Abstract
Microorganisms catalyze the formation of naturally occurring Mn oxides, but little is known about the biochemical mechanisms of this important biogeochemical process. We used tandem mass spectrometry to directly analyze the Mn(II)-oxidizing enzyme from marine Bacillus spores, identified as an Mn oxide band with an in-gel activity assay. Nine distinct peptides recovered from the Mn oxide band of two Bacillus species were unique to the multicopper oxidase MnxG, and one peptide was from the small hydrophobic protein MnxF. No other proteins were detected in the Mn oxide band, indicating that MnxG (or a MnxF/G complex) directly catalyzes biogenic Mn oxide formation. The Mn(II) oxidase was partially purified and found to be resistant to many proteases and active even at high concentrations of sodium dodecyl sulfate. Comparative analysis of the genes involved in Mn(II) oxidation from three diverse Bacillus species revealed a complement of conserved Cu-binding regions not present in well-characterized multicopper oxidases. Our results provide the first direct identification of a bacterial enzyme that catalyzes Mn(II) oxidation and suggest that MnxG catalyzes two sequential one-electron oxidations from Mn(II) to Mn(III) and from Mn(III) to Mn(IV), a novel type of reaction for a multicopper oxidase.
Mn(III,IV) oxides are reactive minerals that play an important role in the global cycling of many major (C and S) and trace elements (Fe, Co, Pb, Cu, Cd, and Cr) in nature (52). Microorganisms are thought to be responsible for the formation of Mn oxides in the environment, catalyzing Mn(II) oxidation rates that are orders of magnitude faster than abiotic Mn(II) oxidation rates (32, 52). This microbially mediated Mn(II) oxidation occurs by an enzymatic pathway (13, 42), whereby biogenic Mn oxides are precipitated on cell surfaces. Although the importance of microbes in driving the oxidative segment of the Mn cycle has been demonstrated in many environments (8, 17, 30, 51, 52), little is known about the biochemical mechanism.
Genetic approaches have elucidated multicopper oxidase (MCO) genes that are required for Mn(II) oxidation in the phylogenetically diverse Mn(II)-oxidizing bacteria Pseudomonas putida strain GB-1 (4), Leptothrix discophora strain SS-1 (7), Bacillus sp. strain SG-1 (55), and Pedomicrobium sp. strain ACM 3067 (38), suggesting a universal mechanism of bacterial Mn(II) oxidation. MCOs are a family of enzymes that use multiple Cu atoms—classified into three types of Cu sites—as cofactors in coupling the oxidation of substrate to the reduction of O2 to H2O (46). MCO substrates include a variety of organic compounds, as well as metal ions such as Fe2+, and it has been hypothesized that bacterial MCOs are in fact the direct catalysts of Mn(II) oxidation. However, a direct link between MCO genes and Mn(II)-oxidizing enzymes has not been made. Repeated efforts to purify native Mn(II) oxidases to completion have failed (1, 22a, 35; D. B. Edwards and B. M. Tebo, unpublished data), as have efforts to produce active Mn(II) oxidase by expressing MCO genes in heterologous hosts (5, 12). Thus, biochemical analysis of the Mn(II) oxidase has not been possible, and uncovering the functional role of MCOs in Mn(II) oxidation has been enigmatic. MCOs are involved in a variety of cellular functions (6, 46), including iron and copper homeostasis (21, 37, 50), siderophore oxidation (16), pigment formation (20), and biopolymerization (47). The loss of such functions could indirectly lead to the non-Mn(II)-oxidizing phenotype that is observed with MCO mutants; therefore, that MCOs directly catalyze Mn(II) oxidation has remained an unproven hypothesis.
In the present study, we investigated the role of an MCO in Mn(II) oxidation by marine Bacillus spores. Phylogenetically diverse Mn(II)-oxidizing Bacillus species have been isolated from a number of environments in which Mn(II) oxidation is prevalent, such as coastal sediments (14), deep-sea hydrothermal vents (11), and the suboxic zone of the Black Sea (G. J. Dick and B. M. Tebo, unpublished results). These Bacillus species oxidize Mn(II) as metabolically dormant spores (not as growing or vegetative cells) via an unidentified Mn(II)-oxidizing enzyme (42) located in the exosporium, the outermost layer of the spore coat (12). In the model organism Bacillus sp. strain SG-1, a cluster of seven genes—designated the mnx genes—are required for Mn oxidation but show very limited similarity to any database sequences (55). One of these genes, mnxG, encodes a predicted protein with Cu-binding motifs that are signatures of MCOs. Mn(II) oxidation by strain SG-1 proceeds through a Mn(III) intermediate, and mnxG is required for both the oxidation of Mn(II) to Mn(III) and of Mn(III) to Mn(IV) (57). The possibility that MnxG is the direct catalyst raises interesting mechanistic questions because all well-characterized MCOs catalyze one-electron transfer reactions (46), whereas the oxidation of Mn(II) to Mn(IV) requires the transfer of two electrons. However, as with all other Mn(II)-oxidizing bacteria, the exact role of the MCO in the Mn(II) oxidation reaction remains unknown.
Recently, the extensive phylogenetic diversity of marine Mn(II)-oxidizing Bacillus spores has been recognized (11, 14). The Mn(II) oxidase of these diverse Mn(II)-oxidizing Bacillus spores can be visualized by a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in-gel activity assay, and its size varies widely among species (14). Here we describe cloning and DNA sequence analysis of the mnx gene cluster from two diverse Bacillus strains—PL-12 and MB-7—and comparative sequence analysis with strain SG-1. DNA sequence of the mnx region also facilitated interpretation of tandem mass spectrometric (MS/MS) analysis of the Mn(II) oxidase, leading to the most prominent finding of our research—the first direct identification of a bacterial Mn(II) oxidase.
MATERIALS AND METHODS
Growth, DNA extraction, cloning, and sequencing.
Bacillus cultures were grown in K medium as described previously (53) with shaking (150 rpm) at room temperature. DNA was isolated by phenol-chloroform extraction (18). The PL-12 mnx region was cloned by a combination of genomic DNA library screening and inverse PCR, and the MB-7 mnx region was cloned entirely by inverse PCR. The PL-12 library was constructed by digestion of genomic DNA with HindIII, fractionation of the HindIII fragments on a 10 to 40% (wt/vol) sucrose density gradient (28), selection of 6- to 8-kb fragments by agarose gel-electrophoresis extraction (Qiagen), ligation with pZERO-2 (Invitrogen), and transformation into Escherichia coli TOP10 competent cells (Invitrogen). Colonies were screened by using a ∼900-bp digoxigenin-labeled mnxG PCR product (amplified with primers MnxGIf and MnxGIr (14) as a probe with the DIG DNA labeling and detection kit (Roche). Hybridizations were done at 42°C, washes at 68°C. A 7-kb fragment, including the PL-12 mnxG gene and ∼3.3 kb downstream, was cloned by this method. Inverse PCR products (typically ∼1.5 kb) were generated as described previously (33) by using the restriction enzyme HaeII and the polymerase Pfu Turbo (Stratagene) and cloned by using a TOPO TA cloning kit (Invitrogen). The PL-12 and MB-7 mnx regions were cloned by primer walking with successive cycles of inverse PCR, cloning, and DNA sequencing.
Purification of the Mn(II) oxidase.
Spores were purified, and exosporium was isolated as described previously (11). Solubilization of crude and trypsin-digested exosporium was tested with various concentrations of deoxycholate, Triton X-100, CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, Tween 20, NP-40, and SDS (0.1 to 5%), NaCl (0 to 0.5 M), and glycerol (0 or 10%). For protein purification, crude exosporium extracts were digested with sequencing-grade trypsin (Promega) in 50 mM HEPES (pH 7.8) for 1 h at 37°C and then solubilized in 2% SDS, 10% glycerol, 1 mM dithiothreitol (DTT), and 50 mM HEPES (pH 7.8) for 2 h. Insoluble complexes were removed by ultracentrifugation at 86,000 × g. Solubilized samples were fractionated at room temperature on a Superose 6HR 10/30 size-exclusion column (Amersham Pharmacia) at a flow rate of 0.3 ml/min using an Akta FPLC (Amersham Pharmacia). Fractions (1 ml) were collected and assayed for Mn(II)-oxidizing activity, and active fractions were concentrated by ultrafiltration (50-kDa molecular mass cutoff; Millipore).
Mn(II) oxidation assays and specific activity determination.
Mn(II) oxidation was assayed in reactions containing 800 μM MnCl2 and 20 mM HEPES (pH 7.8). The concentration of Mn oxidizing equivalents [including Mn(III) and Mn(IV)] expressed as MnO2 was determined by the leukoberbelin blue method (25, 53). For rate determinations for specific activity, time points were analyzed every hour for 4 h. Protein concentrations were determined by using a DC protein assay (Bio-Rad). Based on our standard curve, the protein concentration of the partially purified fraction was negative (and thus below the detection limits of our assay and standard curve). Because the absorbance at 750 nm (the wavelength at which absorbance of the protein assay is read) for the partially purified fraction was greater than the blank (reagents without sample), for purposes of comparing this activity with the activity in the crude exosporium, we present the specific activity per absorption at 750 nm rather than per mg protein.
Transmission electron microscopy (TEM).
Spores were chemically fixed with 2% glutaraldehyde, followed by 1% osmium tetroxide, stained en bloc with 2% uranyl acetate, dehydrated through an ethanol series, and embedded in LR White resin. Thin sections were stained with uranyl acetate and lead citrate before imaging in a Philips EM300 operating under standard conditions at 60 kV.
SDS-PAGE and in-gel digestion.
SDS-PAGE analysis and the in-gel Mn(II) oxidation assay were done as described previously (14). For strain PL-12, the partially purified Mn oxidase was analyzed, whereas crude exosporium extracts were used for strains MB-7 and SG-1. For strain SG-1, a 2.5% stacking gel was required to allow the Mn(II) oxidase to enter the gel. The darkest portions of the Mn(II) oxidase bands were excised from the gel, washed with 200 μl of H2O, and vortexed for 10 min before removal of the water. Destaining solutions A and B (SilverQuest silver staining kit; Invitrogen) were mixed in equal proportions and used to destain the Mn oxide bands (100 μl/band). The sample was then vortex mixed at room temperature for 15 min, and the supernatant was removed and washed again with water. Samples were then taken through two cycles of mixing with 200 μl of 25 mM ammonium bicarbonate-5 mM DTT-50% acetonitrile (ACN), vortex mixing for 10 min, and removal of the supernatant. Finally, the gel piece was dehydrated with 100 μl of ACN (room temperature, 10 min), the ACN was removed, and 400 ng of ice-cold trypsin (Promega) in 25 mM ammonium bicarbonate-5 mM DTT solution was added to the sample and set on ice for 30 min. After complete rehydration, the excess trypsin solution was removed, replaced with fresh 25 mM ammonium bicarbonate-5 mM DTT, and left overnight at 37°C. The peptides were extracted by the addition of 2 μl of 2% trifluoroacetic acid (TFA) and vortex mixing at room temperature for 30 min. The supernatant was removed and saved. A total of 20 μl of 20% ACN-0.1% TFA was added to the sample, which was vortexed again at room temperature for 30 min. The supernatant was removed and combined with the supernatant from the first extraction.
LC-MS/MS analysis.
Trypsin-digested peptides extracted from SDS-PAGE as described above were analyzed by liquid chromatography (LC)-MS/MS with electrospray ionization. All electrospray ionization experiments were performed by using a QSTAR-XL hybrid mass spectrometer (AB/MDS Sciex) interfaced to a nanoscale reversed-phase high-pressure liquid chromatograph (Famos/Ultimate/Switchos; LC Packings) using a 218MS column (10 cm by 75 μm; GraceVydac) packed with 3-μm C18 beads. The buffer compositions were as follows. Buffer A was composed of 98% H2O, 2% ACN, 0.1% formic acid, and 0.01% TFA; buffer B was composed of 98% ACN, 2% H2O, 0.1% formic acid, and 0.01% TFA. A total of 10 μl was injected by the Famos apparatus onto the Switchos C18 precolumn (5 cm by 300 μm, LC Packings) using buffer A at a flow rate of 30 μl/min. After a 3-min wash, the Switchos valve was switched, and the peptides were backflushed onto the analytical column and eluted with a 20-min linear gradient from 10 to 40% buffer B at a flow rate of ∼200 nl/min. Peptides from keratin contamination and trypsin autolysis were identified and excluded from further analyses. Some samples were reanalyzed with exclusion lists of previously observed keratin and/or trypsin autolysis peaks.
Database search and analysis.
Translated mnx gene sequences were used to search MS/MS data for mnx peptides by using Analyst QS 1.1/ProID 1.1 (49) and BioAnalyst 1.1.5/ProBlast 1.1 (43; Applied Biosystems). To ensure statistically meaningful results, the MS/MS spectra were also tested against a database in which the mnx gene sequences were inserted into SwissProt. Each mnx-related MS/MS spectrum was manually inspected to verify the accuracy of the search results. To identify peptides not present in any databases, all 74 MS/MS spectra obtained from strain PL-12 were manually sequenced de novo.
Nucleotide sequence accession numbers.
Nucleotide sequences of mnx genes were deposited in GenBank with the accession numbers EF158106 and EF158107.
RESULTS
Comparative analysis of the ultrastructure, mnx region gene sequence, and Mn(II) oxidase from diverse Bacillus species.
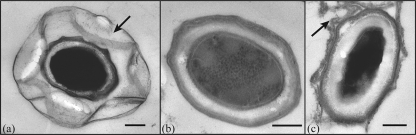
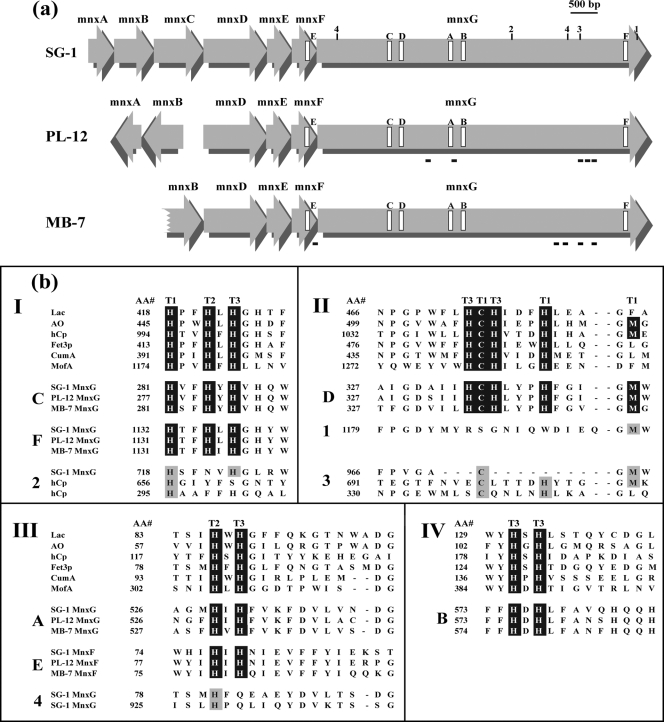
The size of the Mn(II) oxidase varies dramatically among representatives of the three major phylogenetic clusters of Mn(II)-oxidizing Bacillus species, strains PL-12, MB-7, and SG-1 (Fig. 1) (14). TEM shows prominent differences in the extent of exosporium between spores of these three strains (Fig. 2). To investigate the genetic basis for variation of Mn(II) oxidase size, we cloned and sequenced the mnx regions from Bacillus sp. strains PL-12 and MB-7 and compared them with SG-1. The organization of the mnx operon varies between the three strains: mnxC is missing from the mnx region of both PL-12 and MB-7, and mnxA and mnxB are inverted in PL-12 (Fig. 3a). mnxG is the most highly conserved gene (65 to 70% predicted amino acid identity between the three strains), whereas other predicted proteins show 30 to 49% amino acid identity (Table 1). The length of the mnxG gene is nearly identical in all three strains; therefore, the apparent difference in the size of the Mn(II) oxidases (Fig. 1) cannot be explained by differing sizes of the genes that are suspected to encode them.
FIG. 1.
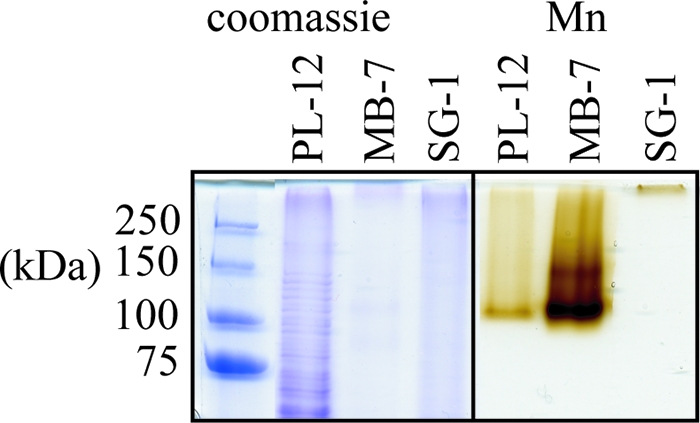
SDS-PAGE of exosporium extracts from three Bacillus strains, stained with Coomassie blue (left) and an in-gel Mn(II) oxidation activity assay (right).
FIG. 2.
TEM of spores from Bacillus species strains PL-12 (a), MB-7 (b), and SG-1 (c). Arrows indicate exosporium, which is not detectable for strain MB-7. Scale bars, 200 nm.
FIG. 3.
(a) Schematic representation of mnx region genes of Bacillus sp. strains SG-1, PL-12, and MB-7. The locations of putative Cu-binding regions are indicated by open rectangles and labeled A to F according to the data in panel b. Additional regions with potential Cu ligands are indicated by numbers (1-4) as in panel b. The locations of peptides detected by MS/MS analysis of the Mn oxide band from the in-gel activity assay are shown with black bars beneath the mnx genes of strains PL-12 and MB-7. (b) Alignment of putative mnx Cu-binding region amino acid sequences to consensus Cu-binding sequence motifs found in all MCOs. The consensus sequences shown (I to IV) are for the well-characterized MCOs laccase (Lac), ascorbate oxidase (AO), ceruloplasmin (hCp), and Fet3p, as well as putative MCOs involved in Mn(II) oxidation in P. putida (CumA) and L. discophora (MofA). “AA#” refers to the number of the first amino acid of each block. Note that the order of the MnxG Cu-binding regions (C, D, A, and B) is different than all other MCOs (III, IV, I, and II). Consensus MCO Cu-binding motifs are highlighted in black, and the type of Cu site for each ligand (based on characterized MCOs) is indicated with T1, T2, or T3. Mnx regions E and F are the fifth and sixth Cu-binding regions that are not found in other MCOs. Additional potential Cu-binding regions present in MnxG (only SG-1 shown here) are shown (sequences 1 to 4), with possible coordinating ligands highlighted with gray. Corresponding regions of hCp are also shown in sequences 2 and 3, with known Cu ligands highlighted in gray.
TABLE 1.
Amino acid sequence identity of Mnx proteins from Bacillus sp. strains SG-1, PL-12, and MB-7
| Predicted protein | Amino acid sequence identitya
|
||
|---|---|---|---|
| SG-1/PL-12 | SG-1/MB-7 | PL-12/MB-7 | |
| MnxA | 46.6 | * | * |
| MnxB | 34.0 | 32.5 | 33.5 |
| MnxC | ** | ** | ** |
| MnxD | 36.1 | 48.7 | 30.7 |
| MnxE | 49.1 | 46.6 | 48.3 |
| MnxF | 43.1 | 37.3 | 43.3 |
| MnxG | 68.6 | 69.7 | 65.4 |
*, Present only in strains PL-12 and SG-1; **, present only in strain SG-1.
The predicted amino acid sequence of MnxG contains the four consensus Cu-binding regions found in all MCOs (46, 55) (labeled A to D in Fig. 3). A fifth putative consensus Cu-binding region near the C terminus of the protein (Fig. 3 region F) (15, 55) is conserved among the three Bacillus spp., a finding consistent with a critical functional role for those amino acids. Another prominent region of conservation occurs at the C terminus of MnxF (Fig. 3a region E), where 12 of 14 amino acids are identical. Intriguingly, this stretch includes a sequence that resembles an MCO Cu-binding region (Fig. 3b, region E). MnxG also has at least seven homologues of MCO Cu-coordinating amino acids in regions that are less similar to the consensus MCO Cu-binding motifs (Fig. 3b, 1 to 4). Based on the homology of MCO Cu-binding regions, the extra Cu-binding amino acids found in MnxF and MnxG are predicted to be ligands at all three types of Cu sites present in MCOs (Fig. 3b, regions E, F, and 1 to 4).
Partial purification of the Mn(II) oxidase from Bacillus sp. strain PL-12.
We attempted to purify the Mn(II) oxidase from strain PL-12. The Mn(II) oxidase is insoluble, so we tested a variety of solubilizing detergents (deoxycholate, Triton X-100, CHAPS, Tween 20, NP-40, SDS, etc.) at various concentrations and conditions (NaOH, NaCl, glycerol, DTT, and EDTA). Only SDS solubilized Mn(II)-oxidizing activity, with high concentrations (up to 2%) being most effective. The Mn(II) oxidase remained active in the presence of SDS, a characteristic observed with some other MCOs (10) and Mn(II) oxidases (13, 35). Efforts to remove the SDS or exchange it for another detergent and maintain activity were unsuccessful, thereby limiting purification options.
The Mn(II)-oxidizing activity from PL-12 is resistant to many proteases in solution. Of the proteases tested (pronase, trypsin, Glu-C, Arg-C, Asp-N, and Lys-C), only pronase was able to disrupt Mn(II)-oxidizing activity as detected by the in-gel assay. We used this trypsin resistance as a purification step. After digestion of crude exosporium extract with trypsin, Mn(II)-oxidizing activity was unaffected or in some cases even enhanced, whereas nearly all other exosporium proteins were digested (Fig. 4). After solubilization and size-exclusion chromatography of the trypsin-digested exosporium, the specific activity increased >50-fold from the solubilized crude exosporium (2 nmol of MnO2 equivalents h−1 A750−1) to the most pure fraction (119 nmol MnO2 equivalents h−1 A750−1) (we report specific activity per A750 rather than per mg of protein because the protein concentration in the partially purified fraction was below our detection limits [see Materials and Methods]). In this partially purified fraction, the Coomassie blue band corresponding to the Mn(II) oxidase is faint, and several other bands are still present (Fig. 4).
FIG. 4.
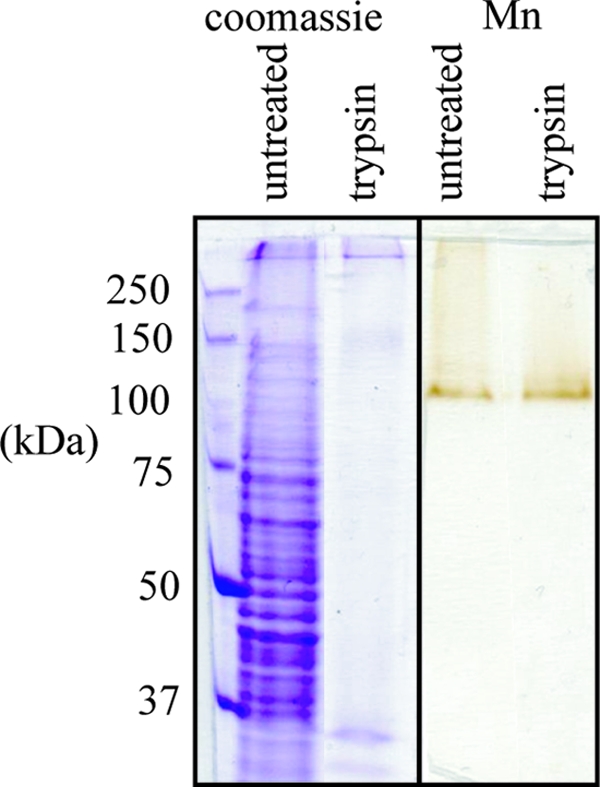
SDS-PAGE of PL-12 exosporium stained with Coomassie blue (left) and in-gel Mn(II) oxidation activity assay (right). Digestion with trypsin degrades most exosporium proteins, but Mn(II)-oxidizing activity is not inhibited.
MS/MS identification of the Mn(II) oxidase.
We analyzed the SDS-PAGE Mn oxide bands of Bacillus spp. strains PL-12, MB-7, and SG-1 by in-gel tryptic digestion, followed by nanoscale LC MS with MS/MS analysis. Although no pertinent peptides were recovered from the crude PL-12 exosporium, five distinct MnxG peptides were repeatedly detected from the partially purified PL-12 Mn(II) oxidase (see Fig. S1 in the supplemental material and Table 2). Four different MnxG peptides and one MnxF peptide (the C-terminal tryptic peptide) were consistently recovered from the Mn oxide band from the crude exosporium of strain MB-7 (Table 2). The MnxG peptides detected from PL-12 and MB-7 cluster toward the C terminus of the predicted protein sequence (Fig. 3a). No other Bacillus proteins were detected in the Mn oxide band of PL-12 or MB-7. A higher-molecular-mass band (∼140 kDa) often present in the in-gel Mn(II) oxidation assay of strain MB-7 (Fig. 1) and occasionally PL-12 (data not shown) did not yield any detectable peptides, and no peptides were recovered from the SG-1 Mn oxide band.
TABLE 2.
Peptides identified by MS/MS analysis of the Mn(II) oxidase
| Strain | Protein | Peptide sequence | Amino acid position |
|---|---|---|---|
| PL-12 | MnxG | AITGENVILR | 1011-1020 |
| MnxG | SFGTFGAFVAESR | 986-998 | |
| MnxG | EFVLLLHDGVR | 1024-1034 | |
| MnxG | MPHILDGDAFQLVTRa | 508-522 | |
| MnxG | APRPPLGIV | 402-410 | |
| MB-7 | MnxG | EFVLVMYDGAR | 1024-1034 |
| MnxG | ANQGDVVEINLTSR | 884-897 | |
| MnxG | ISLHAQLLDYDVK | 927-939 | |
| MnxG | SLGTFGAFIAEPK | 986-998 | |
| MnxF | IPELRDDF | 95-102 |
Only the five C-terminal residues show unambiguous MS/MS sequence for this peptide.
The overall coverage of the MnxG protein sequence with peptides identified by MS/MS was low (Fig. 3a), and there was a high incidence of peptides from trypsin autolysis (trypsin digesting itself) and nonspecific cleavages (having a tryptic cleavage at one end and a nonspecific cleavage at the other). A total of 74 MS/MS spectra obtained from strain PL-12 were de novo sequenced, and 35 spectra were of sufficient data quality for interpretation. Thirty of these peptides were from trypsin, and only five spectra were from MnxG. Two of five MnxG peptides had nonspecific cleavages (L.EFVLLLHDGVR and K.APRPPLGIV), as did one of the MB-7 peptides (Table 2).
DISCUSSION
MCOs are a large family of enzymes with over 500 putative homologs in all three domains of life. Although all MCOs couple the oxidation of substrate to the reduction of O2 to H2O, their substrates (organics versus metals) and specificity (narrow versus broad) vary widely, and their cellular roles are functionally diverse, ranging from trace metal uptake and homeostasis to lignin degradation and antibiotic biosynthesis. MCOs have long been known to be involved in bacterial Mn(II) oxidation, a process of broad environmental importance, but unraveling their role in bacterial Mn(II) oxidation has proven enigmatic. Here we present the first evidence that a bacterial MCO directly catalyzes Mn(II) oxidation; MS/MS analysis of SDS-PAGE purified Mn(II) oxidase from the spores of two Bacillus spp. identified nine different peptides unique to the MCO MnxG and one peptide unique to the small hydrophobic protein MnxF. These peptides were repeatedly detected through multiple analyses, and no other Bacillus peptides were found in the Mn oxide bands from these two strains, indicating that MnxG is the dominant protein in the gel at the site of Mn oxide formation and therefore is the direct catalyst of Mn(II) oxidation in the exosporium. The detection of one MnxF peptide in the MB-7 Mn oxide band from strain MB-7 suggests that this small hydrophobic protein is associated with the Mn(II) oxidase MnxG. Although the nature of this protein-protein interaction is unknown, the presence of a putative copper-binding motif in MnxF suggests that it may be involved either directly in the Mn oxidation reaction or in the delivery of Cu to the MCO as in other Cu proteins (34). Alternatively, the hydrophobic nature of MnxF may reflect a structural role in the spore coat, perhaps in anchoring the Mn(II) oxidase to the exosporium.
MS/MS peptide coverage of the MnxG sequence was low and clustered to the C terminus of the protein (Fig. 3a), perhaps suggesting that the N terminus is either not translated (i.e., an internal start codon is used), posttranslationally modified, or resistant to trypsin digestion. The lack of coverage of MnxG can likely be explained at least in part by the low abundance of the Mn(II) oxidase; there is no Coomassie blue-stainable band corresponding to the Mn oxide band in crude cell extracts, and it is only barely visible in the partially purified fraction. The high incidence of nonspecific cleavages and trypsin autolysis peaks in the MS/MS spectra are also consistent with a low concentration of MnxG protein. These observations, including the absence of a clear Coomassie blue stainable band, suggest that there was at most 10 to 20 ng (ca. 100 to 200 fmol) of MnxG protein present in the gel band. Another possibility is that MnxG is resistant to tryptic digestion; the relatively high occurrence of nonspecific cleavages that we observed suggests that peptides were produced with low efficiency. Although there are many tryptic cleavage sites (arginine or lysine) in MnxG, tryptic digestion of crude exosporium in solution does not inhibit Mn(II)-oxidizing activity. The clustering of peptides detected by MS/MS at the C terminus may indicate that these sites are more accessible, whereas others are buried within the protein. Protease resistance has been observed in other exosporium proteins of other Bacillus spores (23).
No peptides were detected from the Mn oxide band of strain SG-1. This is most likely due to extremely low levels of the Mn(II) oxidase in the gel as a result of its enormous apparent molecular mass, which limits its entry into the SDS-PAGE resolving gel. The larger apparent size of the SG-1 Mn(II) oxidase cannot be accounted for by the size of the predicted protein product of mnxG, which is nearly identical to that of strains PL-12 and MB-7 (138 kDa). Other intrinsic properties of MnxG that might influence the migration by SDS-PAGE, such as isoelectric point and hydrophobicity, are very similar among the three strains (as calculated by amino acid sequence [data not shown]) and thus not expected to be determining factors. The SG-1 Mn(II) oxidase may occur as a high-molecular-mass complex because of the association of additional subunits and/or proteins or because of differences in the overall structure of the exosporium, the outermost layer of the spore coat in which the Mn(II) oxidase is located (12). TEM shows that these three strains have prominent differences in the extent of exosporium that is present: PL-12 has a large exosporium, SG-1's is less prominent, and MB-7's is not detectable (Fig. 2). Differences in exosporium structure such as extent of protein-protein cross-linking or glycosylation could explain the varied migration of the Mn(II) oxidase through SDS-PAGE. Little is known about the composition, structure, or function of the exosporium in diverse Bacillus species, and yet, as the site of Mn oxide deposition and interface between spore and environment, it undoubtedly plays important roles in the ecology and biogeochemical impact of spores.
Taken together with our previous findings that MnxG is required to oxidize Mn(II) to Mn(III) and Mn(III) to Mn(IV) (57), our results indicate that MnxG directly catalyzes Mn(II) to Mn(IV) via a Mn(III) intermediate. MnxG is the first MCO demonstrated to directly catalyze two sequential one-electron oxidations of one substrate molecule. Well-characterized MCOs such as Fet3p, ascorbate oxidase, laccase, and human ceruloplasmin (hCp) oxidize each substrate by a single electron that is accepted by the enzyme at the type 1 Cu site. Electrons are then passed to the trinuclear cluster, consisting of one type 2 and two type 3 Cu sites, where four electrons (from four substrate molecules) reduce O2 to 2H2O (46). The two-electron oxidation reaction catalyzed by MnxG raises interesting mechanistic questions. How does MnxG catalyze two energetically distinct oxidation reactions with products and reactants that demand different ligand chemistry? Does each oxidation step occur at the same site, or is Mn(III) transferred to a different active site prior to oxidation to Mn(IV)? Does MnxG contain extra redox active Cu cofactors to handle two energetically distinct oxidations?
The MCO ferroxidases Fet3p and hCp may provide some insights into the mechanism of Mn oxidation. Fe(II) is oxidized to Fe(III), and the redox potential of the oxidation site is modulated via the relative affinity of the protein for Fe(II) and Fe(III) (36). After oxidation, Fe(III) is translocated to a “holding site,” where it is then donated to transferrin (hCp) or Ftr1p (Fet3p) (3, 26, 31, 56). Such a translocation may occur in the Mn(II) oxidase if the Mn(II) and Mn(III) oxidation sites are distinct, but how that might relate to electron transfer is unclear. Ferroxidases exhibit high specificity for Fe that has been attributed to a substrate-binding pocket (50) and, of the three tested to date (P. aeruginosa MCO, CueO, and Fet3p), none are able to oxidize Mn(II) (21, 24). In contrast, laccases lack a substrate-binding pocket, exhibiting broad substrate specificity that correlates with the substrate's oxidation potential (46). Fungal laccases are capable of oxidizing Mn(II) to Mn(III) (44), in some cases producing Mn(IV) oxide (29), and yet it is currently unclear whether this Mn(IV) is produced enzymatically or by the disproportionation of Mn(III). Thus, MnxG is unique among MCOs in its ability to catalyze the oxidation of Mn(II) to Mn(IV). The only other MCO thought to directly catalyze multiple-electron oxidations of substrate is phenoxazinone synthase, which catalyzes the biosynthesis of actinomycin D via a series of three two-electron oxidations (2). However, the enzymatic mechanism of this multielectron transfer is not understood.
The MnxF/G proteins contain a unique set of Cu-binding regions not present in any characterized MCOs but most similar to hCp (Fig. 3). Homologues of histidines within these regions are required for activity in other MCOs (50), and structural data have confirmed their role in binding Cu (39, 50). Based on sequence homology, the extra Cu-binding ligands of MnxF/G (Fig. 3, regions E, F, and 1 to 4) are predicted to coordinate Cu at all three types of Cu sites. A tally of these putative extra Cu ligands yields 8 type 1, 6 type 2, and 10 type 3 ligands, whereas the typical MCO (with four Cu atoms) has 4 type 1, 2 type 2, and 6 type 3 ligands. It is unclear whether all of these predicted ligands actually bind Cu. Additional Cu ligands that have not yet been detected could lie elsewhere in the MnxG amino acid sequence. Still, an intriguing possibility is that these putative extra Cu ligands are evidence of extra Cu cofactors that are required for the unique two-electron oxidation catalyzed by MnxG. hCp is the only well-characterized MCO known to contain more than four Cu atoms; it has two extra Cu atoms that are both type 1. One is not redox active and therefore catalytically irrelevant (27), and the function of the other type 1 Cu, although it is redox active, is unclear. Other MCOs with extra Cu atoms include CueO (E. coli), where the extra Cu is involved in regulation (40), and phenoxazinone synthase, where the extra Cu is thought to stabilize quaternary structure (45). These cases are distinguished from MnxG because the Cu atoms are coordinated by histidines and methionines that are not homologous to the consensus MCO Cu-binding regions.
Our results identify an enzyme that drives the oxidative segment of the Mn cycle, a biogeochemical process with important environmental consequences. MnxG's unusual complement of putative Cu ligands and the two-electron reaction that it catalyzes suggest that it is a novel MCO that may provide insights into the mechanism and evolution of this important class of enzymes. Determination of the number of Cu atoms bound by MnxG and elucidation of their role in the biochemical mechanism of Mn oxidation awaits purification of active Mn(II) oxidase. The limited abundance and solubility of MnxG make purification of the native enzyme from Bacillus spores difficult, and thus far efforts to express MnxG in E. coli have failed to yield an active Mn(II) oxidase. The myriad obstacles to heterologous expression are well documented; however, many of these difficulties might be overcome by expression in a more closely related host, such as another Bacillus species.
Finally, another major enigma clouding Mn(II) oxidation by marine Bacillus spores is the function that this process serves for the spore. Mn is required for sporulation in Bacillus species (including those not known to oxidize Mn) (22), but a connection between this requirement and Mn oxidation has not been made. Bacterial Mn(II) oxidation has also been suggested as a strategy for accessing refractory organic carbon (48). Other possible functions include storage of an oxidant and protection from predation, ionizing radiation, or reactive oxygen species (54). Mn generally plays an important role in biology in protection against ionizing radiation and/or reactive oxygen species (9, 19, 22), and MCOs have been implicated in melanization for UV protection in the Bacillus subtilis spore coat (20). Whatever the function of Mn(II) oxidation, it is clear that Bacillus spores should be considered reactive catalysts of biogeochemical processes rather than merely dormant/inert structures.
Supplementary Material
Acknowledgments
We thank members of the Tebo laboratory for helpful critical review of the manuscript, Dianne Moyles of T. J. Beveridge's laboratory, and Chris Francis for the preparation of spores for electron microscopy.
This research was supported by grants to B.M.T. from UCSD's Superfund Basic Research Program grant (NIEHS ES10337), NSF Ocean Sciences (OCE-0352081/0635493), and the NSF CRAEMS program (NSF CHE-0089208). G.J.D. was supported in part by a NSF graduate research fellowship. T.J.B.'s research is supported by a US-DOE-NABIR grant. The electron microscopy was performed in the NSERC Guelph Regional Integrated Imaging Facility, which is partially supported through an NSERC Major Facilities Access grant to T.J.B.
Footnotes
Published ahead of print on 28 December 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, L. F., and W. C. Ghiorse. 1987. Characterization of an extracellular Mn2+-oxidizing activity and isolation of Mn2+-oxidizing protein from Leptothrix discophora SS-1. J. Bacteriol. 169:1279-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry, C. E., P. G. Nayar, and T. P. Begley. 1989. Phenoxazinone synthase: mechanism for the formation of the phenoxazinone chromophore of actinomycin. Biochemistry 28:6323-6333. [DOI] [PubMed] [Google Scholar]
- 3.Bonaccorsi di Patti, M. C., S. Pascarella, D. Catalucci, and L. Calabrese. 1999. Homology modeling of the multicopper oxidase Fet3 gives new insights in the mechanism of iron transport in yeast. Protein Eng. 12:895-897. [DOI] [PubMed] [Google Scholar]
- 4.Brouwers, G.-J., J. P. M. de Vrind, P. L. A. M. Corstjens, P. Cornelis, C. Baysse, and E. W. de Vrind-de Jong. 1999. cumA, a gene encoding a multicopper oxidase, is involved in Mn2+-oxidation in Pseudomonas putida GB-1. Appl. Environ. Microbiol. 65:1762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouwers, G.-J., E. Vijgenboom, P. L. A. M. Corstjens, J. P. M. de Vrind, and E. W. de Vrind-de Jong. 2000. Bacterial Mn2+ oxidizing systems and multicopper oxidases: an overview of mechanisms and functions. Geomicrobiol. J. 17:1-24. [Google Scholar]
- 6.Claus, H. 2003. Laccases and their occurrence in prokaryotes. Arch. Microbiol. 179:145-149. [DOI] [PubMed] [Google Scholar]
- 7.Corstjens, P. L. A. M., J. P. M. de Vrind, T. Goosen, and E. W. de Vrind-de Jong. 1997. Identification and molecular analysis of the Leptothrix discophora SS-1 mofA gene, a gene putatively encoding a manganese-oxidizing protein with copper domains. Geomicrobiol. J. 14:91-108. [Google Scholar]
- 8.Cowen, J. P., G. J. Massoth, and E. T. Baker. 1986. Bacterial scavenging of Mn and Fe in a mid- to far-field hydrothermal particle plume. Nature 322:169-171. [Google Scholar]
- 9.Daly, M. J., E. K. Gaidamakova, V. Y. Matrosova, A. Vasilenko, M. Zhai, A. Venkateswaran, M. Hess, M. V. Omelchenko, H. M. Kostrandarithes, K. S. Makarova, L. P. Wackett, J. K. Fredrickson, and D. Ghosal. 2004. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306:1025-1028. [DOI] [PubMed] [Google Scholar]
- 10.Diamantidis, G., A. Effosse, P. Potier, and R. Bally. 2000. Purification and characterization of the first bacterial laccase in the rhizospheric bacterium Azospirillum lipoferum. Soil Biol. Biochem. 32:919-927. [Google Scholar]
- 11.Dick, G. J., Y. E. Lee, and B. M. Tebo. 2006. Manganese(II)-oxidizing Bacillus spores in Guaymas Basin hydrothermal sediments and plumes. Appl. Environ. Microbiol. 72:3184-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis, C. A., K. L. Casciotti, and B. M. Tebo. 2002. Localization of Mn(II)-oxidizing activity and the putative multicopper oxidase, MnxG, to the exosporium of the marine Bacillus sp. strain SG-1. Arch. Microbiol. 178:450-456. [DOI] [PubMed] [Google Scholar]
- 13.Francis, C. A., E.-M. Co, and B. M. Tebo. 2001. Enzymatic manganese(II) oxidation by a marine α-proteobacterium. Appl. Environ. Microbiol. 67:4024-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francis, C. A., and B. M. Tebo. 2002. Enzymatic manganese(II) oxidation by metabolically-dormant spores of diverse Bacillus species. Appl. Environ. Microbiol. 68:874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis, C. A., and B. M. Tebo. 1999. Marine Bacillus spores as catalysts for the oxidative precipitation and sorption of metals. J. Mol. Microbiol. Biotechnol. 1:71-78. [PubMed] [Google Scholar]
- 16.Grass, G., K. Thakali, P. E. Klebba, D. Thieme, A. Muller, G. F. Wildner, and C. Rensing. 2004. Linkage between catecholate siderophores and the multicopper oxidase CueO in Escherichia coli. J. Bacteriol. 186:5826-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvey, J. W., and C. C. Fuller. 1998. Effect of enhanced manganese oxidation in the hyporheic zone on basin-scale geochemical mass balance. Water Resource Res. 34:623-636. [Google Scholar]
- 18.Harwood, C. R., and A. R. Archibald. 1990. Growth, maintenance, and general techniques, p. 1-26. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. Wiley, Chichester, United Kingdom.
- 19.Horsburgh, M. J., S. J. Wharton, M. Karavolos, and S. J. Foster. 2002. Manganese: elemental defense for a life with oxygen? Trends Microbiol. 10:496-501. [DOI] [PubMed] [Google Scholar]
- 20.Hullo, M. F., I. Moszer, A. Danchin, and I. Martin-Verstraete. 2001. CotA of Bacillus subtilis is a copper-dependent laccase. J. Bacteriol. 183:5426-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huston, W. M., M. P. Jennings, and A. G. McEwan. 2002. The multicopper oxidase of Pseudomonas aeruginosa is a ferroxidase with a central role in iron acquisition. Mol. Microbiol. 45:1741-1750. [DOI] [PubMed] [Google Scholar]
- 22.Jakubovics, N. S., and H. F. Jenkinson. 2001. Out of the iron age: new insights into the critical role of manganese homeostasis in bacteria. Microbiology 147:1709-1718. [DOI] [PubMed] [Google Scholar]
- 22a.Johnson, H. A., and B. M. Tebo. 2008. In vitro studies indicate a quinone is involved in bacterial Mn(II) oxidation. Arch. Microbiol. 189:59-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keim, P., M. Mock, J. Young, and T. M. Koehler. 2006. The International Bacillus anthracis, B. cereus, and B. thuringiensis Conference: “Bacillus-ACT05.” J. Bacteriol. 188:3433-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, C., W. W. Lorenz, J. T. Hoopes, and J. F. D. Dean. 2001. Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yakK gene. J. Bacteriol. 183:4866-4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krumbein, W. E., and H. J. Altmann. 1973. A new method for the detection and enumeration of manganese oxidizing and reducing microorganisms. Helgol. Wiss. Meeresunters 25:347-356. [Google Scholar]
- 26.Kwok, E. Y., S. Severance, and D. J. Kosman. 2006. Evidence for iron channeling in the Fet3p-Ftr1p high-affinity iron uptake complex in the yeast plasma membrane. Biochemistry 45:6317-6327. [DOI] [PubMed] [Google Scholar]
- 27.Machonkin, T. E., and E. I. Solomon. 2000. The thermodynamics, kinetics, and molecular mechanism of intramolecular electron transfer in human ceruloplasmin. J. Am. Chem. Soc. 122:12547-12560. [Google Scholar]
- 28.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Miyata, N., Y. Tani, K. Maruo, H. Tsuno, M. Sakata, and K. Iwahori. 2006. Manganese(IV) oxide production by Acremonium sp. strain KR21-2 and extracellular Mn(II) oxidase activity. Appl. Environ. Microbiol. 72:6467-6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moffett, J., and J. Ho. 1996. Oxidation of cobalt and manganese in seawater via a common microbially catalyzed pathway. Geochim. Cosmochim. Acta 60:3415-3424. [Google Scholar]
- 31.Murphy, M. E. P., P. F. Lindley, and E. T. Adman. 1997. Structural comparison of cupredoxin domains: domain recycling to construct proteins with novel functions. Protein Sci. 6:761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nealson, K. H., B. M. Tebo, and R. A. Rosson. 1988. Occurrence and mechanisms of microbial oxidation of manganese. Adv. Appl. Microbiol. 33:279-318. [Google Scholar]
- 33.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Halloran, T. V., and V. Cizewski Culotta. 2000. Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 275:25057-25060. [DOI] [PubMed] [Google Scholar]
- 35.Okazaki, M., T. Sugita, M. Shimizu, Y. Ohode, K. Iwamoto, E. W. de Vrind-de Jong, J. P. M. de Vrind, and P. L. A. M. Corstjens. 1997. Partial purification and characterization of manganese-oxidizing factors of Pseudomonas fluorescens GB-1. Appl. Environ. Microbiol. 63:4793-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quintanar, L., M. Gebhard, T. P. Wang, D. J. Kosman, and E. I. Solomon. 2004. Ferrous binding to the multicopper oxidases Saccharomyces cerevisiae and human ceruloplasmin: contributions to ferroxidase activity. J. Am. Chem. Soc. 126:6579-6589. [DOI] [PubMed] [Google Scholar]
- 37.Rensing, C., and G. Grass. 2003. Escherichia coli mechanisms of copper resistance in a changing environment. FEMS Microbiol. Rev. 27:197-213. [DOI] [PubMed] [Google Scholar]
- 38.Ridge, J. P., M. Lin, E. I. Larsen, M. Fegan, A. G. McEwan, and L. I. Sly. 2007. A multicopper oxidase is essential for manganese oxidation and laccase-like activity in Pedomicrobium sp. ACM 3067. Environ. Microbiol. 9:944-953. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, S. A., A. Weichsel, G. Grass, K. Thakali, J. T. Hazzard, G. Tollin, C. Rensing, and W. R. Montfort. 2002. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:2766-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts, S. A., G. F. Wildner, G. Grass, A. Weichsel, A. Ambrus, C. Rensing, and W. R. Montfort. 2003. A labile regulatory copper ion lies near the T1 copper site in the multicopper oxidase CueO. J. Biol. Chem. 279:31958-31963. [DOI] [PubMed] [Google Scholar]
- 41.Reference deleted.
- 42.Rosson, R. A., and K. H. Nealson. 1982. Manganese binding and oxidation by spores of a marine bacillus. J. Bacteriol. 151:1027-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schevenko, A., S. Sunyaev, A. Loboda, A. Schevenko, P. Bork, W. Ens, and K. G. Standing. 2001. Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 73:1917-1926. [DOI] [PubMed] [Google Scholar]
- 44.Schlosser, D., and C. Höfer. 2002. Laccase-catalyzed oxidation of Mn2+ in the presence of natural Mn3+ chelators as a novel source of extracellular H2O2 production and its impact on manganese peroxidase. Appl. Environ. Microbiol. 68:3514-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, A. W., A. Camara-Artigas, M. Wang, J. P. Allen, and W. A. Franciso. 2006. Structure of phenoxazinone synthase from Streptomyces antibioticus reveals a new type 2 copper center. Biochemistry 45:4378. [DOI] [PubMed] [Google Scholar]
- 46.Solomon, E. I., U. M. Sundaram, and T. E. Machonkin. 1996. Multicopper oxidases and oxygenases. Chem. Rev. 96:2563-2605. [DOI] [PubMed] [Google Scholar]
- 47.Sterjiades, R., J. F. D. Dean, and K.-E. L. Eriksson. 1992. Laccase from sycamore maple (Acer pseudoplatanus) olignols. Plant Physiol. 99:1162-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sunda, W. G., and D. J. Kieber. 1994. Oxidation of humic substances by manganese oxides yields low-molecular-weight organic substrates. Nature 367:62-64. [Google Scholar]
- 49.Tang, W. H., B. R. Halpern, I. V. Shilo, S. L. Seymour, S. P. Keating, A. Loboda, A. A. Patel, D. A. Schaeffer, and L. M. Nuwaysir. 2005. Discovering known and unanticipated protein modifications using MS/MS database searching. Anal. Chem. 77:3931-3946. [DOI] [PubMed] [Google Scholar]
- 50.Taylor, A. B., C. S. Stoj, L. Ziegler, D. J. Kosman, and P. J. Hart. 2005. The copper-iron connection in biology: structure of the metallo-oxidase Fet3p. Proc. Natl. Acad. Sci. USA 102:15459-15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tebo, B. M. 1991. Manganese(II) oxidation in the suboxic zone of the Black Sea. Deep-Sea Res. 38:S883-S905. [Google Scholar]
- 52.Tebo, B. M., J. R. Bargar, B. G. Clement, G. J. Dick, K. J. Murray, D. Parker, R. Verity, and S. M. Webb. 2004. Biogenic manganese oxides: properties and mechanisms of formation. Annu. Rev. Earth Planetary Sci. 32:287-328. [Google Scholar]
- 53.Tebo, B. M., B. G. Clement, and G. J. Dick. 2007. Biotransformations of manganese, p. 1223-1238. In C. J. Hurst, R. L. Crawford, J. L. Garland, D. A. Lipson, A. L. Mills, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 3rd ed. ASM Press, Washington, DC.
- 54.Tebo, B. M., W. C. Ghiorse, L. G. van Waasbergen, P. L. Siering, and R. Caspi. 1997. Bacterially-mediated mineral formation: insights into manganese(II) oxidation from molecular genetic and biochemical studies, p. 225-266. In J. F. Banfield and K. H. Nealson (ed.), Geomicrobiology: interactions between microbes and minerals, vol. 35. Mineralogical Society of America, Washington, DC. [Google Scholar]
- 55.van Waasbergen, L. G., M. Hildebrand, and B. M. Tebo. 1996. Identification and characterization of a gene cluster involved in manganese oxidation by spores of the marine Bacillus sp. strain SG-1. J. Bacteriol. 178:3517-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, T. P., L. Quintanar, S. Severance, E. I. Solomon, and D. J. Kosman. 2003. Targeted suppression of the ferroxidase and iron trafficking activities of the multicopper oxidase Fet3p from Saccharomyces cerevisiae. J. Biol. Inorg. Chem. 8:611-620. [DOI] [PubMed] [Google Scholar]
- 57.Webb, S. M., G. J. Dick, J. R. Bargar, and B. M. Tebo. 2005. Evidence for the presence of Mn(III) intermediates in the bacterial oxidation of Mn(II). Proc. Natl. Acad. Sci. USA 102:5558-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.