Abstract
An insertion mutant of gtcA, responsible for serotype-specific glycosylation of the cell wall teichoic acid in serotype 4b strains of Listeria monocytogenes, was also resistant to both Listeria genus- and serotype 4b-specific phages. The sugar substituents on teichoic acid appeared essential for the adsorption of phages A500 (serotype 4b specific) and A511 (Listeria genus specific) to serotype 4b L. monocytogenes.
In Listeria monocytogenes, serotype designations are based on flagellin antigens (corresponding to letters such as a, b, c, d, and e) as well as specific moieties on the teichoic acid of the cell wall (corresponding to the numerical portion of serotype designations, such as 1/2, 3, or 4). In serotype 4b, teichoic acid harbors N-acetylglucosamine as an integral component of the poly-ribitol phosphate backbone and serotype-specific sugars, specifically galactose and glucose, decorate the N-acetylglucosamine molecule (3).
In earlier studies, gtcA and the gltA-gltB cassette were found to be essential for serotype-specific glycosylation of the teichoic acid of L. monocytogenes serotype 4b with glucose and galactose (8, 12). Within L. monocytogenes, the gltA-gltB cassette is found only in strains of the serotype 4b complex (serotype 4b and the highly similar serotypes 4d and 4e) (8). Originally, gtcA was thought to be unique to serogroup 4 strains (12); however, subsequent studies and genome sequence data revealed that serotype 1/2a strains harbor a divergent gtcA homologue (80% and 82% identity at the nucleotide and amino acid sequence levels, respectively) in a genomically equivalent location (1, 4, 11). A gtcA mutant of the serotype 1/2a strain EGD-e was shown to be resistant to a phage (LMUP121) infecting both serotype 1/2 and 4b strains, but the impact of gtcA inactivation on teichoic acid composition of EGD-e was not investigated (1). In the case of serotype 4b, gtcA was found to be essential for teichoic acid glycosylation with galactose and glucose, and for reactivity of the bacteria with the serotype 4b-, 4d-, and 4e-specific monoclonal antibody c74.22 (5, 12). However, the role of gtcA in infection of serotype 4b strains by serotype 4b-specific and Listeria genus-specific phages has not been described.
In this study, we characterized phage susceptibility of strain M44, a derivative of the serotype 4b strain 4b1 harboring a single Tn916ΔE transposon insertion in gtcA. The construction and teichoic acid glycosylation defects of M44 were described earlier (12). M44 was found to be resistant not only to the serotype 4b-specific phage A500 but also to two different Listeria genus-specific phages, A511 (9, 10) and Φ20422-1, recently isolated from a turkey-processing plant in the United States (6). In contrast, the parental strain 4b1 was fully susceptible to all three phages (Table 1). To determine whether the observed resistance of the mutant to the phages was associated with failure of the phages to adsorb, adsorption assays were done. Adsorption of A500 and A511 onto the M44 cells was indeed impaired in comparison to that of the parental strain 4b1 (Table 2). In contrast, no consistent decrease in adsorption of Φ20422-1 onto M44 versus 4b1 was observed (data not shown).
TABLE 1.
Phage susceptibility and c74.22 reactivity of strains investigated in this study
| Strain | c74.22 reactivitya | Phage susceptibilityb
|
||
|---|---|---|---|---|
| Φ20422-1 | A500 | A511 | ||
| 4b1 | + | S | S | S |
| M44 | − | R | R | R |
| M44::pPL2 | − | R | R | R |
| M44::pPL95 | + | S | S | S |
Determined as described in the legend to Fig. 1.
S and R indicate susceptibility and resistance, respectively, determined on the basis of plaque formation following infection of the bacteria (ca. 1 × 108 CFU/ml) with the phage (ca. 1 × 106 PFU/ml), done as described previously (14). The phage infection assays for each phage were done in duplicate and in at least two independent experiments.
TABLE 2.
Phage A500 and A511 adsorption deficiency of gtcA mutant
| Strain | % Efficiency of phage adsorptiona
|
|
|---|---|---|
| A511 | A500 | |
| 4b1 | 100 | 100 |
| M44 | 26.2 | 7.0 |
| M44::pPL2 | 18.7 | 9.1 |
| M44::pPL95 | 72.9 | 57.4 |
Adsorption of phages was calculated by determining the number of PFU remaining in the supernatant following 30 min of incubation of the phage-bacterium mixture at 37°C and subtracting it from the total PFU/ml of A511 (ca. 1.41 × 105 PFU/ml) or A500 (ca. 1.4 × 104 PFU/ml) added to the bacteria (ca. 1 × 108 CFU/ml). Adsorption efficiency was indicated relative to adsorption on the parental strain 4b1 (set as 100%). The serotype 4b strain F2365 (11) was used as an indicator for the enumeration of total PFU/ml in the inoculum and in the supernatant, and infections were done as described previously (14). The phage adsorption assay for each tested phage was done at least twice. The results shown are from a representative experiment.
To confirm that gtcA is essential for phage infection of L. monocytogenes serotype 4b by serotype-specific as well as by Listeria genus-specific phages, it would be important to examine phage susceptibility profiles of genetically complemented gtcA mutants. Genetic complementation of gtcA in trans (using gtcA cloned in the temperature-sensitive shuttle plasmid vector pKSV7) was described earlier (12). However, that earlier complemented strain is no longer available. In addition, we deemed it important to utilize a complemented strain harboring a chromosomal copy of gtcA, thus obviating copy number and stability issues related to plasmid-borne constructs. To construct such a complemented strain, primers VCpNP95F (5′-ATAAGCGGCCGCTTCAAAGGGACAGGCAACATG, harboring a NotI site [underlined]) and VCpNP95R (5′-ATAACCCGGGGTACTCAGGATGAATTCCAG, harboring an XmaI site [underlined]) were used to amplify a fragment consisting of the coding sequence of gtcA and 300 nucleotides upstream of the start codon of the gene. The amplicon was digested with NotI and XmaI; cloned into the integration shuttle vector pPL2 (7), also digested with NotI-XmaI; and introduced into Escherichia coli S17-1 (13) by electroporation. The resulting recombinant plasmid (pPL95) and the empty vector (pPL2) were transferred from E. coli S17-1 (expressing resistance to chloramphenicol, conferred by these plasmids) to M44 employing the conjugation procedures described before (7), resulting in M44::pPL95 and M44::pPL2, respectively. Transconjugants were selected using chloramphenicol (6 μg/ml) and nalidixic acid (20 μg/ml) (to which L. monocytogenes is naturally resistant). The primer pair gtcA_C_F (5′-TGTGCTTGTACTGAACTACG) and CAT_G+_R (5′-CAAAAGCTTCGGATCTGGAGCTG) amplified a single ∼1.5-kb product, as would be expected for gtcA cloned into pPL2 (data not shown).
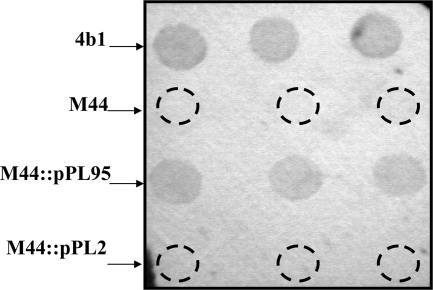
The complemented strain recovered reactivity with the serotype 4b-specific monoclonal antibody c74.22, which recognizes glycosylated teichoic acid and which did not react with M44 (Fig. 1 and Table 1). The c74.22 reactivity of the complemented strain was indistinguishable from that of the parental strain, as was also observed earlier with a complementation derivative harboring gtcA in trans (12). Infection of M44::pPL95 with A500, A511, and Φ20422-1 showed that, in contrast to M44, the complemented strain was now susceptible to these phages (Table 1). Furthermore, adsorption of A511 and A500 to the complemented strain (M44::pPL95) was partially restored (Table 2). The reasons for lack of complete restoration of adsorption are unclear. It is possible that regulation of gtcA expression in the integrated construct may differ from that in the original location. However, the apparently lower adsorption did not result in detectible differences in phage susceptibility and c74.22 reactivity under the conditions employed.
FIG. 1.
cis complementation of surface antigen expression of M44 with wild-type gtcA using colony immunoblotting with monoclonal antibody c74.22. Overnight cultures were spotted on the nitrocellulose membrane in triplicate and processed as described previously (12). From top to bottom are shown 4b1 (parental strain), M44 (transposon-induced gtcA mutant), M44::pPL95 (M44 harboring pLP95), and M44::pPL2 (M44 harboring cloning vector pPL2).
As mentioned earlier, Φ20422-1 did not appear to be impaired in adsorption onto M44 cells; this phage also adsorbed with similar efficiency onto M44 cells harboring pPL2 or pPL95 (data not shown). These findings are in agreement with earlier results that suggested that the receptor for Φ20422-1 was different from that for A511 (2). The findings also suggest that the observed resistance of M44 to Φ20422-1 was due not to failure of this phage to adsorb but to a yet unidentified step that requires intact teichoic acid (and is impaired in M44).
The overall findings with M44 and the genetically complemented mutant (M44::pPL95) constitute strong evidence that gtcA is required for susceptibility of the bacteria to listeriaphage and that glycosylated teichoic acid is required for adsorption of A500 and A511. In the case of the serotype 4b-specific phage A500, our results are in agreement with previous findings indicating that teichoic acid served as receptor for this phage (15). The receptor of the genus-specific phage A511 has been shown to be peptidoglycan (15), and previous studies showed that peptidoglycan was not affected in M44 (12). However, it is possible that the alterations in teichoic acid glycosylation associated with gtcA inactivation impaired topological aspects of cell wall conformation that may be required for proper recognition of peptidoglycan receptors by A511 or for full access of this phage to its receptors. The receptor for the other wide-host-range phage tested here, Φ20422-1, has not yet been determined but appears to be different from those for A500 or A511. Mechanisms underlying the observed resistance of the gtcA mutant to this phage remain unknown. However, restoration of susceptibility to Φ20422-1 in the genetically complemented strain M44::pPL95 (Table 1) suggests that, similarly to A500 and A511, this phage requires glycosylated teichoic acid and intact gtcA for infection of serotype 4b bacteria.
DNA sequence analysis of gtcA in both serotype 1/2a and 4b strains has revealed that the G+C content of this gene is noticeably lower than the average for L. monocytogenes and suggested the possibility that it has been acquired by L. monocytogenes via horizontal transfer, from a currently unidentified source (1, 12). Further studies are needed to elucidate the mechanisms driving the observed sequence diversity of the gene in the different serogroups and underlying the gene's apparent role in the organism's susceptibility to both serotype-specific and genus-specific phages.
Acknowledgments
Funding for this research was partially provided by USDA grant 2006-35201-17377.
We thank R. Calendar for A500 and pPL2 and M. Loessner for the gift of A511. We acknowledge advice and feedback from Driss Elhanafi, the expert technical support of Robin Siletzky, and the encouragement of all members of our laboratory.
Footnotes
Published ahead of print on 11 January 2008.
REFERENCES
- 1.Autret, N., I. Dubail, P. Trieu-Cuot, P. Berche, and A. Charbit. 2001. Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect. Immun. 69:2054-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng, Y., L. Yue, D. Elhanafi, and S. Kathariou. 2007. Absence of serotype-specific surface antigen in laboratory variants of epidemic-associated Listeria monocytogenes strains. Appl. Environ. Microbiol. 73:6313-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiedler, F. 1988. Biochemistry of the cell surface of Listeria strains: a locating general view. Infection 16(Suppl. 2):S92-S97. [DOI] [PubMed] [Google Scholar]
- 4.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 5.Kathariou, S., C. Mizumoto, R. D. Allen, A. K. Fok, and A. A. Benedict. 1994. Monoclonal antibodies with a high degree of specificity for Listeria monocytogenes serotype 4b. Appl. Environ. Microbiol. 60:3548-3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim, J.-W., and S. Kathariou. 2006. Host range of Listeria-specific bacteriophage from the environment of turkey processing plants in the United States, abstr. P2-59. Abstr. Int. Assoc. Food Prot. Conf., Calgary, Alberta, Canada.
- 7.Lauer, P., M. Y. N. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei, X.-H., F. Fiedler, Z. Lan, and S. Kathariou. 2001. A novel serotype-specific gene cassette (gltA-gltB) is required for expression of teichoic acid-associated surface antigens in Listeria monocytogenes of serotype 4b. J. Bacteriol. 183:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loessner, M. J. 1991. Improved procedure for bacteriophage typing of Listeria strains and evaluation of new phages. Appl. Environ. Microbiol. 57:882-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loessner, M. J., and M. Busse. 1990. Bacteriophage typing of Listeria species. Appl. Environ. Microbiol. 56:1912-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Promadej, N., F. Fiedler, P. Cossart, S. Dramsi, and S. Kathariou. 1999. Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serotype-specific gene. J. Bacteriol. 181:418-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 14.Tran, H. L., F. Fiedler, D. A. Hodgson, and S. Kathariou. 1999. Transposon-induced mutations in two loci of Listeria monocytogenes serotype 1/2a result in phage resistance and lack of N-acetylglucosamine in the teichoic acid of the cell wall. Appl. Environ. Microbiol. 65:4793-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wendlinger, G., M. J. Loessner, and S. Scherer. 1996. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetyl-D-glucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology 142:985-992. [DOI] [PubMed] [Google Scholar]