WHY IS ENUMERATION OF SALMONELLA NECESSARY?
Quantitative microbial risk assessment (QMRA) is an important approach for food safety in which risk and factors that influence food safety are identified. The goal is to provide an estimate of the level of illness that a pathogen can cause in a given population (13). For QMRA, there is a need for microbiological methods that generate quantitative data. Furthermore, the sample preparation methods preceding the analytical method itself (e.g., PCR) need to be able to produce quantitative results.
Salmonellosis is one of the most important food-borne disease and causes substantial medical and economic burdens worldwide (9, 30). Food is the main source of infection by Salmonella in humans. Beside eggs, egg products, and poultry meat, pork is one of the most important sources of human salmonellosis (4, 17). Because of this, a number of actions have been taken to reduce the prevalence of Salmonella serovars with public health significance in food-producing animals (3), including a QMRA study of Salmonella in slaughter and breeder pigs (10). QMRA is still hampered by the lack of quantitative data, and often assumptions that generate high degrees of uncertainty have to be included. The generation of appropriate data with high sensitivity is a challenge for microbiologists since currently used bacteriological quantitation methodologies are laborious.
In the past, it was shown that the severity of salmonellosis and the percentage of infected humans after consumption of food are associated with the level of contamination (dose-response relationship [12]). Moreover, the infection dose of Salmonella depends on the food item itself. Salmonellae can enter the food chain at every stage, and the consequences for humans after consumption of the contaminated end product depend on the food-processing conditions. For example, a well-known source of contamination is the lairage environment of slaughterhouses for incoming nonaffected animals (6). Later, salmonellae can multiply to harmful levels due to inappropriate storage conditions. Generally, Salmonella does not grow at temperatures below 6°C for as long as 15 days on chicken meat, while significant growth has been reported at 8°C (27). However, some other reports have indicated that growth at 2 to 7°C might occur (8). Furthermore, quantitative salmonella data for foods associated with severe outbreaks have shown that the type of food plays a major role in the severity of illness. Salmonellae in fatty foods may have an advantage during passage through the acidic environment of the stomach to the intestine, where the cells become invasive regardless of the damage caused by the acids.
Very low numbers of Salmonella cells are typically found in food, feed, and environmental samples (6, 7, 11, 29). Carcasses may be contaminated during transport or slaughtering, resulting in low levels and uneven distribution. However, such contamination may be fatal because of the possibility of multiplication of the cells on the meat, leading to a high risk for consumers. Consequently, to identify critical contamination points and to provide risk modelers with quantitative data for each processing chain, cost-effective methods that can also enumerate low levels of Salmonella are needed.
TRADITIONAL QUANTITATION METHODS
Currently, nearly all quantitative data that have been generated for Salmonella were obtained by traditional bacteriological methods. In principle, there are two culture-based methods. The most-probable-number (MPN) test (5) is particularly useful for determination of low concentrations of Salmonella. Here, triplicate samples or five replicates are prepared from 10-fold serial dilutions. All samples are then tested by using the horizontal culture method (1). The ratio of positive results to negative results in relation to the concentration results in a MPN/g value. The MPN method assumes that bacteria are distributed randomly within the sample and are separated (not clustered together). The growth medium and incubation conditions have been chosen so that one viable cell multiplies and can be detected.
Higher levels of Salmonella (100 to 1,000 CFU/g or 100 to 1,000 CFU/ml) can be determined by direct isolation on selective agar, such as xylose lysine deoxycholate agar. However, depending on the matrix, high levels of background flora can disturb the growth of Salmonella and lead to failures in interpretation of colonies. Consequently, the method for confirming typical colonies is labor-intensive. In addition, selective media may inhibit the growth of stressed cells. Due to the low sensitivity, direct isolation has been combined with concentration of a sample (e.g., by filtration, centrifugation, or immunoconcentration), but even these strategies have limited efficacy (20).
A semiquantitative approach using modified semisolid Rappaport-Vassiliades (MSRV) agar plates has been shown to be useful (21). In this approach a serial dilution of a sample is plated on MSRV agar plates, and growth of salmonellae is recorded and confirmed after 24 and 48 h. It is then possible to obtain a semiquantitative estimate of the salmonella level that was present in the original, nonenriched sample.
The International Standard Organization (ISO) and the European Committee for Standardization have recently decided to include enumeration of Salmonella in their agenda under supervision of the Salmonella Community Reference Laboratory, Bilthoven, The Netherlands. Currently, a new ISO standard is being developed by the TC34/SC9 members. The document comprises enumeration of Salmonella by the MPN technique. An MPN protocol has been developed, as described by Fravalo et al. (15), and it will be validated. This protocol is based on miniaturization of the dilution, preenrichment, and selective enrichment steps in 12-well microwell plates. MSRV agar is used as the selective enrichment medium for detection of motile salmonellae. The mini-MSRV MPN method has the advantage compared to the conventional tube MPN method that a minimal amount of medium is needed in a compact format.
Direct counting might be also performed by immunofluorescence techniques, but these methods are not used yet in practice because of problems with the quality of the antibodies and the choice and linkage of the fluorochrome, etc. In addition, the technology is not sensitive enough for enumeration of Salmonella in food (31).
PCR-BASED QUANTITATION METHODS
Culture-based quantitation methods are both costly and lengthy, and therefore it is necessary to develop easier, high-throughput quantitative methods. PCR has been standardized in the last 5 years by ISO and is now used for food testing (26). The next generation of PCR, real-time PCR, offers the possibility of also estimating the number of bacteria. Quantitation is not based on the end point signal but rather is based on the exponential increase in the initial DNA amount with the number of PCR cycles performed (25). Serial dilution of known numbers of target copies can be used to set up a standard curve which is used to determine an unknown amount of DNA in a sample (absolute quantitation). The automation of DNA sample preparation methods and the real-time PCR setup itself are undoubtedly useful for generating a huge amount of quantitative data at a lower cost than culture methods.
PREENRICHMENT OR NO PREENRICHMENT PRIOR TO ANALYTICAL METHODS
Generally, to detect very low levels of Salmonella in food by molecular methods, the sample preparation step must include a significant time for preenrichment. The time that it takes for cells to multiply depends on various intrinsic and extrinsic factors. Cells can be damaged or stressed (e.g., by heat or cold), and consequently a prolonged adaptation time is necessary. High levels of background flora in the presence of small amounts of Salmonella might also present a challenge. For these reasons, elimination of the preenrichment step would improve the quantitation of salmonellae when the levels of contamination are low. A few studies have described direct quantitation in foods using real-time PCR (16, 38). However, the bottleneck is the use of small volumes of reagents in the PCR, which in turn limits the amount of DNA sample that can be added to the PCR mixture. Consequently, development of a sample preparation method that separates the target pathogen from an appropriate amount of the food sample and concentrates the DNA sample in the small volume that is added to the PCR mixture is a significant challenge.
ONE-STEP OR TWO-STEP DNA ISOLATION PROTOCOLS
Samples for quantitative analyses without prior enrichment of target cells can be prepared by using a single-step approach or a two-step approach. Single-step DNA isolation protocols that start with the transfer of an amount of a foodstuff into a lysis buffer have to overcome the inhibitors present in the food sample and have to yield an amount of DNA resembling the amount of DNA present in the target cells per gram or milliliter of foodstuff. Dilution of the template could circumvent inhibition (18) but creates problems if the quantitation limit of the PCR is very close to the real amount of DNA present in a sample. Two-step extraction strategies separate the target cells from a food matrix prior to isolation of DNA from the capture matrix. Two-step cell extraction can be done using flotation (34, 37), paramagnetic beads (28, 32), or filtration (38).
Flotation is based on density gradient centrifugation. This procedure can separate biological particles and microorganisms with different buoyant densities because their densities are lower than that of the medium, which allows the cells to float. The advantage of using flotation instead of buoyant density centrifugation is that it does not require extra washing steps and the sample can be withdrawn directly from the surface. This method was used to quantify Yersinia enterocolitica in pork (34) and Campylobacter in a chicken rinse (35), as well as to separate living cells from DNA of dead cells (34). Recently, a quantitative multiplex real-time PCR for Campylobacter and Salmonella based on a rather lengthy flotation procedure was described (37). However, there are problems with variation and lack of robustness when this sample preparation method is used, especially when a large number of samples must be processed simultaneously. This method is also too time-consuming to suit the needs of a routine analysis method.
Wolffs et al. (38) developed a two-step filtration protocol to efficiently concentrate salmonellae from chicken carcass rinses and sprouted mung beans. The recovery rate was approximately 100% with a low standard deviation, but it was shown that the filter types and modifications in the conditions used to recover cells from the filters have an important impact on the recovery rate (38). The limit of quantitation was 750 ± 300 CFU per 100-ml sample. Often, commercial DNA extraction kits are also used. It has been shown that the type of kit used can dramatically influence the recovery rate (23). Consequently, for development of a quantification method several kits should be tested to determine their efficiencies for extraction of DNA from the matrix.
Due to the loss of target material during sample preparation and the small volumes analyzed, the limit of quantitation without any enrichment is not less than approximately 100 to 1,000 cells per gram or milliliter of sample. This limit is usually still too high, since, for instance, most samples taken throughout the meat production chain are contaminated with less than 100 salmonellae per g (see above).
To improve the quantitation limit, Josefsen et al. (22) developed a semiquantitative strategy to quantify low numbers of Campylobacter in chicken rinse samples. These workers showed that after a 12-h selective enrichment phase under standardized conditions, the initial amount of cells in a carcass rinse was inversely correlated with the threshold cycle (CT) value. Thus, a larger initial amount of target bacteria may result in a lower CT value. However, the precision of the method needs to be investigated further. Generally, this strategy might be also applicable to Salmonella using adapted enrichment media and times, followed by real-time PCR quantitation (Fig. 1). Thus, careful consideration should be given to the enrichment strategies used for Salmonella cells in combination with subsequent quantitative real-time PCR analysis. An optimal enrichment should inhibit the growth of background flora but simultaneously recover and multiply sublethally damaged Salmonella cells. An advantage of this strategy is also that dead Salmonella cells do not play a major role.
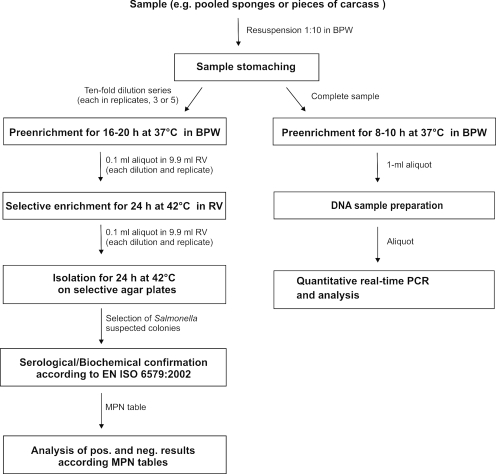
FIG. 1.
Comparison of the procedures of the MPN and real-time PCR-based methods for quantitation of low levels of salmonellae in food and environmental samples. The culture-based MPN method (left) starts with 10-fold serial dilutions of the homogenized sample at least in triplicate. The presence or absence of salmonellae is determined using ISO standard 6579:2002 (1). The pattern of positive and negative results is noted, and an MPN table is consulted for the MPN of organisms per unit of volume of the original sample. The (proposed) real-time PCR-based method (right) starts with a short preenrichment of the homogenized sample in buffered peptone water, followed by DNA sample preparation. The number of copies of DNA in an aliquot is determined using a validated standard curve. BPW, buffered peptone water; RV, Rappaport Vassiliadis medium.
Preliminary results for quantitation of Salmonella from pig carcasses from our laboratories (Federal Institute for Risk Assessment [BFR] and Technical University of Denmark) showed that it is possible to generate robust quantitative data at low contamination levels by this strategy. The data indicate that the real-time PCR-based quantitation method is even more precise than the MPN method. The MPN method can overestimate the number of artificially inoculated Salmonella cells in swab samples, whereas data obtained by using the suggested strategy quantitate cells close to the inoculation level (unpublished data). In addition, the sensitivity of the suggested method is 10 times higher than that of the MPN method because it does not start with a 1:10 dilution. However, the enrichment time has to be adapted to the status of the Salmonella cells usually expected in the specific food matrix investigated. Nevertheless, the precision of this strategy should be elucidated in more detail, and a positive quantitation control measuring the correct enrichment procedure is needed.
STANDARD CURVE SETUP
Absolute quantitation requires a standard curve with exact known amounts of the target. Generally, it is possible to define DNA genome equivalents based on measurement of the DNA (with a spectrophotometer or fluorometer) or to define CFU or cell equivalents based on plating and counting of Salmonella in a suspension. The DNA or cell equivalents used must be chosen with care because interpretation of the results might be different. Our studies have shown that the number of Salmonella DNA equivalents corresponds quite well with the number of CFU taken from the stationary phase. In contrast, cells taken from the exponential phase result in CT values that are 3 or 4 cycles less (BFR, unpublished data). Since different growth phases of Salmonella can lead to different CT values, it might be more objective for standard curve setup to use DNA genome equivalents instead of an indirect measurement based on the number of cells or CFU. However, the recovery of any treated sample before the analytical PCR must be taken into account. The recovery rate of the cells should be determined for each specific food item. In addition, the efficiency of the sample treatment for recovering Salmonella can differ in the presence and in the absence of a food matrix. To determine the recovery rate, replicates of the food item should be artificially contaminated with 10-fold serially diluted cell suspensions with concentrations ranging from approximately 107 to 1 cell per g of food. After the selected DNA purification method is used, an aliquot of the DNA is subjected to the PCR. In parallel, the same 10-fold serially diluted cell suspensions which were used for artificial contamination are used directly in the PCR. The amount of cells in the PCR mixture should be the same as the amount calculated before DNA purification. Consequently, CT values for the two sample preparations can be directly compared. The cell dilutions are defined in the run as standards to generate a standard curve. The numbers of CFU in the cell suspensions are determined by plating the suspensions on an appropriate agar medium, and CFU is used as units for the standard curve. The recovery rate is then defined as the ratio of the number of CFU for the DNA purified from the artificial contamination to the number of CFU for the cell suspension examined.
Another important aspect is how nonlinear areas of the standard curve should be handled. There are several different ways to handle the points that fall outside the linear range of amplification. Some studies extrapolate the standard curve to also include these points, while others divide the standard curve into linear and nonlinear areas (24).
FACTORS INFLUENCING THE ACCURACY OF QUANTITATION
It is important to be aware how precise or accurate a quantitation result is. Two main uncertainties can be expected when quantitative data are generated; the first uncertainty results from the sampling technique, and second results from the analytical method itself (including sample preparation). Heterogeneous distribution of the pathogen in or on the surface of a sample can lead to a false estimate of the pathogen load. Therefore, more accurate results might be obtained if rinses from carcasses or their surfaces are used for quantitation. However, rinses might not always be the most appropriate method. For example, plant seeds might be contaminated with Salmonella, and this can lead to internal colonization of the plant by the pathogen. After sterilization of a surface, salmonellae are still present, which demonstrates that thorough rinsing or washing is not an appropriate technique for separating the pathogen from the matrix (14).
Sampling of salmonellae on red meat carcasses in slaughterhouses is performed by using destructive or nondestructive methods which are described in the ISO 17604:2003 international standard (2). Abrasive sponges are often used with a minimum sampling area of 100 cm2 per site selected. The efficiency of recovery of bacteria from these sponges has been reported to be 82% ± 35% (16), but this value is influenced by whether swabbing is performed before or after the carcasses have hung in a cold room overnight, the actual areas swabbed, the time of sampling after slaughter, and the nature of the sponge material itself. It seems reasonable to recommend that bacterial determinations be done using at least three samples per carcass. Alternatively, it might be useful to investigate the use of pooled samples for this purpose.
DEAD-VIABLE STATUS AND STRESSED CELLS
Since nucleic acids and nonviable cells are detected by PCR, microbiologists are concerned with the significance of a PCR signal. In addition, PCR, which is based on DNA detection, is not able to discriminate between dead and viable cells. For Salmonella in meat we claim that dead cells or free nucleic acids do not play a major role. Studies have shown that DNA is rapidly degraded on meat (36). In addition, raw meat is a reasonable growing medium for Salmonella at ambient temperatures. Consequently, there is little likelihood that stressed cells on raw meat play a major role at temperatures higher than approximately 8°C. Nevertheless, at lower temperatures cells on meat might be stressed. Consequently, special processing conditions in the food chain must be considered. For example, in slaughterhouses in many countries destructive and swab (abrasive sponge) samples are routinely taken from carcasses 1 day after slaughtering and chilling. Experiments performed at the BFR laboratory have shown that chilling (4°C) can reduce the viability of salmonellae and consequently hamper the growth on selective media. As a result, if stressed cells are expected to be present in a sample matrix, the enrichment time and the type of enrichment medium used for sensitive quantitation must be taken into account when the quantitative method shown in Fig. 1 is used.
MPN VERSUS REAL-TIME PCR ENUMERATION: RECOMMENDATIONS
Table 1 summarizes a comparison of the two methods. Real-time PCR-based methods have major advantages compared to the MPN method. PCR can generate much more data in a shorter time, resulting in a higher degree of confidence in the data. The personnel workload is tremendously lower, and consequently the cost of analysis is lower. The MPN method has been used for many years and is familiar to most personnel. Because of this, currently an ISO standard document is being developed for the MPN method, which will be the basis for easier comparison of quantitative data between laboratories. Our experience tells us that modern methodologies are established faster in laboratories if standard documents, such as the document from ISO, are available. Quantitation by real-time PCR has already been established for levels higher than approximately 500 cells per g or 500 cells per ml. Now it is necessary to develop more sensitive methods and to generate data to initiate a standardization process for real-time PCR quantitation. Table 2 summarizes practical recommendations for sensitive Salmonella quantitation by real-time PCR which have been identified and should be considered in the future.
TABLE 1.
Comparison of the MPN method and real-time PCR for enumeration of salmonellae in a food sample
| Parameter | MPN method | Real-time PCR |
|---|---|---|
| Detected units | CFU | Nucleic acid (DNA), genome equivalents, can be correlated to CFU |
| Method | Based on horizontal detection method | Based on CT values of Salmonella DNA |
| Duration | 4-5 days | Approx 12 h |
| Approx cost (including personnel)a | Three-tube MPN (three dilutions), 500 €; five-tube MPN (three dilutions), 750 € | 300 € (three replicates) |
| Quantitation limit | Three-tube MPN (three dilutions), 3.0 MPN/g; five-tube MPN (three dilutions), 1.8 MPN/g | Approx 1 cell in 10 g |
| Precision (confidence limits) | For 1.8 MPN/g, 0.009 (low) to 6.8 (high) | No sufficient data yet available; ranges of 1 to 10, 10 to 100, and 100 to 1,000 cells have been reported |
| Automation possible | No | Yes |
| Standard curve setup necessary | No | Yes |
The cost for the personnel can vary between countries. The higher the cost for personnel, the higher the expected cost for the MPN or real-time PCR method. The calculation is based on the cost in a well-developed European country (e.g., German scale and charges of fees for medical services [http://www.e-bis.de/goae/defaultFrame.htm]).
TABLE 2.
Recommendations for real-time PCR-based quantitation of Salmonella in food and feed
| Recommendation no. | Recommendation |
|---|---|
| 1 | Enrichment is necessary if salmonellae that are present at low levels are to be enumerated (approx <500 cells per g or <500 cells per ml). The time of enrichment has to be adjusted to the expected status of the cells in the food or environmental matrix investigated (approx 8 h for meat samples). For enrichment, a nonselective broth (e.g., buffered peptone water) should be used. |
| 2 | The standard curve setup should consider the loss of nucleic acids due to sample preparation; i.e., samples for the standard curve should be processed in the same way as the test samples. The curve should consist of at least four 10-fold serially diluted data points in duplicate. An additional data point should be included if samples with low levels of contamination are investigated. |
| 3 | Available DNA polymerase enzymes and buffers can differ in robustness. The accuracy and the detection window can vary substantially, which should be considered. It has been shown that Tth polymerase seems to be more appropriate for quantitative PCR than Taq polymerase because of its robustness and accuracy.a |
| 4 | An internal amplification control should be included in the analytical real-time PCR assay.b |
| 5 | The linear range of the standard curve should not be extended below the points included in the standard curve or outside the linear range of amplification. As an alternative approach, the nonlinear part of the standard curve can be modeled.c |
Acknowledgments
The work was supported by the European Union-funded Integrated Project BIOTRACER (contract 036272) under the 6th RTD Framework.
Footnotes
Published ahead of print on 28 December 2007.
REFERENCES
- 1.Anonymous. 2002. Microbiology of food and animal feeding stuffs—horizontal method for the detection of Salmonella (EN ISO 6579:2002). International Organization for Standardization, Geneva, Switzerland.
- 2.Anonymous. 2003. Microbiology of food and animal feeding stuffs—carcass sampling for microbiological analysis (ISO 17604:2003). International Organization for Standardization, Geneva, Switzerland.
- 3.Anonymous. 2003. Regulation (EC) no. 2160/2003 of the European Parliament and of the Council of 17 November 2003 on the control of salmonella and other specified food-borne zoonotic agents. Off. J. Eur. Union L325-1-L325/14:12.12.2003. http://europa.eu.int/eur-lex/pri/en/oj/dat/2003/l_325/l_32520031212en00010015.pdf.
- 4.Berends, B. R., F. van Knapen, D. A. Mossel, S. A. Burt, and J. M. Snijders. 1998. Impact on human health of Salmonella spp. on pork in The Netherlands and the anticipated effects of some currently proposed control strategies. Int. J. Food. Microbiol. 44:219-229. [DOI] [PubMed] [Google Scholar]
- 5.Blodgett, R. February 2006, posting date. Most probable number from serial dilutions. In FDA's bacteriological analytical manual online, appendix 2. http://www.cfsan.fda.gov/∼ebam/bam-a2.html.
- 6.Boughton, C., J. Egan, G. Kelly, B. Markey, and N. Leonard. 2007. Quantitative examination of Salmonella spp. in the lairage environment of a pig abattoir. Foodborne Pathog. Dis. 4:26-32. [DOI] [PubMed] [Google Scholar]
- 7.Boughton, C., F. C. Leonard, J. Egan, G. Kelly, P. O'Mahony, B. K. Markey, and M. Griffin. 2004. Prevalence and number of Salmonella in Irish retail pork sausages. J. Food Prot. 67:1834-1839. [DOI] [PubMed] [Google Scholar]
- 8.D'Aoust, J. Y. 1991. Psychrotrophy and foodborne Salmonella. Int. J. Food Microbiol. 13:207-215. [DOI] [PubMed] [Google Scholar]
- 9.de Jong, B., and K. Ekdahl. 2006. The comparative burden of salmonellosis in the European Union member states, associated and candidate countries. BMC Public Health 6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.EFSA. 2006. Opinion of the scientific panel BIOHAZ related to “risk assessment and mitigation options of Salmonella in pig production.” EFSA J. 341:1-131. [Google Scholar]
- 11.Fegan, N., P. Vanderlinde, G. Higgs, and P. Desmarchelier. 2004. Quantification and prevalence of Salmonella in beef cattle presenting at slaughter. J. Appl. Microbiol. 97:892-898. [DOI] [PubMed] [Google Scholar]
- 12.Food and Agriculture Organization and World Health Organization. 2002. Risk assessments of Salmonella in eggs and broiler chickens. Food and Agriculture Organization and World Health Organization, Rome, Italy. ftp://ftp.fao.org/docrep/fao/005/y4392e/y4392e00.pdf.
- 13.Forsythe, S. J. 2002. The microbiological risk assessment of food. Blackwell Publishing, Oxford, United Kingdom.
- 14.Franz, E., A. A. Visser, A. D. Van Diepeningen, M. M. Klerks, A. J. Termorshuizen, and A. H. van Bruggen. 2007. Quantification of contamination of lettuce by GFP-expressing Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium. Food Microbiol. 24:106-112. [DOI] [PubMed] [Google Scholar]
- 15.Fravalo, P., Y. Hascoet, M. Le Fellic, S. Queguiner, J. Petton, and G. Salvat. 2003. Convenient method for rapid and quantitative assessment of Salmonella enterica contamination: the mini-MSRV MPN technique. J. Rapid Methods Automat. Microbiol. 11:81-88. [Google Scholar]
- 16.Guy, R. A., A. Kapoor, J. Holicka, D. Shepherd, and P. A. Horgen. 2006. A rapid molecular-based assay for direct quantification of viable bacteria in slaughterhouses. J. Food Prot. 69:1265-1272. [DOI] [PubMed] [Google Scholar]
- 17.Hald, T., D. Vose, H. C. Wegener, and T. Koupeev. 2004. A Bayesian approach to quantify the contribution of animal-food sources to human salmonellosis. Risk Anal. 24:255-269. [DOI] [PubMed] [Google Scholar]
- 18.Hein, I., A. Lehner, P. Rieck, K. Klein, E. Brandl, and M. Wagner. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoorfar, J., N. Cook, B. Malorny, P. Rådström, D. De Medici, A. Abdulmawjood, and P. Fach. 2003. Diagnostic PCR: making internal amplification control mandatory. J. Clin. Microbiol. 41:5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humbert, F. 2002. Review of the presentations on quantitative methods, p. 25-27. In M. Raes and A. M. Henken (ed.), Report on the sixth workshop organized by CRL-Salmonella, Bilthoven, 11 and 12 June 2001. RIVM, Bilthoven, The Netherlands.
- 21.Jensen, A. N., G. Sørensen, D. L. Baggesen, R. Bødker, and J. Hoorfar. 2003. Addition of novobiocin in pre-enrichment step can improve Salmonella culture of modified semisolid Rappaport-Vassiliadis. J. Microbiol. Methods 55:249-255. [DOI] [PubMed] [Google Scholar]
- 22.Josefsen, M. H., N. R. Jacobsen, and J. Hoorfar. 2004. Enrichment followed by quantitative PCR both for rapid detection and as a tool for quantitative risk assessment of food-borne thermotolerant campylobacters. Appl. Environ. Microbiol. 70:3588-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klerks, M. M., A. H. van Bruggen, C. Zijlstra, and M. Donnikov. 2006. Comparison of methods of extracting Salmonella enterica serovar Enteritidis DNA from environmental substrates and quantification of organisms by using a general internal procedural control. Appl. Environ. Microbiol. 72:3879-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knutsson, R., C. Löfström, H. Grage, J. Hoorfar, and P. Rådström. 2002. Modeling of 5′ nuclease real-time responses for optimization of a high-throughput enrichment PCR procedure for Salmonella enterica. J. Clin. Microbiol. 40:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackay, I. M. 2004. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 10:190-212. [DOI] [PubMed] [Google Scholar]
- 26.Malorny, B., P. T. Tassios, P. Rådström, N. Cook, M. Wagner, and J. Hoorfar. 2003. Standardization of diagnostic PCR for the detection of foodborne pathogens. Int. J. Food Microbiol. 83:39-48. [DOI] [PubMed] [Google Scholar]
- 27.Pintar, K., A. Cook, F. Pollari, A. Ravel, S. Lee, and J. A. Odumeru. 2007. Quantitative effect of refrigerated storage time on the enumeration of Campylobacter, Listeria, and Salmonella on artificially inoculated raw chicken meat. J. Food Prot. 70:739-743. [DOI] [PubMed] [Google Scholar]
- 28.Rudi, K., H. K. Hoidal, T. Katla, B. K. Johansen, J. Nordal, and K. S. Jakobsen. 2004. Direct real-time PCR quantification of Campylobacter jejuni in chicken fecal and cecal samples by integrated cell concentration and DNA purification. Appl. Environ. Microbiol. 70:790-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seo, K. H., I. E. Valentin-Bon, and R. E. Brackett. 2006. Detection and enumeration of Salmonella Enteritidis in homemade ice cream associated with an outbreak: comparison of conventional and real-time PCR methods. J. Food Prot. 69:639-643. [DOI] [PubMed] [Google Scholar]
- 30.Voetsch, A. C., T. J. Van Gilder, F. J. Angulo, M. M. Farley, S. Shallow, R. Marcus, P. R. Cieslak, V. C. Deneen, and R. V. Tauxe. 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin. Infect. Dis. 38(Suppl. 3):S127-S134. [DOI] [PubMed] [Google Scholar]
- 31.Wang, X., and M. F. Slavik. 1999. Rapid detection of Salmonella in chicken washes by immunomagnetic separation and flow cytometry. J. Food Prot. 62:717-723. [DOI] [PubMed] [Google Scholar]
- 32.Warren, B. R., H. G. Yuk, and K. R. Schneider. 2007. Detection of Salmonella by flow-through immunocapture real-time PCR in selected foods within 8 hours. J. Food Prot. 70:1002-1006. [DOI] [PubMed] [Google Scholar]
- 33.Wolffs, P., H. Grage, O. Hagberg, and P. Rådström. 2004. Impact of DNA polymerases and their buffer systems on quantitative real-time PCR. J. Clin. Microbiol. 42:408-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolffs, P., R. Knutsson, B. Norling, and P. Rådström. 2004. Rapid quantification of Yersinia enterocolitica in pork samples by a novel sample preparation method, flotation, prior to real-time PCR. J. Clin. Microbiol. 42:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolffs, P., B. Norling, J. Hoorfar, M. Griffiths, and P. Rådström. 2005. Quantification of Campylobacter spp. in chicken rinse samples by using flotation prior to real-time PCR. Appl. Environ. Microbiol. 71:5759-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolffs, P., B. Norling, and P. Rådström. 2005. Risk assessment of false-positive quantitative real-time PCR results in food, due to detection of DNA originating from dead cells. J. Microbiol. Methods 60:315-323. [DOI] [PubMed] [Google Scholar]
- 37.Wolffs, P. F., K. Glencross, B. Norling, and M. W. Griffiths. 2007. Simultaneous quantification of pathogenic Campylobacter and Salmonella in chicken rinse fluid by a flotation and real-time multiplex PCR procedure. Int. J. Food Microbiol. 117:50-54. [DOI] [PubMed] [Google Scholar]
- 38.Wolffs, P. F., K. Glencross, R. Thibaudeau, and M. W. Griffiths. 2006. Direct quantitation and detection of salmonellae in biological samples without enrichment, using two-step filtration and real-time PCR. Appl. Environ. Microbiol. 72:3896-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]