inoculation, 10 20-μl drops of a suspension containing 107 or 5 × 105 conidia/ml, respectively, were placed onto the primary leaf. In treatments subjected to stress of mechanical injury, a part of the primary leaf (about 50% of the leaf area) was cut off using a scalpel. After infection with fungal pathogens, plants were placed in the dark at 22°C and a relative humidity of 100% for 2 days, prior to resuming the standard light and temperature conditions. Eight days after inoculation with bacteria and pathogens, plants were removed from the flasks, adhering soft agar was discarded, and roots were transferred to 10 ml autoclaved distilled water in a sterilized 50-ml Erlenmeyer flask. Flasks were vigorously shaken at 400 rpm for 20 min. After a final filtration step to remove all trace of agar particles (5-μm-pore-size Minisart filters; Sartorius, Goettingen, Germany), samples of 2 ml were taken from the resulting suspensions, transferred on ice, and immediately analyzed by FACS. Roots were assessed for fresh weight, and the number of culturable bacteria was determined by plating serial dilutions of root washes onto KMB agar containing tetracycline (125 μg/ml) and subsequent counting of CFU. All treatments were performed in triplicate, and the experiment was conducted twice.
FACS analysis.
Monitoring of antifungal gene expression was performed by flow cytometry on samples of root washes obtained from the experiments described above using a FACSCalibur flow cytometer equipped with a 15-mW, air-cooled argon ion laser excitation light source (488 nm) (Becton Dickinson, San Jose, CA). The flow cytometer collects data about each single cell flowing in a thin capillary, including its size and granularity (internal complexity), determined with the forward scatter (FSC) detector and the side scatter (SSC) detector, respectively, and its green or red fluorescence emissions are recorded with the FL1-H and FL3-H fluorescence detectors, respectively. FSC signals were collected using a photodiode with an amplification factor of 10 and a threshold set to 72 and were processed in log gain. SSC signals were detected in log gain using a photomultiplier tube set at 350 V and a threshold of 72. Green fluorescence was detected in the range of 515 to 545 nm with the FL1-H detector set at a photomultiplier tube voltage of 505 V with logarithmic gain. Red fluorescence was detected in the range of 670 nm by the FL3-H detector in log gain using a photomultiplier tube set at 690 V. Data were collected using CellQuest software (Becton Dickinson, San Jose, CA) and analyzed with WinMDI software, version 2.8 (Joseph Trotter; available at http://facs.scripps.edu/software.html). Gating (subtraction of background) was done by setting a marker M1 on the histogram of green fluorescence above the background fluorescence noise (i.e., the autofluorescence emitted by root-derived particles, bacterial cell fragments, and bacterial cells not expressing GFP) on the histogram of green fluorescence. For all experiments presented in this study, the marker M1 used to define the geometric mean fluorescence was limited from 137 to 1,023 on the histogram set location. The geometric mean of the fluorescence emitted by the gated events was calculated with WinMDI software. This mean reflects the mean fluorescence per gated event and corresponds to the average GFP expression per expressing bacterial cell. Red fluorescent microspheres (Flow Check Ruby Red Fluorescent Microspheres; Polysciences, Inc., Warrington, PA) were added at a final concentration of 2.5 × 105 per ml to each sample prior to analysis. These microspheres of 6 μm in diameter are suitably excited at 488 nm and easily detected with the FL3-H detector. The analysis of each sample was stopped after 500 red microspheres (gated by size) were counted, standardizing in this way the volume of the measured sample (at 2 μl).
Detection limits.
CHA0 cells carrying the phlA-gfp or prnA-gfp reporter were analyzed by FACS at different cell concentrations, ranging from 50 to 50,000 cells in a total injection volume of 2 μl. For both constructs, 500 gfp-expressing cells were necessary to obtain a measurable fluorescence peak that was clearly distinguishable from background fluorescence. In order to determine the minimal bacterial concentration on roots necessary to obtain a clear gfp expression signal in the FACS analysis, the roots of 1-week-old cucumber and wheat plants were incubated for 15 min in differentially concentrated suspensions of CHA0 cells carrying either the phlA-gfp or the prnA-gfp reporter. Subsequently, bacterial cells were extracted from the roots as described above, and root washes were analyzed by FACS. For both plant species and both reporter constructs, around 107 gated events (active, phlA- or prnA-expressing cells) per g of root were necessary to obtain a clearly measurable gfp expression peak. A total of 107 gated events per g of root reflected about 8 × 106 CFU per g of root for cucumber and around 106 CFU per g of root for wheat.
Statistical analysis.
Statistical analyses were performed using the statistics program Systat, version 10.0 (Systat Inc., Evanston, IL). Results of independent repetitions over time were first analyzed by two-way analysis of variance. This analysis revealed for all experiments (Tables 2, 3, and 4) significant repetition versus treatment interactions. Therefore, data collected from individual repetitions over time could not be pooled and are thus presented separately. Each individual repetition was analyzed first by one-way analysis of variance and then by Fisher's protected least significant difference (LSD) test (P ≤ 0.05).
TABLE 2.
Expression of phlA-gfp or prnA-gfp reporter fusions in P. fluorescens CHA0 in the rhizosphere of different plant speciesa
| Reporter fusion | Trial | No. of gated events (108 per g of root)b | No. of CFUc
|
Relative fluorescence
|
||
|---|---|---|---|---|---|---|
| % of gfp-expressing cells | Per g of root (107) | Per gated eventd | Per g of root (1010)e | |||
| phlA-gfp | Wheat | 7.9 ABe | 14 AB | 7.9 A | 33.1 A | 2.7 AB |
| Triticale | 22.3 AB | 7 A | 11.6 AB | 32.2 A | 7.2 A | |
| Barley | 23.8 AB | 6 A | 14.8 AB | 26.0 B | 6.1 AB | |
| Maize | 4.7 AB | 4 A | 2.3 A | 41.0 C | 2.0 AB | |
| Turnip | 16.0 AB | 18 AB | 31.9 B | 22.3 B | 3.7 AB | |
| Cress | 26.9 A | 10 A | 16.3 AB | 21.2 B | 5.8 AB | |
| Tomato | 2.1 AB | 28 B | 6.2 A | 28.9 AB | 0.6 B | |
| Cucumber | 1.0 B | 87 C | 9.1 AB | 28.0 AB | 0.7 B | |
| prnA-gfp | Wheat | 4.1 A | 8 A | 2.3 B | 34.1 A | 1.4 B |
| Triticale | 2.7 A | 15 A | 3.7 AB | 34.2 A | 0.9 B | |
| Barley | 4.5 A | 22 A | 9.7 AB | 30.6 AB | 1.4 B | |
| Maize | 1.1 A | 7 A | 4.2 AB | 48.3 C | 0.7 B | |
| Turnip | 10.8 AB | 22 A | 12.1 A | 36.6 A | 3.8 AB | |
| Cress | 21.2 B | 6 A | 11.7 A | 31.8 AB | 6.4 A | |
| Tomato | 0.6 A | 34 AB | 2.6 B | 17.8 D | 0.1 B | |
| Cucumber | 0.8 A | 63 B | 4.4 AB | 23.8 BD | 2.7 AB | |
Plant seedlings were inoculated with CHA0/pME7100 carrying a phlA-gfp fusion or with CHA0/pME7116 carrying a prnA-gfp fusion and grown in growth pouches as described in Materials and Methods. Gene expression in root washes was measured after 10 days by FACS-based flow cytometry. Means of three replicates per treatment are given. The experiment has been repeated once with similar results (data not shown). Values in the same column for the same reporter fusion followed by different letters are significantly different according to Fisher's protected LSD (P ≤ 0.05).
Cells expressing reporter fusions.
The number of CFU corresponding to culturable cells was determined by serial plating on KMB agar containing tetracycline.
Relative fluorescence per gated event (cell) reflects the average phlA-gfp or prnA-gfp expression per active CHA0 cell.
Relative fluorescence per root weight reflects the total phlA-gfp or prnA-gfp expression per root weight.
TABLE 3.
Expression of phlA-gfp in P. fluorescens CHA0 on the roots of healthy wheat and wheat infected with the foliar pathogen P. triticinaa
| Trial | P. triticina infection | Total no. of events per g of root (109)b | No. of gated eventsc
|
No. of CFUd
|
Relative fluorescence
|
|||
|---|---|---|---|---|---|---|---|---|
| % of total events | Per g of root (109) | % of phlA-gfp-expressing cells | Per g of root (107) | Per gated evente | Per g of root (1010)f | |||
| 1 | − | 2.8 | 56 | 1.5 | 8 | 12.1 | 41.7 | 6.5 |
| + | 1.7 | 66 | 1.1*g | 21* | 18.4 * | 40.4 | 4.5* | |
| 2 | − | 4.0 | 62 | 2.5 | 6 | 14.4 | 36.5 | 9.0 |
| + | 4.0 | 61 | 1.1 | 16* | 16.1 | 37.3 | 9.2 | |
| 3 | − | 2.7 | 65 | 1.8 | 6 | 10.5 | 41.1 | 7.3 |
| + | 2.9 | 71 | 2.0 | 3 | 5.9 | 42.1 | 8.6 | |
Wheat seedlings were inoculated with CHA0/pME7100 carrying a phlA-gfp fusion and were grown in growth pouches. After 6 days, plants were infected with spores of P. triticina. Nine days later, root washes were collected and 2-μl samples were analyzed by flow cytometry, as described in Materials and Methods. Three independent trials are presented.
Total analyzed particles.
phlA-gfp-expressing cells.
The number of CFU corresponding to culturable cells was determined by serial plating on KMB agar containing tetracycline.
Relative fluorescence per gated event (cell) reflects the average phlA-gfp expression per active CHA0 cell.
Relative fluorescence per root weight reflects the total phlA-gfp expression per root weight.
Values derived from P. triticina infection treatments which are marked with an asterisk are significantly different from values derived from the corresponding control treatment without pathogen infection (Fisher's protected LSD, P ≤ 0.05).
TABLE 4.
Influence of leaf pathogen infections or mechanical injury on the expression of a phlA-gfp fusion (pME7100) carried by strain CHA0 on cucumber rootsa
| Trial | No. of gated events per g of root (108)b | No. of CFUc
|
Relative fluorescence
|
||
|---|---|---|---|---|---|
| % of phlA-gfp expressing cells | Per g of root (107) | Per gated eventd | Per g of root (1010)e | ||
| Control | 4.5 A | 106 A | 46.8 A | 82.9 A | 3.7 A |
| Botrytis cinerea | 2.1 B | 108 A | 21.5 B | 63.1 B | 1.3 B |
| Glomerella lagenaria | 2.8 B | 85 A | 23.3 B | 65.4 B | 1.8 B |
| Mechanical injury | 2.6 B | 69 A | 17.3 B | 61.5 B | 1.6 B |
Cucumber plants were root-inoculated with CHA0/pME7100 and leaves were infected with B. cinerea or G. lagenaria or were mechanically injured. Eight days later, root washes were analyzed by FACS. Means of four replicates per treatment are given. The experiment was repeated once with similar results. Values in the same column followed by different letters are significantly different according to Fisher's protected LSD (P ≤ 0.05).
phlA-gfp expressing cells.
The number of CFU corresponding to culturable cells was determined by serial plating on KMB agar containing tetracycline.
Relative fluorescence per gated event (cell) reflects the average phlA-gfp expression per active CHA0 cell.
Relative fluorescence per root weight reflects the total phlA-gfp expression per root weight.
RESULTS
Calibration of a FACS-based method for monitoring antifungal gene expression in the rhizosphere.
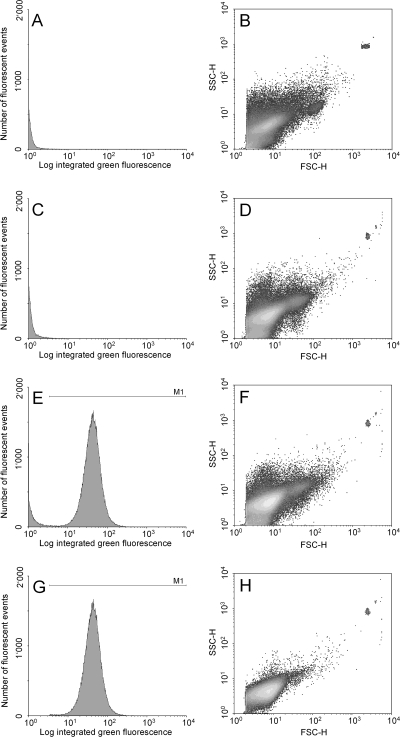
In a set of method calibration experiments, the expression of a phlA-gfp reporter fusion on pME7100 was tested in a CHA0 background in the rhizosphere of wheat grown in growth pouches for 8 days. Root washes containing bacterial cells were analyzed with a FACSCalibur. Illustrations of the FACS analysis are presented in Fig. 1 showing density plots and green fluorescence histograms.
FIG. 1.
FACS analysis of the expression of phlA-gfp in P. fluorescens CHA0 on the roots of wheat grown in growth pouches for 12 days. Histograms show green fluorescence detected by the FL1-H detector (A, C, E, and G) and dot plots show the size and internal granularity of analyzed particles (events) detected by the FSC-H and the SSC-H detector, respectively (B, D, F, and H). Red fluorescent microspheres which were used to determine the volume of the analyzed sample are visualized on the right top corner of each dot plot. (A and B) Analysis of a treatment without added bacteria. Total analyzed particles (events) in 2 μl, 288,166. (C and D) Analysis of a treatment inoculated with wild-type CHA0. Total analyzed particles (events) in 2 μl, 343,185. (E and F) Analysis of a treatment inoculated with CHA0 carrying a phlA-gfp fusion on pME7100. Total analyzed particles (events) in 2 μl, 334,647. (G and H) The same analysis as in panels E and F after subtraction of the background fluorescence by gating with M1. Total gated particles (events) in 2 μl, 195,929 (58.5% of the total events are cells expressing phlA-gfp). Mean fluorescence value per gated particle, 41.5.
Samples from plants grown without added bacteria emitted low background fluorescence detected by the FL1-H detector, as shown in the histogram in Fig. 1A. As the growth pouches were exposed to ambient air, contaminations with external microorganisms could not be prevented, although they did not result in any significant green fluorescence. Serial dilution of the root washes on nutrient agar plates exhibited a maximal bacterial contamination of 6 × 104 CFU/g of root. In the analyzed 2-μl sample derived from plants grown without added bacteria, a total of 288,000 particles (events) were detected by the FSC-H and SSC-H detectors which measured size and internal granularity of the particles, as shown in the dot plot in Fig. 1B. Inoculation of wheat seedlings with wild-type strain CHA0 resulted in an increase of analyzed particles to 343,000 (Fig. 1D). The emitted fluorescence, however, did not increase (Fig. 1C). The data suggest that neither root-derived particles nor CHA0 cells or other bacteria contaminating the growth pouch system emit any considerable autofluorescence. Analysis of a sample derived from plants cultivated with P. fluorescens harboring the phlA-gfp fusion reporter gene on plasmid pME7100 revealed a clear fluorescent peak (Fig. 1E). A total of 334,000 particles were detected (Fig. 1F). To calculate the mean fluorescence of CHA0 cells expressing phlA-gfp, background fluorescence resulting from particles was subtracted. For this purpose, the total detected events were gated with the marker M1 (Fig. 1E and G) which was defined based on autofluorescence of root particles, contaminating bacteria, and cells of wild-type strain CHA0 which did not carry a GFP-based reporter. A specific M1 setting allowed us to eliminate the background noise emitted by these cells and particles. Figures 1G and H finally show fluorescence, size, and granularity of the 195,000 particles remaining after gating with M1. These particles corresponded to cells of CHA0/pME7100 expressing the phlA-gfp reporter fusion and represented about 58% of the total detected events. The mean fluorescence (geometric mean) per gated cell reflecting the mean phlA-gfp expression per CHA0/pME7100 cell could then be calculated by the software WinMDI 2.8. In this example, the value is 41.5. After gating with M1, each detected event (Fig. 1H, dot plot) represents an active Pseudomonas cell producing GFP. The histogram in Fig. 1G shows the total fluorescence emitted by these cells.
In summary, this representative experiment illustrates that the newly developed method is suitable to measure the expression of an antifungal gene by P. fluorescens in the rhizosphere, and cells expressing the GFP-based reporter construct can be easily separated from other bacteria and root-derived particles also present in the analyzed samples.
FACS-based evaluation of plant species-dependent expression of antifungal genes in the rhizosphere.
Several experiments were performed to test the efficiency of the developed method to monitor differences in antifungal gene expression due to plant-induced variations in the rhizosphere. In a first series of experiments, P. fluorescens CHA0 was grown on the roots of several monocotyledonous (wheat, triticale, barley, and maize) and dicotyledonous (turnip, cress, tomato, and cucumber) plants in order to evaluate the impact of the plant species on the level of antifungal gene expression. Plants were cultivated in growth pouches and inoculated with P. fluorescens CHA0 harboring phlA-gfp, pltA-gfp, or prnA-gfp reporter fusions. Plants were harvested after 10 days, and bacterial cells separated in root washes were analyzed with the FACSCalibur flow cytometer. Background fluorescence was corrected as described above. Results of one representative experiment are presented in Table 2. Significant differences in expression levels of antifungal genes between the plant species could be recorded. The highest expression of the phlA-gfp reporter fusion per individual cell was observed in bacteria harvested from maize roots (Table 2) with a mean GFP-derived fluorescence value of 41.0. The lowest expression levels were recorded in bacteria colonizing roots of turnip and cress, with mean fluorescence values of 22.3 and 21.2, respectively, which is about half the value for maize. Intermediate phlA expression levels occurred in the rhizosphere of the other five plant species. The highest phlA-gfp expression per gram of root was found for triticale, with a significantly higher relative fluorescence value than that for tomato and cucumber (Table 2).
Distinct expression patterns were obtained when a prnA-gfp reporter fusion was used to monitor the expression of PRN biosynthetic genes (Table 2). As for phlA, the highest prnA-gfp expression per individual cell was found in the rhizosphere of maize with a mean fluorescence value of 48.3. In contrast to phlA expression, the lowest prnA expression was detected in cells isolated from roots of tomato and cucumber plants with values of 17.8 and 23.8, respectively, i.e., about half the maize rhizosphere value. The highest prnA-gfp expression per gram of root was found for cress, with a significantly higher relative fluorescence value than that found for most of the other plant species tested. Analysis of root washes derived from plants inoculated with CHA0 carrying a pltA-gfp fusion did not reveal detectable levels of PLT gene expression (data not shown).
The number of gated cells (i.e., cells expressing the antifungal gene reporter) varied between 6 × 107 and 2.7 × 109 cells per g of root (Table 2), with cress roots showing a significantly higher root colonization than most of the other plant species tested. When the proportion of culturable cells (CFU) per gram of root and CHA0 cells detected by FACS per gram of root was calculated, a significantly higher percentage of cells capable of forming colonies on KMB agar was found in samples derived from cucumber plants and to some extent also from tomato plants than from the other plant species tested (Table 2).
To summarize, the experiment confirms that the FACS-based flow cytometric approach is suitable for the quantification of differences in antifungal gene expression in the rhizosphere of different plant species.
DAPG gene expression and root colonization by P. fluorescens CHA0 on healthy and leaf rust-infected wheat.
A second set of experiments aimed at testing the influence of leaf rust infection on antifungal gene expression and root colonization of P. fluorescens CHA0 in the wheat rhizosphere. Results of three individual experiments are presented in Table 3. In all experiments, treatment of wheat with spores of P. triticina resulted in severe leaf rust symptoms with about 50 to 300 rust pustules per leave (data not shown). The number of total events determined by FACS ranged between 1.7 × 109 and 4.0 × 109 particles per g of root (Table 3) in all experiments. In all analyzed bacterial populations, gating with M1 (subtracting the particles emitting background) indicated that 56 to 71% of the total detected events were active phlA-expressing CHA0 cells. No difference between rust-infected and healthy plants was observed. When calculating the number of phlA-expressing cells (gated events) per g of root, only experiment 1 revealed a significant difference between healthy and rust-infected wheat plants. On roots of healthy plants, 1.5 × 109 phlA-expressing cells per g of root were detected whereas on the roots of rust-infected plants the number of phlA-expressing cells was significantly lower, i.e., 1.1 × 109 (Table 3).
The most pronounced differences were found when the numbers of cells capable of forming colonies on KMB agar plates were quantified. With respect to the number of CFU per root weight, in only experiment 1 was there a significant 50% increase (1.8 × 108 CFU/g of root) in samples derived from rust-attacked plants compared to healthy control plants (1.2 × 108 CFU/g of root). When the proportion of culturable cells (CFU) per gram of root and total active CHA0 cells (gated cells detected by FACS) per gram of root was calculated, interestingly, in two out of three experiments a significantly higher percentage of CFU was found in samples derived from diseased plants (Table 3). On the roots of healthy plants, only 6 to 8% of the total FACS-detected active CHA0 cells formed colonies on KMB agar plates. In contrast, on the roots of rust-attacked plants the culturability of CHA0 cells increased to 21 and 16% of the gated events in experiments 1 and 2, respectively. The proportion of CFU remained as low as in the rhizosphere of healthy plants only in experiment 3.
No significant differences between the rhizosphere of healthy and rust-attacked plants were found with regard to mean phlA expression per actively expressing CHA0 cell (mean fluorescence per gated event). The phlA expression levels in experiments 1, 2, and 3 ranged between mean fluorescence values of 36.5 and 42.1 (Table 3). Expression of phlA calculated per root weight revealed that the mean fluorescence per gram of root varied between values of 4.5 × 1010 and 9.2 × 1010 in the three independent experiments. It could, however, be shown in one case (experiment 1) that a decreased number of phlA-expressing cells present on the roots of diseased plants also resulted in a significantly lower total phlA expression in the rhizosphere.
In summary, the experiments indicate that leaf rust infections do not have a major effect on phlA expression levels in individual CHA0 cells growing on wheat roots. However, leaf rust infections had an impact on the culturability of CHA0 cells, inasmuch as they significantly increased the percentage of culturable cells in two of three experiments.
Effect of leaf pathogen attack and mechanical injury on the expression of phlA on the roots of cucumber.
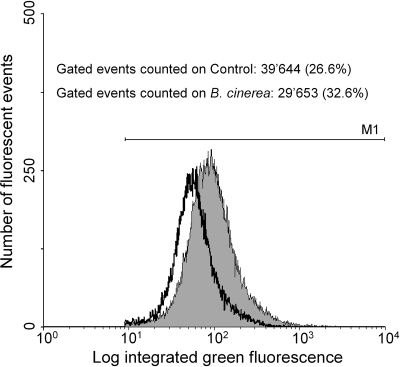
In a last set of experiments, the influence of leaf pathogen infection on phlA-gfp expression in strain CHA0 in the rhizosphere of cucumber was investigated. In contrast to the previous experiments, cucumber plants were cultivated in a gnotobiotic assay. Cucumber seedlings were planted into sterile flasks containing Knop nutrient agar. Ten days later, the root system was inoculated with CHA0/pME7100, and cucumber leaves were either mechanically injured or inoculated with G. lagenaria or B. cinerea. Eight days later, disease symptoms had fully developed, and about 15% (treatments with G. lagenaria) or 20% (treatments with B. cinerea) of the area of infected leaves was covered by necrotic lesions (data not shown). At this time point, root washes containing CHA0 cells were analyzed by FACS, and data obtained are presented in Table 4 and Fig. 2. Mean phlA expressions levels per actively expressing CHA0 cell (mean fluorescence intensities per gated event) were calculated for each sample of root washes, and averages per treatment are shown in Table 4. For all treatments, the total number of phlA-gfp-expressing cells represented 27 to 38% of the total particles present in the samples (gated events). A significant decrease in phlA expression per individual cell was observed on the roots of all cucumber plants that had received a pathogenic or mechanic stress treatment, compared to healthy control plants. The mean fluorescence value obtained for the control treatment was 82.9, whereas values for treatments with B. cinerea, G. lagenaria, or mechanical injury ranged between 61.5 and 65.4. The stress-induced decrease in phlA expression calculated per gram of root was even more pronounced (Table 4). In Fig. 2, two representative histograms of FACS analyses derived from a control plant and a plant infected with B. cinerea (one of each) are overlaid. On this graph, the fluorescence peak detected in the Botrytis-treated sample is clearly shifted to the left on the axis of the fluorescence intensity detected by FL1-H, indicating the decrease in phlA expression. Moreover, pathogen infection as well as mechanical injury led to a significant reduction of root colonization by P. fluorescens CHA0 (Table 4). Healthy plants were colonized by 4.7 × 108 CFU/g of root whereas root colonization of stressed plants ranged only between 1.7 × 108 and 2.3 × 108 CFU/g of root. In contrast to the wheat-leaf rust experiments, no significant difference between the number of culturable cells (CFU) per g of root and total active CHA0 cells (gated cells detected by FACS) per g of root was observed for any of the treatments (Fisher's protected LSD, P ≤ 0.05). The number of gated cells per gram of root detected by FACS was 4.5 × 108 for healthy plants and 2.1 × 108 to 2.8 × 108 for injured plants (Table 4).
FIG. 2.
Influence of leaf pathogen infections or mechanical injury on the expression of a phlA-gfp fusion in pME7100 carried by strain CHA0 on cucumber roots. Cucumber plants were root inoculated with CHA0/pME7100, and leaves were infected with B. cinerea. Eight days later, root washes were analyzed by flow cytometry. The histogram shows the green fluorescence emitted by strain CHA0/pME7100 grown on the roots of healthy cucumber plants (gray peak) or cucumber plants attacked by B. cinerea (black peak).
Taken together, these data illustrate that physiological stress of cucumber plants induced by leaf pathogens or mechanical injury can result in a significant reduction of DAPG gene expression on roots.
DISCUSSION
In this study, we have developed a new application technique for FACS-based flow cytometry that allows in combination with GFP-based reporter constructs the study of the expression of antifungal genes by plant-beneficial P. fluorescens in the rhizosphere. So far, the use of fluorescence-based flow cytometry to measure bacterial gene expression in the environment was restricted to a few studies in which gfp-expressing whole-cell biosensors were employed for example to monitor bacterial quorum-sensing molecules in soil (16) or tetracycline production in soil microcosms (31). Previously, the main use of flow cytometry in combination with GFP was to investigate proliferation, behavior, and viability of GFP-tagged bacteria in pure culture or in specific environments such as plant surfaces and soil (4, 25, 45, 64, 68). Recently, flow cytometry has also been applied to monitor horizontal gene transfer between soil bacteria (62) and to quantify interactions between protists and their bacterial prey (26).
Here, we extended the use of flow cytometry in combination with recently developed reporter fusions based on stable variants of GFP (5) to study the expression of specific genes. The method was used to monitor changes in DAPG and PRN biosynthetic gene expression in P. fluorescens CHA0 in response to host species variation (Table 2) and the physiological stress status of the plant host (induced by pathogen attack or mechanical injury) (Tables 3 and 4 and Fig. 2). To our best knowledge, this is the first report of in situ analysis of antifungal gene expression in a root-associated bacterium monitored by flow cytometry.
A consistent problem with the use of flow cytometry in environmental applications is the background noise originating from basal autofluorescence of bacteria or from soil, plant, substrate, and other contaminating particles emitting fluorescence signals that may interfere with analysis of relevant data. One method of separating the monitored signal from the background noise consists in the gating of specific regions on density plots by selecting only particles of a certain size and shape or displaying particular fluorescence properties for data analysis. This gating approach for specifying populations of bacterial cells within a sample has been used by several research groups, e.g., in studies on the simultaneous detection of GFP- and DsRed-tagged Escherichia coli cells (44) and on the physiological state of GFP-tagged P. fluorescens SBW25 under different environmental conditions (45).
Gating particles by size was not appropriate for our purposes because some root-derived particles or other bacteria present on the roots might have had a similar size as the Pseudomonas cells of interest. We have, therefore, gated the total events by fluorescence in that we have analyzed only particles emitting fluorescence higher than a certain threshold. This threshold was determined in samples derived from plants inoculated with CHA0 cells not carrying a GFP-based reporter construct (background fluorescence). The method allowed us to distinguish between bacterial cells expressing phlA or prnA from other particles present. In wheat, the fraction of CHA0 cells expressing the phlA-gfp fusion was about 60% of the total present particles (Table 3 and Fig. 1) showing that the majority of particles in our root washes were indeed active CHA0 cells. A major drawback of the method is that CHA0 cells which are alive but do not express phlA cannot be detected and are thus not included in the analysis. One solution to overcome this problem would be to additionally tag the bacteria with a variant of the red fluorescent protein DsRed (9, 20, 61). In this way, it would be possible to quantify the total number of cells of a bacterial inoculant and the fraction expressing the gene of interest. Previous studies have successfully illustrated the strength of dual- or multicolor applications of fluorescent proteins for microscopic observations of the behavior of bacterial cells in mixed populations on plant surfaces (12, 14).
The data presented in Tables 2 and 3 show that for CHA0 the numbers of CFU counted on agar plates were about one magnitude lower than the numbers of cells expressing phlA-gfp counted by flow cytometer. Our findings are in accordance with those of Unge et al. (68) and Gamalero et al. (27), who also reported that the number of GFP-tagged bacteria counted by flow cytometry was higher than the number of bacteria capable of forming colonies when they analyzed P. fluorescens populations isolated from roots or artificial medium. Similar results were obtained by Tombolini et al. (65) when they compared samples from barley seeds for numbers of GFP-tagged Pseudomonas cells counted by microscopy and numbers of culturable cells. Taken together, these results suggest that a significant fraction of Pseudomonas cells isolated from in situ assays and detected by GFP fluorescence cannot grow on agar plates. For P. fluorescens CHA0, it has been shown that, in particular, under conditions of abiotic stress, the majority of cells persist in a nonculturable state in soil (47, 66). However, the culturability of cells seems to be strongly dependent on the experimental assay and on the conditions plants and bacteria have been exposed to. Interestingly, a significantly larger fraction of cells isolated from the rhizosphere of cucumber and tomato than from the rhizosphere of the other plant species tested was able to grow on agar plates (Tables 2 and 4). Yet further experiments would be needed to clarify whether the plant species can influence the culturability of CHA0 cells grown in the rhizosphere.
Unge et al. (68) showed for P. fluorescens SBW25 soil populations tagged with GFP and a luciferase marker that GFP fluorescence remained stable during 1 month of observation while luciferase activity corresponding to cell activity rapidly declined. Unge and Jansson (67) proposed that GFP tags can be used only to monitor total cells in a sample but, in contrast to luciferase tags, do not give any information about the metabolic activity of bacteria isolated from environmental samples. The authors argued that the GFP crystal structure is highly stable and might still be detectable in inactive or dead cells as long as the cell membrane remains intact. In the experiments presented here, however, detected GFP fluorescence clearly corresponds to the expression of antifungal genes which is only performed by metabolically active cells (5). We cannot completely exclude the possibility that some of the detected fluorescence was emitted by cells which once had been active but were not any longer at the time of sampling though they still contained active GFP. The quantification of antifungal gene expression by our GFP-based reporters, though, is rather cumulative and gives information about the total amount of antifungal metabolites produced by bacterial cells in the rhizosphere and does not reflect antifungal activity at an exact time point. However, the stable GFP reporters used in this study are still sensitive enough to monitor changes in antifungal gene expression in response to physiological changes in the host plant, as illustrated in Table 4 and Fig. 2.
A possible way to avoid cumulative effects is to use unstable GFP variants. Such unstable variants have been used in several studies to observe bacterial growth (1) and metabolic activity (43). As recently proposed by G. Bloemberg (10), the unstable GFP variants constructed by Andersen et al. (1) could facilitate the analysis of transient gene expression in the rhizosphere. In fact, we have tested such unstable variants for monitoring of antifungal metabolite production by the biocontrol agent P. fluorescens CHA0 (5). Such unstable GFP variants were suitable to quantify antifungal gene expression in vitro; however, they were not expressed strongly enough under in situ conditions (our unpublished observations).
The direct quantification of antifungal metabolites in soil is possible but difficult and labor-intensive, especially if numerous samples have to be analyzed. DAPG has been extracted from the rhizosphere of different plant species and quantified by high-performance liquid chromatograph as described in several studies (8, 13, 37, 49, 55). Notz et al. (55) showed that increased levels of DAPG produced in the rhizosphere of wheat in comparison with the rhizosphere of cucumber also correlated with increased rhizosphere expression levels of a phlA-lacZ reporter fusion. The use of reporter genes for monitoring the expression of antifungal genes certainly represents a much easier and quicker way to monitor antifungal compound production in the rhizosphere although it has to be kept in mind that this measurement is indirect and that reporter gene expression levels cannot be translated directly into amounts of antifungal compounds. Advantages of the FACS-based flow cytometric analysis of cells expressing gfp-reporter constructs are that the analysis of gene expression can be performed in intact cells, without any addition of substrates or cofactors and at a single-cell level, which is not the case when bacterial gene expression is quantified using lacZ or lux reporters in combination with enzymatic assays. Enzymatic assays give only global information on gene expression of the whole analyzed population, and furthermore bacterial cells derived from environmental samples have to be enumerated by conventional methods such as colony plating. In contrast, direct enumeration of individual cells by flow cytometry provides statistical data on specific population fractions, is rapid, and does not rely on culturability for detection (68). A drawback of the method is that it does not give information about the exact localization of cells. To investigate the site of gene expression, bacterial cells carrying GFP-based reporter constructs can be visualized on plant surfaces by epifluorescence or confocal microscopy. For example, Brandl et al. (15) monitored the spatial expression pattern of a plant-inducible gene (involved in indoleacetic acid biosynthesis) in E. herbicola on plant surfaces. Using whole-cell GFP-based bacterial biosensors, Joyner and Lindow (34) examined the iron bioavailability on plants, and Axtell and Beattie (3) visualized bacterial water deprivation on bean leaves using various bacterial species. A further method to monitor antifungal gene expression would be to quantify the mRNA of antifungal biosynthetic genes by real time reverse transcription-PCR. However, such a method for quantifying antifungal gene expression in fluorescent pseudomonads in the rhizosphere has never been published so far.
The main aim of this study was to examine whether flow cytometric analysis is sensitive enough to monitor changes in antifungal gene expression in response to environmental factors. In the experiment presented in Table 2, we show that phlA-gfp and prnA-gfp gene expression levels monitored by FACS were significantly different in CHA0 cells harvested from the roots of different plant species. The highest expression of antifungal genes was observed in the rhizosphere of maize. In our experiments, we could not find similar clear-cut differences in phlA expression between the rhizosphere of monocotyledons and dicotyledons as described by Notz et al. (55), who used a phlA-lacZ reporter for the in situ monitoring of DAPG gene expression in CHA0. However, we were able to observe similar trends in that the highest antifungal gene expressions were always found in cells colonizing the rhizosphere of a monocotyledon, and the lowest expression levels were always in cells isolated from the rhizosphere of a dicotyledon (Table 2). Our data show that not only DAPG but also PRN biosynthetic gene expression can vary substantially between individual plant species (Table 2), which could result in variation of biocontrol activity for different species. We suggest that differences in DAPG and PRN biosynthetic gene expression between plant species may be due to differences in the quantity and/or composition of root exudates which are known to modulate gene expression in root-associated bacteria (7, 24, 32, 46, 70). However, high antifungal gene expression levels in individual cells as observed for bacteria isolated from maize roots do not necessarily lead to high gene expression levels in the rhizosphere. The total amount of an antifungal metabolite produced in the rhizosphere is also dependent on the amount of bacteria colonizing the roots. The bacterial root colonization levels for triticale and cress, for example (Table 2), were higher than those for maize. Therefore, the highest phlA-gfp (triticale) and prnA-gfp (cress) expression levels per gram of root were measured in the rhizosphere of these two plant species, although the gene expression levels per individual cell were significantly lower than the level for maize (Table 2). In the experimental setups used here, it was not possible to measure the expression of a GFP-based pltA reporter gene fusion. One possible explanation is that in the growth pouch assays, pltA expression might have been too low to be detected by FACS analysis.
Since a plant host can strongly influence bacteria colonizing its roots, the assumption that the physiological condition of the plant can also have an impact on rhizobacteria is self-evident. We have investigated the influence of leaf pathogen infections or physical stress on bacterial phlA expression on wheat and cucumber roots (Tables 3 and 4 and Fig. 2). Infection of wheat leaves with P. triticina did not result in an altered antifungal gene expression (Table 3). An interesting result, however, was that in two out of three experiments, the number of culturable cells in comparison with total number of active CHA0 cells detected by FACS was significantly higher for bacteria harvested from rust-infected plants than for bacteria from healthy plants. Yet whether the observed effect really was due to pathogen infection remains unclear and would need to be further clarified. For cucumber, infection with a leaf pathogen (B. cinerea or G. lagenaria) as well as physical stress (leaf cut) resulted in significantly lowered phlA expression (Table 4 and Fig. 2). In a previous study performed by Notz et al. (55), it was shown that infection of maize and cucumber with the root pathogen Pythium ultimum resulted in an enhanced expression of a phlA-lacZ reporter construct in strain CHA0 in the rhizosphere. It could not, however, be specified whether the increase in DAPG expression can be attributed to direct signaling from the pathogen or to an indirect effect of the root disease, for example, a modification in root exudate composition. Here, in contrast, spatial separation of the beneficial rhizobacteria and the foliar pathogen permits us to attribute the observed difference in phlA expression to a direct signaling from the pathogen-attacked plant. A similar decrease in phlA expression was observed independently of whether plant stress was induced by different leaf pathogens or mechanical injury (Table 4). It is known that plant stress and infection with leaf pathogens lead to an alteration in root exudate composition and phenolic compound production (6, 7, 41, 50, 53, 60). It is tempting to speculate that the rhizobacterial population detects a change in root exudate composition or a specific plant stress signal, resulting in modification of antifungal gene expression. The nature of this signal remains unknown. Since we have observed that certain plant phenolics strongly influence the expression of antifungal genes in P. fluorescens CHA0 (5, 23, 59), we speculate that plant phenolics might play a role in the regulation of antifungal compound production in the rhizosphere.
Taken together, our results show that the combined use of flow cytometry with GFP-based reporters is a powerful and promising method for studying the in situ expression of biocontrol traits in root-colonizing pseudomonads. The method proved to be suitable for quantitative monitoring of changes in antifungal gene expression in response to plant-derived signals. The novel approach opens up new possibilities for studying the interaction of plant-beneficial bacteria with their plant host and their biotic and abiotic soil environments.
Acknowledgments
We thank Fabio Mascher, research station Agroscope Changins-Wädenswil, for providing wheat seeds and the leaf rust pathogens and Caroline Loutre, Institute of Plant Biology, University of Zurich, for appreciable advice on the leaf rust infection protocol. We also thank DSP Delley for providing us with different cereals. Finally, we thank Matthias Lutz, Michele Frapolli, and Caroline Loutre for correcting the manuscript.
We gratefully acknowledge financial support from the Swiss National Science Foundation (project 3100A0-105881) and from the State Secretariat for Education and Research (project C04.0201).
Footnotes
Published ahead of print on 28 December 2007.
REFERENCES
- 1.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjørn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artursson, V., R. D. Finlay, and J. K. Jansson. 2006. Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environ. Microbiol. 8:1-10. [DOI] [PubMed] [Google Scholar]
- 3.Axtell, C. A., and G. A. Beattie. 2002. Construction and characterization of a proU-gfp transcriptional fusion that measures water availability in a microbial habitat. Appl. Environ. Microbiol. 68:4604-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backman, A., N. Maraha, and J. K. Jansson. 2004. Impact of temperature on the physiological status of a potential bioremediation inoculant, Arthrobacter chlorophenolicus A6. Appl. Environ. Microbiol. 70:2952-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baehler, E., M. Bottiglieri, M. Péchy-Tarr, M. Maurhofer, and C. Keel. 2005. Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. J. Appl. Microbiol. 99:24-38. [DOI] [PubMed] [Google Scholar]
- 6.Bais, H. P., B. Prithiviraj, A. K. Jha, F. M. Ausubel, and J. M. Vivanco. 2005. Mediation of pathogen resistance by exudation of antimicrobials from roots. Nature 434:217-221. [DOI] [PubMed] [Google Scholar]
- 7.Bais, H. P., T. L. Weir, L. G. Perry, S. Gilroy, and J. M. Vivanco. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57:233-266. [DOI] [PubMed] [Google Scholar]
- 8.Bergsma-Vlami, M., M. E. Prins, and J. M. Raaijmakers. 2005. Influence of plant species on population dynamics, genotypic diversity and antibiotic production in the rhizosphere by indigenous Pseudomonas spp. FEMS Microbiol. Ecol. 52:59-69. [DOI] [PubMed] [Google Scholar]
- 9.Bevis, B. J., and B. S. Glick. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20:83-87. [DOI] [PubMed] [Google Scholar]
- 10.Bloemberg, G. V. 2007. Microscopic analysis of plant-bacterium interactions using auto fluorescent proteins. Eur. J. Plant Pathol. 119:301-309. [Google Scholar]
- 11.Bloemberg, G. V., A. H. M. Wijfjes, G. E. M. Lamers, N. Stuurman, and B. J. J. Lugtenberg. 2000. Simultaneous imaging of Pseudomonas fluorescens WCS365 populations expressing three different autofluorescent proteins in the rhizosphere: new perspectives for studying microbial communities. Mol. Plant-Microbe Interact. 13:1170-1176. [DOI] [PubMed] [Google Scholar]
- 12.Bolwerk, A., A. L. Lagopodi, A. H. M. Wijfjes, G. E. M. Lamers, T. F. C. Chin-A-Woeng, B. J. J. Lugtenberg, and G. V. Bloemberg. 2003. Interactions in the tomato rhizosphere of two Pseudomonas biocontrol strains with the phytopathogenic fungus Fusarium oxysporum f. sp radicis-lycopersici. Mol. Plant-Microbe Interact. 16:983-993. [DOI] [PubMed] [Google Scholar]
- 13.Bonsall, R. F., D. M. Weller, and L. S. Thomashow. 1997. Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonas spp. in vitro and in the rhizosphere of wheat. Appl. Environ. Microbiol. 63:951-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandl, M. T., and R. E. Mandrell. 2002. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl. Environ. Microbiol. 68:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandl, M. T., B. Quiñones, and S. E. Lindow. 2001. Heterogeneous transcription of an indoleacetic acid biosynthetic gene in Erwinia herbicola on plant surfaces. Proc. Natl. Acad. Sci. USA 98:3454-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burmølle, M., L. H. Hansen, G. Oregaard, and S. J. Sørensen. 2003. Presence of N-acyl homoserine lactones in soil detected by a whole-cell biosensor and flow cytometry. Microb. Ecol. 45:226-236. [DOI] [PubMed] [Google Scholar]
- 17.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene-expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 18.Chin-A-Woeng, T. F. C., G. V. Bloemberg, A. J. van der Bij, K. van der Drift, J. Schripsema, B. Kroon, R. J. Scheffer, C. Keel, P. Bakker, H. V. Tichy, F. J. de Bruijn, J. E. Thomas-Oates, and B. J. J. Lugtenberg. 1998. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol. Plant-Microbe Interact. 11:1069-1077. [Google Scholar]
- 19.Cook, R. J. 1993. Making greater use of introduced microorganisms for biological control of plant pathogens. Annu. Rev. Phytopathol. 31:53-80. [DOI] [PubMed] [Google Scholar]
- 20.Dandie, C. E., E. Larrainzar, G. L. Mark, F. O'Gara, and J. P. Morrissey. 2005. Establishment of DsRed.T3_S4T as an improved autofluorescent marker for microbial ecology applications. Environ. Microbiol. 7:1818-1825. [DOI] [PubMed] [Google Scholar]
- 21.Daunert, S., G. Barrett, J. S. Feliciano, R. S. Shetty, S. Shrestha, and W. Smith-Spencer. 2000. Genetically engineered whole-cell sensing systems: coupling biological recognition with reporter genes. Chem. Rev. 100:2705-2738. [DOI] [PubMed] [Google Scholar]
- 22.DeAngelis, K. M., P. S. Ji, M. K. Firestone, and S. E. Lindow. 2005. Two novel bacterial biosensors for detection of nitrate availability in the rhizosphere. Appl. Environ. Microbiol. 71:8537-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Werra, P., A. Huser, E. Baehler, C. Keel, and M. Maurhofer. 2006. Using flow cytometry for in situ monitoring of antimicrobial compound production in the biocontrol bacteria Pseudomonas fluorescens CHA0, p. 117-121. In J. M. Raaijmakers and R. A. Sikora (ed.), Multitrophic interactions in soil and integrated control. Proceedings of a Meeting at Wageningen, The Netherlands. International Organisation for Biological Control/West Palaearctic Regional Section.
- 24.Duffy, B. K., and G. Défago. 1999. Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 65:2429-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elväng, A. M., K. Westerberg, C. Jernberg, and J. K. Jansson. 2001. Use of green fluorescent protein and luciferase biomarkers to monitor survival and activity of Arthrobacter chlorophenolicus A6 cells during degradation of 4-chlorophenol in soil. Environ. Microbiol. 3:32-42. [DOI] [PubMed] [Google Scholar]
- 26.Fu, Y. T., C. O'Kelly, M. Sieracki, and D. L. Distel. 2003. Protistan grazing analysis by flow cytometry using prey labeled by in vivo expression of fluorescent proteins. Appl. Environ. Microbiol. 69:6848-6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamalero, E., G. Lingua, F. G. Capri, A. Fusconi, G. Berta, and P. Lemanceau. 2004. Colonization pattern of primary tomato roots by Pseudomonas fluorescens A6RI characterized by dilution plating, flow cytometry, fluorescence, confocal and scanning electron microscopy. FEMS Microbiol. Ecol. 48:79-87. [DOI] [PubMed] [Google Scholar]
- 28.Gantner, S., M. Schmid, C. Dürr, R. Schuhegger, A. Steidle, P. Hutzler, C. Langebartels, L. Eberl, A. Hartmann, and F. B. Dazzo. 2006. In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol. Ecol. 56:188-194. [DOI] [PubMed] [Google Scholar]
- 29.Haas, D., and G. Défago. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3:307-319. [DOI] [PubMed] [Google Scholar]
- 30.Haas, D., and C. Keel. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41:117-153. [DOI] [PubMed] [Google Scholar]
- 31.Hansen, L. H., B. Ferrari, A. H. Sørensen, D. Veal, and S. J. Sørensen. 2001. Detection of oxytetracycline production by Streptomyces rimosus in soil microcosms by combining whole-cell biosensors and flow cytometry. Appl. Environ. Microbiol. 67:239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James, D. W., and N. I. Gutterson. 1986. Multiple antibiotics produced by Pseudomonas fluorescens HV37a and their differential regulation by glucose. Appl. Environ. Microbiol. 52:1183-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jansson, J. K. 2003. Marker and reporter genes: illuminating tools for environmental microbiologists. Curr. Opin. Microbiol. 6:310-316. [DOI] [PubMed] [Google Scholar]
- 34.Joyner, D. C., and S. E. Lindow. 2000. Heterogeneity of iron bioavailability on plants assessed with a whole-cell GFP-based bacterial biosensor. Microbiology 146:2435-2445. [DOI] [PubMed] [Google Scholar]
- 35.Kamath, R., J. L. Schnoor, and P. J. J. Alvarez. 2004. Effect of root-derived substrates on the expression of nah-lux genes in Pseudomonas fluorescens HK44: implications for PAH biodegradation in the rhizosphere. Environ. Sci. Technol. 38:1740-1745. [DOI] [PubMed] [Google Scholar]
- 36.Keel, C., and G. Défago. 1997. Interactions between beneficial soil bacteria and root pathogens: mechanisms and ecological impact, p. 27-46. In A. C. Gange and V. K. Brown (ed.), Multitrophic interactions in terrestrial systems. Blackwell Science, London, England.
- 37.Keel, C., U. Schnider, M. Maurhofer, C. Voisard, J. Laville, U. Burger, P. Wirthner, D. Haas, and G. Défago. 1992. Suppression of root diseases by Pseudomonas fluorescens CHA0—importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol. Plant Microbe Interact. 5:4-13. [Google Scholar]
- 38.Keel, C., C. Voisard, C. H. Berling, G. Kahr, and G. Défago. 1989. Iron sufficiency, a prerequisite for the suppression of tobacco black root-rot by Pseudomonas fluorescens strain CHA0 under gnotobiotic conditions. Phytopathology 79:584-589. [Google Scholar]
- 39.Larrainzar, E., F. O'Gara, and J. P. Morrissey. 2005. Applications of autofluorescent proteins for in situ studies in microbial ecology. Annu. Rev. Microbiol. 59:257-277. [DOI] [PubMed] [Google Scholar]
- 40.Laville, J., C. Blumer, C. Von Schroetter, V. Gaia, G. Défago, C. Keel, and D. Haas. 1998. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J. Bacteriol. 180:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, L., Z. K. Punja, and J. E. Rahe. 1997. Altered root exudation and suppression of induced lignification as mechanisms of predisposition by glyphosate of bean roots (Phaseolus vulgaris L.) to colonization by Pythium spp. Physiol. Mol. Plant Pathol. 51:111-127. [Google Scholar]
- 42.Loper, J. E., and M. D. Henkels. 1997. Availability of iron to Pseudomonas fluorescens in rhizosphere and bulk soil evaluated with an ice nucleation reporter gene. Appl. Environ. Microbiol. 63:99-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowder, M., and J. D. Oliver. 2001. The use of modified GFP as a reporter for metabolic activity in Pseudomonas putida. Microb. Ecol. 41:310-313. [DOI] [PubMed] [Google Scholar]
- 44.Maksimow, M., K. Hakkila, M. Karp, and M. Virta. 2002. Simultaneous detection of bacteria expressing gfp and dsred genes with a flow cytometer. Cytometry 47:243-247. [DOI] [PubMed] [Google Scholar]
- 45.Maraha, N., A. Backman, and J. K. Jansson. 2004. Monitoring physiological status of GFP-tagged Pseudomonas fluorescens SBW25 under different nutrient conditions and in soil by flow cytometry. FEMS Microbiol. Ecol. 51:123-132. [DOI] [PubMed] [Google Scholar]
- 46.Mark, G. L., J. M. Dow, P. D. Kiely, H. Higgins, J. Haynes, C. Baysse, A. Abbas, T. Foley, A. Franks, J. Morrissey, and F. O'Gara. 2005. Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc. Natl. Acad. Sci. USA 102:17454-17459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mascher, F., C. Hase, Y. Moënne-Loccoz, and G. Défago. 2000. The viable-but-nonculturable state induced by abiotic stress in the biocontrol agent Pseudomonas fluorescens CHA0 does not promote strain persistence in soil. Appl. Environ. Microbiol. 66:1662-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maurhofer, M., E. Baehler, R. Notz, V. Martinez, and C. Keel. 2004. Cross talk between 2,4-diacetylphloroglucinol-producing biocontrol pseudomonads on wheat roots. Appl. Environ. Microbiol. 70:1990-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maurhofer, M., C. Keel, D. Haas, and G. Defago. 1995. Influence of plant species on disease suppression by Pseudomonas fluorescens strain CHA0 with enhanced antibiotic production. Plant Pathol. 44:40-50. [Google Scholar]
- 50.McCrady, J. K., and C. P. Andersen. 2000. The effect of ozone on below-ground carbon allocation in wheat. Environ. Pollut. 107:465-472. [DOI] [PubMed] [Google Scholar]
- 51.Miller, W. G., M. T. Brandl, B. Quiñones, and S. E. Lindow. 2001. Biological sensor for sucrose availability: relative sensitivities of various reporter genes. Appl. Environ. Microbiol. 67:1308-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nebe-von-Caron, G., P. J. Stephens, C. J. Hewitt, J. R. Powell, and R. A. Badley. 2000. Analysis of bacterial function by multi-colour fluorescence flow cytometry and single cell sorting. J. Microbiol. Methods 42:97-114. [DOI] [PubMed] [Google Scholar]
- 53.Nicholson, R. L., and R. Hammerschmidt. 1992. Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30:369-389. [Google Scholar]
- 54.Normander, B., N. B. Hendriksen, and O. Nybroe. 1999. Green fluorescent protein-marked Pseudomonas fluorescens: localization, viability, and activity in the natural barley rhizosphere. Appl. Environ. Microbiol. 65:4646-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Notz, R., M. Maurhofer, U. Schnider-Keel, B. Duffy, D. Haas, and G. Défago. 2001. Biotic factors affecting expression of the 2,4-diacetylphloroglucinol biosynthesis gene phlA in Pseudomonas fluorescens biocontrol strain CHA0 in the rhizosphere. Phytopathology 91:873-881. [DOI] [PubMed] [Google Scholar]
- 56.Péchy-Tarr, M., M. Bottiglieri, S. Mathys, K. B. Lejbølle, U. Schnider-Keel, M. Maurhofer, and C. Keel. 2005. RpoN (σ54) controls production of antifungal compounds and biocontrol activity in Pseudomonas fluorescens CHA0. Mol. Plant Microbe Interact. 18:260-272. [DOI] [PubMed] [Google Scholar]
- 57.Raaijmakers, J. M., M. Vlami, and J. T. de Souza. 2002. Antibiotic production by bacterial biocontrol agents. Antonie van Leeuwenhoek. 81:537-547. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 59.Schnider-Keel, U., A. Seematter, M. Maurhofer, C. Blumer, B. Duffy, C. Gigot-Bonnefoy, C. Reimmann, R. Notz, G. Défago, D. Haas, and C. Keel. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182:1215-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shaw, L. J., P. Morris, and J. E. Hooker. 2006. Perception and modification of plant flavonoid signals by rhizosphere microorganisms. Environ. Microbiol. 8:1867-1880. [DOI] [PubMed] [Google Scholar]
- 61.Sörensen, M., C. Lippuner, T. Kaiser, A. Miβlitz, T. Aebischer, and D. Bumann. 2003. Rapidly maturing red fluorescent protein variants with strongly enhanced brightness in bacteria. FEBS Lett. 552:110-114. [DOI] [PubMed] [Google Scholar]
- 62.Sørensen, S. J., A. H. Sørensen, L. H. Hansen, G. Oregaard, and D. Veal. 2003. Direct detection and quantification of horizontal gene transfer by using flow cytometry and gfp as a reporter gene. Curr. Microbiol. 47:129-133. [DOI] [PubMed] [Google Scholar]
- 63.Steidle, A., K. Sigl, R. Schuhegger, A. Ihring, M. Schmid, S. Gantner, M. Stoffels, K. Riedel, M. Givskov, A. Hartmann, C. Langebartels, and L. Eberl. 2001. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67:5761-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tombolini, R., A. Unge, M. E. Davey, F. J. de Bruijn, and J. K. Jansson. 1997. Flow cytometric and microscopic analysis of GFP-tagged Pseudomonas fluorescens bacteria. FEMS Microbiol. Ecol. 22:17-28. [Google Scholar]
- 65.Tombolini, R., D. J. van der Gaag, B. Gerhardson, and J. K. Jansson. 1999. Colonization pattern of the biocontrol strain Pseudomonas chlororaphis MA 342 on barley seeds visualized by using green fluorescent protein. Appl. Environ. Microbiol. 65:3674-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Troxler, J., M. Zala, Y. Moënne-Loccoz, C. Keel, and G. Défago. 1997. Predominance of nonculturable cells of the biocontrol strain Pseudomonas fluorescens CHA0 in the surface horizon of large outdoor lysimeters. Appl. Environ. Microbiol. 63:3776-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Unge, A., and J. Jansson. 2001. Monitoring population size, activity, and distribution of gfp-luxAB-tagged Pseudomonas fluorescens SBW25 during colonization of wheat. Microb. Ecol. 41:290-300. [DOI] [PubMed] [Google Scholar]
- 68.Unge, A., R. Tombolini, L. Mølbak, and J. K. Jansson. 1999. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl. Environ. Microbiol. 65:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Valverde, C., S. Heeb, C. Keel, and D. Haas. 2003. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 50:1361-1379. [DOI] [PubMed] [Google Scholar]
- 70.van Overbeek, L. S., and J. D. van Elsas. 1995. Root exudate-induced promoter activity in Pseudomonas fluorescens mutants in the wheat rhizosphere. Appl. Environ. Microbiol. 61:890-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Voisard, C., C. Bull., C. Keel, J. Laville, M. Maurhofer, U. Schnider, G. Défago, and D. Haas. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches, p. 67-89. In F. O'Gara, D. Dowling, and B. Boesten (ed.), Molecular ecology of rhizosphere microorganisms. VCH Publishers, Weinheim, Germany.
- 72.Weingart, H., S. Stubner, A. Schenk, and M. S. Ullrich. 2004. Impact of temperature on in planta expression of genes involved in synthesis of the Pseudomonas syringae phytotoxin coronatine. Mol. Plant Microbe Interact. 17:1095-1102. [DOI] [PubMed] [Google Scholar]
- 73.Weller, D. M., J. M. Raaijmakers, B. B. M. Gardener, and L. S. Thomashow. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309-348. [DOI] [PubMed] [Google Scholar]