Abstract
The thick wall of gram-positive bacteria is a polymer meshwork composed predominantly of peptidoglycan (PG) and teichoic acids, both of which have a critical function in maintenance of the structural integrity and the shape of the cell. In Bacillus subtilis 168 the major teichoic acid is covalently coupled to PG and is known as wall teichoic acid (WTA). Recently, PG insertion/degradation over the lateral wall has been shown to occur in a helical pattern. However, the spatial organization of WTA assembly and its relationship with cell shape and PG assembly are largely unknown. We have characterized the localization of green fluorescent protein fusions to proteins involved in several steps of WTA synthesis in B. subtilis: TagB, -F, -G, -H, and -O. All of these localized similarly to the inner side of the cytoplasmic membrane, in a pattern strikingly similar to that displayed by probes of nascent PG. Helix-like localization patterns are often attributable to the morphogenic cytoskeletal proteins of the MreB family. However, localization of the Tag proteins did not appear to be substantially affected by single disruption of any of the three MreB homologues of B. subtilis. Bacterial and yeast two-hybrid experiments revealed a complex network of interactions involving TagA, -B, -E, -F, -G, -H, and -O and the cell shape determinants MreC and MreD (encoded by the mreBCD operon and presumably involved in the spatial organization of PG synthesis). Taken together, our results suggest that, in B. subtilis at least, the synthesis and export of WTA precursors are mediated by a large multienzyme complex that may be associated with the PG-synthesizing machinery.
In most bacteria, the rigid cell wall (CW) is responsible for providing shape and structural integrity to the cell. The thick CW of gram-positive bacteria is a multilayered structure composed predominantly of peptidoglycan (PG) (also called murein) and anionic polymers, particularly teichoic acids (TA) (for recent reviews, see references 5, 47, and 54). The highly cross-linked PG polymer (poly-N-acetylglucosamine and N-acetylmuramic acid) network is an essential determinant of cell shape and is responsible for protection from the cellular turgor pressure. Many roles for TA have been proposed, including cell shape maintenance (61, 68), resistance to antimicrobial peptides (1, 35, 36), biofilm formation (27), acid tolerance (9) and, efficient release of secreted proteins into the culture medium (49).
TA are either covalently bound to the PG (wall TA [WTA]) or anchored to the cytoplasmic membrane (lipo-TA). In the gram-positive model organism Bacillus subtilis, WTA is present in quantities roughly equal to those of PG and constitutes the major class of anionic polymers (26). The type of WTA polymer varies between strains. In B. subtilis 168, the major WTA consists of a poly-glycerol-phosphate [poly-(Gro-P)] chain of 45 to 60 subunits and a “PG linkage unit” of N-acetylglucosamine-β-(1-4)-N-acetylmannosamine (GlcNAc-ManNAc). WTA is covalently linked to the PG through a phosphodiester bond between the anomeric carbon of GlcNAc in the PG linkage unit and the 6-hydroxyl of MurNAc in the PG chain.
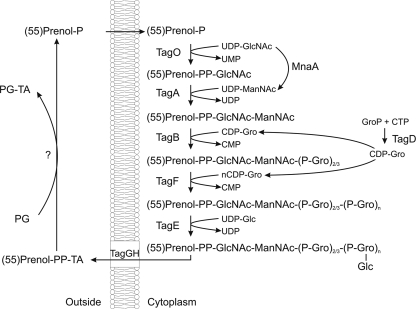
In B. subtilis 168 the genes responsible for WTA synthesis are tagABDEFGHO and mnaA (Fig. 1). According to proposed models, WTA biosynthesis occurs predominantly at the cytoplasmic side of the membrane (5). Initially, TagO is thought to couple GlcNAc to the membrane-embedded lipid undecaprenyl-pyrophosphate. The next step in the pathway, thought to be catalyzed by TagA, is the addition of ManNAc, which is made from the epimerization of GlcNAc by MnaA (59, 60), to produce the lipid-linked GlcNAc-ManNAc disaccharide. Recent in vitro biochemical studies from the Brown laboratory have established that TagB then functions as the “Tag primase,” which couples the first glycerol-phosphate to ManNAc (7), followed by TagF acting as the “Tag polymerase,” which adds the additional glycerol-phosphates extending the poly-(Gro-P) chain (56). TagD activates glycerol-phosphate through linkage to CTP, making CDP-glycerol, the substrate for both TagB and TagF (50), and TagE glucosylates glycerol-phosphate subunits in the poly-(Gro-P) chain (51). Finally, TagG and TagH are thought to constitute a two-component ABC-type transporter that mediates the translocation of the TA polymer across the membrane. TagH constitutes the nucleotide binding domain (NBD), and TagG is the membrane-spanning domain and presumed translocase component of the ABC-type transporter TagGH (40). After translocation, the WTA polymer is covalently attached to the PG through an unknown mechanism (40). As all of these synthetic reactions are believed to take place at the membrane in an ordered fashion (5), it could be envisaged that these proteins form a complex in which the lipid-linked WTA precursor molecule is passed through the chain of biosynthetic steps, but evidence for such a complex has not yet been provided.
FIG. 1.
Overview of WTA biosynthesis. The biosynthetic steps in the formation of WTA precursors are shown. Arrows indicate catalytic steps, and the enzymes responsible are shown. The precursor is assembled at the cytoplasmic membrane prior to transport by TagGH and linkage to the PG. The question mark indicates an unknown enzymatic reaction.
Until recently it was thought that WTA was an indispensable component of the CW, as mutations in any of the tag genes, except tagE, were lethal in B. subtilis (see, e.g., references 6, 40, and 59). However, careful analysis has revealed that in fact WTA is dispensable for growth in B. subtilis and Staphylococcus aureus, provided that the first committed step catalyzed by TagO is deleted (20, 21). The cause for the lethality of downstream mutations probably lies in the sequestration of metabolites such as undecaprenyl-pyrophosphate, which is also required as a lipid linker for the translocation of PG precursors (20). Nevertheless, loss of WTA in B. subtilis is associated with loss of rod shape and swelling, nonuniform thickening of the PG layer, aberrant placement of the division septum, and reduced growth rate, indicating that WTA plays an important role in maintaining the shape and structural integrity of the bacterial cell (20).
A number of shape mutants (namely, Rod mutants) that show a rounded phenotype in B. subtilis have been identified. Two of the early rod genes to be identified were mapped to tag genes. rodC1 was a temperature sensitive mutation in tagF, and rodD was a mutation in tagE (29). The other two classes of Rod mutations were mapped to rodA and mreD (rodB), which are both believed to play a role in PG synthesis (28, 67). mreB morphogenes have come to the fore since the discovery that they encode actin homologues. B. subtilis contains three MreB isoforms (MreB, Mbl, and MreBH) that assemble into helical filaments around the cell periphery (19, 32, 65) and colocalize in what appears to be a single helical structure (11, 18). Use of fluorescent derivatives of the antibiotics vancomycin and ramoplanin, which label newly incorporated PG precursors, has revealed an underlying helical pattern of PG synthesis in B. subtilis (15, 63). Whether this pattern is influenced by the MreB cytoskeleton is at present controversial. Nevertheless, it has been suggested that the MreB isoforms are involved in the positioning of PG synthases known as penicillin-binding proteins (PBPs) (24), a PG hydrolase (LytE) (11), and/or other membrane-associated cell morphogenesis proteins, such as MreC and MreD (22, 23, 38, 41, 66). Conversely, it has been suggested that MreB requires MreC/D for proper localization in B subtilis (17). MreC and MreD (encoded by the mreBCD operon) are essential membrane proteins that also localize in a helix-like pattern and that are believed to function in the same morphogenetic pathway as MreBs. MreC has been recently shown to interact with high-molecular-weight PBPs in B. subtilis (66) and in Escherichia coli (38) by bacterial two-hybrid (B2H) and yeast two-hybrid (Y2H) experiments, respectively, and in Caulobacter crescentus by affinity chromatography (22). Altogether, it is currently believed that MreBCD form an essential membrane-bound complex involved in the spatial organization of the PG synthesis machinery along the cylindrical CW (5, 54).
Several studies have attempted to determine the localization of WTA on the surface of B. subtilis cells (12, 52, 57, 58). It has been suggested that they localize at division septa and either in patches or uniformly along the cell cylinder, but none of these studies were conclusive. One recent study addressed the location of the “Tag primase” TagB and showed that it localizes to the cytoplasmic membrane in a disperse pattern (7). Since a considerable body of recent work has suggested that CW biogenesis may occur in a helical manner governed by the MreB (actin-like) proteins, we decided to make a more systematic study of Tag protein localization and of its relationship with the MreB cytoskeleton. Imaging of functional green fluorescent protein (GFP) fusions to TagO, TagB, TagF, TagG, and TagH revealed a similar set of membrane-associated localization patterns. Fluorescence was present at the division sites and in a pattern consistent with a helix along the sidewall. For TagG and TagH at least, it was reduced at mature cell poles. We then addressed the protein-protein interactions of Tag proteins by using B2H and Y2H assays (30, 34). Extensive interactions between the WTA biosynthetic proteins as well as with the putative translocase TagGH were revealed. Interactions between several of the Tag proteins and the shape determinants MreC and MreD were also detected. Our findings suggest that the synthesis and translocation of WTA precursors are mediated by a large multienzyme complex (consisting of at least TagO, TagA, TagB, TagF, TagG, and TagH) that localizes at sites of the cytoplasmic membrane similar to those where insertion of nascent PG takes place, possibly through interactions with the bacterial cytoskeleton proteins MreC and MreD.
MATERIALS AND METHODS
General methods.
The strains and plasmids used in this study are listed in Table 1 (for a full list of plasmids used in the two-hybrid studies, see Table S2 in the supplemental material). B. subtilis cells were made competent for transformation with DNA either by the method of Kunst and Rapoport (39) or by the method of Anagnostopoulos and Spizizen (2) as modified by Jenkinson (31). DNA manipulations and E. coli DH5α or XL1-Blue transformations were carried out using standard methods (53). Solid medium used for growing B. subtilis was nutrient agar (Oxoid), and liquid medium was either casein hydrolysate (CH) medium (62) or S medium (33) supplemented with 1% (vol/vol) CH (S+ medium) and with xylose (0.5%). Where SMM was added, the above media were made at 2× concentration and diluted 50:50 with a 2× SMM solution (1 M sucrose, 33.7 mM maleic acid, 40 mM MgCl2, pH 7.0). Antibiotics were used at the following concentrations: kanamycin, 5 μg/ml; chloramphenicol, 5 μg/ml; erythromycin, 1 μg/ml; lincomycin, 25 μg/ml; and/or spectinomycin, 50 μg/ml. Media used for growing E. coli strains were 2× TY (53) and nutrient agar supplemented with ampicillin (100 μg/ml) or kanamycin (5 μg/ml), as required.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source, reference, or constructionb |
|---|---|---|
| B. subtilis strains | ||
| 168 | trpC2 | Laboratory collection |
| EB247 | tagF1 (rodC1), S644F ts mutant | 56 |
| 2501 | trpC2 Ω(mbl::pSG4503, lacZ lacI bla erm Pspac-′mbl′) | 10 |
| 2505 | trpC2 Ω(mbl::spc) | 32 |
| 2535 | trpC2 Ω(mreBH::spc) | 11 |
| 2580J | trpC2 Ω(amyE::spc Pxyl-gfp-tagB) | pSG4537J→168 (Spc) |
| 3110 | trpC2 tagH::pSG5050 (cat Pxyl-gfp-tagH1-648) | pSG5050→168 (Cm) |
| 3506 | trpC2 Ω(mbl::spc) tagG::pSG5322 (cat Pxyl-gfp-tagG1-346) | 2505→3512 (Spc) |
| 3512 | trpC2 tagG::pSG5322 (cat Pxyl-gfp-tagG1-346) | pSG5322→168 (Cm) |
| 3530 | trpC2 Ω(mbl::spc) tagH::pSG5050 (cat Pxyl-gfp-tagH1-648) | 2505→3110 (Spc) |
| 3725 | trpC2 Ωneo3427 ΔmreB | 25 |
| 3741 | trpC2 Ωneo3427 ΔmreB tagG::pSG5322 (cat Pxyl-gfp-tagG1-346) | 3725→3512 (Neo) |
| 3742 | trpC2 Ωneo3427 ΔmreB tagH::pSG5050 (cat Pxyl-gfp-tagH1-648) | 3725→3110 (Neo) |
| 3746 | trpC2 Ω(mreBH::spc) tagG::pSG5322 (cat Pxyl-gfp-tagG1-346) | 2535→3512 (Spc) |
| 3747 | trpC2 Ω(mreBH::spc) tagH::pSG5050 (cat Pxyl-gfp-tagH1-648) | 2535→3110 (Spc) |
| 3798 | trpC2 tagB::pSG5879 (tagB-gfp cat) | pSG5879→168 (Cm) |
| 3799 | trpC2 tagF::pSG5880 (tagF-gfp cat) | pSG5880→168 (Cm) |
| 3800 | trpC2 tagO::pSG5881 (tagO-gfp cat) | pSG5881→168 (Cm) |
| E. coli strains | ||
| DH5α | F−endA1 hsdR17 supE44 thi-1 λ-recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 φ80 dlacZ ΔM15 | GIBCO-BRL |
| BTH101 | F−glnV44 recA1 endA gyrA96 thi-1 hsdR17 spoT1 rfbD1 cya-854 | 34 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene Ltd. |
| Plasmids | ||
| pSG1151 | bla cat gfp | 43 |
| pSG1729 | bla amyE::spc Pxyl-gfp | 43 |
| pSG4537J | bla amyE::spc Pxyl-gfp-tagB | This work |
| pSG4902 | bla cat Pxyl-gfp | 69 |
| pSG5050 | bla cat Pxyl-gfp-tagH1-648 | This work |
| pSG5322 | bla cat Pxyl-gfp-tagG1-346 | This work |
| pSG5879 | bla cat tagB-gfp | This work |
| pSG5880 | bla cat tagF-gfp | This work |
| pSG5881 | bla cat tagO-gfp | This work |
gfp, F64L S65T variant of GFP gene (13); ′mbl′ denotes 5′-3′ gene truncation.
X→Y indicates that strain Y was transformed with DNA from source X, with the selected marker in parentheses: Cm, chloramphenicol; Sp, spectinomycin; Neo, neomycin.
Construction of GFP fusions.
Primers and restriction endonucleases used are listed in Table S1 in the supplemental material. For the gfp-tagG and gfp-tagH fusions, approximately one-third of each of the promoter-proximal parts of the tagG and tagH genes was amplified by PCR and cloned into pSG4902 (69). Transformation of the resulting plasmids (Table 1) into B. subtilis, with selection for chloramphenicol resistance, resulted in strains that carried a gfp fusion to tagG or tagH at the chromosomal locus as the only copy of the gene of interest and under the control of the Pxyl promoter. Correct integration at the chromosomal locus was confirmed by PCR.
For tagB-gfp, tagF-gfp, and tagO-gfp fusions, approximately one-third of the 3′-proximal parts of each of the tagB, tagF, and tagO genes was amplified by PCR and cloned into pSG1151 (43), generating plasmids pSG5879, pSG5880, and pSG5881, respectively. The plasmids were checked by sequencing and transformed into B. subtilis 168, with selection for chloramphenicol resistance to produce strains carrying a gfp fusion to tagB (strain 3798), tagF (strain 3799), and tagO (strain 3800) at the chromosomal locus under control of the native promoter.
For the gfp-tagB and gfp-tagF fusions, the full-length coding regions of tagB and tagF were PCR amplified and cloned into plasmid pSG1729 (43) to generate plasmids pSG4535J and pSG4537J. The resulting inserts were sequenced, and the plasmids pSG4535J and pSG4537J were transformed into wild-type B. subtilis 168 cells, with selection for spectinomycin resistance, to give strains 2575J and 2580J, which contain both the natural copy of tagF and tagB and a second, xylose-dependent copy of tagF or tagB, respectively, fused to gfp at the ectopic amyE locus.
To test the functionality of the GFP-TagF fusion, B. subtilis strain EB247 (56), harboring the tagF1 (rodC1) temperature-sensitive mutation, was transformed by plasmid pSG4535J to produce strain 2577J, which contains the tagF1 mutation at the chromosomal locus and the xylose-inducible gfp-tagF fusion at amyE. Disruption of amyE was confirmed for all pSG1729 derivates using a starch plate assay (14), and the correct integration of the inserts at the amyE locus was confirmed by PCR.
To disrupt the native mbl gene in the strains containing the GFP fusions, chromosomal DNA of strain 2505 [Ω(mbl::spc)] (32) was used to transform strains 3110 and 3512, to produce strains 3530 and 3506, respectively. For mreB disruptions, chromosomal DNA of strain 3725 (Ωneo3427 ΔmreB) (25) was used to transform strains 3110 and 3512, to produce strains 3742 and 3741, respectively. For mreBH disruptions, chromosomal DNA of strain 2535 [Ω(mreBH::spc)] (11) was used to transform strains 3110 and 3512, to produce strains 3747 and 3746, respectively.
Microscopy.
Microscopy was performed essentially as described previously (55). Image acquisition was done as described previously (42) using Metamorph v 6.0 software (Universal Imaging Corp., Downingtown, PA). Images from a single focal plane were deconvolved using the “No Neighbors” algorithm from the Metamorph software package. Overlays of micrographs were assembled using Metamorph before exporting the images to Adobe Photoshop v 6.0.
B2H plasmid construction and assay.
The method used was that of Karimova et al. (34), adapted as described by Daniel et al. (16). The coding sequence of each gene was amplified by PCR from the wild-type strain 168 genomic DNA using the primers listed in Table S1 in the supplemental material and inserted into pKT25, pUT18C, and pUT18. For a full list of plasmids used in this study, see Table S2 in the supplemental material.
Y2H plasmid construction and assay.
Coding sequences of each gene were amplified by PCR from the wild-type strain 168 genomic DNA using the primers listed in Table S1 in the supplemental material and cloned as translational fusions with the Gal4 DNA-binding (BD) and Gal4 activation (AD) domains in the recipient vectors pGBDU-C1 (bait, Ura+) and pGAD-C1 (prey, Leu+), respectively (30). Two-hybrid bait and prey constructions were introduced into the Saccharomyces cerevisiae haploid strains PJ69-4a and PJ69-4α, respectively, by gap repair (44). The two-hybrid assays were performed according to a previously described mating strategy (48). Interaction phenotypes were scored by replica plating the diploids onto plates selecting for the expression of the HIS3 and ADE2 interaction reporters. Control matings with empty pGBDU and pGAD vectors were used to detect self-activation and as negative controls for interaction.
RESULTS
Localization of the putative WTA transporter TagGH in a helix-like pattern.
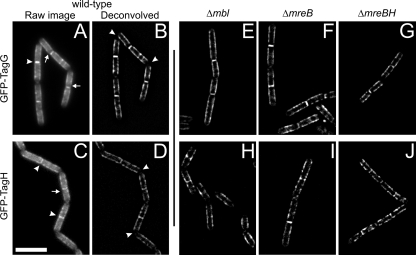
Lipid-linked WTA precursors are thought to be transported across the membrane by a dedicated ABC-type transporter, TagGH. TagG and TagH are predicted to have 6 transmembrane spans and 1 transmembrane span, respectively, with both N termini in the cytosol (TMHMM v 2.0; http://www.cbs.dtu.dk/services/TMHMM/) (37). To examine the sites of WTA export, i.e., the sites of nascent WTA insertion, N-terminal gfp fusions to tagG and tagH were constructed. The gfp-tagG and gfp-tagH fusions were integrated into the chromosome at the tagGH locus, replacing the respective wild-type copy of the gene. This generated two strains containing either gfp-tagG (strain 3512) or gfp-tagH (strain 3110) under the control of the xylose-inducible promoter Pxyl. Both strains displayed xylose-dependent growth (not shown), indicating that expression of gfp-tagG and gfp-tagH could replace that of the essential tagG and tagH genes, respectively. In the presence of inducer, these strains showed no morphological defects, further indicating that the GFP fusions were functional. GFP-TagG and GFP-TagH were both expressed as full-length fusion proteins, as shown by Western blotting using a specific anti-GFP antibody (data not shown), and showed similar localization patterns (Fig. 2A and C). They localized to ongoing division sites, and a weak, uneven fluorescence signal was detected over the cell cylinder. Strikingly, little fluorescence was detected in old cell poles, and, consistent with this, a gap between the fluorescence signal at the division site and that in the cylinder emerged as cells completed division (Fig. 2A and C). Out-of-focus light in the GFP fluorescence images was reduced by two-dimensional (2D) deconvolution (see Materials and Methods), which was applied to single images of cells expressing either GFP-TagG or GFP-TagH. Interestingly, in both the raw and the deconvolved images of GFP-TagG (Fig. 2A and B, respectively) or GFP-TagH (Fig. 2C and D), the cylindrical staining was reminiscent of the helix-like patterns seen in B. subtilis cells stained with fluorescent probes for nascent PG (15, 63) or expressing a GFP fusion to the CW hydrolase LytE (11), which were shown to be dependent on the MreB isoforms Mbl and MreBH, respectively, and of the helical configuration displayed at the membrane by GFP fusions to MreC and MreD (41). To test if the helix-like distribution of TagG and/or TagH (and thus WTA export) was also influenced by the actin-like cytoskeleton, the GFP-TagG and GFP-TagH fusions were examined in mreB, mreBH, and mbl single mutant strains. These strains were grown under conditions (CH medium supplemented with SMM) previously shown to allow these mutants to grow with near-wild-type morphology (11, 25). Under these conditions, the localization pattern of either GFP-TagG or GFP-TagH in any of the three mreB-like single mutants (Fig. 2E to J) was not significantly different from that displayed in wild-type cells (Fig. 2B and D). mreC and mreD are essential under normal growth conditions, but viability can also be rescued by high concentrations of magnesium (41). However, mreC and mreD mutants are unstable and display severe shape defects under these conditions (41), unlike the mreB-like mutants, in which the rod shape is restored by Mg2+ (11, 25). Thus, a possible effect of MreC and/or MreD on the positioning of the WTA transporter TagGH could not be tested. Indeed, in a ΔmreC or a ΔmreD background, any change in the localization patterns of TagGH could be attributed either to the lack of MreCD proteins or to a secondary effect resulting from the shape defects.
FIG. 2.
Localization of the ABC transporter TagGH. Fluorescence images of cells expressing GFP-TagG (A and B), GFP-TagH (C and D) in a wild type background or in the absence of Mbl (E and H), MreB (F and I), or MreBH (G and J) are shown. Cells were grown to mid-exponential stage at 30°C in CH medium (or CH supplemented with SMM for the mutants [see text]) with 5 mM K2HPO4/KH2PO4 (pH 7.5) and 0.5% xylose and imaged on wet agarose slides. Panels B and D show 2D deconvolutions of panels A and C, respectively. Panels E to J show 2D deconvolutions of raw images that are not included in the figure. In panels A and C, white arrowheads represent ongoing division sites and white arrows indicate regions of reduced fluorescence between the division septum and the lateral CW. White arrowheads in B and D indicate the absence of TagGH from polar regions. Bar, 5 μm.
Localization of the WTA biosynthetic enzymes TagO, TagB, and TagF to the membrane.
The first step of WTA biosynthesis is thought to be catalyzed by TagO (Fig. 1), which is predicted to have 11 transmembrane spans with its C terminus located in the cytosol (TMHMM v 2.0) (37). To examine the subcellular localization of TagO, a gfp fusion was made to the C terminus of the gene. The fusion was integrated into the chromosome at the tagO locus, under control of the native tagO promoter (strain 3800). The TagO-GFP fusion, which was the only copy of TagO present in the cells, was judged to be functional, as cells displayed a wild-type morphology whereas cells with tagO disrupted become spherical (20). TagO-GFP localized to the membrane, in a nonuniform pattern along the cylindrical region, and also to the division sites (Fig. 3A). Deconvolution (Fig. 3D) revealed that this pattern was also consistent with a helix-like organization, similar to that displayed by the TagG- and TagH-GFP fusions (Fig. 2B and D, respectively). Again, the fluorescence intensity appeared to be reduced at older cell poles (Fig. 3A), although this effect was less clear than for the TagGH proteins.
FIG. 3.
Localization of TagO, TagB, and TagF. Fluorescence images of cells expressing TagO-GFP (A), TagB-GFP (B), and TagF-GFP (C) are shown. Panels D to F show 2D deconvolutions of the images in panels A to C, respectively. Cells were grown to mid-exponential stage at 30°C in CH medium with 5 mM K2HPO4/KH2PO4 (pH 7.5) and imaged on wet agarose slides. Expression of the fusion proteins was under control of the native promoters. Bar, 5 μm.
TagB, the “Tag primase,” was previously reported to localize to the cell periphery and septa using a TagB-GFP fusion cloned at an ectopic locus, under control of an inducible promoter and in the presence of the wild-type tagB copy (7). To test whether a similar localization of TagB was observed at its endogenous levels of expression, we integrated a tagB-gfp fusion in place of wild-type tagB at the chromosomal locus (strain 3798). Cells expressing the tagB-gfp fusion as the only copy of the essential tagB gene in the cell were viable and displayed normal growth and morphology, indicating that the fusion protein was functional (data not shown). Expression of TagB-GFP from the native tagB promoter produced a very weak fluorescence signal, the majority of which was associated with the division sites in either raw or deconvolved images (Fig. 3B and, E, respectively). Here deconvolution underscored the weakness of the signal and did not improve the resolution as much as for the other GFP fusions described. A fluorescence signal at the membrane along the cell cylinder was occasionally seen, but it was not as strong and/or uniform as that previously reported for the (overexpressed) inducible TagB-GFP fusion (7). Thus, at endogenous expression levels and under the growth conditions tested, TagB mainly localizes to division sites, with perhaps only a few molecules present in the sidewall.
The polymerization of the poly-(Gro-P) chain, catalyzed by the “Tag polymerase” TagF, is a key step in WTA biosynthesis (56). To establish the sites of WTA polymerization in B. subtilis and their relationship with the sites of WTA nucleation (i.e., TagB localization) and of WTA export (i.e., TagGH localization), N- and C-terminal GFP fusions were constructed for tagF. The tagF-gfp fusion was integrated into the chromosome at the tagF locus, placing it under control of the native promoter (strain 3799). This fusion was judged to be functional, as it could replace the essential tagF gene in the cell without any morphological defects (not shown). Despite the fact that TagF has no predicted membrane-spanning sequences, the TagF-GFP fusion (Fig. 3C) displayed a pattern of localization similar to those of TagO-GFP (Fig. 3A), GFP-TagG (Fig. 2A), and GFP-TagH (Fig. 2C). Again, deconvolution revealed a pattern that could be consistent with a helix-like organization along the sidewall (Fig. 3F), although the possible helicity of TagF was not as striking as that for the membrane proteins TagG and TagH (see above). As for the TagO-GFP fusion, a possible reduction in fluorescence at old cell poles was not especially evident. The N-terminal gfp-tagF fusion was integrated at an ectopic locus (amyE) under the control of Pxyl, in addition to the wild-type copy of tagF (strain 2575J). This fusion was judged to be functional, as it could complement the temperature-sensitive rodC1 (tagF1) mutation (strain 2577J; data not shown). The inducible GFP-TagF fusion gave a very bright signal that localized uniformly around the cell periphery (not shown), in a pattern similar to that described by Bhavsar et al. for the inducible TagB-GFP fusion (7). Thus, both TagB and TagF localize nonuniformly along the cell cylinder at their endogenous levels but are more uniformly distributed around the membrane when overexpressed.
Interaction network of the WTA biosynthetic machinery.
The similarity between the localization patterns of TagO, TagF, TagG, and TagH, which act at different steps of the WTA biosynthetic pathway (see Fig. 1), suggested common sites of action for all these WTA-synthesizing enzymes and prompted us to address the possibility that WTA might be synthesized by a membrane-associated multienzyme complex. Potential binary interactions between all the Tag proteins involved in WTA synthesis (TagABDEFGHO) (Fig. 1) were tested in vivo using both the Y2H (30) and the B2H (34) systems.
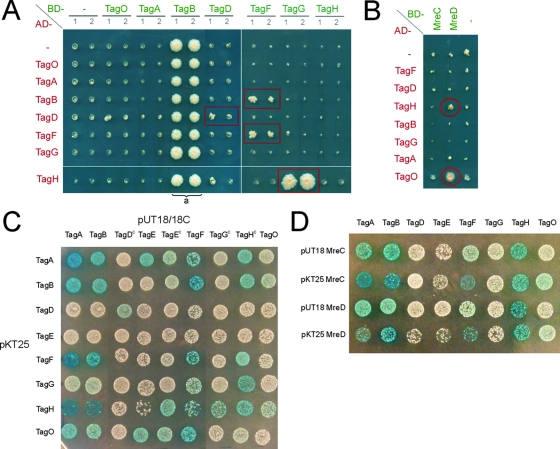
For the Y2H assay, fusions of the Gal4 BD (“bait”) and the Gal4 AD (“prey”) to full-length copies of the Tag proteins were constructed. Bait and prey fusions were combined in yeast diploid cells, and their ability to interact was tested (Fig. 4A). TagB expressed as bait (BD-TagB) exhibited a self-activation phenotype (a in Fig. 4A) and so was uninformative in these assays. A strong interaction was detected between the two components of the putative ABC-type transporter, TagG and TagH, in the orientation BD-TagH/AD-TagG although not in the reverse orientation of the AD and BD fusions. This lack of reciprocal interaction, which might be due to conformational changes in the proteins fused to the AD and BD domains, is not unusual in Y2H assays (for instance, see Table 2 in reference 64). The “Tag primase” TagB interacted with the “Tag polymerase” TagF. Self-interactions were detected with TagD and TagF, suggesting that these proteins dimerize or oligomerize in vivo. Tests such as that of AD-TagD/BD-TagO in Fig. 4A, in which only one of the two independent matings reporting an interaction showed a positive result and where the interaction was not seen in the reciprocal experiment, were disregarded.
FIG. 4.
Two-hybrid interaction assays. (A and B) Y2H interactions. Matrices of Y2H interactions occurring between TagABDFGHO (A) and between TagABDFGHO and MreCD (B) are shown. The indicated proteins were expressed as baits (BD, Gal4 BD fusion) and/or as preys (AD, Gal4 AD fusion). Pairs of independent diploid yeast cells (1 and 2) expressing the various BD/AD combinations were arrayed as indicated and subjected to selection for expression of the HIS3 interaction reporter as described in Materials and Methods. The empty bait and prey vectors (BD and AD) were used to detect self-activation and as negative controls for interaction (−). The BD-TagB fusion protein exhibited a strong self-activation phenotype (a). Positive interactions are boxed or circled in red. (C and D) B2H interactions. Matrices of bacterial two-hybrid interactions occurring between TagABDEFGHO (C) and crosses of TagABDEFGHO with MreC and MreD (D) are shown. C refers to a fusion to the C terminus of T18 in the high-copy pUT18C vector. Other fusions are made to the C terminus of T25 (low-copy pKT25 vector) or to the N terminus of T18 (high-copy pUT18 vector). Transformations were carried out and plates incubated at 30°C for a maximum of 36 h.
To further investigate the interactions between the WTA proteins, the B2H system was used. In the B2H system, the T25 and the T18 fragments of the catalytic domain of Bordetella pertussis adenylate cyclase are fused to full-length copies of the Tag proteins. One potential advantage of this system is that it does not require the interaction partners to be associated with the DNA for the interaction to be reported and is therefore more suited for membrane proteins (34) (see Discussion). As shown in Fig. 4C, TagA, TagB, TagH, and TagF all showed strong self-interactions. TagD also showed self-interaction, albeit with a slightly weaker readout. TagO, TagA, TagB, TagF, and TagH all gave positive interaction signals with each other, and these interactions were reciprocal, with the exception of TagO, for which only the T25-TagO fusion gave a positive result. In previous observations with the FtsW protein of E. coli (34) it was suggested that T18 fusions to proteins that contain 10 or more transmembrane spans (FtsW and TagO contain 10 and 11, respectively) cannot insert correctly into the cytoplasmic membrane. TagH also showed a strong and reciprocal interaction with TagG. TagE, the last enzyme acting on the WTA precursor prior to export, showed interactions only when fused to T18. TagE interacted with TagA and TagH, as well as TagB and TagO, but for the latter two only when TagE was fused to the C-terminal end of T18. The lack of reciprocity means that the TagE results should be interpreted with caution.
In B. subtilis, MreC and MreD proteins localize in a helix-like pattern (41), and MreC was recently shown to interact with several high-molecular-weight PBPs, including PbpA and PbpH (66), suggesting that MreCD may help to position the PG machinery (22-24, 41, 66). To address whether they might also be involved in positioning the WTA biosynthetic machinery, the Y2H and B2H systems were used to test for interactions between MreCD and the Tag proteins. In the Y2H assay, interactions between MreD and TagH and TagO were detected (Fig. 4B). In the B2H, interactions between both MreC and MreD and TagH, TagO, TagA, TagB, and TagF were detected (Fig. 4D). The MreB proteins of B. subtilis do not seem to be functional in standard two-hybrid experiments (our unpublished results) and were therefore excluded from the test matrices.
DISCUSSION
Subcellular sites of synthesis and export of WTA.
We have characterized the localization of functional GFP fusions to TagB, TagF, TagG, TagH, and TagO. All of the fusions showed localization at division sites and, to a greater or lesser extent, along the lateral sides of the cells. In general, the fluorescence intensity for fusions under native control was relatively low, making imaging difficult, with TagB being weakest and TagGH strongest. Since TagGH comprises the putative ABC-type transporter for WTA, the GFP fusions to TagGH probably identify the sites of export of WTA precursors. Both TagG and TagH localized to new septa in dividing cells and unevenly, in a helix-like pattern (see below), along the length of the cell (Fig. 2A to D). The fluorescence signal at the poles was reduced in separating cells (Fig. 2A and C) and was essentially absent from poles after cell separation (Fig. 2B and D). We note that the helix-like pattern is best observed for GFP fusions to proteins containing at least one transmembrane span. The virtual absence of TagGH in the newly completed cell pole suggests that the export and hence the incorporation of WTA into the crosswalls occurs during division (septum formation) and is absent after cell separation and formation of the cell poles. This may provide the molecular basis for the observation that turnover of WTA at the cell pole is significantly slower than that in the cell cylinder, as detected using both phage binding assays (3, 4, 12) and concanavalin A binding (46). If WTA and PG synthesis are coupled, this is also consistent with the observation that the turnover of PG is very slow at cell poles (12, 57). Interestingly, the fusions to the other proteins, of which that of TagO was strongest, did not show such a striking reduction in signal at mature cell poles (Fig. 3A and D). It is therefore possible that TA synthesis can occur at the old cell poles, uncoupled from export.
All of the localization patterns, especially those of TagGH and TagO, were highly reminiscent of the staining seen with fluorescent probes that label the topology of nascent PG insertion, which has been shown to occur at the septum and in a helix-like pattern over the lateral wall (15, 63). This suggested that nascent WTA might also be incorporated into the lateral wall following an underlying helical tract. Early studies using radiolabeling suggested that in B. subtilis new WTA polymers are incorporated only into newly synthesized PG strands (45). Although we cannot provide conclusive evidence that the sites of export of PG precursors correspond exactly to the sites of export of WTA precursors, this seems plausible in the light of our observations and would allow for rapid incorporation of the WTA into the CW, without the long WTA polymers having to diffuse laterally along the outer surface of the membrane to reach the sites where nascent PG strands are about to be inserted.
MreB proteins control cell shape determination, and they are believed to achieve this by directing the spatial organization of the sites of PG insertion. This prompted us to analyze whether the helix-like pattern of localization displayed by the WTA transporter TagGH was also directed by the MreB cytoskeleton. No significant difference in the localization of either TagG or TagH was found in the mreB, mbl, or mreBH single mutant backgrounds (Fig. 2E to J) relative to the wild-type background (Fig. 2A and B). Although this raises the possibility that the actin-like cytoskeleton does not position the sites of WTA export, it seems more likely that the system is redundant and that more than one MreB homologue can act as a helical guide for WTA export.
Evidence for a WTA-synthesizing multienzyme complex at the membrane.
Using the B2H and Y2H systems, a complex network of interactions has been found between the WTA synthetic enzymes (TagO, TagA, TagB, and TagF), all of which also interacted with TagH, the NBD of the ABC-type transporter. Furthermore, TagA, TagB, TagD, TagH, and TagF all showed self-interactions, suggesting that they dimerize or oligomerize in vivo. All of these protein-protein interactions are believed to be physiologically relevant in view of the WTA biosynthetic pathway (Fig. 1) and of the similar localization patterns displayed by the Tag proteins in vivo (Fig. 2 and 3). Such a multienzyme complex would allow WTA to be rapidly synthesized at the cell membrane prior to (or simultaneously with) export through the TagGH transporter. The B2H system detected more interactions than the Y2H system. Both systems rely on the reconstitution of function by the interaction of two independent protein domains fused to candidate interacting proteins. In the Y2H system an additional requirement is that the interacting fusion proteins are correctly targeted to the yeast nucleus, where they have to bind DNA in order for an interaction to be recorded. In the B2H system the readout is provided by the response to a cyclic AMP-activated signaling cascade, which precludes the need for the interacting partners to be near the transcription machinery. Hence, the B2H system appears more suited for membrane (and membrane-associated) proteins such as the Tag proteins, which could explain the larger number of interactions detected in our B2H assays compared to our Y2H assays (34).
TagG and TagH, the respective membrane-spanning domain and NBD of the predicted ABC transporter that translocates TA precursors, displayed a strong interaction in both the Y2H and the B2H assays. Interestingly, TagG did not show a positive interaction with any of the other proteins tested, supporting the idea that the interactions detected by these systems reflect a physiologically relevant specificity. TagH also showed a self-interaction, whereas TagG did not, which was unexpected since dimerization is a common feature in the structure of ABC transporters (8). However, it is important to note that a lack of interaction in two-hybrid assays does not exclude a possible interaction in the native cellular system. Thus, it is possible that TagG forms a dimer in B. subtilis, which either cannot be detected in our assays or requires other factors (such as TagH) to be formed. Our analysis suggests that TagH plays a key role in WTA synthesis, presumably by linking the enzymes involved in the synthesis of the WTA precursors to the translocase TagG and providing the energy for active export of WTA.
TagA, TagB, TagF, TagH, and TagO were found to interact with MreC and MreD in the B2H system (Fig. 4D). The interactions between MreD and both TagH and TagO were further confirmed by the Y2H system (Fig. 4B). Interestingly, interactions of MreC with high-molecular-weight PBPs were recently detected using the same B2H system (66). This suggests that, in B. subtilis at least, MreC and MreD may have a role in positioning (or coupling) both the WTA-synthesizing machinery and the PG-synthesizing machinery along the cylindrical wall. In E. coli, MreC has been shown to interact with both MreB and MreD, and MreC and MreB both showed self-interactions in a B2H assay (38). In B. subtilis, MreB, Mbl and MreBH are currently not functional in standard two-hybrid experiments (i.e., the expected self- and cross-interactions between them are not detected in either Y2H or B2H assays [our unpublished results]), but the three isoforms colocalize in a single helical structure (11), and direct interactions between MreC and Mbl and between MreB, Mbl, and MreBH were recently detected at the cell membrane by bimolecular fluorescence complementation (18). Thus, if MreC and MreD are positioned by an interaction with MreB and/or MreBH and/or Mbl and if they are in turn positioning the PG and the WTA synthetic machineries as suggested by the direct interactions detected between MreC/D and both PBPs (66) and Tag proteins (Fig. 4B and D), then lack of a single MreB isoform would not lead to a loss of localization of either PBPs (55) or Tag proteins (Fig. 2E to J). It is tempting to speculate that through (co-)localizing all these proteins along an organized helical path, the bacterial actin cytoskeleton spatially coordinates PG and WTA synthesis to ensure that new tracts of PG containing WTA are inserted in the lateral wall. Obviously, the interactions suggested here should be tested biochemically in due course, e.g., via coimmunoprecipitation or affinity chromatography experiments.
In summary, the results presented here suggest that WTA, like PG (i.e., all major CW polymers) is laid down in helical tracts and that WTA is incorporated into the PG at the same place and at the same time that the PG precursors are inserted into the CW. This may define discrete and highly organized sites on the membrane where PG and WTA coupled synthesis takes place, allowing the balanced composition and structure of the CW that is necessary to ensure controlled CW growth in rod-shaped bacteria.
Supplementary Material
Acknowledgments
We thank Eric Brown for providing B. subtilis strain EB247.
R.C.-L. was funded by a long-term fellowship from the Human Frontier Science Programme Organization (HFSPO) during part of this work. R.C-L. and P.N. are supported by the Institut National de la Recherche Agronomique (INRA). Work in the Errington lab was supported by a grant from the HFSPO. D.-J.S. was funded by a VENI fellowship from the Research Council for Earth and Life Sciences from The Netherlands Organization for Scientific Research (NWO).
Footnotes
Published ahead of print on 21 December 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abachin, E., C. Poyart, E. Pellegrini, E. Milohanic, F. Fiedler, P. Berche, and P. Trieu-Cuot. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 431-14. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, A. J., R. S. Green, A. J. Sturman, and A. R. Archibald. 1978. Cell wall assembly in Bacillus subtilis: location of wall material incorporated during pulsed release of phosphate limitation, its accessibility to bacteriophages and concanavalin A, and its susceptibility to turnover. J. Bacteriol. 136886-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archibald, A. R., and H. E. Coapes. 1976. Bacteriophage SP50 as a marker for cell wall growth in Bacillus subtilis. J. Bacteriol. 1251195-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhavsar, A. P., and E. D. Brown. 2006. Cell wall assembly in Bacillus subtilis: how spirals and spaces challenge paradigms. Mol. Microbiol. 601077-1090. [DOI] [PubMed] [Google Scholar]
- 6.Bhavsar, A. P., L. K. Erdman, J. W. Schertzer, and E. D. Brown. 2004. Teichoic acid is an essential polymer in Bacillus subtilis that is functionally distinct from teichuronic acid. J. Bacteriol. 1867865-7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhavsar, A. P., R. Truant, and E. D. Brown. 2005. The TagB protein in Bacillus subtilis 168 is an intracellular peripheral membrane protein that can incorporate glycerol phosphate onto a membrane-bound acceptor in vitro. J. Biol. Chem. 28036691-36700. [DOI] [PubMed] [Google Scholar]
- 8.Biemans-Oldehinkel, E., M. K. Doeven, and B. Poolman. 2006. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 5801023-1035. [DOI] [PubMed] [Google Scholar]
- 9.Boyd, D. A., D. G. Cvitkovitch, A. S. Bleiweis, M. Y. Kiriukhin, D. V. Debabov, F. C. Neuhaus, and I. R. Hamilton. 2000. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 1826055-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carballido-López, R., and J. Errington. 2003. The bacterial cytoskeleton. In vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev. Cell 419-28. [DOI] [PubMed] [Google Scholar]
- 11.Carballido-López, R., A. Formstone, Y. Li, S. D. Ehrlich, P. Noirot, and J. Errington. 2006. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev. Cell 11399-409. [DOI] [PubMed] [Google Scholar]
- 12.Clarke-Sturman, A. J., A. R. Archibald, I. C. Hancock, C. R. Harwood, T. Merad, and J. A. Hobot. 1989. Cell wall assembly in Bacillus subtilis: partial conservation of polar wall material and the effect of growth conditions on the pattern of incorporation of new material at the polar caps. J. Gen. Microbiol. 135657-665. [DOI] [PubMed] [Google Scholar]
- 13.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 17333-38. [DOI] [PubMed] [Google Scholar]
- 14.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular bological methods for Bacillus. John Wiley and Sons Ltd., Chichester, United Kingdom.
- 15.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113767-776. [DOI] [PubMed] [Google Scholar]
- 16.Daniel, R. A., M. F. Noirot-Gros, P. Noirot, and J. Errington. 2006. Multiple interactions between the transmembrane division proteins of Bacillus subtilis and the role of FtsL instability in divisome assembly. J. Bacteriol. 1887396-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Defeu Soufo, H. J., and P. L. Graumann. 2005. Bacillus subtilis actin-like protein MreB influences the positioning of the replication machinery and requires membrane proteins MreC/D and other actin-like proteins for proper localization. BMC Cell Biol. 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Defeu Soufo, H. J., and P. L. Graumann. 2006. Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB. Mol. Microbiol. 621340-1356. [DOI] [PubMed] [Google Scholar]
- 19.Defeu Soufo, H. J., and P. L. Graumann. 2004. Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep. 5789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Elia, M. A., K. E. Millar, T. J. Beveridge, and E. D. Brown. 2006. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J. Bacteriol. 1888313-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Elia, M. A., M. P. Pereira, Y. S. Chung, W. Zhao, A. Chau, T. J. Kenney, M. C. Sulavik, T. A. Black, and E. D. Brown. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 1884183-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Divakaruni, A. V., R. R. Loo, Y. Xie, J. A. Loo, and J. W. Gober. 2005. The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 10218602-18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dye, N. A., Z. Pincus, J. A. Theriot, L. Shapiro, and Z. Gitai. 2005. Two independent spiral structures control cell shape in Caulobacter. Proc. Natl. Acad. Sci. USA 10218608-18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figge, R. M., A. V. Divakaruni, and J. W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 511321-1332. [DOI] [PubMed] [Google Scholar]
- 25.Formstone, A., and J. Errington. 2005. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol. Microbiol. 551646-1657. [DOI] [PubMed] [Google Scholar]
- 26.Foster, S. J., and D. L. Popham. 2002. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-41. In L. Sonenshein, R. Losick, and J. A. Hoch (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, DC.
- 27.Gross, M., S. E. Cramton, F. Gotz, and A. Peschel. 2001. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 693423-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28235-247. [DOI] [PubMed] [Google Scholar]
- 29.Honeyman, A. L., and G. C. Stewart. 1988. Identification of the protein encoded by rodC, a cell-division gene from Bacillus subtilis. Mol. Microbiol. 2735-741. [DOI] [PubMed] [Google Scholar]
- 30.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1441425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkinson, H. F. 1983. Altered arrangement of proteins in the spore coat of a germination mutant of Bacillus subtilis. J. Gen. Microbiol. 1291945-1958. [DOI] [PubMed] [Google Scholar]
- 32.Jones, L. J. F., R. Carballido-López, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104913-922. [DOI] [PubMed] [Google Scholar]
- 33.Karamata, D., and J. D. Gross. 1970. Isolation and genetic analysis of temperature-sensitive mutants of Bacillus subtilis defective in DNA synthesis. Mol. Gen. Genet. 108277-287. [DOI] [PubMed] [Google Scholar]
- 34.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 1872233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovacs, M., A. Halfmann, I. Fedtke, M. Heintz, A. Peschel, W. Vollmer, R. Hakenbeck, and R. Bruckner. 2006. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 1885797-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kristian, S. A., V. Datta, C. Weidenmaier, R. Kansal, I. Fedtke, A. Peschel, R. L. Gallo, and V. Nizet. 2005. d-Alanylation of teichoic acids promotes group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 1876719-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305567-580. [DOI] [PubMed] [Google Scholar]
- 38.Kruse, T., J. Bork-Jensen, and K. Gerdes. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 5578-89. [DOI] [PubMed] [Google Scholar]
- 39.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 1772403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazarevic, V., and D. Karamata. 1995. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids Mol. Microbiol. 16345-355. [DOI] [PubMed] [Google Scholar]
- 41.Leaver, M., and J. Errington. 2005. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol. Microbiol. 571196-1209. [DOI] [PubMed] [Google Scholar]
- 42.Lewis, P. J., and J. Errington. 1997. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol. Microbiol. 25945-954. [DOI] [PubMed] [Google Scholar]
- 43.Lewis, P. J., and A. L. Marston. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227101-109. [DOI] [PubMed] [Google Scholar]
- 44.Ma, H., S. Kunes, P. J. Schatz, and D. Botstein. 1987. Plasmid construction by homologous recombination in yeast. Gene 58201-216. [DOI] [PubMed] [Google Scholar]
- 45.Mauck, J., and L. Glaser. 1972. On the mode of in vivo assembly of the cell wall of Bacillus subtilis. J. Biol. Chem. 2471180-1187. [PubMed] [Google Scholar]
- 46.Mobley, H. L. T., A. L. Koch, R. J. Doyle, and U. N. Streips. 1984. Insertion and fate of the cell wall in Bacillus subtilis. J. Bacteriol. 158169-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noirot-Gros, M. F., E. Dervyn, L. J. Wu, P. Mervelet, J. Errington, S. D. Ehrlich, and P. Noirot. 2002. An expanded view of bacterial DNA replication. Proc. Natl. Acad. Sci. USA 998342-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nouaille, S., J. Commissaire, J. J. Gratadoux, P. Ravn, A. Bolotin, A. Gruss, Y. Le Loir, and P. Langella. 2004. Influence of lipoteichoic acid d-alanylation on protein secretion in Lactococcus lactis as revealed by random mutagenesis. Appl. Environ. Microbiol. 701600-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park, Y. S., T. D. Sweitzer, J. E. Dixon, and C. Kent. 1993. Expression, purification, and characterization of CTP:glycerol-3-phosphate cytidylyltransferase from Bacillus subtilis. J. Biol. Chem. 26816648-16654. [PubMed] [Google Scholar]
- 51.Pooley, H. M., F. X. Abellan, and D. Karamata. 1991. A conditional-lethal mutant of Bacillus subtilis 168 with a thermosensitive glycerol-3-phosphate cytidylyltransferase, an enzyme specific for the synthesis of the major wall teichoic acid. J. Gen. Microbiol. 137921-928. [DOI] [PubMed] [Google Scholar]
- 52.Pooley, H. M., J. M. Schlaeppi, and D. Karamata. 1978. Localised insertion of new cell wall in Bacillus subtilis. Nature 274264-266. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 54.Scheffers, D.-J. 2007. The cell wall of Bacillus subtilis, p. 331-373. In P. L. Graumann (ed.), Bacillus: cellular and molecular biology. Horizon Scientific Press, Norwich, United Kingdom.
- 55.Scheffers, D.-J., L. J. F. Jones, and J. Errington. 2004. Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 51749-764. [DOI] [PubMed] [Google Scholar]
- 56.Schertzer, J. W., and E. D. Brown. 2003. Purified, recombinant TagF protein from Bacillus subtilis 168 catalyzes the polymerization of glycerol phosphate onto a membrane acceptor in vitro. J. Biol. Chem. 27818002-18007. [DOI] [PubMed] [Google Scholar]
- 57.Schlaeppi, J.-M., and D. Karamata. 1982. Cosegregation of cell wall and DNA in Bacillus subtilis. J. Bacteriol. 1521231-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schlaeppi, J.-M., O. Schaefer, and D. Karamata. 1985. Cell wall and DNA cosegregation in Bacillus subtilis studied by electron microscope autoradiography. J. Bacteriol. 164130-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soldo, B., V. Lazarevic, and D. Karamata. 2002. tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology 1482079-2087. [DOI] [PubMed] [Google Scholar]
- 60.Soldo, B., V. Lazarevic, H. M. Pooley, and D. Karamata. 2002. Characterization of a Bacillus subtilis thermosensitive teichoic acid-deficient mutant: gene mnaA (yvyH) encodes the UDP-N-acetylglucosamine 2-epimerase. J. Bacteriol. 1844316-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steen, A., G. Buist, K. J. Leenhouts, M. El Khattabi, F. Grijpstra, A. L. Zomer, G. Venema, O. P. Kuipers, and J. Kok. 2003. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J. Biol. Chem. 27823874-23881. [DOI] [PubMed] [Google Scholar]
- 62.Sterlini, J. M., and J. Mandelstam. 1969. Committment to sporulation in Bacillus subtilis and its relationship to the development of actinomycin resistance. Biochem. J. 11329-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tiyanont, K., T. Doan, M. B. Lazarus, X. Fang, D. Z. Rudner, and S. Walker. 2006. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. USA 10311033-11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403623-627. [DOI] [PubMed] [Google Scholar]
- 65.van den Ent, F., L. A. Amos, and J. Löwe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 41339-44. [DOI] [PubMed] [Google Scholar]
- 66.van den Ent, F., M. Leaver, F. Bendezu, J. Errington, P. de Boer, and J. Lowe. 2006. Dimeric structure of the cell shape protein MreC and its functional implications. Mol. Microbiol. 621631-1642. [DOI] [PubMed] [Google Scholar]
- 67.Varley, A. W., and G. C. Stewart. 1992. The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (MinCD) and cell shape (MreBCD) determinants. J. Bacteriol. 1746729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wecke, J., M. Perego, and W. Fischer. 1996. d-Alanine deprivation of Bacillus subtilis teichoic acids is without effect on cell growth and morphology but affects the autolytic activity. Microb. Drug Resist. 2123-129. [DOI] [PubMed] [Google Scholar]
- 69.Wu, L. J., and J. Errington. 2003. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol. Microbiol. 491463-1475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.