Abstract
The DNA sequence of the genome of Staphylococcus haemolyticus JCSC1435 revealed a putative capsule operon composed of 13 genes in tandem. The first seven genes (capABCDEFGSh) showed ≥57% similarity with the Staphylococcus aureus cap5 or cap8 locus. However, the capHIJKLMSh genes are unique to S. haemolyticus and include genes encoding a putative flippase, an aminotransferase, two glycosyltransferases, and a transcriptional regulator. Capsule-like material was readily apparent by immunoelectron microscopy on bacteria harvested in the postexponential phase of growth. Electron micrographs of a JCSC1435 mutant with a deleted cap region lacked the capsule-like material. Both strains produced small amounts of surface-associated material that reacted with antibodies to polyglutamic acid. S. haemolyticus cap genes were amplified from four of seven clinical isolates of S. haemolyticus from humans, and three of these strains produced a serologically cross-reactive capsular polysaccharide. In vitro assays demonstrated that the acapsular mutant strain showed greater biofilm formation but was more susceptible to complement-mediated opsonophagocytic killing than the parent strain. Structural characterization of capsule purified from S. haemolyticus strain JCSC1435 showed a trisaccharide repeating unit: −3-α-l-FucNAc-3-(2-NAc-4-N-Asp-2,4,6-trideoxy-β-d-Glc)-4-α-d-GlcNAc-. This structure is unique among staphylococcal polysaccharides in that its composition includes a trideoxy sugar residue with aspartic acid as an N-acyl substituent.
Among the coagulase-negative staphylococci (CoNS), Staphylococcus haemolyticus plays an important role in hospital-acquired opportunistic infections. S. haemolyticus is second only to Staphylococcus epidermidis in its frequency of isolation from human blood cultures (7, 9), and it may also cause peritonitis, otitis, urinary tract infections, and septicemia. Methicillin-resistant S. haemolyticus strains have been associated with foreign body infections (7), and S. haemolyticus is notorious for its multidrug-resistant phenotype, with decreased susceptibility to methicillin, teicoplanin, and vancomycin (24). S. haemolyticus was the first species of CoNS sharing teicoplanin and vancomycin resistance (23).
The genome of the human-pathogenic S. haemolyticus strain JCSC1435 was sequenced in 2005 (25). At least 57 open reading frames (ORFs) associated with virulence were reported. As suggested by its species name, S. haemolyticus carries three candidate ORFs encoding hemolysins; other putative virulence factors include adhesins, exonucleases, proteases, and genes encoding a capsular polysaccharide (CP). Although slime production by S. haemolyticus has been reported (4), biofilm formation and adherence to acrylic by S. haemolyticus are significantly reduced compared to those of Staphylococcus epidermidis (2, 3). Moreover, the biofilm-associated ica locus, present in S. epidermidis and Staphylococcus aureus, is absent from strain JCSC1435 and other clinical isolates of S. haemolyticus (2, 6).
Previous reports have indicated that certain bovine strains of S. haemolyticus (21, 26) produced a CP that was serologically cross-reactive with the S. aureus type 5 CP (CP5). However, probes specific for the S. aureus cap5 or cap8 locus showed little hybridization to chromosomal or plasmid DNA prepared from the bovine strains of S. haemolyticus (26).
The purpose of this study was to purify and characterize the CP produced by JCSC1435, an S. haemolyticus strain of human origin that is resistant to multiple antibiotics (25). We have visualized the CP by immunoelectron microscopy and shown that it is produced by other clinical isolates of S. haemolyticus. Antibodies specific for the CP neutralized the antiphagocytic effect of the capsule in a complement-dependent opsonophagocytic killing assay.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. haemolyticus strains used in this study are listed in Table 1. Strains JCSC1435 and 8HT4 were previously described (25); mutant 8HT4 has a 91.9-kb chromosomal deletion within the oriC environ that includes the entire capsule operon. Unless otherwise indicated, S. haemolyticus strains were cultivated in tryptic soy broth (TSB) with 1% (wt/vol) glucose at 37°C with vigorous shaking.
TABLE 1.
Presence of S. haemolyticus capsule genes in strains from humans and cows
| Strain | Source (reference) | Presence of capsule gene as determined by PCR assay
|
|||
|---|---|---|---|---|---|
| capABCDSh | capEFGSh | capHIJSh | capKLMSh | ||
| JCSC1435 | Clinical isolate from Japan (25) | + | + | + | + |
| 8HT4 | JCSC1435 mutant with a 91.9-kb chromosomal deletion (25) | ||||
| NRS9a | Peritoneal fluid | − | − | − | − |
| NRS50 | Wound/skin/soft tissue | + | + | + | + |
| NRS62 | Dialysis fluid | − | − | − | − |
| NRS69 | Bloodstream | + | + | + | + |
| NRS115 | Wound/skin/soft tissue | + | + | + | + |
| NRS116 | Wound/skin/soft tissue | + | + | +b | +b |
| M176 | Peritoneal fluid (3) | − | − | − | − |
| 8005-3 | Bovine strain from Norway (26) | − | − | − | − |
| 8622-1 | Bovine strain from Norway (26) | − | − | − | − |
| 8690-3 | Bovine strain from Norway (26) | − | − | − | − |
| 8733-3 | Bovine strain from Norway (26) | − | − | − | − |
| 8785-4 | Bovine strain from Norway (26) | − | − | − | − |
| 8690-3 | Bovine strain from Norway (26) | − | − | − | − |
NRS strains were clinical isolates from the United States that were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus.
Unlike the other clinical isolates that carried the capSh genes, strain NRS116 showed negative RT-PCRs with primers derived from capHISh and capKLSh (see text).
CP antibodies and serotyping.
Antiserum to the S. haemolyticus CP was prepared by immunizing a rabbit with heat-killed (70°C for 1 h) cells of strain JCSC1435 as described previously (30). The antiserum was adsorbed with the acapsular mutant 8HT4 to render the serum capsule specific. The reactivity of S. haemolyticus clinical isolates with JCSC1435 CP antiserum was determined by a colony immunoblot method (16). In addition, capsule extracts (26) from S. haemolyticus strains were reacted with polyclonal antibodies specific for the S. haemolyticus CP and S. aureus CP1, CP2, CP5, and CP8 by double immunodiffusion.
DNA and RNA manipulations.
S. haemolyticus strains were harvested from TSB cultures incubated overnight with shaking at 37°C. The bacterial cells were lysed with lysozyme and lysostaphin (1 mg/ml each) for 1 h at 37°C, and genomic DNA was isolated with the Wizard Genomic Purification kit (Promega Corp., Madison, WI). PCRs were performed with Taq DNA polymerase (Invitrogen Corp., Carlsbad, CA) and 1.5 mM MgCl2. Primers for PCR and reverse transcription-PCR (RT-PCR) and the lengths of the amplicons are indicated in Table 2. Amplification of the S. haemolyticus recA gene served as a positive PCR control.
TABLE 2.
Primers used in this study
| Primer name | Position in Fig. 1 | Amplicon (length in bp) | Sequence (5′-3′) |
|---|---|---|---|
| SH0383-f | 1 | SH0383 (889) | TTTTAAGCGCAGGAACAAGG |
| SH0383-r | 2 | SH0383 (889) | CCAATTTCACTTTTATTAGTTTTGC |
| SH0384-f | 3 | SH0384 (391) | TTCCCAGTTAGTAAGACTGAAATAGC |
| SH0384-r | 4 | SH0384 (391) | TCTTCTCCATTATATTAACGTCTATCC |
| SH0385-f | 5 | SH0385 (586) | TTATTACCATTAAACCTTTCG |
| SH0385-r | 6 | SH0385 (586) | TGAAAAGAATATTTGATATTGG |
| acpD-f | 7 | acpD (551) | ATTTCTTCAGTTTTCTCAGG |
| acpD-r | 8 | acpD (551) | CATCCTTTAGATGAATTAGG |
| capA-f | 9 | TTCCTGTAAATATTGCGGTAGC | |
| capC-r | 10 | capABCSh (1,477) | GTCTTTGAGATTCATGATGAGC |
| capD-r | 11 | capABCDSh (3,184) | CGAATACTTCTCCACCTTTAGC |
| capE-f | 12 | ACCTTCTTGCGAATTCTTCC | |
| capF-r | 13 | capEFSh (1,603) | CCGAAACTTGACCTCTGTC |
| capG-r | 14 | capEFGSh (2,927) | TCGATAGCGTCAATCATAGG |
| capH-f | 15 | GTATTGATCATGCGGAATCG | |
| capI-r | 16 | capHISh (1,497) | AAATGGAAACAAGAAGAAAGC |
| capJ-r | 17 | capHIJSh (3,201) | AAGCCCAAACTTTCTACTTGG |
| capK-f | 18 | CAGAGCTAATGGCTTATTTGACC | |
| capL-r | 19 | capKLSh (1,539) | CACTTCCATTTGTCCTCTTCC |
| capM-r | 20 | capKLMSh (2,800) | AGATTGTCATCACCCAGTGC |
| phoB-f | 21 | phoB (1,319) | TGACCATAAGTAGTCCAACC |
| phoB-r | 22 | phoB (1,319) | TTTGGTAAAACAACAGTAGC |
| recA-f | Not shown | recA (935) | TCAACATCTCCATCGAAAATACC |
| recA-r | Not shown | recA (935) | GGTGCCGTTATGAAACTAGGC |
| icaR-f | Not shown | icaR-icaA (770) | GAAAATAAGGTTATTGCGTTATCA |
| icaA-r | Not shown | icaR-icaA (770) | TTAATACAGCCAATTAAACTTGCATATTC |
Total RNA was prepared from S. haemolyticus cells collected in the exponential, postexponential, and stationary phases of growth. The bacteria were lysed with 0.5 ml zirconia-silicon beads (Fisher Scientific, Pittsburgh, PA) in a dental amalgamator (32), and RNA was purified with the RNeasy Mini kit (Qiagen, Valencia, CA). RNA was treated with 80 U RNase OUT (Invitrogen), and contaminating DNA was removed by treatment for 30 min with 10 U DNase I (Invitrogen). RT-PCR amplification was carried out with the Access RT-PCR System kit (Promega) following the manufacturer's recommendations. A control sample without reverse transcriptase was included for each reaction to confirm the absence of contaminating DNA.
Transmission electron microscopy.
S. haemolyticus cells were harvested from TSB cultures incubated with shaking until the postexponential growth phase. The bacterial cells were mixed for 2 h at room temperature with 0.2 ml of preimmune rabbit serum or JCSC1435 CP antiserum. Samples were fixed and sectioned as described previously (29).
Immunogold labeling of S. haemolyticus surface antigens was performed on Formvar carbon-coated copper grids inoculated with ∼107 CFU in phosphate-buffered saline (PBS). The samples were blocked for 10 min in 0.5% fish skin gelatin in PBS with 0.1% Tween 20. A 1:10 dilution of JCSC1435 CP antiserum or antibodies to poly-γ-d-glutamic acid (PGA) (1) (kindly provided by Julia Wang, Channing Laboratory) was added, and the samples were incubated for 30 min at ambient temperature. The grids were washed four times in PBS and incubated with protein A-gold particles (20 nm). After four washes in deionized water, the samples were air dried and examined with a JEOL 1200EX microscope.
In vitro opsonophagocytic killing assay.
The opsonophagocytic killing assay was performed as described by Xu et al. (30) with the following modifications. S. haemolyticus strains were cultivated with shaking for 16 h at 37°C in TSB. The bacterial cells were washed once and diluted in a minimal essential medium (Invitrogen) containing 1% bovine serum albumin (MEM-BSA; Sigma Chemical Co., St. Louis, MO). The assay was performed in polypropylene tubes containing ∼3.2 × 106 human polymorphonuclear leukocytes (PMNs), ∼7.2 × 105 CFU S. haemolyticus, 0.25% heat-inactivated rabbit antiserum, and 1% guinea pig serum (as a complement source) in a total volume of 500 μl MEM-BSA. Control samples contained S. haemolyticus and PMNs with either complement or antiserum or serum and complement with no added PMNs. The tubes were rotated end over end (12 rpm) for 2 h at 37°C. Samples were vortexed and diluted in sterile deionized water, and bacterial killing was estimated by plating the diluted samples in duplicate on tryptic soy agar. The percentage killing was defined as the reduction in CFU/ml after 2 h compared with that at time zero (30).
Biofilm formation.
S. haemolyticus biofilm formation was measured using a standard microtiter assay, as described previously (2, 6). Bacteria were grown overnight in TSB plus 1% glucose and diluted 1:100 in the same medium, and 200-μl aliquots were transferred into wells of tissue culture-treated polystyrene microtiter plates. The plates were incubated for 24 h at 37°C without shaking, and culture turbidities were measured in each well at 650 nm. To remove the medium and nonadherent bacterial cells, the microtiter plates were washed twice with 0.9% NaCl, and 200 μl of 0.15% safranin per well was added. After 10 min at ambient temperature, the safranin solution was removed by washing, and the biofilms were air dried for 45 min at 50°C. One hundred microliters H2O was added to each well before measurement of optical density at 540 nm. The absorbance reading of each sample was normalized by dividing the optical density at 540 by the culture turbidity at 650 nm. Differences in biofilm production were evaluated by the Student t test.
CP purification.
CP was purified by methods similar to those we have described previously (27). Briefly, S. haemolyticus JCSC1435 was cultivated in 20 liters of TSB to an optical density at 600 nm of 2.0. Autoclaved aqueous bacterial extracts were treated with DNase and RNase at 37°C overnight, followed by treatment for 8 h with pronase. The dialyzed material was passed over a column of DEAE-Sephacel equilibrated with 0.05 M sodium acetate-0.05 M NaCl, pH 6.0. Fractions that eluted in a 2-liter gradient (0.05 M sodium acetate with 0.05 to 0.5 M NaCl) were analyzed for absorbance at 206 nm and for serologic activity with antibodies to S. haemolyticus CP. Serologically active fractions were pooled, dialyzed, and lyophilized. The CP was chromatographed on a Sephacryl S-300 column equilibrated in 0.2 M NaCl, and the fractions were monitored as described above. Serologically active fractions were pooled, dialyzed, and lyophilized. The purity of the polysaccharide was assessed by UV (260- and 280-nm) absorption spectra, chemical assays for protein and phosphate, and nuclear magnetic resonance (NMR) spectroscopy.
Structural characterization of the S. haemolyticus CP.
1H and 13C NMR spectra were recorded using a Varian Inova 500 MHz spectrometer in D2O solutions at 60°C with an acetone standard (2.225 ppm for 1H and 31.5 ppm for 13C) using standard pulse sequences for correlation spectroscopy, total correlation spectroscopy (mixing time, 120 ms), nuclear Overhauser effect spectroscopy (NOESY) (mixing time, 200 ms), and heteronuclear single quantum correlation and heteronuclear multiple-bond correlation (HMBC) (optimized for an 8-Hz coupling constant). Spectra in 10% D2O were recorded at 25°C with WET water suppression (water suppression enhanced through T1 effects).
For monosaccharide analysis and identification of the aspartic acid residue, the S. haemolyticus CP (0.5 mg) was hydrolyzed (0.2 ml of 3 M trifluoroacetic acid, 120°C, 2 h) and evaporated to dryness under a stream of air. The residue was dissolved in water (0.5 ml), reduced with NaBH4 (∼5 mg, 1 h), neutralized with acetic acid (0.3 ml), and dried, and methanol (1 ml) was added. The mixture was dried twice with the addition of methanol, and the residue was acetylated with acetic anhydride (0.5 ml, 100°C, 30 min), dried, and analyzed by gas-liquid chromatography (GLC) on an HP1 capillary column (30 m by 0.25 mm) with a flame ionization detector (Agilent 6850 chromatograph) in a temperature gradient of 170 (4 min) to 260°C at 4°C/min and by gas chromatography (GC)-mass spectrometry on a Varian Saturn 2000 system with the same column and an ion-trap mass spectral detector.
For determination of the absolute configuration of the monosaccharides, CP (1 mg) was treated with (S)-2-butanol/AcCl (0.25 ml, 10:1 [vol/vol], 2 h, 85°C), dried under a stream of air, acetylated, and analyzed by GC in comparison with authentic standards prepared from respective monosaccharides with (S)- and (R)-2-butanol. The absolute configuration of l-aspartic acid was determined in the hydrolysate of S. haemolyticus CP by high-pressure liquid chromatography on a penicillamine chiral column (Phenomenex) in 2 mM CuSO4 in 15% methanol eluent.
RESULTS
Expression of the S. haemolyticus capsule genes.
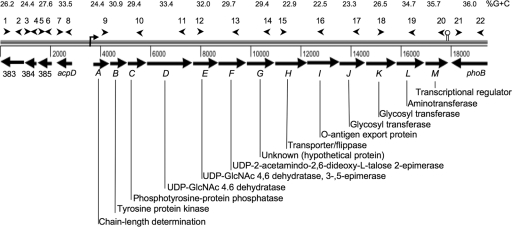
The capSh operon in the genome of S. haemolyticus JCSC1435 (25) is comprised of 13 ORFs (SH0388 to SH0400) in a 14,652-bp region (Fig. 1) spanning nucleotides 402008 to 416209. The first seven genes (capASh through capGSh) are homologous to S. aureus cap5A (or cap8A) through cap5G (or cap8G), with DNA homologies between 65% and 81% and amino acid identities between 57% and 89%. CapHSh through CapKSh share only limited homology with other gene products and probably include putative glycosyltransferases, as well as genes involved in CP transport and polymerization. CapLSh is homologous to bacterial aminotransferases, and CapMSh is a putative transcriptional regulator (Fig. 1). The overall G+C content in the coding regions of the S. haemolyticus cap region is 28.8%, which is lower that that of the total genome (32.8%). The G+C content of the individual capSh genes is shown in Fig. 1. The capHIJSh genes in the center of the capSh locus showed the lowest G+C content, ranging from 22.5% to 23.3% G+C. This suggests that this central DNA region, specific to S. haemolyticus, represents the genes most recently acquired by the bacterium via horizontal gene transfer. A putative Rho-dependent transcriptional terminator was located at the end of the capMSh gene.
FIG. 1.
S. haemolyticus capsule locus and flanking genes. Arrowheads denote sites where primers anneal. Primer names and sequences are shown in Table 2. The percent G+C content for individual genes is shown above each ORF. The predicted functions of each gene product based on protein homologies are shown.
Previous serologic studies indicated that certain bovine strains of S. haemolyticus produce a CP that reacts serologically with polyclonal and monoclonal antibodies to S. aureus CP5 (21, 26). We prepared capsule extracts from strain JCSC1435 and seven S. haemolyticus clinical isolates of human origin (Table 1). None of the extracts reacted with antibodies specific to S. aureus CP1, CP2, CP5, or CP8 (not shown).
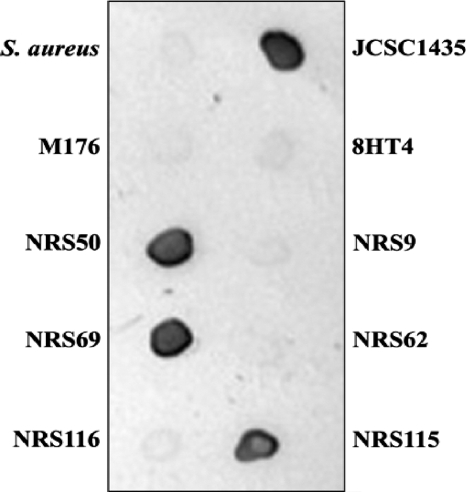
To determine whether clinical isolates of S. haemolyticus carried capsule genes similar to those within the JCSC1435 genome, we designed primers (Table 2) to amplify specific regions of the S. haemolyticus capsule locus: capABCDSh, capEFGSh, capHIJSh, and capKLMSh. Genomic DNA from S. haemolyticus JCSC1435 yielded amplicons of the expected size with each primer pair, whereas none of the genomic DNA preparations from six CP5+ bovine isolates (26) or mutant 8HT4 were positive by PCR. Of the seven clinical S. haemolyticus isolates from humans, four (NRS50, NRS69, NRS115, and NRS116) produced amplicons identical in size to those of JCSC1435 (Table 1). PCR analysis indicated that three clinical isolates (NRS9, NRS62, and M176) did not carry capsule genes similar to those of JCSC1435, nor did they react with S. haemolyticus CP-specific antiserum by immunodiffusion or immunoblotting (Fig. 2). Likewise, six bovine isolates of S. haemolyticus and deletion mutant 8HT4 failed to react by immunoblotting or immunodiffusion with the JCSC1435 CP antiserum (not shown). Strains NRS9 and NRS62, but not M176, were positive by PCR for the phoB gene just downstream of the capSh locus (Fig. 1), whereas all three strains were negative by PCR for four genes (SH0383 to SH0386) upstream of the capSh locus. All of the S. haemolyticus isolates, but not an S. aureus strain, yielded a positive amplicon with primers specific for S. haemolyticus recA (not shown).
FIG. 2.
Colony immunoblotting of staphylococcal strains reacted with S. haemolyticus CP-specific antiserum. The strains tested are described in Table 1. A capsule serotype 5 S. aureus strain was used as a negative control.
To determine whether the capsule genes were expressed by the clinical S. haemolyticus isolates, we performed RT-PCR experiments with primers that yielded products ≤1.6 kb in size and spanned the length of the capSh locus (Table 2). Our results revealed that S. haemolyticus strains JCSC1435, NRS50, NRS69, and NRS115 expressed the capSh genes since they were positive by RT-PCR with primer pairs with homology to capABCSh, capEFSh, capHISh, and capKLSh. Furthermore, the clinical isolates NRS50, NRS69, and NRS115 each produced a serologically detectable CP when tested with antiserum specific for the JCSC1435 CP (Fig. 2). Although strain NRS116 was positive for transcription of the upstream capSh genes, it was negative for the expected bands corresponding to the downstream genes (capHISh and capKLSh), suggesting that strain NRS116 may carry a defective promoter downstream of capFSh. This observation is consistent with our inability to detect CP production by this strain (Fig. 2).
Influence of culture medium and bacterial growth phase on S. haemolyticus CP production.
To investigate environmental influences on CP production, we cultivated S. haemolyticus JCSC1435 on different growth media, including TSB, TSB plus 1% glucose, brain heart infusion broth, Columbia broth, or Columbia broth or agar supplemented with 2% NaCl. CP, visualized by electron microscopy and immunogold labeling, was abundant when strain JCSC1435 was grown in brain heart infusion broth, TSB plus 1% glucose, or Columbia broth plus 2% NaCl (data not shown). In contrast to that observed with S. aureus (19, 20), cultivation on Columbia salt agar plates was suboptimal for CP production by S. haemolyticus.
Similar experiments were performed to determine the influence of the bacterial growth phase on capsule expression by strain JCSC1435. Similar to CP expression by S. aureus, little CP was detected on the surface of S. haemolyticus cells until late in the exponential growth phase. Maximal CP was observed on the bacterial cells during the postexponential and early-stationary phases of growth (Fig. 3 and 4). This finding was consistent with RT-PCR experiments performed with RNA prepared from strain JCSC1435. S. haemolyticus capSh transcripts were detected once the culture reached an optical density at 600 nm of 0.5 but were absent from cells harvested from 24-h cultures (data not shown).
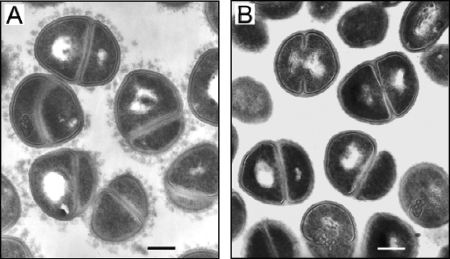
FIG. 3.
Transmission electron micrographs of S. haemolyticus JCSC1435. To visualize the capsule, the bacteria were incubated with S. haemolyticus CP-specific antiserum (A). Bacteria incubated with preimmune serum showed no evidence of CP (B). Magnification, ×22,500. Bars, 0.2 μm.
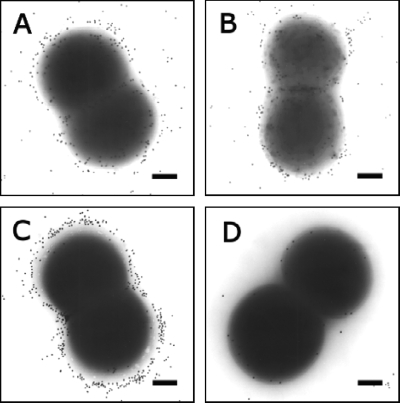
FIG. 4.
Detection of S. haemolyticus PGA or CP by immunogold staining and visualization by transmission electron microscopy. Samples (A and B) incubated with PGA antiserum were harvested from Columbia salt agar plates, since salt enhances PGA expression (13). Samples (C and D) incubated with S. haemolyticus CP-specific antiserum were harvested from postexponential-phase cultures in TSB. (A and C) S. haemolyticus JCSC1435; (B and D) cap deletion mutant 8HT4. Magnification, ×45,000. Bars, 0.1 μm.
To better visualize the S. haemolyticus CP, we prepared ultrathin sections of strain JCSC1435 harvested from overnight TSB plus 1% glucose cultures. Electron micrographs of S. haemolyticus JCSC1435 that were incubated with CP-specific antibodies showed abundant surface-associated CP (Fig. 3A) that was lacking on bacterial cells incubated with preimmune serum (Fig. 3B).
Differentiation between PGA and CP production by S. haemolyticus.
PGA is a surface-associated polymer produced by Bacillus anthracis and members of the S. epidermidis group, and PGA has been shown to play a role in staphylococcal resistance to high osmolarity (13). Because the genes encoding PGA biosynthesis are present within the S. haemolyticus genome (25), we wanted to ensure that the capsule-like material visualized on the surface of JCSC1435 was CP and not PGA. To accomplish this, we performed immunogold labeling experiments of JCSC1435 with antibodies to either PGA or the S. haemolyticus CP. Both S. haemolyticus JCSC1435 and acapsular mutant 8HT4 harvested from Columbia salt agar plates showed immunogold labeling with PGA antibodies; the gold particles were distributed in small clusters closely associated with the bacterial cells (Fig. 4A and B). Similar findings were observed on bacteria cultivated in TSB (data not shown). Our data corroborate those of Kocianova et al., who reported that PGA production by S. epidermidis is sparse, i.e., 1.2 μg PGA/liter (13). Our experimental results indicate that PGA synthesis and CP production by S. haemolyticus are not linked, since PGA was produced by wild-type strain JCSC1435 and the acapsular mutant 8HT4 (Fig. 4A and B), whereas CP was apparent on the wild-type strain (Fig. 4C) but not on mutant 8HT4 (Fig. 4D).
S. haemolyticus biofilm formation.
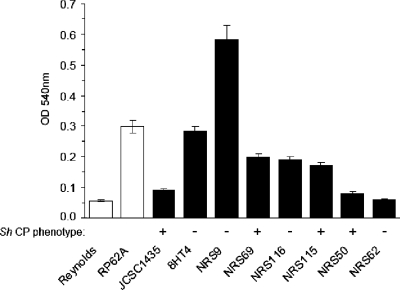
Adherence of bacteria to polystyrene microtiter plates is a convenient assay for biofilm formation (2, 6). S. haemolyticus JCSC1435 is a poor biofilm producer (Fig. 5), and it lacks the ica genes implicated in biofilm formation (25). PCR analysis of the S. haemolyticus clinical isolates revealed that they also lacked the ica genes (not shown). However, other cell wall structures, such as wall teichoic acid and protein, have been shown to mediate staphylococcal biofilm formation in strains lacking or not expressing the ica locus (8, 14). Several of the S. haemolyticus clinical isolates produced biofilms in vitro that were comparable to that of the control strain S. epidermidis RP62A (Fig. 5). Although the acapsular mutant 8HT4 showed strongly enhanced biofilm production (P < 0.001) compared to the wild-type strain (Fig. 5), there were no other obvious correlations between CP production and biofilm formation among the clinical isolates.
FIG. 5.
Biofilm production by S. haemolyticus strains. Each bar represents the mean ± standard deviation of 12 separate determinations. S. aureus Reynolds and S. epidermidis RP62A are control negative and positive biofilm-producing strains, respectively. The S. haemolyticus CP phenotype of each strain is indicated.
Effect of CP on in vitro opsonophagocytic killing of S. haemolyticus.
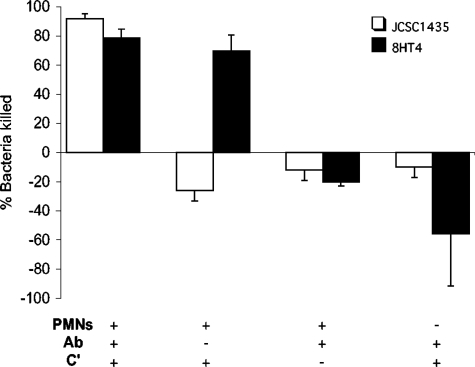
To determine whether there were differences in resistance to phagocytosis attributable to CP production by S. haemolyticus, we measured the killing of strains JCSC1435 and the acapsular mutant 8HT4 by human PMNs. As shown in Fig. 6, >78% of the JCSC1435 and 8HT4 inocula were killed by PMNs in the presence of both CP-specific antibodies and complement. Similarly, ∼70% of the mutant 8HT4 (CP−) inoculum was effectively opsonized for phagocytic killing by complement alone. The addition of CP antibodies to the mutant strain did not improve the killing observed by PMNs and complement. In contrast, the JCSC1435 inoculum bacteria increased in number when incubated with PMNs and complement but no antibodies. In serum with no complement activity, rabbit CP-specific antibodies were not opsonic for either strain. No killing was observed in control samples lacking PMNs or in samples with PMNs but no opsonin. These results confirm the predicted antiphagocytic activity of the capsule produced by S. haemolyticus JCSC1435.
FIG. 6.
Opsonophagocytic killing of S. haemolyticus JCSC1435 and acapsular mutant 8HT4 by human PMNs. The percentage of bacteria killed (mean ± standard error of the mean) was calculated as a reduction in CFU/ml after 2 h at 37°C compared with the CFU/ml in each sample at time zero. The data were pooled from experiments performed two to six times. Ab, S. haemolyticus CP-specific antibodies; C', guinea pig serum with complement activity.
Purification of S. haemolyticus JCSC1435 CPs.
Capsule was extracted and purified from 20 liters of a JCSC1435 TSB culture. The extract (∼1 g), released into the supernatant by autoclaving the bacterial pellet, was clarified by DNase, RNase, and protease treatments. The crude CP preparation was then separated from contaminating teichoic acids by ion-exchange chromatography. The CP (∼10% of the crude extract) eluted from the DEAE column with 0.14 to 0.18 M NaCl and was detected by immunodiffusion assays with rabbit S. haemolyticus antiserum. The serologically active fractions were pooled, dialyzed, concentrated, and further purified by size-exclusion chromatography. The purified CP (13 mg) eluted near the void volume of an S-300 Sephacryl column with a Kav of 0.01. The final CP yield was ∼0.65 mg of capsule per liter of culture. The capsule preparations used for biochemical characterization contained <2% protein, 0.5% nucleic acid, and <0.2% phosphorus.
Structural characterization of S. haemolyticus CP.
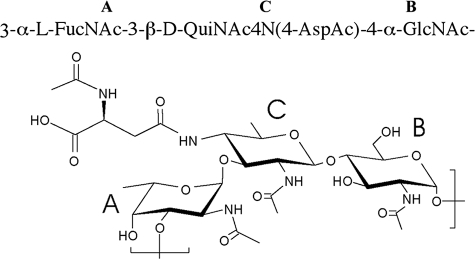
A set of the NMR spectra—correlation spectroscopy, total correlation spectroscopy, NOESY, heteronuclear single quantum correlation, and HMBC—was recorded for the S. haemolyticus CP. Spectra contained signals of spin systems of three monosaccharides—α-fucosamine (α-FucN, unit A), α-glucosamine (α-GlcN, unit B), and 2,4-diamino-2,4,6-trideoxy-β-glucose (bacillosamine or QuiN4N, unit C) in pyranose form and an aspartic acid residue (Table 3). Relative configurations of the constituent monosaccharides were identified on the basis of vicinal proton coupling constants and 13C NMR chemical shifts. Anomeric configurations were deduced from the J1,2 coupling constants and chemical shifts of H-1, C-1, and C-5 signals and by observations of intraresidual NOE connectivities (H-1:H-3, H-1:H-5) characteristic of the β-pyranosides for β-bacillosamine unit C. The sequence of the monosaccharides was determined from NOE and HMBC data, which showed correlations from anomeric proton to transglycosidic proton: A1:C3, B1:A3, and C1:B4, establishing the sequence [A-C-B]n (Fig. 7).
TABLE 3.
NMR data for the S. haemolyticus CP (δ, ppm)
| Residue | Nucleus | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|---|
| FucN A | 1H | 5.10 | 4.32 | 3.84 | 3.85 | 3.91 | 1.27 |
| 13C | 97.9 | 49.0 | 74.1 | 71.6 | 67.9 | 16.5 | |
| GlcN B | 1H | 5.02 | 3.96 | 3.87 | 3.59 | 3.73 | 3.65/3.77 |
| 13C | 98.9 | 54.1 | 70.6 | 80.6 | 71.9 | 61.1 | |
| BacN C | 1H | 4.51 | 3.98 | 3.87 | 3.72 | 3.70 | 1.24 |
| 13C | 101.9 | 57.2 | 75.5 | 56.4 | 71.6 | 17.9 | |
| Asp | 1H | 4.54 | 2.62/2.86 | ||||
| 13C | 173.6 | 52.5 | 39.7 | 176.9 |
FIG. 7.
Structure of the S. haemolyticus JCSC1435 CP.
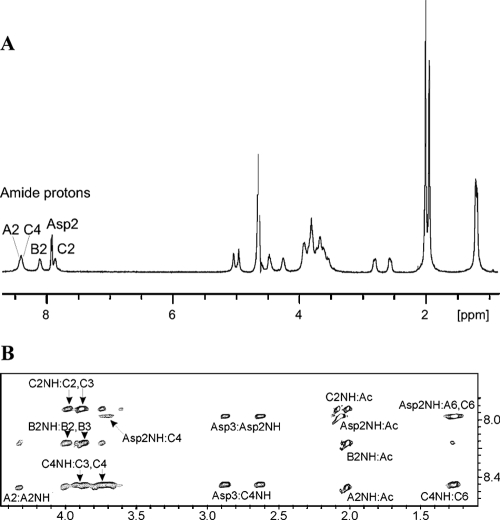
For determination of the position of the aspartic acid residue, a NOESY spectrum of the polysaccharide was recorded in 10% D2O-90% H2O in order to reveal correlations for NH protons. The 1H NMR spectrum contained five amide signals (Fig. 8A), corresponding to four amino groups of the amino sugars (A2NH, B2NH, C2NH, and C4NH) and the amino group of the aspartic acid (Asp2NH). They showed NOE correlations (Fig. 8B) with the protons at the site of the amide location (A2NH:A2; B2NH:B2; C2NH:C2; C4NH:C4), to the ones of the neighboring protons (B2NH:B3; C2NH:C3; C4NH:C3, C4NH:C6), and to the CH3 of the acetyl groups (A2NH:Ac; B2NH:Ac; C2NH:Ac). The proton of the aspartic acid amino group showed a NOE correlation with a CH3 of an acetyl group (Asp2NH:Ac) due to N acetylation of the aspartic acid residue. Aspartate H-3 gave strong NOE correlation with the C4NH (Asp3:C4NH, Fig. 8B), whereas Asp2 gave no correlation with C4NH; this indicated the attachment of the aspartate to the bacillosamine 4-amino group through the C-4 carboxyl of the aspartate. Thus, it could be determined that amino groups at A2, B2, and C2 and the amino group of the aspartic acid are acetylated and the amino group at position 4 of the bacillosamine residue is acylated with the 4-carboxyl group of the N-acetyl aspartate (Fig. 8B). Similar structures with N-acetyl-aspartate were described as constituents of the O-antigen polysaccharides of enterobacteria of the genera Providencia and Proteus (31).
FIG. 8.
Structural analysis of CP produced by S. haemolyticus JCSC1435. (A) 1H NMR spectrum of the S. haemolyticus CP in 10% D2O with WET water suppression. (B) Fragment of the NOESY spectrum of the polysaccharide showing correlations from amide protons.
The NMR data were confirmed by composition analyses that showed the presence of GlcN, FucN, and Asp. BacN was not detected after hydrolysis. The absolute configurations of GlcN and FucN were determined by GC of (S)-2-butylglycoside acetates. The absolute d-configuration of the BacN residue was deduced from 13C NMR chemical shift effects from l-FucN and d-GlcN, as described by Lipkind et al. (18). The structure of the CP purified from S. haemolyticus strain JCSC1435 was a trisaccharide repeating unit (Fig. 7): −3-α-l-FucNAc-3-(2-NAc-4-N-Asp-2,4,6-trideoxy-β-d-Glc)-4-α-d-GlcNAc-.
DISCUSSION
CP production is an important determinant of bacterial virulence, and it is a trait common to many invasive bacterial pathogens. Although clinical isolates of S. aureus were recognized as encapsulated by the early 1980s (10), the CoNS were considered nonencapsulated species. However, when the S. haemolyticus JCSC1435 genome was sequenced by Takeuchi et al. (25), they reported a capsule operon located within the “oriC environ” of the chromosome. The oriC environ includes genes specific to each staphylococcal species, and this region of the S. haemolyticus genome is particularly subject to frequent rearrangements and deletions (25, 28). We sought to determine whether a CP with antiphagocytic properties was actually expressed by S. haemolyticus JCSC1435 and to elucidate the structure of the CP encoded by the capSh genes.
Previous reports indicated that at least some bovine strains of S. haemolyticus produced an uncharacterized surface antigen that was serologically cross-reactive with S. aureus CP5 (21, 26). Poutrel et al. reported that 13 of 19 S. haemolyticus strains from six bovine herds in France reacted with antibodies to CP5 (21), and Tollersrud et al. indicated that all seven Norwegian bovine isolates of S. haemolyticus reacted with both monoclonal and polyclonal antibodies to S. aureus CP5 (26). DNA prepared from the encapsulated bovine isolates showed weak hybridization with S. aureus cap5 DNA probes under low-stringency conditions. Our studies indicate that the CP produced by bovine S. haemolyticus isolates is unrelated at the genetic and serologic levels to that produced by the human isolate JCSC1435. In contrast, four of seven S. haemolyticus clinical isolates from humans carried the capASh to capMSh genes, and three of the four produced a CP that reacted with JCSC1435 CP antiserum. Whether the strains lacking the capSh genes carry genes encoding a different capsule type has not yet been addressed.
Biochemical characterization of the purified CP from S. haemolyticus JCSC1435 revealed a new polysaccharide structure: −3-α-l-FucNAc-3-(2-NAc-4-N-Asp-2,4,6-trideoxy-β-d-Glc)-4-α-GlcNAc-. This structure is consistent with the predicted functions of the genes within the capSh locus. The first seven genes of the S. haemolyticus JCSC1435 cap operon showed high amino acid identity to S. aureus cap5ABCDEFG or cap8ABCDEFG. The capABCD genes are conserved among the cap loci of S. haemolyticus, Staphylococcus saprophyticus, and S. aureus (cap1, cap5, and cap8) (15, 17, 22, 25). The capA gene product may be involved in chain length determination, capB encodes a putative tyrosine protein kinase, and capC encodes a putative phosphotyrosine-protein phosphatase. Cap5D is a putative 4,6-dehydratase, converting UDP-GlcNAc to UDP-2-acetamido-2,6 dideoxy-d-xylo-4-hexulose, a precursor for S. aureus UDP-d-FucNAc or, in the case of S. haemolyticus, 2-NAc-4-N-Asp-2,4,6-trideoxy-d-Glc. The presence of l-FucNAc in the S. haemolyticus CP was predicted by the presence of the capEFGSh genes, which show 89%, 71%, and 87% identity, respectively, to the cap5EFG or cap8EFG genes responsible for l-FucNAc synthesis in S. aureus (11). The S. haemolyticus CP is the first example of a staphylococcal polysaccharide that includes a trideoxy sugar residue, and it is also unique in containing aspartic acid as an N-acyl substituent of QuiNAc4N, which has been described only in the O polysaccharides of Proteus and Providencia spp. (12). The putative aminotransferase capLSh gene likely adds the amino group to C-4 of UDP-QuiNAc, which is then acylated by the aspartic acid residue, possibly involving capHSh.
Despite the similarity between the S. aureus and S. haemolyticus CP biosynthetic genes, there is little nucleotide homology upstream of the cap5A gene, suggesting that regulation of CP biosynthesis may differ for the two species. Furthermore, the putative transcriptional regulator at the 3′ end of the capSh locus is absent from the S. aureus cap5 or cap8 gene cluster. The effects of other regulators of S. aureus CP expression, such as agr, arlS, mgrA, and sigB, on S. haemolyticus CP production are unknown. Although CP production by both S. aureus and S. haemolyticus was maximal during postexponential growth, cultivation of S. haemolyticus cells in a liquid medium resulted in CP production greater than that by bacteria grown on solid medium with added salt, a condition that promotes S. aureus CP production (19, 20).
The S. haemolyticus CP demonstrated biologic properties consistent with those produced by other encapsulated bacterial pathogens. Strain JCSC1435 was resistant to complement-mediated opsonophagocytic killing by human neutrophils, whereas the acapsular mutant 8HT4 was susceptible. The addition of anticapsular antibodies neutralized the antiphagocytic effect of the S. haemolyticus capsule. In addition to CP, S. haemolyticus produces PGA, a surface polymer that has been shown to have antiphagocytic properties when expressed by S. epidermidis (13). PGA is produced by many CoNS, including S. haemolyticus ATCC 29970. We visualized PGA on the surface of JCSC1435 and acapsular mutant 8HT4 by immunogold labeling of bacteria examined by electron microscopy, indicating that the biological differences that we observed between the two bacterial strains were due to production of CP and not PGA.
Biofilm formation is an important characteristic of opportunistic pathogens, such as CoNS. Some S. haemolyticus isolates are associated with foreign body infections (7), but there is no evidence that members of this species carry the ica genes that are implicated in biofilm production (2, 6). The influence of CP production on the development of staphylococcal biofilm production has received little attention. We evaluated the biofilm-forming ability of S. haemolyticus strains cultivated under growth conditions favorable for capsule production. The observation that the capsule deletion mutant strain showed greater biofilm formation than did the parental strain suggested that CP might partially mask or inhibit surface factors critical for biofilm production, as has been shown for other encapsulated pathogens (5, 31). However, there was no correlation among the clinical S. haemolyticus isolates between CP production and biofilm production, suggesting that the biofilm formation is strain dependent and mediated by factors that remain poorly understood.
In conclusion, we have isolated, purified, and characterized CP from the first CoNS shown to be encapsulated. Similar studies characterizing the CP of an S. saprophyticus strain (15) are in progress. CoNS are important agents of opportunistic infections in compromised patients, and S. haemolyticus is notorious for its resistance to antibiotics. CP produced by strain JCSC1435 cross-reacted serologically with CP detected on other clinical isolates of S. haemolyticus. The capSh locus bears some homology to the S. aureus cap5 or cap8 locus, but it is unique since it carries genes that encode enzymes to synthesize a trideoxy sugar residue that is N acylated by aspartic acid. We have visualized the bacterial polysaccharide by electron microscopy and shown that it protects the bacterium from uptake and killing by human neutrophils.
Acknowledgments
This work was supported by Public Health Service grant AI29040 (J.C.L.). S.F. was supported by a Fulbright-Nord-Pas de Calais (France) grant from the Fulbright Franco-American commission.
We thank Irina Sadovskaya for her critical review of the manuscript and Robert Solinga for technical assistance. Clinical isolates of S. haemolyticus were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) program supported under NIAID, NIH contract no. N01-AI-95359. Tore Tollersrud kindly provided the bovine S. haemolyticus isolates, and Julia Wang generously donated the antiserum to PGA.
Footnotes
Published ahead of print on 28 December 2007.
REFERENCES
- 1.Aulinger, B. A., M. H. Roehrl, J. J. Mekalanos, R. J. Collier, and J. Y. Wang. 2005. Combining anthrax vaccine and therapy: a dominant-negative inhibitor of anthrax toxin is also a potent and safe immunogen for vaccines. Infect. Immun. 733408-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerca, N., S. Martins, S. Sillankorva, K. K. Jefferson, G. B. Pier, R. Oliveira, and J. Azeredo. 2005. Effects of growth in the presence of subinhibitory concentrations of dicloxacillin on Staphylococcus epidermidis and Staphylococcus haemolyticus biofilms. Appl. Environ. Microbiol. 718677-8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerca, N., G. B. Pier, R. Oliveira, and J. Azeredo. 2004. Comparative evaluation of coagulase-negative staphylococci (CoNS) adherence to acrylic by a static method and a parallel-plate flow dynamic method. Res. Microbiol. 155755-760. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, G. D., J. T. Parisi, A. L. Bisno, W. A. Simpson, and E. H. Beachey. 1983. Characterization of clinically significant strains of coagulase-negative staphylococci. J. Clin. Microbiol. 18258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davey, M. E., and M. J. Duncan. 2006. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J. Bacteriol. 1885510-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Silva, G. D., M. Kantzanou, A. Justice, R. C. Massey, A. R. Wilkinson, N. P. Day, and S. J. Peacock. 2002. The ica operon and biofilm production in coagulase-negative staphylococci associated with carriage and disease in a neonatal intensive care unit. J. Clin. Microbiol. 40382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falcone, M., M. Giannella, G. Raponi, C. Mancini, and M. Venditti. 2006. Teicoplanin use and emergence of Staphylococcus haemolyticus: is there a link? Clin. Microbiol. Infect. 1296-97. [DOI] [PubMed] [Google Scholar]
- 8.Fitzpatrick, F., H. Humphreys, and J. P. O'Gara. 2005. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 431973-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ing, M. B., L. M. Baddour, and A. S. Bayer. 1997. Bacteremia and infective endocarditis: pathogenesis, diagnosis, and complications, p. 331-354. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, NY.
- 10.Karakawa, W. W., and W. F. Vann. 1982. Capsular polysaccharides of Staphylococcus aureus. Semin. Infect. Dis. 4285-293. [Google Scholar]
- 11.Kneidinger, B., K. O'Riordan, J. Li, J.-R. Brisson, J. C. Lee, and J. S. Lam. 2003. Three highly conserved proteins catalyze the conversion of UDP-N-acetyl-d-glucosamine to precursors for the biosynthesis of O antigen in Pseudomonas aeruginosa O11 and capsule in Staphylococcus aureus type 5—implications for the UDP-N-acetyl-l-fucosamine biosynthetic pathway. J. Biol. Chem. 2783615-3627. [DOI] [PubMed] [Google Scholar]
- 12.Kocharova, N. A., S. N. Senchenkova, A. N. Kondakova, A. I. Gremyakov, G. V. Zatonsky, A. S. Shashkov, Y. A. Knirel, and N. K. Kochetkov. 2004. D- and L-aspartic acids: new non-sugar components of bacterial polysaccharides. Biochemistry (Moscow) 69103-107. [DOI] [PubMed] [Google Scholar]
- 13.Kocianova, S., C. Vuong, Y. Yao, J. M. Voyich, E. R. Fischer, F. R. DeLeo, and M. Otto. 2005. Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J. Clin. Investig. 115688-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kogan, G., I. Sadovskaya, P. Chaignon, A. Chokr, and S. Jabbouri. 2006. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol. Lett. 25511-16. [DOI] [PubMed] [Google Scholar]
- 15.Kuroda, M., A. Yamashita, H. Hirakawa, M. Kumano, K. Morikawa, M. Higashide, A. Maruyama, Y. Inose, K. Matoba, H. Toh, S. Kuhara, M. Hattori, and T. Ohta. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. USA 10213272-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, J. C., M. J. Liu, J. Parsonnet, and R. D. Arbeit. 1990. Expression of type 8 capsular polysaccharide and production of toxic shock syndrome toxin 1 are associated among vaginal isolates of Staphylococcus aureus. J. Clin. Microbiol. 282612-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, W. S., T. Cunneen, and C. Y. Lee. 1994. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J. Bacteriol. 1767005-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipkind, G. M., A. S. Shashkov, Y. A. Knirel, E. V. Vinogradov, and N. K. Kochetkov. 1988. A computer-assisted structural analysis of regular polysaccharides on the basis of 13C-n.m.r. data. Carbohydr. Res. 17559-75. [DOI] [PubMed] [Google Scholar]
- 19.Pohlmann-Dietze, P., M. Ulrich, K. B. Kiser, G. Doring, J. C. Lee, J. M. Fournier, K. Botzenhart, and C. Wolz. 2000. Adherence of Staphylococcus aureus to endothelial cells: influence of the capsular polysaccharide, the global regulator agr, and the bacterial growth phase. Infect. Immun. 684865-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poutrel, B., F. B. Gilbert, and M. Lebrun. 1995. Effects of culture conditions on production of type 5 capsular polysaccharide by human and bovine Staphylococcus aureus strains. Clin. Diagn. Lab. Immunol. 2166-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poutrel, B., C. Mendolia, L. Sutra, and J. M. Fournier. 1990. Reactivity of coagulase-negative staphylococci isolated from cow and goat milk with monoclonal antibodies to Staphylococcus aureus capsular polysaccharide type 5 and type 8. J. Clin. Microbiol. 28358-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sau, S., N. Bhasin, E. R. Wann, J. C. Lee, T. J. Foster, and C. Y. Lee. 1997. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology 1432395-2405. [DOI] [PubMed] [Google Scholar]
- 23.Schwalbe, R. S., J. T. Stapleton, and P. H. Gilligan. 1987. Emergence of vancomycin resistance in coagulase-negative staphylococci. N. Engl. J. Med. 316927-931. [DOI] [PubMed] [Google Scholar]
- 24.Sieradzki, K., P. Villari, and A. Tomasz. 1998. Decreased susceptibilities to teicoplanin and vancomycin among coagulase-negative methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 42100-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 1877292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tollersrud, T., K. Kenny, A. J. Reitz, and J. C. Lee. 2000. Genetic and serologic evaluation of capsule production by bovine mammary isolates of Staphylococcus aureus and other Staphylococcus spp. from Europe and the United States. J. Clin. Microbiol. 382998-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzianabos, A. O., J. Y. Wang, and J. C. Lee. 2001. Structural rationale for the modulation of abscess formation by Staphylococcus aureus capsular polysaccharides. Proc. Natl. Acad. Sci. USA 989365-9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe, S., T. Ito, Y. Morimoto, F. Takeuchi, and K. Hiramatsu. 2007. Precise excision and self-integration of a composite transposon as a model for spontaneous large-scale chromosome inversion/deletion of the Staphylococcus haemolyticus clinical strain JCSC1435. J. Bacteriol. 1892921-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watts, A., D. Ke, Q. Wang, A. Pillay, A. Nicholson-Weller, and J. C. Lee. 2005. Staphylococcus aureus strains that express serotype 5 or serotype 8 capsular polysaccharides differ in virulence. Infect. Immun. 733502-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu, S., R. D. Arbeit, and J. C. Lee. 1992. Phagocytic killing of encapsulated and microencapsulated Staphylococcus aureus by human polymorphonuclear leukocytes. Infect. Immun. 601358-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi, K., A. W. Rasmussen, S. K. Gudlavalleti, D. S. Stephens, and I. Stojiljkovic. 2004. Biofilm formation by Neisseria meningitidis. Infect. Immun. 726132-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yim, H. H., and C. E. Rubens. 1997. Use of a dental amalgamator to extract RNA from the gram-positive bacterium Streptococcus agalactiae. BioTechniques 23229-231. [DOI] [PubMed] [Google Scholar]