Abstract
In Escherichia coli, YaeT, together with four lipoproteins, YfgL, YfiO, NlpB, and SmpA, forms a complex that is essential for β-barrel outer membrane protein biogenesis. Data suggest that YfgL and YfiO make direct but independent physical contacts with YaeT. Whereas the YaeT-YfiO interaction needs NlpB and SmpA for complex stabilization, the YaeT-YfgL interaction does not. Using bioinformatics, genetics, and biochemical approaches, we have identified three residues, L173, L175, and R176, in the mature YfgL protein that are critical for both function and interactions with YaeT. A single substitution at any of these sites produces no phenotypic defect, but two or three simultaneous alterations produce mild or yfgL-null phenotypes, respectively. Interestingly, biochemical data show that all YfgL variants, including those with single substitutions, have weakened in vivo YaeT-YfgL interaction. These defects are not due to mislocalization or low steady-state levels of YfgL. Cysteine-directed cross-linking data show that the region encompassing L173, L175, and R176 makes direct contact with YaeT. Using the same genetic and biochemical strategies, it was found that altering residues D227 and D229 in another region of YfgL from E221 to D229 resulted in defective YaeT bindings. In contrast, mutational analysis of conserved residues V319 to H328 of YfgL shows that they are important for YfgL biogenesis but not YfgL-YaeT interactions. The five YfgL mutants defective in YaeT associations and the yfgL background were used to show that SurA binds to YaeT (or another complex member) without going through YfgL.
The discovery of Omp85 in Neisseria meningitidis represents a major piece of the outer membrane biogenesis puzzle (39). Omp85, an essential, highly conserved, integral β-barrel outer membrane protein (OMP), is required for the assembly of OMPs (40). Recently, it was shown that the Omp85 homolog in Escherichia coli, YaeT, forms a multiprotein complex comprised of the OMP YaeT (BamA) and four lipoproteins, YfgL (BamB), YfiO (BamC), NlpB (BamD), and SmpA (BamE) (32, 43) (the names in the parentheses are newly standardized designations [22], but the old computer-assigned names are used in this paper due to their prevalence in the literature). Like the N. meningitidis counterpart, the YaeT complex is required for the assembly of OMPs in E. coli (4, 42, 43). The recently solved crystal structure of the N-terminal polypeptide transport-associated (POTRA) domains of YaeT provided a possible clue for the mechanism by which YaeT interacts with OMPs and YfgL (15, 22).
OMPs are synthesized in the cytoplasm as precursor polypeptides with N-terminal signal sequences that direct them to the SecYEG complex for translocation across the inner membrane (26). The signal sequences are cleaved during translocation, and the mature polypeptides are released into the periplasm, where they are thought to interact with chaperones, foldases, and lipopolysaccharide (LPS) to avoid aggregation and acquire the proper folding status required for the eventual assembly and insertion into the outer membrane at the YaeT complex site (31).
The two essential members of the multicomponent OMP assembly complex, YaeT and YfiO, make direct contact with each other (18). Moreover, the YaeT-YfiO interaction is stabilized by NlpB and SmpA (32). YfgL also interacts directly with YaeT, but in contrast to YfiO, this interaction is independent of NlpB and SmpA (18, 32). Although YfgL is nonessential, it is a highly conserved protein found in many (but not all) gram-negative bacteria, and its absence produces a pleiotropic phenotype. An E. coli strain with a deletion of yfgL has reduced levels of many OMPs (2, 25, 30). Consistent with a role for YfgL in OMP biogenesis, a strain lacking YfgL shows slow LamB monomer folding (36), while a strain lacking both YfgL and the major periplasmic protease DegP displays a conditional lethal phenotype (2). Additionally, a yfgL strain displays hypersensitivity to vancomycin, bacitracin, novobiocin, and other antibiotics, reflecting a compromised outer membrane permeability barrier (30). A deletion of yfgL even attenuates some pathogenic bacterial strains. For example, in a Salmonella enterica serovar Enteritidis yfgL mutant, the transcription of genes encoding many type III secretion system proteins involved in virulence is downregulated (8). Similarly, transposon disruption of yfgL in the invasive pathogenic E. coli strain LF82 isolated from chronic lesions of Crohn's disease patients markedly reduced its invasive ability in intestinal epithelial cells (28). Finally (and unexpectedly), YfgL in E. coli was reported to have activities unrelated to OMP biogenesis, namely, DNA break repair and homologous recombination (14).
The pleiotropic phenotype of ΔyfgL could be due solely to the disruption of critical interactions with YaeT, resulting in compromised OMP biogenesis, or it could be due to the absence of YfgL from the outer membrane in addition to the lack of YaeT interactions, causing broader structural defects in the outer membrane. At present, it is unclear which is the case. If it is the former, then alterations at specific YfgL residues that disrupt the function of YfgL and the interaction with YaeT should produce the same pleiotropic phenotype as the absence of YfgL. Using bioinformatics, genetics, and biochemical techniques, we found that altering just three amino acids of the mature YfgL, L173, L175, and R176, resulted in a yfgL-null phenotype and severely weakened YaeT interactions without significant defects in the biogenesis of the mutant YfgL protein. Substitutions in another region of YfgL, E221 to D229, also resulted in YaeT binding defects. In contrast, alterations of conserved residues near the C terminus of YfgL, from V319 to H328, affected the structural stability of the lipoprotein. Finally, data from the YfgL mutants provide evidence that SurA, presumably with its OMP cargo bound to it, interacts with YaeT directly or indirectly via another complex member but not via YfgL.
MATERIALS AND METHODS
Bacterial strains.
All E. coli strains used in this study are derived from MC4100 [F− araD139 Δ(argF-lac)U169 rpsL150 relA1 flb5301 deoC1 ptsF25 rbsR thi-1]. Three of the strains, Δara ΔyfgL-scar, Δara ΔyfgL-scar ΔdegP::Tn10, and ΔacrA-scar ΔtolC::Km, were created in previous studies (1, 2). A fourth strain, Δara ΔyaeT/λPBAD-yaeT bla, contains an arabinose-inducible copy of yaeT introduced to replace the disrupted native yaeT. The strain also harbors plasmid pZS21 with a hexahistidine-tagged yaeT insert (32). Without arabinose induction, the expression of the chromosomally integrated copy of yaeT is negligible, and the expression of the plasmid-borne yaeT is comparable to that of the wild-type strain (J. Malinverni, personal communication). From this strain, we constructed two more strains for this study by moving a yfgL+ or ΔyfgL-scar allele with a linked Cmr marker that is 90% linked to yfgL by P1 transduction.
DNA manipulations.
Wild type E. coli yfgL was cloned with a 3′ extension coding for a C-terminal six-histidine tag into plasmid pBAD24 (12) and expressed under the control of an arabinose-inducible promoter. Chromosomal yfgL was amplified using the Phusion high-fidelity DNA polymerase kit (Finnzymes) and forward primer 5′-ATCTGCTAGCCTGAGAGGGACCCGATG-3′, containing an NheI (underlined) restriction site, and reverse primer 5′-TCTAGAAGCTTAGTGATGGTGATGGTGATGACGGGTAATAGAGT ACACGGTTCCG-3′, containing a HindIII (underlined) restriction site and a six-histidine tail (italicized). The yfgL PCR product and the pBAD24 plasmid were cut with NheI and HindIII restriction enzymes (New England Biolabs) and ligated with T4 DNA ligase (Fermentas). In a similar fashion, yfgL homologs from Pseudomonas aeruginosa and Vibrio cholerae were cloned into vector pTrc99A, which had been digested with NcoI and HindIII. P. aeruginosa yfgL was amplified using forward primer 5′-CCTGCCATGGTGCAATGGAAACACGCGGCGC-3′ (the NcoI restriction site is underlined) and reverse primer 5′-ATCATAAGCTTAGTGATGGTGATGGTGATGGCGGATGGTGTAGGCGACGAGC-3′ (the HindIII cut site is underlined, and the six-histidine tag is italicized). V. cholerae yfgL was amplified using forward primer 5′-CTCCCTCATGAAGAAGCTGTTCAATCAAGTG-3′ (the BspHI restriction site is underlined) and reverse primer 5′-ATCATAAGCTTAGTGATGGTGATGGTGATGTTGTTGAATCGTCAGCTTCTTTATCTGGC-3′ (the HindIII site is underlined, and the six-histidine tail is italicized). E. coli chromosomal acrA was amplified with forward primer 5′-GCAGGTACCGGACACTCGAGGTTTACATATG-3′ (the KpnI cut site is underlined) and reverse primer 5′-GCTCTAGAAGCTTAGTGATGGTGATGGTGATGAGACTTGGACTGTTCAGGCTGACG-3′ (the HindIII cut site is underlined, and the six-histidine tail is italicized). The amplified acrA gene and plasmid pBAD33 were restricted with KpnI and HindIII and ligated. yaeT and tolC were cloned into vectors pBAD33 and pTrc99A (Pfizer-Pharmacia), respectively, in our previous works (41, 42).
The YfgL-His6 variants were created using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions and the pBAD24-yfgL-His6 construct as the template. The two YfgL-His6 scramble alleles were generated via two rounds of site-directed mutagenesis (SDM). In the first round, a guanine was inserted between the second and third bases of the P171 codon (scramble 1), and the first nucleotide of the S172 codon was deleted (scramble 2). In the second round of SDM, the third nucleotide of the 180th codon was deleted (scramble 1), and a thymine was inserted between the 180th and 181st codons (scramble 2) to restore the downstream reading frame. SDM primer sequences are available upon request.
Growth conditions.
The growth of strain expressing a plasmid-borne yfgL allele in a ΔyfgL-scar ΔdegP::Tn10 background was tested by incubating the mutant at 30°C for 20 h on an LB agar plate with arabinose (0.2% [wt/vol], final). MIC determination was performed by inoculating 106 cells in 1 ml LB containing 0.2% arabinose, followed by incubation at 37°C for 18 h on a roller drum.
Immunoprecipitation.
Twenty-five milliliters of LB was inoculated with a 1:100 dilution of a culture grown overnight and incubated to an optical density at 600 nm (OD600) of approximately 0.3 at 37°C before the cells were induced with arabinose (0.2%, final) for 2 h to overexpress both YaeT and His-tagged YfgL. Cells were pelleted and lysed by freeze-thawing in 0.1 ml lysis buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.1 mM EDTA, 0.35 μM lysozyme, 0.1% Triton). After incubation on ice for 30 min, proteins in the cell lysates were solubilized under gentle conditions using 1 ml BugBuster protein extraction reagent (Novagen), 0.5 μl Benzonase nuclease (Novagen), and 0.1 mM (final) phenylmethylsulfonyl fluoride for 20 min at room temperature on a rotating mixer. The mixtures were centrifuged for 25 min at 20,000 × g, and supernatants containing the solubilized proteins were saved. Proteins in the clarified extract were precipitated using the IP50 protein G immunoprecipitation kit (Sigma-Aldrich) according to the manufacturer's protocol, with the following optimizations. Incubation of 300 μl extract with 0.4 μg Penta-His antibody (QIAGEN) was done overnight at 4°C on a rocker. Following incubation with protein G beads, seven washes were performed, the first two of which contained 0.5 M NaCl. Precipitates were eluted from immunoprecipitate columns after boiling for 5 min in 50 μl sodium dodecyl sulfate (SDS) sample buffer containing β-mercaptoethanol. Eluates were separated on an 11% SDS-polyacrylamide gel. Proteins were visualized using the SilverQuest silver staining kit (Invitrogen) according to the manufacturer's instructions. Proteins were also subjected to Western analysis as described below.
Cross-linking.
Cells harboring mutant yfgL-His6 and wild-type yaeT alleles under the control of the arabinose-inducible promoter PBAD on compatible plasmids were grown in 25 ml LB to an OD600 of approximately 0.3 at 37°C before they were induced with 0.2% arabinose for 2 h. For cells expressing plasmid-borne yaeT-His6 in a yfgL+ or yfgL mutant background, 25-ml cultures were grown to an OD600 of approximately 0.8 without induction. In either case, cells were then harvested, washed twice with cross-linking buffer (20 mM Na-phosphate [pH 7.5], 150 mM NaCl, 1 mM EDTA), and resuspended in 5 ml cross-linking buffer. Resuspended cells were incubated in the absence or presence of a DSP [dithiobis(succinimidylpropionate)] (0.5 mM; Pierce) or SPDP [N-succinimidyl 3-(2-pyridyldithio) propionate] (0.2 mM; Pierce) cross-linker for 30 min at room temperature on a rotating mixer. The reaction was quenched with Tris-HCl (pH 7.5) (for DSP [40 mM]) or l-cysteine (for SPDP [25 mM]). Cells were pelleted and washed with cross-linking buffer. Cells cross-linked with DSP were made into extracts for immunoprecipitation as described above.
Proteins in cells cross-linked with SPDP were solubilized and denatured in PUTTS (100 mM NaH2PO4, 8 M urea, 10 mM Tris-HCl [pH 7.5], 1% Triton X-100, 0.2% Sarkosyl) with 10 mM imidazole for 1 h at room temperature on a rotating mixer. Cellular debris was spun down, and supernatants containing cross-linked soluble proteins were saved. Cross-linked proteins were purified using Ni-nitrilotriacetic acid (NTA) spin columns (QIAGEN). Briefly, 600 μl of sample was passed through a Ni-NTA spin column preequilibrated with PUTTS containing 10 mM imidazole. Next, the column was washed three times with PUTTS containing 100 mM imidazole. Bound proteins were eluted with 130 μl PUTTS containing 500 mM imidazole. All spins were done for 2 min at 400 × g or 800 × g. Eluates were mixed with SDS sample buffer, boiled for 5 min, and resolved on an 11% SDS-polyacrylamide gel. Purified proteins were visualized by silver staining.
Western blot analysis.
Cultures were grown to an OD600 of approximately 0.3 before induction with arabinose (0.2% [wt/vol], final) at 37°C for 2 h. The amount of His-tagged YfgL in 1 ml of whole cells was detected using HisProbe-HRP as described previously (19). LamB in 0.25 ml of whole cells was probed using antibody raised against LamB (1:10,000) as described previously (2). In a similar manner, AcrA, SurA, and YaeT were detected using antibodies raised against these proteins (AcrA [1:10,000] and SurA [1:5,000]) or a 15-residue N-terminal mature peptide (YaeT [1:2,000]).
Bioinformatics.
Homologs of E. coli YfgL in other bacterial species were found by submitting the entire E. coli YfgL protein sequence to the Blastp protein-protein BLAST (Basic Local Alignment Search Tool) program via the Swiss Institute of Bioinformatics interface at http://au.expasy.org/tools/BLAST/ (10). The database option was set to “bacteria,” and the number of best-scoring sequences to show was set to 250. The mature protein sequence alignment of YfgL homologs was generated by ClustalW (3) (http://www.ebi.ac.uk/Tools/clustalw/). ClustalW was also used to align TolC-VceC, AcrA-MexA, and AcrB-MexB and to determine their sequence identities. Putative binding domains in E. coli YfgL were found by submitting the GenBank accession number of the lipoprotein, P77774, to the Simple Modular Architecture Research Tool (SMART) Web application (17) (http://smart.embl.de/smart/show_motifs.pl?ID=P77774) for analysis.
RESULTS
Identification of structurally and functionally significant YfgL residues.
Clusters of conserved residues of homologous, functionally exchangeable proteins from evolutionarily distant species often reflect the importance of those regions to the proteins' structures and functions. This reasoning was used to identify structurally and functionally important residues of E. coli YfgL. The BLAST program was run to search for YfgL homologs in other bacteria. Initially, YfgL homologs from many different bacterial species were selected and submitted to ClustalW for sequence alignment against E. coli YfgL. The output from ClustalW showed only one cluster of conserved residues toward the C terminus of the protein. When we selected fewer YfgL homologs to align with E. coli YfgL, more (but not many) patches of conserved residues appeared. While it is reasonable to expect these new regions to be less critical, they may nevertheless have important functions.
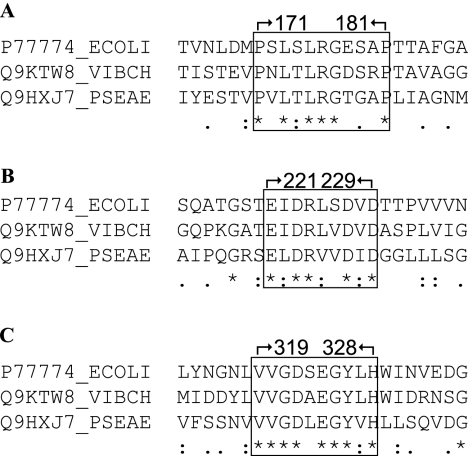
We were drawn to YfgL homologs from Vibrio cholerae and Pseudomonas aeruginosa, two bacterial species that are evolutionarily distant from E. coli. The YfgL amino acid sequences of V. cholerae and P. aeruginosa share only 43% and 31% sequence identity with E. coli YfgL, respectively. Nevertheless, alignments of the two YfgL homolog sequences to that of E. coli YfgL showed several conserved regions (see Fig. S1 in the supplemental material). Moreover, expression of the cloned V. cholerae and P. aeruginosa yfgL genes from an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible pTrc99A vector complemented a ΔyfgL allele of E. coli (data not shown). This is significant since other proteins from V. cholerae and P. aeruginosa do not complement their E. coli counterparts despite having higher sequence identities. For instance, VceC, the outer membrane component of the VceABC multidrug-resistant efflux pump of V. cholerae, cannot complement TolC, the outer membrane member of the AcrAB-TolC efflux pump in E. coli (38), even though VceC and TolC are 45% identical at the protein sequence level. MexA and MexB of P. aeruginosa and AcrA and AcrB of E. coli share 53% and 70% (34) sequence identities, respectively, and yet MexAB is nonfunctional in E. coli (34). The finding that V. cholerae and P. aeruginosa YfgL can still function in E. coli, despite sharing lower sequence identity, gives credence to the conserved regions. This study focuses on three regions in the mature E. coli YfgL sequence: P171 to P181, E221 to D229, and V319 to H328 (Fig. 1).
FIG. 1.
Homologs of E. coli YfgL in V. cholerae and P. aeruginosa were found by the BLAST program. Alignment of the three mature YfgL protein sequences by the ClustalW application reveals several conserved regions. (A) One conserved region spans residues 171 to 181 of the mature protein sequence. (B) Another region encompasses residues 221 to 229. (C) A third region encompasses residues 319 to 328.
The region at P171 to P181 of YfgL has functional significance.
Initially, to assess the overall relevance of this P171-to-P181 region to the structure and function of YfgL, wild-type residues S172 to A180 of the histidine-tagged yfgL clone were changed to amino acids that are overall bulkier, aromatic, and nonpolar (scramble 1) and those that have overall properties similar to those of the wild type (scramble 2) via two rounds of SDM. (The two flanking conserved proline residues, P171 and P181, were not altered because doing so might adversely affect the stability of the YfgL variant.)
The two scramble yfgL mutants exhibited a pleiotropic phenotype very similar to that of a ΔyfgL strain; that is, the scrambled yfgL strains had reduced LamB levels, displayed vancomycin hypersensitivity, and formed extremely small, nonhomogeneous colonies in a ΔdegP background at 30°C (Table 1). The levels of the two mutant YfgL proteins were very similar to that of the wild type; hence, the yfgL mutant phenotype was not due to low YfgL levels (Table 1). The scramble 1 and scramble 2 alleles were recessive to the wild-type yfgL allele (data not shown), indicating that the pleiotropic phenotype exhibited by the YfgL mutants was due to the synthesis of nonfunctional YfgL proteins. We surmise from these results that wild-type residues S172 to A180 are indispensable for the proper functioning of YfgL.
TABLE 1.
Phenotypes of strains producing plasmid-borne YfgL variants
| Proteina | Mean YfgL level ± SDb | Mean LamB level ± SDb | MIC of vancomycin (μg/ml) | Growth in ΔdegP backgroundc |
|---|---|---|---|---|
| YfgL− | 0.00 | 0.46 ± 0.12 | 40 | −/+ |
| YfgL-His6 wild type | 1.00 | 1.00 | 120 | +++ |
| YfgL-His6 scramble 1 | 0.95 ± 0.03 | 0.69 ± 0.09 | 40 | −/+ |
| YfgL-His6 scramble 2 | 1.05 ± 0.01 | 0.41 ± 0.22 | 40 | −/+ |
| YfgL(L173S,L175S,R176A)-His6 | 0.82 ± 0.12 | 0.61 ± 0.09 | 40 | +/− |
| YfgL(L175S,R176A)-His6 | 0.89 ± 0.03 | 0.79 ± 0.10 | 60 | ++ |
| YfgL(L173S,R176A)-His6 | 0.95 ± 0.02 | 0.72 ± 0.13 | 80 | ++ |
| YfgL(L173S,L175S)-His6 | 0.94 ± 0.04 | 1.02 ± 0.27 | 120 | +++ |
| YfgL(R176A)-His6 | 0.92 ± 0.10 | 0.90 ± 0.04 | 120 | +++ |
| YfgL(L175S)-His6 | 0.86 ± 0.04 | 1.08 ± 0.03 | 120 | +++ |
| YfgL(L173S)-His6 | 0.92 ± 0.05 | 0.94 ± 0.14 | 120 | +++ |
| YfgL(S172A)-His6 | 0.96 ± 0.05 | 1.14 ± 0.16 | 120 | +++ |
| YfgL(S172C)-His6 | 0.72 ± 0.16 | 0.78 ± 0.26 | 80 | ++ |
| YfgL(E221A)-His6 | 0.84 ± 0.04 | 0.77 ± 0.05 | 120 | +++ |
| YfgL(D223A)-His6 | 0.94 ± 0.08 | 0.89 ± 0.13 | 120 | +++ |
| YfgL(R224A)-His6 | 0.78 ± 0.23 | 0.97 ± 0.01 | 120 | +++ |
| YfgL(D227A)-His6 | 0.65 ± 0.07 | 0.80 ± 0.06 | 80 | ++ |
| YfgL(D229A)-His6 | 0.95 ± 0.15 | 0.90 ± 0.15 | >120 | +++ |
| YfgL(S226C)-His6 | 0.86 ± 0.17 | 0.90 ± 0.03 | 120 | +++ |
Plasmid pBAD24 served as the negative control (YfgL−). All experiments were done with full arabinose induction (0.2% [wt/vol], final). Residue numbers are relative to the beginning of the mature YfgL protein sequence.
Steady-state YfgL or LamB protein levels were analyzed by Western analysis. Three independent LB cultures were quantified and averaged relative to the wild-type level.
Growth of a strain producing YfgL-His6 or a variant in a DegP− background at 30°C on an LB agar plate with 0.2% arabinose. −/+ indicates the strain formed extremely tiny heterogeneous colonies. +/− indicates that the strain formed small heterogeneous colonies.
L173, L175, and R176 are functionally important residues in the P171-to-P181 region of YfgL.
Having established a functional significance of the region at S172 to A180 of YfgL, we proceeded to pinpoint the specific residues in this region that are important for YfgL's function. As shown in Fig. 1A, three conserved residues, L173, L175, and R176, stood out as good candidates for mutagenesis analysis. (The conserved G177 was not included because, like P171 and P181, it probably plays a structural role.) Seven yfgL alleles were created from a yfgL-His6 construct via SDM. Of the resulting mutant proteins, three had a single alteration each (L173S, L175S, or R176A), three had two alterations each (L173S and L175S, L173S and R176A, or L175S and R176A), and one had all three alterations (L173S, L175S, and R176A).
All three YfgL mutants with a single-amino-acid substitution produced the lipoprotein at levels close to that of the wild type, and they were phenotypically identical to the wild type in their production of LamB, sensitivity to vancomycin, and ability to form single colonies in a degP background at 30°C (Table 1). Even when L173S and L175S are paired together, a strain expressing the double mutant yfgL allele still showed wild-type-like phenotypes (Table 1). However, cells expressing YfgL with R176A and either L173S or L175S started to exhibit phenotypic defects. In particular, they had lower LamB levels, were sensitive to vancomycin, and could not form robust colonies in the absence of DegP (Table 1). These defects were exacerbated and, with respect to vancomycin hypersensitivity and growth in the degP background, became yfgL-null-like in a strain expressing the yfgL allele with all three alterations (Table 1). Note that all three strains exhibiting phenotypic defects produced YfgL(L173S,R176A)-His6, YfgL(L175S,R176A)-His6, and YfgL(L173S,L175S,R176A)-His6 at roughly 80% to 90% of wild-type levels (Table 1). The phenotypes were unlikely to be due to reduced YfgL levels since another mutant described below, YfgL(Y326A)-His6, did not display vancomycin hypersensitivity or growth defects in a ΔdegP background even when YfgL was expressed at only 31% of the wild-type level. Taken together, these data showed that residues L173, L175, and R176 of YfgL are functionally relevant, with R176 being the most important, followed by L175 and then L173.
Altering the L173, L175, or R176 residue of YfgL results in defective YaeT interactions.
The fact that strains expressing the yfgL scramble 1, scramble 2, or triple mutant exhibit a yfgL-null phenotype even though the variant proteins are produced at levels close to that of wild-type YfgL suggests that the mutant lipoproteins might have lost their ability to interact with YaeT. Sucrose density gradient fractionation analysis of inner and outer membranes of strains expressing the mutant yfgL alleles found the YfgL proteins in the outer membrane fractions, just as the OMP LamB control did (data not shown). Since it is difficult to imagine how alterations in the mature region at P171 to P181 of YfgL could have interfered with the outer membrane anchoring of YfgL, a process mediated solely by the addition of acyl chains to the N-terminal cysteine residue of lipoproteins (35), it is reasonable to expect that YfgL variants were localized properly to the outer membrane. Thus, the apparent lack of interactions is unlikely to be due to the mislocalization of YfgL.
Previous studies showed that each member of the YaeT complex makes physical and direct contacts with at least one other member (18, 32). These interactions were demonstrated by His tagging of a component protein at the C terminus followed by coimmunoprecipitation using anti-His-tag antibody (18, 32). Similar copurification experiments were performed to assay the physical interactions between YaeT and the YfgL-His6 variants in this study.
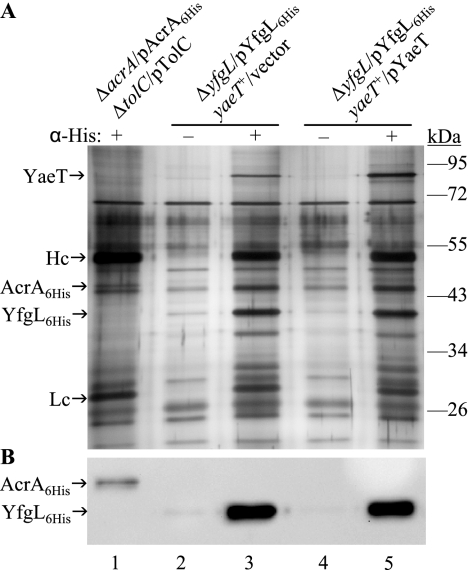
To assess the specificity of the coimmunoprecipitation procedure, cell extracts from two strains expressing wild-type YfgL-His6 from a plasmid construct and either chromosomal or chromosomal plus plasmid-borne YaeT were incubated with and without His tag antibody. Additionally, cell extracts from another culture expressing an inner membrane lipoprotein, AcrA-His6, were used as a negative control. Eluted immunoprecipitates were separated on an SDS-polyacrylamide gel electrophoresis (PAGE) gel and silver stained (Fig. 2A). In the presence of anti-His-tag antibodies, YfgL-His6 coprecipitated with a protein that migrated at an apparent molecular mass of ≈90 kDa, which is the reported size of YaeT (43) (Fig. 2A, lanes 3 and 5). The protein was confirmed to be YaeT by antibodies raised against it (data not shown). In addition, more of YaeT was pulled down in the strain harboring a multicopy yaeT plasmid than in the strain expressing chromosomal yaeT only (Fig. 2A, lanes 3 and 5). Without His tag antibody, YfgL-His6 and YaeT were not precipitated in the samples (Fig. 2A, lanes 2 and 4). As for the negative control, His tag antibody was able to precipitate AcrA-His6 but not YfgL or YaeT, thus further demonstrating the specificity of the antibody and the entire immunoprecipitation procedure (Fig. 2A, lane 1). The presence of AcrA-His6 and YfgL-His6 in the corresponding immunoprecipitates was verified by HisProbe-HRP (Fig. 2B). There are other proteins of approximately 47, 38, and 32 kDa that coprecipitated with YfgL-His6 (Fig. 2, lanes 3 and 5). The 38-kDa band could be NlpB, a 35-kDa lipoprotein that is a member of the YaeT complex (43). The other bands are likely contaminant proteins; they are not consistently brought down by YfgL-His6 (compare Fig. 2A and 3A).
FIG. 2.
Coimmunoprecipitation of YaeT with histidine-tagged YfgL to establish the specificity of the histidine antibody and the coimmunoprecipitation procedure. (A) Whole-cell lysates were immunoprecipitated with (+) or without (−) monoclonal anti-His antibody. The collected immunoprecipitates were separated by SDS-PAGE and visualized by silver staining. Lane 1, production of plasmid-borne histidine-tagged AcrA (AcrA6His), a component of the TolC-AcrAB tripartite efflux system (9) in E. coli; lanes 2 and 3, YfgL-His6 (YfgL6His) was produced from plasmid pBAD24 and YaeT from the chromosome; lanes 4 and 5, yfgL-His6 was expressed from pBAD24, and yaeT was expressed from both the chromosome and plasmid pBAD33 (12). Hc, immunoglobulin G heavy chain; Lc, immunoglobulin G light chain. (B) Histidine-tagged AcrA or YfgL in the corresponding immunoprecipitate eluates was confirmed by Western analysis using HisProbe-HRP.
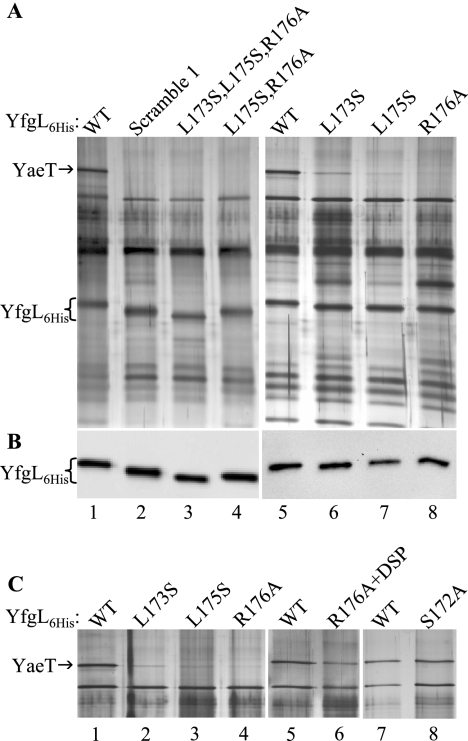
FIG. 3.
Alterations in the first conserved region of YfgL (residues 171 to 181) result in weakened YaeT interactions. (A) Coimmunoprecipitation of YaeT with histidine-tagged YfgL variants. Wild-type (WT) YfgL-His6 (YfgL6His) (lanes 1 and 5) and variants (lanes 2 to 4 and 6 to 8) were produced from pBAD24. All the strains carried a copy of yaeT on the chromosome and on pBAD33. Proteins immunoprecipitated from cell lysates incubated with Penta-His antibody were resolved on an 11% SDS-polyacrylamide gel and silver stained. (B) HisProbe-HRP detection of histidine-tagged YfgL in the same immunoprecipitate samples from A. (C) Silver-stained SDS-polyacrylamide gel of YaeT coimmunoprecipitated with YfgL-His6 variants. The left panel (lanes 1 to 4) is a repeat of the right panel in A (lanes 5 to 8) but without the two 0.5 M NaCl washes. The middle panel (lanes 5 and 6) shows YaeT precipitated by YfgL(R176A)-His6. Prior to immunoprecipitation, cells expressing YfgL(R176A)-His6 were incubated with the DSP cross-linker. The right panel (lanes 7 and 8) shows YaeT coprecipitated with YfgL(S172A)-His6 without cross-linking treatment.
Coimmunoprecipitation experiments were performed on strains producing wild-type YfgL-His6, YfgL-His6 scramble 1, YfgL(L173S,L175S,R176A)-His6, or YfgL(L175S,R176A)-His6 using His tag antibody. YfgL-His6 was precipitated from all four strain extracts (Fig. 3A, lanes 1 to 4), as verified by Western blotting using HisProbe-HRP (Fig. 3B, lanes 1 to 4). YaeT coprecipitated only from the extract obtained from the strain producing wild-type YfgL (Fig. 3A, lane 1) and not from those producing any of the three YfgL mutant proteins (Fig. 3A, lanes 2 to 4). These results showed that YaeT could no longer stably associate with YfgL-His6 scramble 1, YfgL-His6(L173S,L175S,R176A), or YfgL(L175S,R176A)-His6. Thus, there is a positive correlation between the phenotypic defects of the three YfgL mutants and their inabilities to interact with YaeT.
Next, we checked the association between YaeT and the three YfgL-His6 mutants that do not display an obvious phenotype, YfgL(L173S)-His6, YfgL(L175S)-His6, and YfgL(R176A)-His6. Coimmunoprecipitation experiments showed that while all three YfgL-His6 variants were effectively pulled down, as confirmed by Western blotting using HisProbe-HRP (Fig. 3B, lanes 6 to 8), little YaeT was coprecipitated (Fig. 3A, lanes 6 to 8). Based on the amount of YaeT copurified, it appears that in terms of interactions with YaeT in vivo, YfgL(R176A)-His6 and YfgL(L175S)-His6 are weaker than YfgL(L173S)-His6.
We noted that the SDS-denatured YfgL mutant proteins migrated slightly differently from each other and wild-type YfgL-His6 (Fig. 3B). The reason for this behavior is not known, but it could be due to changes in the secondary structure of the protein. Changes in gel mobility have been observed previously in some OMP mutants carrying single-amino-acid substitutions (21, 23, 37).
It is surprising that only little YaeT could be precipitated in strains producing YfgL with a single substitution (L173S, L175S, or R176A), considering that these strains are phenotypically wild type. An explanation could be that the YfgL-YaeT interaction in vivo is compromised but not to an extent that produces a phenotype. It is conceivable that if the YfgL-YaeT interaction in vivo is already weakened due to single substitutions in YfgL, the association may have been further disrupted by the two stringent 0.5 M NaCl washes of the coimmunoprecipitation procedure or by detergents present in the BugBuster protein extraction reagent. To test the first possibility, we replaced the two harsh washes with ones without salt and found no difference in the amount of YaeT coprecipitated (Fig. 3C, lanes 1 to 4, versus A, lanes 5 to 8). If weak YfgL-YaeT complexes get dissociated during the protein solubilization/extraction step, then cross-linking the complex to stabilize it before cell lysis should yield more YfgL-YaeT coprecipitate. This was tested by incubating cells with the amine-reactive DSP cross-linker prior to protein extraction. Cell extract from the cross-linked culture was then used in a coimmunoprecipitation reaction. Silver staining of immunoprecipitated proteins separated on an SDS-PAGE gel revealed that a significantly increased amount of YaeT was brought down (Fig. 3C, lane 6, versus A, lane 8), thus demonstrating that YfgL(R176A)-His6, and also presumably YfgL(L173S)-His6 and YfgL(L175S)-His6, has a defective association with YaeT in vivo. As described below in the section on YfgL-YaeT-SurA interactions, a similar experiment involving DSP cross-linking and immunoprecipitation confirmed that YfgL(L173S)-His6 and YfgL(L175S)-His6 do have defective in vivo YaeT binding (see Fig. 5A).
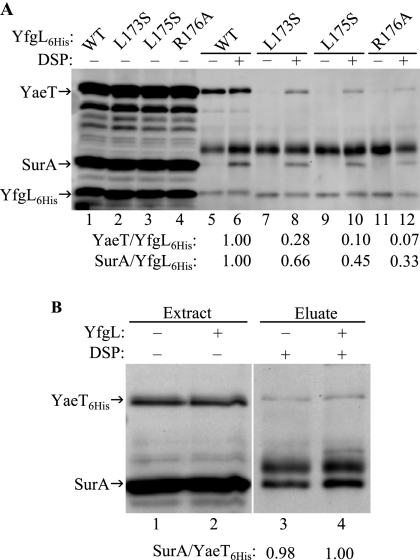
FIG. 5.
(A) Western blot analysis of the YfgL-YaeT-SurA complex. Cells producing wild-type (WT) YfgL-His6 (lanes 5 and 6), YfgL(L173S)-His6 (lanes 7 and 8), YfgL(L175S)-His6 (lanes 9 and 10), or YfgL(R176A)-His6 (lanes 11 and 12) were treated with (+) or without (−) the DSP cross-linker before they were gently lysed. Extracts were incubated with His antibody, and protein complexes were immunoprecipitated. Immunoprecipitated proteins (lanes 5 to 12), along with those in whole-cell extracts (lanes 1 to 4), were resolved by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. YfgL-His6, YaeT, and SurA protein bands were probed using HisProbe-HRP and antibodies against YaeT and SurA, respectively, and the protein levels were quantified. For the immunoprecipitated samples, the ratios of YaeT to YfgL-His6 were calculated and then normalized to 1. The normalized ratios of SurA to YfgL-His6 were calculated in an analogous manner. Only the ratios for the cross-linked samples are shown. (B) Effect of YfgL on YaeT machinery-SurA binding. Plasmid-borne histidine-tagged yaeT and chromosomal surA were expressed in a yfgL+ or yfgL mutant background. Whole-cell extracts of YaeT-His6 and SurA (lanes 1 and 2) and coimmunoprecipitation of YaeT-His6-SurA complexes stabilized by DSP (lanes 3 and 4) were subjected to Western analysis using antibodies against YaeT and SurA. The ratio of SurA to YaeT-His6 precipitated in the yfgL+ or yfgL eluate is shown normalized to 1 (lanes 3 and 4).
To recap, we have shown that cells producing YfgL-His6 with single substitutions at residue L173, L175, and R176 exhibit no apparent phenotypic defects, but they do display weakened YaeT-YfgL interactions. In order for the destabilized YaeT-YfgL association to survive gentle extraction, cross-linker-mediated stabilization is necessary. It is possible that an alteration to any residue in the P171-to-P181 region of YfgL and not just the conserved L173, L175, and R176 residues would produce the same results. To eliminate this remote possibility, the nonconserved S172 residue was mutagenized to alanine. Cells expressing YfgL(S172A)-His6 had the wild-type phenotype, just like those producing YfgL-His6 with L173, L175, or R176 (Table 1). However, the amount of YaeT coimmunoprecipitated (without cross-linking treatment) was the same as that pulled down by wild-type YfgL-His6 (Fig. 3C, lanes 7 to 8), reflecting an in vivo intact YaeT association. These data show that residues L173, L175, and R176 of YfgL are specifically involved in the interaction with YaeT. Moreover, the inability of YfgL(L173S)-His6, YfgL(L175S)-His6, and YfgL(R176A)-His6 to coprecipitate YaeT is indicative of genuine defective in vivo interaction and not an experimental artifact.
The region of P171 to P181 of YfgL makes direct contacts with YaeT.
Our data suggest that the region of P171 to P181 of YfgL interacts with YaeT since altering the conserved residue L173, L175, or R176 in this region resulted in weakened associations with YaeT in vivo. However, our data do not indicate whether the interaction is direct or indirect. (The region at P171 to P181 of YfgL might indirectly interact with YaeT by influencing another YfgL region that comes into direct contact with YaeT.) If the region at P171 to P181 directly interacts with YaeT, it is reasonable to expect this YfgL region and YaeT to be in very close proximity.
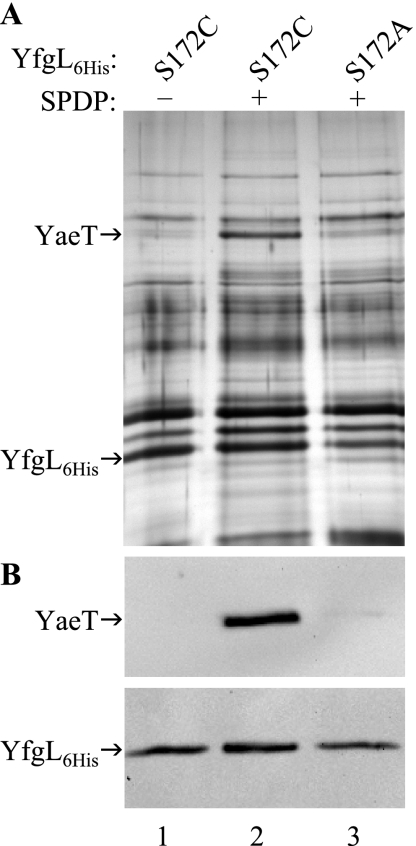
This hypothesis was tested by using SPDP, a chemical cross-linker that has a spacer arm length of only 6.8 Å and forms amine-to-sulfhydryl cross-links. Since cells producing YfgL(S172A)-His6 have a wild-type phenotype and no defect in interactions with YaeT, as discussed above, the same S172 residue was replaced by a cysteine residue. Cells expressing this new yfgL allele exhibited only a slight phenotype (Table 1). Since YfgL(S172A)-His6 has no phenotype, the mild phenotype of YfgL(S172C)-His6 is due to the presence of the sulfhydryl group. Since the only cysteine residue in the wild-type YfgL lipoprotein gets modified during and immediately after translocation across the inner membrane (26), the SPDP cross-linker could conjugate only YfgL-His6 at the S172C residue with its sulfhydryl-reactive end and YaeT with its amine-reactive end. We noted that YaeT has two cysteine residues in the β-barrel domain, and therefore, it is possible that SPDP forms sulfhydryl bonds with these cysteines and amine bonds with lysine residues of YfgL in a region other than P171 to P181, which lacks lysine. If this indeed happens, SPDP should cross-link YfgL-His6(S172A) and YaeT as efficiently as it does YfgL(S172C)-His6 and YaeT.
Cells producing YfgL(S172C)-His6 or YfgL(S172A)-His6 were grown and then incubated in the presence or absence of SPDP. Cross-linked protein complexes were isolated under denaturing conditions, affinity purified, separated on an SDS-PAGE gel, and visualized by silver staining (Fig. 4A). Proteins were denatured prior to affinity purification so that only chemically cross-linked interactions were examined. As expected, mutant YfgL-His6 was present in all three samples, as verified by HisProbe-HRP (Fig. 4B, bottom). Western blot analysis using anti-YaeT antibody showed that significant amounts of YaeT copurified with YfgL(S172C)-His6 but only when SPDP was present (Fig. 4B, top, lanes 1 to 2). In contrast, the amount of YaeT copurified with YfgL(S172A)-His6 was dramatically lower than the amount of YaeT copurified with YfgL(S172C)-His6 (Fig. 4B, top, lanes 2 to 3). Thus, SPDP could efficiently conjugate YfgL(S172C)-His6 with its sulfhydryl-reactive end and YaeT with its amine-reactive end but not the other way around. These results show that residues within the P171-to-P181 region of YfgL make direct contact with YaeT.
FIG. 4.
YfgL residue S172 makes direct contact with YaeT. (A) Cells producing YfgL-His6 (YfgL6His) harboring an S172C (lanes 1 and 2) or S172A (lane 3) alteration were treated with (lanes 2 and 3) or without (lane 1) the cysteine-directed cross-linker SPDP. YfgL-His6 and conjugated proteins were extracted under denaturing conditions and purified by nickel affinity spin columns. Purified protein complexes were separated on an SDS-PAGE gel and visualized by silver staining. (B) The presence of purified YfgL-His6 (bottom) and YaeT (top) was confirmed by Western blot analysis using HisProbe-HRP and YaeT antibodies, respectively.
YfgL-YaeT-SurA interaction.
After translocation across the inner membrane, periplasmic chaperones such as SurA and Skp are thought to bind to nascent OMP peptides to keep them in a proper folding state and to traffic them across the periplasm to the YaeT assembly complex (31). Based on the kinetic analysis of LamB assembly, it was proposed that SurA delivers LamB to the YaeT complex via YfgL (36). If SurA does deliver its cargo to YaeT via YfgL, then YfgL mutants defective in YaeT binding should still be able to pull down the same amount of SurA as wild-type YfgL does, independent of the amount of YaeT coprecipitated.
To test this hypothesis, we performed coimmunoprecipitation experiments using four strains producing wild-type YfgL-His6 and the YfgL(L173S)-His6, YfgL(L175S)-His6, and YfgL(R176A)-His6 variants. Prior to lysis, cells were treated with the DSP cross-linker to stabilize the weakened association between YaeT and the YfgL(R176A)-His6 mutant and to lock in the presumably transient interactions between SurA and the YaeT assembly apparatus. Recall that YfgL(L173S)-His6 and YfgL(L175S)-His6 presumably exhibit weaker YaeT interactions in vivo as well, so DSP also served to enable the two YfgL-His6 variants and YaeT to withstand the immunoprecipitation protocol.
The amounts of YaeT and SurA coprecipitated with histidine-tagged YfgL were assessed by Western analysis. The levels of YaeT, SurA, and YfgL in the whole-cell extracts of the four strains were very similar (Fig. 5A, lanes 1 to 4). As expected, whereas the amounts of YaeT pulled down by wild-type YfgL-His6 were similar with or without DSP (Fig. 5A, lanes 5 and 6), the amounts of YaeT coprecipitated by the three YfgL-His6 variants were significantly greater when stabilized by DSP (Fig. 5A, lanes 8, 10, and 12 versus 7, 9, and 11). This demonstrates that in addition to YfgL(R176A)-His6, YfgL(L173S)-His6 and YfgL(L175S)-His6 also have defects in YaeT interactions in vivo. Moreover, altering residues L173, L175, and R176 of YfgL resulted in decreasing amounts of copurified YaeT (Fig. 5A, lane 8, 10, 12), showing that R176 is the most important residue in YaeT interactions, followed by L175 and then L173. Note that this corroborates the finding in the phenotype analysis section above that R176 is the most functionally relevant residue and L173 is the least functionally relevant amino acid (Table 1).
Not surprisingly, significantly more SurA could be detected when cells received DSP treatment (Fig. 5A, lanes 6, 8, 10, and 12 versus lanes 5, 7, 9, and 11). In contrast to the large amount of YaeT, a barely visible amount of SurA coprecipitated with wild-type YfgL-His6 without DSP (Fig. 5A, lane 5). These observations show that the interactions between SurA and the YaeT assembly complex are not as strong as those between wild-type YfgL-His6 and YaeT. Coincidentally, and consistent with our findings, recent work also detected SurA bound to the YaeT complex and found that DSP stabilization is required (33).
More importantly, the amounts of SurA precipitated in the wild-type YfgL-His6 background and in YfgL-His6 variant backgrounds differed (Fig. 5A, lane 6 versus 8, 10, and 12). Specifically, the more defective the YfgL variant is in YaeT binding (that is, the less YaeT that is coprecipitated), the less SurA was brought down (Fig. 5A, lanes 6, 8, 10, and 12). The data suggest that the SurA species was not directly pulled down by YfgL-His6; rather, YfgL-His6 precipitated the YaeT machinery with SurA complexed to it. Thus, SurA does not appear to deliver its OMP cargo to the YaeT machinery via YfgL.
Even though our data provide evidence that incoming SurA does not bind to YfgL, they do not preclude the possibility that YfgL may still exert an effect on SurA binding. For instance, YfgL's binding to YaeT may enhance YaeT's ability to receive SurA-OMP complexes. This possibility seems to be supported by the YaeT/YfgL-His6 and SurA/YfgL-His6 ratios shown in Fig. 5A. If YfgL has no influence on SurA reception, one would predict the amount of SurA pulled down per YfgL-His6 to be less than (or the same as) the amount of YaeT precipitated for each bait molecule, since DSP has to first conjugate YfgL to YaeT and then conjugate the YaeT complex to SurA. However, this is not the case: YfgL(L173S)-His6, YfgL(L175S)-His6, and YfgL(R176A)-His6 brought down relatively more SurA than YaeT (Fig. 5A, lanes 8, 10, and 12). To test if YfgL has any influence on SurA docking, we performed DSP conjugation and immunoprecipitation on strains expressing hexahistidine-tagged yaeT in a yfgL+ or yfgL mutant background. Precipitated YaeT-His6-SurA complexes were subjected to Western analysis (Fig. 5B, lanes 3 and 4) along with YaeT-His6 and SurA in whole cells (Fig 5B, lanes 1 and 2). As shown in Fig. 5B, the normalized ratios of SurA to YaeT-His6 precipitated in a yfgL+ or yfgL mutant background were very similar (1.00 versus 0.98, respectively) (Fig. 5B, lanes 3 and 4). These data suggest that the absence of YfgL has little or no impact on SurA docking to the YaeT machinery.
As for the SurA/YfgL-His6 ratio being higher than the YaeT/YfgL-His6 ratio, it is likely due to the YaeT antibody, which was raised against the first 15 residues of the mature YaeT peptide (32), and as such, it may not be sensitive enough for accurate detection when the level of YaeT falls below a certain threshold. The insensitive nature of the YaeT antiserum also surfaced when it was observed that YaeT immunoprecipitated by YfgL(L173S)-His6 without DSP treatment was not detected by the YaeT antibody (Fig. 5A, lane 7), even though the YaeT band was clearly visible in a silver-stained gel (gel not shown, but for comparison purposes, see Fig. 3A, lane 6, and C, lane 2).
The YfgL region of E221 to D229 also makes contact with YaeT.
We also investigated the significance of a second conserved region, which encompasses residues 221 to 229, of the YfgL protein by mutagenesis (Fig. 1B). Residues that are identical in E. coli, V. cholerae, and P. aeruginosa were altered to alanine: E221A, D223A, R224A, D227A, and D229A. In addition, a yfgL allele containing the alteration S226C was created to gauge the proximity of this region to YaeT.
The steady-state levels of YfgL(E221A)-His6, YfgL(D223A)-His6, YfgL(R224A)-His6, YfgL(S226C)-His6, and YfgL(D229A)-His6 were at least 78% of the wild-type YfgL-His6 level (Table 1). Cells expressing these five yfgL alleles exhibited a slight decrease in LamB levels but no vancomycin hypersensitivity or growth defect in a degP background at 30°C (Table 1). The YfgL(D227A)-His6 mutant expressed YfgL at 65% of the wild-type level, but even so, it showed only a slight drop in the LamB level, a slight increase in vancomycin sensitivity, and a small growth defect in the absence of DegP at 30°C (Table 1).
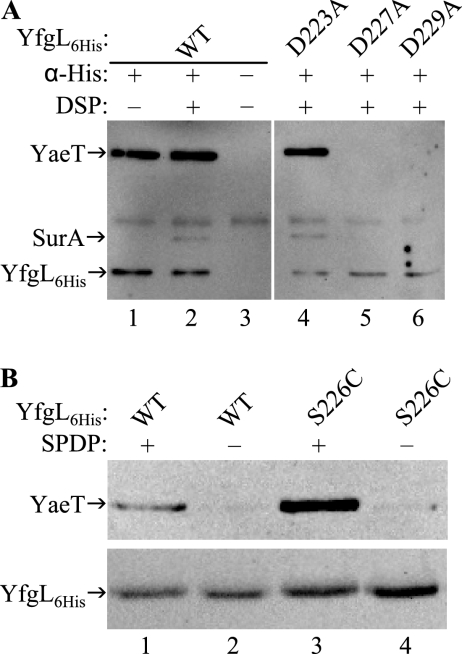
Although altering individual YfgL residues in the E221-to-D229 region did not result in gross phenotypic defects, we nevertheless checked the mutants for defects in YaeT binding. Immunoprecipitation followed by Western blotting found that YfgL(E221A)-His6, YfgL(D223A)-His6, and YfgL(R224A)-His6 could copurify YaeT but that YfgL(D227A)-His6 and YfgL(D229A)-His6 could not (data not shown). These results showed that YfgL(D227A)-His6 and YfgL(D229A)-His6 exhibit YaeT association defects. The immunoprecipitation experiment was repeated, with DSP cross-linking prior to cell lysis this time, on strains producing wild-type YfgL-His6, YfgL(D223A)-His6, YfgL(D227A)-His6, or YfgL(D229A)-His6. As expected, Western analysis on the eluates found that YaeT was brought down by both wild-type YfgL-His6 and YfgL(D223A)-His6 with or without DSP treatment (Fig. 6A, lanes 1, 2, and 4). In contrast, YfgL(D227A)-His6 and YfgL(D229A)-His6 still failed to copurify YaeT even with DSP-mediated stabilization (Fig. 6A, lanes 5 and 6). Parenthetically, we also probed the eluates for SurA. Consistent with the above-described data on the YfgL-YaeT-SurA interaction using YfgL(L173S)-His6, YfgL(L175S)-His6, and YfgL(R176A)-His6, SurA could be detected only if YaeT was copurified and SurA was stabilized by DSP (Fig. 6A, lanes 1, 2, and 4 to 6).
FIG. 6.
(A) Testing YfgL-His6 variants with a D223A, D227A, or D229A alteration for defective YaeT associations. Cells expressing wild-type (WT) YfgL-His6 (YfgL6His), YfgL(D223A)-His6, YfgL(D227A)-His6, or YfgL(D229A)-His6 were incubated in the presence or absence of the cross-linker DSP prior to lysis under nondenaturing conditions. His-tagged YfgL complexes were precipitated by Penta-His antibody (α-His) and analyzed by Western blotting. YfgL-His6, YaeT, and SurA were probed using HisProbe-HRP, anti-YaeT antibody, and anti-SurA antibody, respectively. (B) Residue S226 of the YfgL region at E221 to D229 comes in close contact with YaeT. Cells expressing wild-type YfgL-His6 or YfgL(S226C)-His6 were incubated with or without the cysteine-directed cross-linker SPDP before they were lysed under denaturing conditions. YfgL-His6 and conjugated proteins were purified with an Ni-NTA spin column and subjected to Western analysis. HisProbe-HRP and YaeT antibody were used to probe YfgL-His6 and YaeT.
Next, we tested if the E221-to-D229 region of YfgL forms another contact point with YaeT using the same approach and logic described above to show that the P171-to-P181 region makes direct contact with YaeT. Cells producing wild-type YfgL-His6 or YfgL(S226C)-His6 were incubated with the cysteine-directed cross-linker SPDP before protein complexes were extracted under harsh conditions, affinity purified, and analyzed by Western blotting. Without SPDP treatment, the amounts of YaeT copurified in the YfgL-His6 and YfgL(S226C)-His6 eluates were insubstantial (Fig. 6B, lanes 2 and 4). With SPDP incubation, the amount of YaeT copurified by YfgL(S226C)-His6 was significantly greater than that coprecipitated by wild-type YfgL-His6 (Fig. 6B, lane 3 versus 1). These results show that the S226 residue and therefore the E221-to-D229 region of YfgL are in very close proximity to YaeT.
The region AT V319 to H328 has structural importance but no apparent functional significance.
Thus far, we have investigated two regions of YfgL and found that both regions interact with YaeT. We analyzed a third conserved region, from V319 to H328, by mutagenesis to see if it also contacts YaeT (Fig. 1C). Of the eight conserved residues in the region, G321 and G325 were not analyzed because they likely play a significant role in the structure of YfgL. The remaining six conserved amino acids were altered to serine or alanine: V319S, V320S, D322A, E324A, Y326A, and H328A. In addition, the serine at nonconserved residue 323 was altered to a cysteine. Western analysis revealed that steady-state levels of plasmid-borne YfgL-His6 harboring a V319S, V320S, D322A, or H328A alteration were at or below 12% of wild-type YfgL levels. The severe biogenesis defects suggest that these four single alterations most likely prevented the proper assembly of the YfgL protein, leading to its degradation. Thus, residues V319, V320, D322, and H328 have an important role in the structure of the YfgL protein.
In contrast, mutants producing YfgL(S323C)-His6, YfgL(E324A)-His6, or YfgL(Y326A)-His6 exhibited modest defects in the biogenesis of the lipoprotein: steady-state protein levels were at 62%, 61%, and 31% of wild-type YfgL levels, respectively. As such, each of the three YfgL mutants had no increased sensitivity to vancomycin and could grow at 30°C in a DegP− background as well as a strain expressing wild-type yfgL. Coimmunoprecipitation experiment revealed that the amounts of YaeT precipitated by wild-type YfgL-His6, YfgL(S323C)-His6, YfgL(E324A)-His6, and YfgL(Y326A)-His6 were very similar (data not shown), thus demonstrating that the YfgL variants have no apparent defect in YaeT interactions. In summary, these results show that unlike the previous two YfgL regions, P171 to P181 and E221 to D229, this region is not involved in YaeT binding. However, the residues in this region, in particular, V319, V320, D322, and H328, are important for the structural integrity of the YfgL protein.
DISCUSSION
In this study, we used bioinformatics, genetics, and biochemical approaches and identified five residues in the mature YfgL protein, L173, L175, R176, D227, and D229, involved in the interaction with YaeT. Cysteine-directed cross-linking experiments demonstrated that the two YfgL regions encompassing these residues are in close contact with YaeT. Individual substitutions at L173, L175, and R176 in one region of YfgL destabilized interactions with YaeT, but together, they abolished these interactions and produced a yfgL-null phenotype. These results show that it is not the absence of YfgL per se but rather the disrupted YfgL functions and destabilized YaeT interactions that give rise to the pleiotropic phenotype of ΔyfgL.
A single substitution at D227 or D229 in another region of YfgL resulted in defective YaeT associations, but an alteration at any of the three nearby residues E221, D223, and R224 did not. Furthermore, YfgL with an alteration at D227 or D229 failed to precipitate YaeT even with DSP-mediated stabilization. The inability of DSP to conjugate YfgL to YaeT could be due to reactive amine groups on YfgL and YaeT not in the correct orientation or in the close proximity required for cross-linking. All five yfgL alleles, including the two with the YaeT binding problem, produced little or no phenotype. These findings do not necessarily suggest that the region from E221 to D229 plays a role only in YaeT associations and no other function. Indeed, it has been reported in the literature that deleting the four residues V233 to N236 downstream of D229 results in a stable YfgL variant that confers antibiotics hypersensitivity and decreased OMP levels (30). Also, recall that cells expressing yfgL with a single L173S, L175S, or R176A substitution were phenotypically wild type. They started to exhibit defects when YfgL(L173S,R176A)-His6 was produced. Thus, in order for the more severe phenotype to appear, YfgL variants may need to harbor multiple alterations in the regions at E221 to D229 and V233 to N236.
We showed that two regions of YfgL, P171 to P181 and E221 to D229, make direct contact with YaeT, but where they make contact in YaeT is unknown. A clue can be found in the recently published paper on the structure and function of the N-terminal POTRA domains P1 to P5 of YaeT (15). Kim et al. noted a singular β-bulge in P3 formed by residues I240 and D241 (15). When the significance of the bulge was tested by shifting I240 and D241 by two and four residues, it was found that both altered YaeT proteins were produced at wild-type levels and complemented a YaeT depletion strain (15). However, the two- and four-residue shifts resulted in either severely weakened or a loss of interactions between the mutant YaeT and YfgL, respectively (15). Since moving residues I240 and D241 of YaeT impacted YfgL binding but not its essential functions (15), it is tempting to propose that the bulge region of YaeT's P3 domain and the YfgL region at P171 to P181 or E221 to D229 make direct contact with each other. However, these may not be the only two sites where YfgL interacts with YaeT since Kim et al. also demonstrated that YfgL did not copurify with YaeT if the P2, P3, P4, or P5 domain was deleted (15). It is unclear at this point whether the effect of a P2, P4, or P5 deletion on the YfgL interaction results from (i) the direct loss of interacting surfaces of YaeT, (ii) an indirect effect on P3's conformation, or (iii) shifting P3's bulge region away from YfgL.
The exact role of YfgL in OMP biogenesis is not known, but several lines of evidence suggest that YfgL and the periplasmic chaperone SurA perform a related and maybe even somewhat overlapping function(s) in OMP biogenesis. Study on the kinetics of LamB assembly found that cells with a deletion of yfgL and those without surA both had the same defective assembly step; namely, the rate of conversion of unfolded mature LamB monomers into folded ones was reduced (29, 36). Previous works showing that folded LamB monomer intermediates localized to the outer membrane (5, 24), coupled with the kinetic data on the yfgL and surA mutants (36), suggest that both YfgL and SurA expedite the delivery of LamB to the YaeT complex in the outer membrane for final assembly and insertion.
Despite the fact that a strain with deletions of both yfgL and surA exhibits a cold sensitivity synthetic phenotype (25) and has significantly lower LamB levels than strains with deletions of one of the genes (30), the available data suggest that YfgL and SurA are not functionally equivalent. A ΔyfgL ΔdegP double mutant displays a conditional lethal phenotype that cannot be rescued by the overexpression of surA (2). Moreover, the surA mutant has significantly reduced levels of OmpA, OmpC, OmpF, and LamB compared to those of the yfgL mutant (2, 16). Although YfgL and SurA may both function in the delivery of LamB to YaeT, they most likely are involved in different aspects or steps of that process.
Coimmunoprecipitation experiments using YfgL(L173S)-His6, YfgL(L175S)-His6, YfgL(R176A)-His6, YfgL(D227A)-His6, and YfgL(D229A)-His6 variants as the bait and DSP cross-linking showed that the amount of SurA precipitated was commensurate with the amount of YaeT precipitated. The data suggest that SurA, presumably with its OMP cargo, binds directly to YaeT or another complex member but not to YfgL. In this model, the mutant YfgL proteins, with their weakened interactions with YaeT, cannot coprecipitate YaeT effectively, and subsequently, less SurA was brought down. The weakened associations between the YfgL variants and YaeT could stem from conformation changes to the YfgL proteins introduced by alterations to the L173, L175, and R176 residues, as evidenced by the mobility shift on SDS-PAGE gels.
However, our coimmunoprecipitation data do not preclude a second model in which SurA docks to YfgL before the delivery of its OMP cargoes to YaeT. For this to happen, our alterations to YfgL would have to interfere simultaneously with YaeT interactions and SurA bindings. Since it is highly unlikely for the same P171-to-P181 region of YfgL to simultaneously engage in both YaeT and SurA bindings, the YfgL mutants would have to exert their effects on SurA docking secondarily. Because this second model is more complicated, we do not favor it. Also, it is unlikely that YfgL would function as a receptor for SurA or SurA-OMP complexes for at least two reasons. First, if YfgL does function as a docking site for SurA, then the yfgL mutant should exhibit OMP biogenesis defects as severe as those observed in the surA mutant, but as discussed above, it does not. Consistent with this view, there is a SurA homolog but no YfgL counterpart in N. meningitidis. Second, it has been reported that Omp85 and its homologs in mitochondria and chloroplasts, Sam50 and Toc75, respectively, directly receive incoming OMPs (7, 13, 27), rendering membrane protein receptor activity in YfgL somewhat redundant.
Although it is very unlikely that YfgL would serve as a docking site for SurA-OMP complexes, it is conceivable that YfgL binding to YaeT may enhance the YaeT complex to receive SurA and its OMP cargo. Preliminary assessment of YfgL's influence on SurA reception using histidine-tagged YaeT in a yfgL+ and yfgL-null background found that the amounts of SurA cross-linked to YaeT-His6 were almost identical in both backgrounds. However, more experiments need to be performed, perhaps using histidine-tagged NlpB or SmpA as bait, to confidently conclude that YfgL has no influence on SurA binding to the YaeT complex.
If YfgL's interaction with YaeT indeed does not enhance YaeT's ability to receive SurA-bound OMPs for assembly, then we think that the YfgL-YaeT association facilitates the subsequent steps of OMP assembly and insertion. In the absence of YfgL, YaeT still receives OMPs via SurA and assembles and inserts them, albeit not as efficiently as it does in the presence of YfgL. Recent structure-function analyses of YaeT revealed a potential YfgL binding site within one of the POTRA domains (P3) of YaeT. Since YaeT's POTRA domains are also thought to be binding sites for OMPs, it is possible that YfgL binding alters the conformation of the POTRA domain so as to make it more receptive for interactions with OMPs. Clearly, more work is needed to pinpoint the molecular nature of the YaeT-YfgL interaction and its functional outcome.
Curiously, a recent paper reported that the YfgL protein in E. coli has binding motifs for pyrroloquinoline-quinone (PQQ) (14), a redox cofactor for dehydrogenases (11). The biological relevance of these PQQ-binding motifs in E. coli is unknown since it lacks the gene encoding PQQ synthase (20). Nevertheless, computational analysis of the E. coli YfgL amino acid sequence using the SMART Web application found seven PQQ-binding motifs. The YfgL region at P171 to P181, which we have shown to directly interact with YaeT, resides outside the predicted PQQ-binding domains. In contrast, the YfgL region at V319 to H328, which is involved in the lipoprotein's biogenesis but not in YaeT binding, lies in the last predicted PQQ-binding domain at residues 310 to 342 (E value of 1.39 × 10−4).
It was also reported that the yfgL-null mutant exhibits a dramatically low homologous recombination frequency. However, we found no difference in the homologous recombination frequency as measured by P1 transductional crosses in MC4100 yfgL+ and yfgL mutant strains (data not shown). It is also difficult to reconcile the reported in vitro kinase activity of YfgL because there is no evidence that the periplasmic space of bacteria contains high-energy phosphates.
The yfgL-null mutant displays a pleiotropic phenotype, affecting functions associated with the bacterial envelope. The genetic and biochemical data showing YfgL and YaeT interactions unambiguously establish a direct role of YfgL in OMP biogenesis. However, yfgL-null mutants are also hypersensitive to inhibitors that normally cannot penetrate the outer membrane, indicating a gross breach in the outer membrane permeability barrier. Moreover, yfgL mutations were first isolated as suppressors of a mutant allele of the imp gene, whose product is involved in LPS transport, establishing a genetic link between YfgL and LPS biogenesis (6). Taken together, these findings suggest a broader role for YfgL in cell envelope biogenesis.
Supplementary Material
Acknowledgments
This work was supported by grant R01-GM048167 from the National Institute of General Medical Sciences.
We thank Muriel Masi for critical reading of the first draft of the manuscript. We especially thank Roberto Kolter for generously providing us with anti-SurA antiserum. We are also grateful to Thomas Silhavy's laboratory for sharing their antibody against YaeT, the pZS21-yaeT-His construct, and the ΔyaeT/λPBAD-yaeT strain.
Footnotes
Published ahead of print on 28 December 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Augustus, A. M., T. Celaya, F. Husain, M. Humbard, and R. Misra. 2004. Antibiotic-sensitive TolC mutants and their suppressors. J. Bacteriol. 1861851-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charlson, E. S., J. N. Werner, and R. Misra. 2006. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J. Bacteriol. 1887186-7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 313497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doerrler, W. T., and C. R. Raetz. 2005. Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J. Biol. Chem. 28027679-27687. [DOI] [PubMed] [Google Scholar]
- 5.Duguay, A. R., and T. J. Silhavy. 2002. Signal sequence mutations as tools for the characterization of LamB folding intermediates. J. Bacteriol. 1846918-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggert, U. S., N. Ruiz, B. V. Falcone, A. A. Branstrom, R. C. Goldman, T. J. Silhavy, and D. Kahne. 2001. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science 294361-364. [DOI] [PubMed] [Google Scholar]
- 7.Ertel, F., O. Mirus, R. Bredemeier, S. Moslavac, T. Becker, and E. Schleiff. 2005. The evolutionarily related β-barrel polypeptide transporters from Pisum sativum and Nostoc PCC7120 contain two distinct functional domains. J. Biol. Chem. 28028281-28289. [DOI] [PubMed] [Google Scholar]
- 8.Fardini, Y., K. Chettab, O. Grepinet, S. Rochereau, J. Trotereau, P. Harvey, M. Amy, E. Bottreau, N. Bumstead, P. A. Barrow, and I. Virlogeux-Payant. 2007. The YfgL lipoprotein is essential for type III secretion system expression and virulence of Salmonella enterica serovar Enteritidis. Infect. Immun. 75358-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 1785803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 313784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin, P. M., and C. Anthony. 1998. The biochemistry, physiology and genetics of PQQ and PQQ-containing enzymes. Adv. Microb. Physiol. 401-80. [DOI] [PubMed] [Google Scholar]
- 12.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habib, S. J., T. Waizenegger, A. Niewienda, S. A. Paschen, W. Neupert, and D. Rapaport. 2007. The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial β-barrel proteins. J. Cell Biol. 17677-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khairnar, N. P., V. A. Kamble, S. H. Mangoli, S. K. Apte, and H. S. Misra. 2007. Involvement of a periplasmic protein kinase in DNA strand break repair and homologous recombination in Escherichia coli. Mol. Microbiol. 65294-304. [DOI] [PubMed] [Google Scholar]
- 15.Kim, S., J. C. Malinverni, P. Sliz, T. J. Silhavy, S. C. Harrison, and D. Kahne. 2007. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317961-964. [DOI] [PubMed] [Google Scholar]
- 16.Lazar, S. W., and R. Kolter. 1996. SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 1781770-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, J. Schultz, C. P. Ponting, and P. Bork. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malinverni, J. C., J. Werner, S. Kim, J. G. Sklar, D. Kahne, R. Misra, and T. J. Silhavy. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 6151-64. [DOI] [PubMed] [Google Scholar]
- 19.Masi, M., P. Vuong, M. Humbard, K. Malone, and R. Misra. 2007. Initial steps of colicin E1 import across the outer membrane of Escherichia coli. J. Bacteriol. 1892667-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsushita, K., J. C. Arents, R. Bader, M. Yamada, O. Adachi, and P. W. Postma. 1997. Escherichia coli is unable to produce pyrroloquinoline quinone (PQQ). Microbiology 1433149-3156. [DOI] [PubMed] [Google Scholar]
- 21.Misra, R. 1993. A novel ompC mutation of Escherichia coli K-12 that reduces OmpC and OmpF levels in the outer membrane. Mol. Microbiol. 101029-1035. [DOI] [PubMed] [Google Scholar]
- 22.Misra, R. 2007. First glimpse of the crystal structure of YaeT's POTRA domains. ACS Chem. Biol. 2649-651.18041813 [Google Scholar]
- 23.Misra, R., and S. A. Benson. 1988. Genetic identification of the pore domain of the OmpC porin of Escherichia coli K-12. J. Bacteriol. 1703611-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misra, R., A. Peterson, T. Ferenci, and T. J. Silhavy. 1991. A genetic approach for analyzing the pathway of LamB assembly into the outer membrane of Escherichia coli. J. Biol. Chem. 26613592-13597. [PubMed] [Google Scholar]
- 25.Onufryk, C., M. L. Crouch, F. C. Fang, and C. A. Gross. 2005. Characterization of six lipoproteins in the σE regulon. J. Bacteriol. 1874552-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 5750-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robert, V., E. B. Volokhina, F. Senf, M. P. Bos, P. Van Gelder, and J. Tommassen. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 41984-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rolhion, N., N. Barnich, L. Claret, and A. Darfeuille-Michaud. 2005. Strong decrease in invasive ability and outer membrane vesicle release in Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 with the yfgL gene deleted. J. Bacteriol. 1872286-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouviere, P. E., and C. A. Gross. 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 103170-3182. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz, N., B. Falcone, D. Kahne, and T. J. Silhavy. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121307-317. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz, N., D. Kahne, and T. J. Silhavy. 2006. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 457-66. [DOI] [PubMed] [Google Scholar]
- 32.Sklar, J. G., T. Wu, L. S. Gronenberg, J. C. Malinverni, D. Kahne, and T. J. Silhavy. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA 1046400-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sklar, J. G., T. Wu, D. Kahne, and T. J. Silhavy. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 212473-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tikhonova, E. B., Q. Wang, and H. I. Zgurskaya. 2002. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 1846499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tokuda, H., and S. Matsuyama. 2004. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 16935-13. [DOI] [PubMed] [Google Scholar]
- 36.Ureta, A. R., R. G. Endres, N. S. Wingreen, and T. J. Silhavy. 2007. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J. Bacteriol. 189446-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vakharia, H., and R. Misra. 1996. A genetic approach for analysing surface-exposed regions of the OmpC protein of Escherichia coli K-12. Mol. Microbiol. 19881-889. [DOI] [PubMed] [Google Scholar]
- 38.Vediyappan, G., T. Borisova, and J. A. Fralick. 2006. Isolation and characterization of VceC gain-of-function mutants that can function with the AcrAB multiple-drug-resistant efflux pump of Escherichia coli. J. Bacteriol. 1883757-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299262-265. [DOI] [PubMed] [Google Scholar]
- 40.Voulhoux, R., and J. Tommassen. 2004. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res. Microbiol. 155129-135. [DOI] [PubMed] [Google Scholar]
- 41.Werner, J., A. M. Augustus, and R. Misra. 2003. Assembly of TolC, a structurally unique and multifunctional outer membrane protein of Escherichia coli K-12. J. Bacteriol. 1856540-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werner, J., and R. Misra. 2005. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol. 571450-1459. [DOI] [PubMed] [Google Scholar]
- 43.Wu, T., J. Malinverni, N. Ruiz, S. Kim, T. J. Silhavy, and D. Kahne. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121235-245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.