Abstract
The bacterial flagellum is important for motility and adaptation to environmental niches. The sequence of events required for the synthesis of the flagellar apparatus has been extensively studied, yet the events that dictate where the flagellum is placed at the onset of flagellar biosynthesis remain largely unknown. We addressed this question for alphaproteobacteria by using the polarly flagellated alphaproteobacterium Caulobacter crescentus as an experimental model system. To identify candidates for a role in flagellar placement, we searched all available alphaproteobacterial genomes for genes of unknown function that cluster with early flagellar genes and that are present in polarly flagellated alphaproteobacteria while being absent in alphaproteobacteria with other flagellation patterns. From this in silico screen, we identified pflI. Loss of PflI function in C. crescentus results in an abnormally high frequency of cells with a randomly placed flagellum, while other aspects of cell polarization remain normal. In a wild-type background, a fusion of green fluorescent protein (GFP) and PflI localizes to the pole where the flagellum develops. This polar localization is independent of the flagellar protein FliF, whose oligomerization into the MS ring is thought to define the site of flagellar synthesis, suggesting that PflI acts before or independently of this event. Overproduction of PflI-GFP often leads to ectopic localization at the wrong, stalked pole. This is accompanied by a high frequency of flagellum formation at this ectopic site, suggesting that the location of PflI is a sufficient marker for a site for flagellar assembly.
Flagellar arrangement has long been used as a defining characteristic in bacterial taxonomy. Bacteria can have flagella emerging from a specific site, such as the cell pole, or from seemingly random positions along the cell body (as in peritrichously flagellated bacteria). Differences in flagellar distribution can be important for how various species adapt to specific environments. While the biosynthesis of the flagellum and the temporal regulation of its assembly have been extensively studied (5, 32), we know little about what controls the cellular position of flagellar assembly. The distribution of flagella in peritrichous bacteria may be random and stochastically driven, but spatial mechanisms must exist in polarly flagellated bacteria to ensure that flagellar assembly takes place at the right location.
It has been shown that in several polarly flagellated species of gammaproteobacteria, the flhF gene product is necessary for restricting the flagellum to a polar position (38, 41). flhF encodes a GTP-binding protein homologous to components of the signal recognition particle (SRP) pathway; in its absence, Pseudomonas putida and Pseudomonas aeruginosa have reduced motility and randomly placed flagella on the cell surface (25, 38, 41). Surprisingly, flhF homologs are also found in species with multiple, randomly placed flagella, indicating that FlhF is not a defining component of polarly flagellated bacteria. Furthermore, many polarly flagellated bacteria lack flhF homologs, indicating the existence of other mechanisms for flagellar placement. A large number of these species belong to the vast, understudied class of alphaproteobacteria, which includes species found in a variety of environmental niches, from aquatic environments to soils to plant and animal cells. Caulobacter crescentus belongs to this bacterial group. Its flagellation state varies with respect to cell cycle stage. The newly born “swarmer” daughter cells are motile and have a single flagellum and several pili at one pole. Before initiating DNA replication, swarmer cells differentiate into stalked cells by ejecting the flagellum and forming a thin extension of the cell envelope (the stalk, which is involved in nutrient uptake and surface attachment) at the pole where the flagellum once was. The sessile stalked cells then begin expressing flagellar genes in a stepwise, hierarchical manner, and a new flagellum is formed at the pole opposite the stalk (28, 35, 52). Division yields a flagellated swarmer progeny for dispersal and a sessile stalked progeny for local colonization.
Defects in flagellar placement have been observed in several C. crescentus mutants, but the mutations lie in genes that have pleiotropic functions in cell polarization and polar morphogenesis (1, 20, 21, 27, 47, 49). In this study, we identify CC_2060, renamed pflI (the polar-flagellum-linked gene), as one of the genes specifically associated with flagellar positioning. In polarly flagellated alphaproteobacteria, pflI homologs cluster with flagellar genes involved in the initial step of flagellar assembly, whereas pflI homologs are absent in nonflagellated and peritrichous alphaproteobacteria, suggesting a conserved function.
MATERIALS AND METHODS
Strains, plasmids, media, and growth conditions.
C. crescentus cells were grown at 30°C in peptone yeast extract (PYE) or M2G+ medium supplemented with antibiotics and sugars (0.3% xylose or 0.2% glucose) as described previously (14, 26). Escherichia coli cells were grown at 37°C in Luria-Bertani medium supplemented with appropriate antibiotics. The methods by which the strains and plasmids used in this study were constructed can be found in the supplemental material. The strains and plasmids used are listed in Table 1.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant genotype and/or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5 | Cloning strain | Invitrogen |
| S17-1 | M294::RP4-2 (Tc::Mu)(Km::Tn7); for plasmid mobilization | 46 |
| C. crescentus | ||
| CJW27 | CB15N | 15 |
| CJW686 | CB15N pleC::pBGENTpleCmyfp | 34 |
| CJW816 | CB15N ΔpleC/pMR20 | P. Angelastro (gift) |
| CJW1208 | CB15N ΔcreS | 6 |
| CJW1298 | CB15N/pMR20 | P. Angelastro (gift) |
| CJW1406 | CB15N tipN::pBGENT{tipNgfp | 27 |
| CJW1524 | CB15N ΔfliM | This study |
| CJW1757 | CB15N ΔpflI | This study |
| CJW2179 | CB15N/pMR20pxylpflIgfp | This study |
| CJW2181 | CB15N pflI::pHL32{pflIgfp | This study |
| CJW2182 | CB15N ΔfliF pflI::pHL32{pflIgfp | This study |
| CJW2184 | CB15N ΔpflI pleC::pBGENTpleCmyfp | This study |
| CJW2185 | CB15N pleC::pBGENTpleCmyfp/pMR20pxylpflI | This study |
| CJW2186 | CB15N ΔpflI tipN::pBGENT{tipNgfp | This study |
| CJW2187 | CB15N tipN::pBGENT{tipNgfp/pMR20pxylpflI | This study |
| CJW2188 | CB15N ΔpflI cpaE::cpaE-yfp | This study |
| CJW2189 | CB15N cpaE::cpaE-yfp/pMR20pxylpflI | This study |
| CJW2192 | CB15N/pMR20pxylpflI | This study |
| LS 1218 | CB15N ΔfliF | 22 |
| PV418 | CB15N cpaE::cpaE-yfp | 49 |
| UJ506 | CB15N ΔpleC | 2 |
| Plasmids | ||
| pBGENT | Gmr variant of pBGST18 integration vector | 34 |
| pBGENT{tipNgfp | Integration plasmid with 3′ end of tipN fused to gfp | 27 |
| pBGENTpleC-myfp | Integration plasmid with full-length pleC fused to monomeric yfp | 34 |
| pBGENTpxylpflIgfp | Integration plasmid with pflI-gfp under the xylose-inducible promoter | This study |
| pBluescript KS(+) | Apr cloning vector | Stratagene |
| pEGFP-N2 | Kmr cloning vector harboring gfp | Clontech |
| pHL32 | Kmr variant of pBGST18 with pBluescript KS multiple cloning site | 27 |
| pHL32{pflIgfp | Integration plasmid with 3′ end of pflI fused to gfp | This study |
| pHPV227 (pNPTScpaEyfp) | Sucrose suicide vector harboring cpaE fused to yfp | 49 |
| pJS14 | Cmr medium-copy-no. pBBR1-derived broad-host-range vector | Jeff Skerker |
| pJS14pflIgfp | pJS14 carrying pflI fused to gfp | This study |
| pJS14pxylpflI | pJS14 carrying pflI under the control of the xylose-inducible promoter | This study |
| pJS14pxylpflIgfp | pJS14 carrying pflI fused to gfp under the control of the xylose-inducible promoter | This study |
| pMR20 | Tcr low-copy-no. broad-host-range vector | 44 |
| pMR20pxylpflI | pMR20 carrying pflI under the control of the xylose-inducible promoter | This study |
| pMR20pxylpflIgfp | pMR20 carrying pflI-gfp under the control of the xylose-inducible promoter | This study |
| pNPTS138 | Litmus 38-derived vector; oriT sacB Kmr | MRK Alley |
| pNPTSfliMko | Sucrose suicide vector with sequences upstream and downstream of fliM | This study |
| pNPTSpflIko | Sucrose suicide vector with sequences upstream and downstream of pflI | This study |
Tc, tetracycline; Km, kanamycin; Gm, gentamicin; Ap, ampicillin; Cm, chloramphenicol.
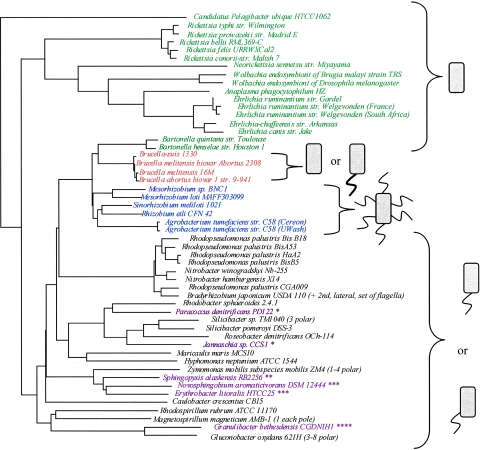
Small-subunit rRNA phylogenetic tree of sequenced alphaproteobacteria.
A list of all 51 alphaproteobacteria whose genomes had been completely sequenced was obtained by looking at the Complete Microbial Genomes section of the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). The 16S rRNA sequence was obtained from the website associated with the genome project for each organism (see Table S1 in the supplemental material). These sequences were aligned via the computer program ClustalX (http://www-igbmc.u-strasbg.fr/BioInfo/ClustalX/Top.html). Finally, a bootstrapped neighbor-joining phylogenetic tree was created using the same program. The locations and numbers of flagella of all of these species were determined by a literature search (see Table S1 in the supplemental material).
Screen for genes involved in polar flagellar placement in alphaproteobacteria.
The chromosomal loci including and surrounding class II flagellar genes (flhA, fliD, fliE, fliF, fliG, fliL, fliM, fliN, fliP, fliQ, and fliR) in the 21 polarly flagellated alphaproteobacterial species were determined by searching for the gene names at the websites associated with the genome project for each organism (see Table S1 and Table S2 in the supplemental material). A “class II genome region” was defined as the set of genes surrounding the class II genes listed above until a gene with a known function unrelated to flagella was seen. The open reading frames (ORFs) of unknown function (e.g., ORFs identified solely by systematic gene numbers) in the class II genome regions of all species examined were used as queries for the Basic Local Alignment Search Tool (BLAST) offered by NCBI (3, 4). An Excel file was created to group ORFs with their homologs found in any of the 51 fully sequenced alphaproteobacteria (see Table S3 in the supplemental material). A homolog was defined as an ORF of similar size with an E value less than or equal to 0.002. Each homolog group was placed into a row of this database. Using this Excel database, the ORFs that were prevalent in class II flagellar genome regions of polarly flagellated alphaproteobacteria and absent from all other species were identified. Additionally, homologs not limited to polarly flagellated bacteria (e.g., genes found not only in polarly flagellated bacteria but also in nonflagellated or peritrichous bacteria) were also found.
In silico analysis of the CC_2060 protein (PflI).
Localization and general topology were determined based on the information from the pSORTb algorithm (17). The coiled-coil content was determined by the COILS program (29). It should be noted that performing a BLAST search on the CC_2060 protein did not find all 20 homologs mentioned in Table S3 in the supplemental material. Due to the repetitive patterns of the coiled-coil repeat found in the CC_2060 protein, it was possible that there might be other proteins with similar function and structure that were not found during a standard BLAST. To account for this, a position-specific iterative BLAST (4, 45) was performed with the CC_2060 protein as a query. After three iterations, it was found that the CC_2060 protein has homologs in 20 species of polarly flagellated alphaproteobacteria. It is likely that these genes are indeed homologs of CC_2060, as they all have similar operon structures (see below). Also, when each of the direct homologs of CC_2060 was used as a query of a BLAST search, the total list of homologs was the same as the results of the position-specific iterative BLAST search using the CC_2060 protein as a query. Finally, the C-terminal portion of the CC_2060 protein was determined to be proline-rich based on the fact that 24 of the last 102 amino acid residues are proline.
Swarm assays.
As previously described (8), bacterial overnight cultures were stabbed into PYE soft (0.3%) agar (with xylose where indicated) and allowed to swim away from the point of inoculation for 3 days. The diameter of the swarm was then measured.
Blind EM.
To avoid biases, bacterial cultures were prepared and labeled by a colleague so as to hide the identity of strains used in the experiments until after the quantification of flagellation phenotypes observed by electron microscopy (EM). Exponential-phase cells were then adhered to glow-discharged carbon-coated EM grids and negatively stained with 2% uranyl acetate solution. A total of three (for the analysis of the ΔpflI strain) or nine (for the analysis of the pflI-overexpressing strain) grids, each grid made from a separate culture, were made for each strain and condition tested. Grids were then viewed using a Zeiss EM10C transmission electron microscope (80 kV), noting the position and number of flagella on each cell. After this, the identities of the cells on the grid were revealed so that further statistical analysis could be used to determine if any of the strains tested had a flagellar positioning defect.
Fluorescence microscopy.
For immobilization, cells were placed on a 1% agarose pad containing growth medium and viewed on a Nikon E1000 microscope through a 100× differential interference contrast objective using a Hamamatsu Orca-ER liquid crystal display camera. For the quantitation of fluorescence intensity, a 100× phase-contrast objective was used. Image analysis was performed using MetaMorph (Universal Imaging, PA). For time-lapse experiments, cells were grown in M2G+ and synchronized via ludox density centrifugation as previously described (15). Images were taken every 15 to 20 min during the course of the cell cycle.
Optical sectioning and three-dimensional (3D) deconvolution experiments were performed as described previously (6). Briefly, z series were obtained using MetaMorph software, taking a total of 33 images with 100-nm increments in the z direction. Out-of-focus light was removed from these images by using 3D deconvolution of z-series stacks with an Image J plug-in (U.S. National Institutes of Health).
Flagellar staining was performed as previously described (27). Briefly, cells from a mid-log-phase cell culture grown in PYE were fixed with a solution of 2.5% paraformaldehyde and 30 mM NaPO4 (pH 7.5) for 30 min at room temperature. Samples were treated with DAPI (4′,6′-diamidino-2-phenylindole; 1.5 μg/ml), incubated for 15 min at room temperature, and observed by fluorescence microscopy using an appropriate filter set.
Quantitation of fluorescence.
To quantify the cellular levels of PflI-green fluorescent protein (GFP) fluorescence during the cell cycle, cell populations were synchronized and several images were taken at each 15-min interval. The average fluorescence of each cell was determined by setting the threshold for the GFP images in MetaMorph, drawing regions around objects (cells), and calculating the average fluorescence per object with the Region Measurements tool. The average fluorescence and standard deviation of the average fluorescence per object at each time point were determined with Excel. Since the localization pattern of PflI-GFP was either at the new pole or diffuse, cells were placed into either category. This was determined by using the Region Measurements tool in MetaMorph to find the maximum fluorescence intensity in each cell. A cell was considered to have a polar focus if the difference between the maximum and average intensity of the cell divided by the average intensity was greater than or equal to 0.1.
Immunoblotting experiments.
Immunoblot analysis was performed using anti-FliF (1/10,000), anti-FliM (1/5,000), antiflagellin (1/10,000), anti-GFP JL-8 (1/1,000) (Clontech, CA), anti-CtrA (1/10,000), or affinity-purified anticrescentin (1/20,000). An equal amount of protein was loaded in each lane based on protein assays (Bio-Rad, CA).
RESULTS
The alphaproteobacterial class includes species with diverse patterns of flagellar positioning.
For the identification of candidate genes with a potential role in polar flagellar placement in alphaproteobacteria in general (and C. crescentus in particular), we hypothesized that these genes would be present in the genomes of polarly flagellated alphaproteobacteria but absent in those of species with other flagellation patterns. We first built a 16S rRNA phylogenetic tree for the 51 alphaproteobacteria with complete genome sequences and surveyed the literature and public databases to determine the flagellation pattern of each one of them (Fig. 1; see Table S1 in the supplemental material). From the phylogenetic tree, it became apparent that these species that share a long evolutionary history together have evolved various flagellation types. Organisms possessing polar or subpolar flagella, such as those shown in Fig. 1, must tightly regulate the expression and localization of their flagellar proteins to ensure that future generations have a flagellum at the correct position. In C. crescentus, this is done in part by coordinating the timing of flagellar gene expression with the cell cycle (7, 9, 39). Several alphaproteobacterial species are obligate intracellular parasites and do not possess flagellar genes due to genome reductions that are characteristically associated with this type of lifestyle (Fig. 1) (48). Brucella species possess a sheathed polar flagellum, which is believed to play a role in the persistence of these organisms within host cells (Fig. 1) (16). It should be noted that because of the sheathed nature of the flagellum in these organisms, there are probably many differences in how flagella are formed compared to species with nonsheathed flagella. Rhizobial species have peritrichous flagella. Other organisms shown in Fig. 1 have conflicting data as to their flagellation state. For example, Novosphingobium aromaticivorans has been described as polarly flagellated, yet genes required for flagellar synthesis are not found in the genome (http://genome.jgi-psf.org/finished_microbes/novar/novar.home.html). More detailed explanations as to the flagellation state of each organism and the references from which this information was obtained can be found in Table S1 in the supplemental material.
FIG. 1.
The alphaproteobacteria include species with diverse types of flagellation. The 16S rRNA sequences of alphaproteobacteria with completely sequenced genomes were aligned to produce the phylogenetic tree shown. On the right are cartoon depictions of what is known about the flagellation state of these organisms, as determined by literature searches (see Table S1 in the supplemental material). Organisms without flagella are shown in green. Brucella species, which have a sheathed polar flagellum under certain conditions (shown as thicker flagellum) but are often thought of as nonflagellated, are shown in orange. Species with peritrichous flagella are shown in blue. Organisms with a single polar or subpolar flagellum are shown in black. There are some examples of sequenced alphaproteobacteria with conflicting data concerning flagellar placement. These organisms, shown in purple, can be classified into four categories. The first group consists of organisms in which many or all flagellar genes are present but for which reports indicate that either a polar tuft of flagella exists or no flagella exist (*). For this study, they were considered polarly flagellated (18, 40, 50; http://genome.jgi-psf.org/draft_microbes/parde/parde.home.html and http://genome.jgi-psf.org/finished_microbes/jan_c/jan_c.home.html). The second group consists of organisms in which some flagellar genes are present and the species is thought to be nonflagellated (**) (13; http://genome.jgi-psf.org/draft_microbes/sphal/sphal.home.html). The third group consists of organisms whose flagellar genes are not annotated, but the organisms are listed as having a single polar flagellum (***). Since the organisms have no flagellar genes, these organisms were considered nonflagellated (18; http://genome.jgi-psf.org/draft_microbes/sphal/sphal.home.html and http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org=ntel01). The fourth group consists of organisms for whom no flagellar genes are annotated but for which there is no confirmation of the lack of a flagellum (****) (http://www.expasy.ch/sprot/hamap/GRABC.html and http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=17111). str., strain.
Strikingly, there are examples of species that show great similarity to each other with respect to 16S rRNA sequence but do not have similar flagellation patterns. For example, Rhodopseudomonas palustris shares more sequence similarity with Mesorhizobium loti than it does with C. crescentus, yet M. loti has peritrichous flagella whereas R. palustris and C. crescentus both have a single polar flagellum. This observation lends credence to the possibility that there may be genes or a gene system dedicated to the control of flagellar positioning in polarly (or subpolarly) flagellated species of alphaproteobacteria.
Screen for genes involved in polar flagellar positioning.
Using computer-based analysis of the 51 completed genome sequences of alphaproteobacteria, we conducted a screen to identify genes that are present in all or most of the 21 polarly flagellated species but absent in species with either peritrichous flagella or no flagella. Rather than use a genome-wide approach, we took advantage of the fact that genes involved in flagellar function and synthesis tend to be clustered together in chromosomal clusters or operons (37). Since the first structural components formed during flagellar biosynthesis are class II flagellar proteins (5, 32), we restricted our search to genes of unknown function that are found in chromosomal regions surrounding class II flagellar genes. Using the genome page associated with each of the 21 polarly flagellated alphaproteobacterial species, we determined the chromosomal loci adjacent to each of 11 class II genes: flhA, fliD, fliE, fliF, fliG, fliL, fliM, fliN, fliP, fliQ, and fliR (see Table S2 in the supplemental material).
These chromosomal loci included a total of 227 ORFs of unknown function (e.g., ORFs given only systematic gene numbers rather than a gene name). These ORFs are indicated in Table S3 in the supplemental material. By performing BLAST analysis with each of the 227 ORFs of unknown function as queries, we identified 127 homolog groups (see Table S3 in the supplemental material). We were most interested in homologs that were prevalent in species with polarly localized flagella and absent in nonflagellated and peritrichous species. Only three genes most closely matched these criteria. In C. crescentus, they correspond to CC_2058, CC_2059, and CC_2060, which have homologs in 13, 20, and 20 polarly flagellated alphaproteobacteria, respectively.
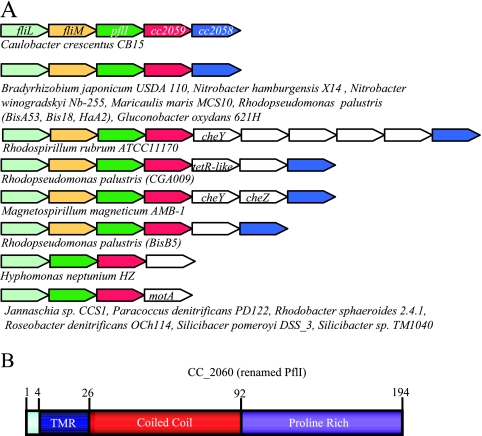
When they are present, the three candidate genes are found in similar genome regions (Fig. 2A). In C. crescentus, they are found downstream of the operonic genes fliL and fliM (53). There is a 4-base overlap at the fliM-CC_2060 and CC_2060-CC_2059 junctions, and 11 bases separate CC_2059 and CC_2058. This spacing suggests that these genes may share an operon. We chose to study the gene CC_2060, hereafter referred to as pflI (the polar-flagellum-linked gene) because pflI is the first gene following the class II flagellar gene fliM and it encodes features that make its gene product a good candidate for a potential scaffold involved in localizing the flagellum to a polar location (Fig. 2B). PflI is a short protein of 194 amino acids with a predicted transmembrane region at the N terminus, suggesting an inner-membrane location. Most of the protein is predicted to face the cytoplasmic side, and about half of its sequence consists of a coiled-coil motif (as predicted by the COILS program), which has been shown to be an important structure for initiating and maintaining protein-protein interactions (12, 29, 33). The last half of the protein has high proline content (24%), raising the possibility that this region forms a polyproline helix, which can also serve as a protein-protein interaction surface (11, 43).
FIG. 2.
The three genes identified as candidates for a putative role in polar localization of flagella are found in similar chromosomal loci. (A) Genome regions of candidate genes. There are three genes that are largely present in polarly flagellated alphaproteobacteria (including C. crescentus) and absent from all other types of bacteria. Shown here are schematic diagrams of the genome regions of these genes. Homologs of fliL, fliM, CC_2060, CC_2059, and CC_2058 (pflI) are shown in light green, yellow, green, red, and blue, respectively. Other genes are shown in white. The directions of the arrows indicate the relative directions of transcription. The organisms listed under each schematic are the species with that particular genome organization. (B) Domain architecture of PflI. In silico analysis of PflI reveals three domains: a transmembrane region from residues 4 through 25 (TMR) shown in blue, a coiled-coil region from residues 26 through 92 shown in red, and an uncharacterized proline-rich region from residues 93 through 194, in which 24 of 102 amino acids are prolines, shown in purple.
Deletion of pflI impacts flagellar location.
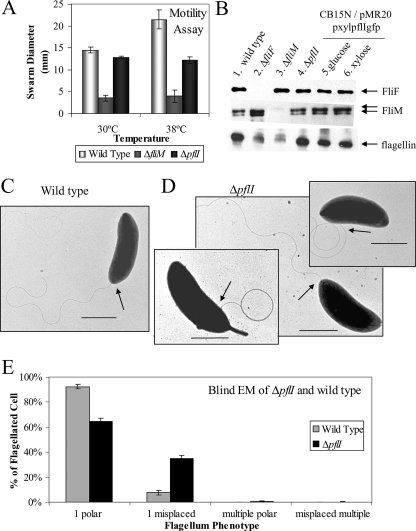
We generated an in-frame pflI deletion strain (CJW1757) to avoid polar effects on CC_2059 and CC_2058 expression. We showed that the mutant strain retains the ability to swim as determined by light microscopy (data not shown) and swarm assays at 30°C (Fig. 3A). Interestingly, there was a noticeable swarming defect in the ΔpflI mutant at 38°C (Fig. 3A), which corresponds to a heat shock condition for C. crescentus. The higher temperature may exacerbate any slight motility defect that ΔpflI has, perhaps by destabilizing protein-protein interactions that may be weakened in the absence of pflI.
FIG. 3.
PflI affects the localization of the flagellum but not its biosynthesis. (A) Swarm motility assay performed at 30°C or 38°C. The swarming abilities of wild-type (positive control; light gray bars), nonmotile ΔfliM (negative control; dark gray bars), and ΔpflI (black bars) strains were compared to determine if loss of pflI affects swimming ability. Swarm diameter was measured after 3 days of incubation at 30°C or 38°C. Growth at 30°C showed that ΔpflI cells are as motile as wild-type cells. When under heat shock at 38°C, ΔpflI cells are still motile but have a decreased swarm diameter compared to that of the wild type. (B) Flagellar protein levels are unchanged when PflI is absent or overproduced. Immunoblots were performed on the following strains: wild-type CB15N (CJW27; lane 1), ΔfliF (CJW1373; lane 2), ΔfliM (CJW1524; lane 3), ΔpflI (CJW1757; lane 4), and CB15N/pMR20pxylpflI (CJW2179) grown in glucose (lane 5) and xylose (lane 6). Immunoblots show that levels of flagellar proteins FliF, FliM, and flagellins are similar in wild-type, ΔpflI, and pflI overexpression strains (compare lanes 1, 2, 5, and 6). (C) Representative electron micrograph of wild-type CB15N cell showing a polar flagellum (arrow pointing to the base of the flagellum). Scale bar, 1 μm. (D) Electron micrographs of ΔpflI cells with a flagellum at the seemingly normal polar location (middle) or at aberrant locations (left and right panels). Scale bars, 1 μm. (E) Loss of pflI increases the prevalence of misplaced flagella. A blind EM study was performed to quantify the percentage of cells with flagellar placement defects. Flagellar position phenotypes of flagellated bacteria were grouped together into the following categories: those with one correctly placed flagellum, those with one misplaced flagellum, those with multiple correctly placed flagella, and those with multiple misplaced flagella. Cells with a correctly placed flagellum included small (presumably swarmer or early stalked) cells with a polar flagellum or predivisional cells with a flagellum found at the pole opposite the stalk. Cells identified as having a single misplaced flagellum included cells with a flagellum found at the stalked pole or not found at a pole. Cells with multiple correctly placed flagella included small cells with nonvisible stalks with two flagella coming out of the same pole or predivisional cells with two flagella at the pole opposite the stalk. Cells with multiple misplaced flagella were cells that had at least one flagellum found in a nonpolar position or at the stalked pole. For example, cells with bipolar flagella were classified as having multiple misplaced flagella.
EM confirmed that the pflI mutant can produce a flagellum under normal growth conditions. ΔpflI and wild-type cell populations had similar percentages of flagellated cells (65% and 56%, respectively) (see Table S4 in the supplemental material). The C. crescentus cell cycle includes several nonflagellated cell types (corresponding to the period between flagellum ejection and the assembly of a new flagellum at the opposite pole), explaining why only 50 to 65% of both wild-type and PflI mutant populations are flagellated. Immunoblot analysis showed that the abundances of several flagellar proteins, including FliF, FliM, and flagellin, were not significantly affected by the loss of pflI (Fig. 3B). These data indicate that PflI does not affect the biosynthetic pathways of flagellar components.
However, EM also revealed that a considerable fraction of ΔpflI cells had flagella emanating from aberrant positions, unlike wild-type cells, for which flagella were consistently found at a single pole (Fig. 3C). While there were still many cells that had a polar flagellum (Fig. 3D, center panel), a considerable fraction of ΔpflI cells showed flagella protruding near the stalked pole (Fig. 3D, left panel) or from sites along the lateral sides of the cell (Fig. 3D, right panel). To quantify this defect in flagellar placement, we performed an EM study (Fig. 3E; see Table S4 in the supplemental material). To avoid biases, this study was performed blind via a colleague labeling bacterial cultures such that the identities of the samples were unknown to us until after flagellar phenotypes were tabulated. Thirty-five percent (±2%) of flagellated pflI mutant cells were scored as carrying a flagellum at a nonpolar location, compared to only 8% (±2%) of wild-type flagellated cells. Thus, while the flagellum can assemble at the pole in the absence of PflI, its absence results in four- to fivefold more mistakes. It should be noted that young cells lacked a visible stalk, which prevented us from distinguishing between the two poles; in these cells, a flagellum located at the pole was considered to be correctly placed, despite the fact that the flagellum could be coming from the wrong pole, thus potentially underestimating the number of cells with misplaced flagella. In any case, the abnormal frequency of flagella at aberrant locations in the absence of PflI is consistent with a role for PflI in flagellar placement.
PflI localizes to the new pole where the flagellum develops.
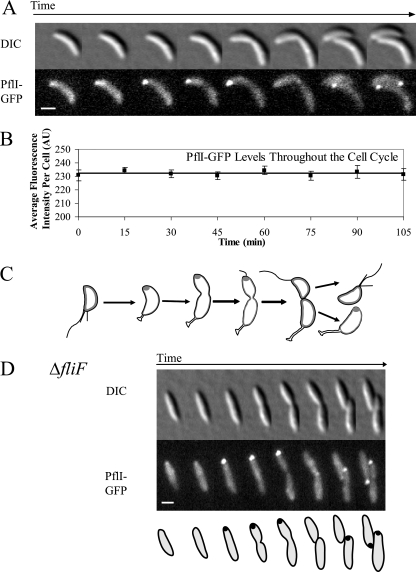
Since the anomalous rate of flagellum mispositioning in the ΔpflI background suggested a role for PflI in the proper polar placement of the flagellum, we examined the subcellular distribution of PflI by using a pflI-gfp fusion integrated at the pflI chromosomal locus as the only copy (CJW2181). Immunoblot analysis showed that full-length PflI-GFP was produced and GFP was not cleaved in this strain (data not shown). The PflI-GFP fusion exhibited some functionality based on the swarming ability of strain CJW2181 at 38°C (see Fig. S1A in the supplemental material). Time-lapse experiments showed that PflI-GFP localized as a fluorescent focus at the new pole (opposite the pole where the stalk develops) early in the stalked-cell stage and remained there during much of the cell cycle (Fig. 4A). At the late predivisional cell stage, the polar focus disappeared then reappeared at the new pole of the stalked progeny first and of the swarmer cell later (after it reached the stalked-cell stage). This pattern was verified by quantitation of 15-min-interval time-course experiments starting with synchronized swarmer-cell populations (see Fig. S1B in the supplemental material); under these conditions, the cell cycle took about 90 min.
FIG. 4.
PflI-GFP localizes at the pole where the flagellum forms in a FliF-independent manner. DIC, differential interference contrast. (A) PflI-GFP changes position during the cell cycle. Strain CJW2181 (carrying pflI-gfp) was synchronized and visualized using time-lapse fluorescence microscopy to determine the localization pattern of PflI-GFP. PflI-GFP is present as a focus in stalked cells at the pole opposite the stalk and remains at that pole for much of the cell cycle. This is the time and place at which the flagellum is assembled. In late predivisional cells, the focus disappears, only to reappear at the new pole of stalked-cell progeny. Scale bar, 1 μm. (B) The average fluorescence intensity of the whole cell does not change during the cell cycle. From the time course experiments whose results are shown in Fig. S1B in the supplemental material, the average fluorescence of each cell was determined at each time point. AU, arbitrary units. (C) Schematic representation of the prevalent localization of PflI-GFP during the cell cycle. Cells are outlined in black, and PflI-GFP is shown in dark gray. (D) PflI localization does not require assembly of the flagellar MS ring. Strain CJW2182 was visualized by time-lapse fluorescence microscopy to examine the localization of PflI-GFP in the absence of FliF. Scale bar, 1 μm.
In order to determine whether the absence of a polar focus of PflI-GFP in swarmer and predivisional cells was due to proteolysis during those parts of the cell cycle, we determined the average PflI-GFP fluorescence in the entire cell for several hundred cells at each time point and found similar average cellular fluorescence intensities throughout the cell cycle (Fig. 4B). Since the fluorescence intensity of cells producing PflI-GFP was well above the level of cellular autofluorescence intensity (see Fig. S1C in the supplemental material) and levels of PflI-GFP protein remain largely steady during the cell cycle (see Fig. S1D in the supplemental material), it is unlikely that the protein is proteolyzed when no polar localization is discernible. Rather, it is more likely that the protein is redistributed to a more diffuse localization. Altogether, our data argue that PflI condenses from a mostly dispersed distribution in the swarmer-cell stage to a focus at the new pole in the stalked-cell and predivisional cell stages and then delocalizes from the pole at the late predivisional cell stage (Fig. 4C).
PflI is predicted to be located in the inner membrane, based on computer algorithms, yet (as with other low-abundance membrane-bound proteins in C. crescentus [51]) PflI-GFP localization appears diffuse. In order to confirm membrane location, optical sectioning followed by 3D deconvolution was performed on cells overproducing PflI-GFP. This resulted in a halo of fluorescence around the cell, in addition to the polar focus, consistent with a membrane localization (see Fig. S2A in the supplemental material). At a normal expression level, PflI predominantly localizes at the pole where the flagellum develops, which is consistent with the hypothesis that PflI plays a role in localizing the flagellum to the proper cellular location.
Polar localization of PflI does not require the assembly of the flagellar MS ring.
The first flagellar protein believed to assemble at the site of flagellar assembly is FliF, which oligomerizes into the MS ring and initiates the assembly of the flagellum (23, 24). We therefore examined the localization of PflI-GFP in a ΔfliF background (strain LS1218). We noticed that the ΔfliF strain had a straight rod shape instead of the normal crescent shape (19). This shape anomaly was attributed to the lack of crescentin, based on immunoblot analysis (see Fig. S3 in the supplemental material), suggesting the presence of an independent mutation affecting the crescentin-encoding gene. Regardless, the polar localization pattern of PflI-GFP was unaffected by the ΔfliF mutation (or the loss of cell curvature) (Fig. 4D). This is consistent with the notion that PflI helps to specify the correct site for flagellar assembly.
Overproduction of PflI affects PflI localization pattern and flagellum placement.
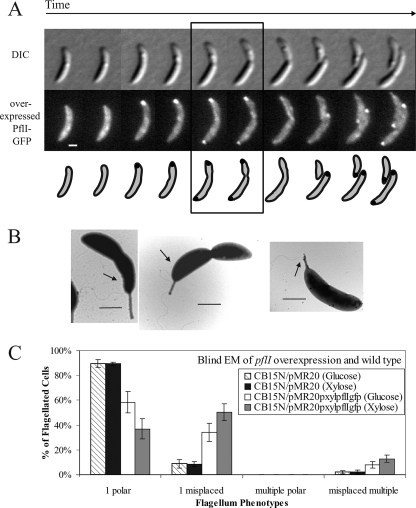
We reasoned that if polar localization of PflI in wild-type cells is involved in correct polar flagellar positioning, ectopic localization of PflI may lead to aberrant flagellar placement. Since overexpression of a gene can lead to ectopic localization of its gene product, we placed pflI-gfp under the xylose-inducible promoter on a low-copy plasmid (pMR20) and examined how wild-type cells carrying this plasmid (CB15N/pMR20pxylpflIgfp; CJW2179) localize PflI-GFP when it is overproduced. These CJW2179 cells, when grown in the presence of xylose, produced about 4.5-fold more PflI-GFP than did CJW2181 cells expressing pflI-gfp from the native promoter on the chromosome, based on relative whole-cell fluorescence intensities (see Fig. S1C in the supplemental material). Under these overexpressing conditions, PflI-GFP displayed a normal localization profile except for an additional accumulation of PflI-GFP at the stalked pole in 36% of predivisional cells (Fig. 5A; see Fig. S2B in the supplemental material for the quantitative analysis). The xylose-inducible promoter is not tightly repressed in the absence of the xylose inducer (36), especially when present on a multicopy plasmid (presumably due to the titration of the repressor). Consequently, when the same CJW2179 strain was grown in glucose (instead of xylose), there was a ∼2.5-fold increase in the PflI-GFP protein level compared to the endogenous level (see Fig. S1C in the supplemental material), and 24% of predivisional cells had an extra PflI-GFP focus at the stalked pole (see Fig. S2B in the supplemental material), in addition to the normal pattern. Thus, the degree of PflI-GFP overproduction correlates with the percentage of cells with PflI-GFP accumulating at the ectopic, stalked-pole location.
FIG. 5.
Overproduction of PflI-GFP results in ectopic localization of the protein and the flagellum. (A) Overexpression of PflI-GFP alters its localization pattern. Strain CJW2179 was synchronized and observed by fluorescence timelapse microscopy. Localization is dispersed in swarmer cells, at the new pole in stalked cells, bipolar in early-predivision cells, diffuse throughout the cytoplasmic membrane in late-predivision cells, and at the new pole of stalked-cell progeny. Scale bar, 1 μm. DIC, differential interference contrast. (B) Aberrant flagellar placement is evident upon overexpression of pflI-gfp. Strain CJW2179 was observed by EM. Many of the cells, including those in the electron micrographs shown in this panel, had flagella growing from improper locations. Arrows point to the base of the flagellum. Scale bar, 1 μm. (C) Increased abundance of PflI-GFP increases the percentage of cells with misplaced flagella at the stalked pole, where PflI-GFP accumulates ectopically. A blind EM study was performed on CB15N/pMR20 (CJW1298) and CB15N/pMR20pxylpflIgfp (CJW2179) cells grown in glucose or xylose to examine flagellar-placement phenotypes. Descriptions of these phenotypes are found in the legend of Fig. 3 (panel E).
Strains overexpressing pflI or pflI-gfp, whether grown in glucose or xylose, had motility (see Fig. S2C in the supplemental material) and levels of flagellar proteins FliF, FliM, and flagellin (Fig. 3B) similar to the wild type levels. The percentages of flagellated cells also did not change considerably upon overexpression of pflI; about 53% of wild-type cells carrying an empty plasmid (CB15N/pMR20; CJW1298) are flagellated when grown in glucose or xylose compared to 62% or 67% of pflI overexpressing (CB15N/pMR20pxylpflIgfp; CJW2179) cells grown in glucose or xylose, respectively.
The frequent ectopic localization of PflI at the stalked pole caused by overproduction was, however, accompanied by a frequent misplacement of flagella (Fig. 5B). Using a fluorescence microscopy technique that visualizes flagella (27), we found cells with bipolar PflI-GFP fluorescence as well as a flagellum emerging from the stalked pole (see Fig. S2D in the supplemental material). It should be noted that this method, which employs formaldehyde-induced fixation and DAPI staining, is not quantitative for two major reasons. First, fixation considerably disrupts polar protein localization, resulting in reduction or loss of PflI-GFP signal at the poles. Second, the flagellum staining can be difficult to observe because of the bright cellular staining of the DNA. We therefore performed a blind EM study to quantify the flagellar positioning phenotypes in cells overproducing PflI-GFP. Roughly 90% of flagellated, wild-type cells carrying an empty plasmid (CB15N/pMR20; CJW1298) had the expected unipolar flagellum; only about 60% of flagellated CB15N/pMR20pxylpflIgfp (CJW2179) cells grown in glucose (mild pflI-gfp overexpression) and about 36% of CJW2179 cells grown in xylose (stronger pflI-gfp overexpression) had this phenotype (Fig. 5C). Thirty-four percent of flagellated CJW2179 cells in glucose (with mild pflI-gfp overexpression) and 50% of flagellated CJW2179 cells in xylose (with stronger pflI-gfp overexpression) had a misplaced flagellum, compared to less than 10% of flagellated wild-type CJW1298 cells grown in glucose or xylose (see Table S5 in the supplemental material). Most of the CJW2179 cells with misplaced flagella had a single flagellum at or near the stalked pole, which is the site of ectopic PflI-GFP localization in this strain (Fig. 5A; see Table S5 in the supplemental material). These observations suggest that ectopic localization of PflI at the stalked pole is accompanied by inappropriate flagellar assembly at that ectopic site.
The role of PflI in cell polarization appears specific to the flagellum.
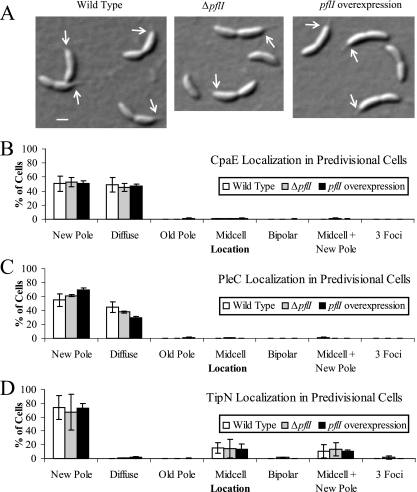
Our data argue that the presence and proper localization of PflI are important for positioning the flagellum at the correct pole. It is known that mutations in genes involved in multiple aspects of cell polarity or polar morphogenesis can result in flagellar placement defects (1, 20, 21, 27, 47, 49). Unlike some of these genes with pleiotropic functions, pflI does not affect cell division or stalk biogenesis, as judged by the normal cell and stalk morphology of ΔpflI and pflI-overexpressing cells (Fig. 3D, 5B, and 6). Both ΔpflI and pflI-overexpressing strains were, like wild-type strains, sensitive to the bacteriophage φCbK (data not shown), indicating the presence of pili, which are used as receptors for this phage. Furthermore, the pilus assembly protein CpaE exhibited the normal new-pole localization (49) in ΔpflI (CJW2188) and pflI-overexpressing (CJW2189 plus xylose) predivisional cells carrying cpaE-yfp (Fig. 6B), suggesting that PflI does not affect pilus function or localization.
FIG. 6.
Localization of polar markers CpaE, PleC, and TipN is not significantly affected by altered PflI abundance. (A) Polar morphogenesis does not appear to be affected by pflI. Differential interference contrast images of wild-type (CJW1388; left panel), ΔpflI (CJW2188; center panel), and pflI-overexpressing (CJW2189; right panel) cells show normal cellular asymmetry and stalk formation. Arrows point to the base of the stalk. Scale bar, 1 μm. (B) Localization of CpaE-yellow fluorescent protein was determined in synchronized predivisional cells of strains PV418 (wild type), CJW2188 (ΔpflI), and CJW2189 (with pflI overexpression in xylose-containing medium) by fluorescence microscopy. The localization of a fluorescent focus, if any, was assigned to the category of diffuse, new pole, old pole, midcell, bipolar, midcell plus new pole, or three foci. Cells were considered to have diffuse localization if no focus was present. Cells were determined to have a focus at the new pole if the focus was present at the pole opposite the stalk or at the pole closer to the division site (when no stalk was visible) because C. crescentus cells divide off-center, slightly closer to the new pole. Cells with old-pole localization had a focus at the stalked pole or at the pole farther from the division site. Cells with midcell localization had a focus at the division site. Cells with foci at both poles were called bipolar. Cells with two foci (one at the division plane and the other at the new pole, as described above) were assigned to the midcell-plus-new-pole category. Cells with foci at both poles plus the division plane were identified as having three foci. (C) Localization of PleC-monomeric yellow fluorescent protein was visualized by fluorescence microscopy using synchronized predivisional-cell populations of CJW686 (wild type), CJW2184 (ΔpflI), and CJW2185 grown in xylose (with pflI overexpression). Fluorescent foci were categorized as described for panel B. (D) Predivisional cells of CJW1406 (wild type), CJW2186 (ΔpflI), and CJW2187 grown in xylose (with pflI overexpression) were observed by fluorescence microscopy to determine TipN-GFP localization. Fluorescent foci were categorized as described for panel B.
We also examined whether PflI plays any role in the localization of polar markers, such as PleC and TipN. PleC is a signal transduction protein involved in polar morphogenesis that normally localizes to the flagellated pole in predivisional cells (51). We found no major differences in the localization of a PleC-monomeric yellow fluorescent protein fusion among wild-type (CJW686), ΔpflI (CJW2184), and pflI-overexpressing (CJW2185 plus xylose) predivisional cells (Fig. 6C). TipN is a cell polarity determinant that marks the new pole (where the flagellum assembles) for most of the cell cycle and relocates to the division site during the predivisional cell stage (21, 27). TipN-GFP showed similar localization profiles in wild-type (CJW1406), ΔpflI (CJW2186), and pflI-overexpressing (CJW2187 plus xylose) backgrounds (Fig. 6D).
These data, together with the clustering of pflI with class II flagellar genes and the segregation of pflI homologs in polarly flagellated alphaproteobacteria, strongly suggest that the role of pflI is specific for flagellar positioning, rather than being involved in overall cell polarization or polar morphogenesis.
DISCUSSION
Here we identify a factor that is important for flagellar placement in C. crescentus. PflI localizes to the new pole and is required for accurate and consistent positioning of the flagellar apparatus at that site. Oligomerization of FliF into the MS ring in the cytoplasmic membrane is the first recognized step in flagellar biogenesis. Without FliF, cytoplasmic components of the flagellum (such as FliM) are no longer associated with the membrane, and extracellular components (such as the flagellins), if made, can no longer be secreted (23, 24, 31). Strikingly, PflI localizes at the correct time and place in C. crescentus cells lacking FliF, thereby identifying PflI as an early or independent component marking the flagellum site. When PflI is overproduced, it localizes ectopically to the stalked pole, and this correlates with a considerable increase in ectopic flagellar synthesis at that site. C. crescentus populations carrying tipN, podJ, pleC, or pleD mutations also have a proportion of cells with a flagellum at the stalked pole or at the stalk itself, but these mutations also alter many aspects of cell polarity and polar morphogenesis (1, 20, 21, 27, 47, 49). On the other hand, we found no evidence of defects in cell polarization or polar morphogenesis in the ΔpflI or pflI-overexpressing strains, apart from a high frequency of flagellum misplacement. Furthermore, pflI and its orthologs are exclusively found in the genomes of polarly flagellated alphaproteobacteria, where they cluster with class II flagellar genes, most likely in operon structures. Collectively, these findings argue that PflI acts specifically on the flagellum.
Based on swarm assays, which provide information on cell populations rather than individual cells, PflI does not seem to affect motility. The flagellar misplacement phenotype is found in only 30% of flagellated ΔpflI cells and 34% or 50% of cells overproducing PflI (in glucose or xylose, respectively). The assay may not be sensitive enough to find motility defects in a subset of the population.
PflI appears to function differently from FlhF, which has been identified for its role in polar flagellar placement in several species (38, 41). How FlhF affects flagellum positioning is not well understood, but the sequence similarity between FlhF and GTP-binding SRP proteins suggests a possible role in translocation of early flagellar proteins. PflI has no predicted GTP-binding motifs and has no similarity to SRP proteins, but it has several potential protein-protein interaction domains (defined by the coiled-coil and proline-rich regions), suggesting a potential role in scaffolding flagellar proteins. We propose that PflI, by localizing at the new pole, works in concert with other, possibly redundant, localization factors to recruit early flagellar proteins to the proper site, thus ensuring high fidelity of flagellum assembly at the correct pole. In the absence of pflI, these other localization factors could still function, but fidelity would be compromised, resulting in a flagellar placement defect, because of which a portion of the cell population would have randomly placed flagella. When overproduced, PflI localizes ectopically at the stalked pole (in addition to its normal position) in 36% of cells, which would be sufficient to mark this site for flagellum assembly.
Based on its location next to early flagellar genes, its prevalence in polarly flagellated alphaproteobacteria, and its absence in others, pflI was predicted to be a potential mediator of flagellar positioning. Experimental data confirming this hypothesis show that these criteria can be successfully used for finding potential genes involved in flagellar positioning. Therefore, other genes fitting these criteria could be interesting candidates for involvement in this process as well (see Table S3 in the supplemental material). Of particular interest is CC_2059, which is downstream of pflI in 20 species of polarly flagellated alphaproteobacteria.
Based on our analysis of available genomic information, pflI appears limited to polarly flagellated alphaproteobacteria; only one species in this group, Zymomonas mobilis, does not have a recognizable pflI homolog (or CC_2059 homolog). Z. mobilis is, however, unique among this group of bacteria because its only set of flagellar genes comes from a horizontal gene transfer event with a gammaproteobacterium (10, 30, 42). Rhodobacter sphaeroides (which has a pflI homolog) has also acquired this horizontally transferred DNA but has two sets of flagellar genes: one of gammaproteobacterial origin and one inherited throughout the alphaproteobacteria. Thus, Z. mobilis would be more likely to act more similarly to gammaproteobacteria in terms of flagellation. Consequently, the presence of PflI is strongly correlated with the polar flagellar arrangement of alphaproteobacteria, suggesting a conserved and defining function for PflI in flagellar placement in this class. Interestingly, while the majority of polarly flagellated alphaproteobacteria bacteria do not possess an flhF homolog, Magnetospirillum magneticum, R. sphaeroides, Hyphomonas neptunium, Maricaulis maris, and Rhodospirillum rubrum do. In these five species, flhF and pflI may be acting on parallel pathways to achieve polar flagellation.
Supplementary Material
Acknowledgments
We are grateful to Patrick Viollier, Urs Jenal, and Lucy Shapiro for providing strains and antibodies, Barry Piekos for support with the EM, and the members of the Jacobs-Wagner laboratory for critical reading of the manuscript. We also thank Matthew Cabeen and Paula Montero Llopis for assistance with the blind EM experiments.
This work was supported by a National Institutes of Health predoctoral training grant in cellular and molecular biology (T32 GM07223) (to P.L.O.), National Institutes of Health grant GM065835 (to C.J.-W.), and the Pew Scholars Program in the Biological Sciences sponsored by the Pew Charitable Trust (to C.J.-W.).
Footnotes
Published ahead of print on 28 December 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aldridge, P., and U. Jenal. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32379-391. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 471695-1708. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apel, D., and M. G. Surette. 24 July 2007, posting date. Bringing order to a complex molecular machine: the assembly of the bacterial flagella. Biochim. Biophys. Acta. doi: 10.1016/j.bbamem.2007.07.005. [DOI] [PubMed]
- 6.Ausmees, N., J. R. Kuhn, and C. Jacobs-Wagner. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115705-713. [DOI] [PubMed] [Google Scholar]
- 7.Bryan, R., M. Purucker, S. L. Gomes, W. Alexander, and L. Shapiro. 1984. Analysis of the pleiotropic regulation of flagellar and chemotaxis gene expression in Caulobacter crescentus by using plasmid complementation. Proc. Natl. Acad. Sci. USA 811341-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton, G. J., G. B. Hecht, and A. Newton. 1997. Roles of the histidine protein kinase PleC in Caulobacter crescentus motility and chemotaxis. J. Bacteriol. 1795849-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Champer, R., A. Dingwall, and L. Shapiro. 1987. Cascade regulation of Caulobacter flagellar and chemotaxis genes. J. Mol. Biol. 19471-80. [DOI] [PubMed] [Google Scholar]
- 10.Choudhary, M., Y. X. Fu, C. Mackenzie, and S. Kaplan. 2004. DNA sequence duplication in Rhodobacter sphaeroides 2.4.1: evidence of an ancient partnership between chromosomes I and II. J. Bacteriol. 1862019-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creamer, T. P., and M. N. Campbell. 2002. Determinants of the polyproline II helix from modeling studies. Adv. Protein Chem. 62263-282. [DOI] [PubMed] [Google Scholar]
- 12.Delahay, R. M., and G. Frankel. 2002. Coiled-coil proteins associated with type III secretion systems: a versatile domain revisited. Mol. Microbiol. 45905-916. [DOI] [PubMed] [Google Scholar]
- 13.Eguchi, M., M. Ostrowski, F. Fegatella, J. Bowman, D. Nichols, T. Nishino, and R. Cavicchioli. 2001. Sphingomonas alaskensis strain AFO1, an abundant oligotrophic ultramicrobacterium from the North Pacific. Appl. Environ. Microbiol. 674945-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204372-384. [DOI] [PubMed] [Google Scholar]
- 15.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fretin, D., A. Fauconnier, S. Kohler, S. Halling, S. Leonard, C. Nijskens, J. Ferooz, P. Lestrate, R. Delrue, I. Danese, J. Vandenhaute, A. Tibor, X. DeBolle, and J. Letesson. 2005. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol. 7687-698. [DOI] [PubMed] [Google Scholar]
- 17.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. L. Brinkman. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21617-623. [DOI] [PubMed] [Google Scholar]
- 18.Garrity, G. M., et al. (ed.). 2005. Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, part C. Springer-Verlag, New York, NY.
- 19.Grünenfelder, B., S. Gehrig, and U. Jenal. 2003. Role of the cytoplasmic C terminus of the FliF motor protein in flagellar assembly and rotation. J. Bacteriol. 1851624-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinz, A. J., D. E. Larson, C. S. Smith, and Y. V. Brun. 2003. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol. Microbiol. 47929-941. [DOI] [PubMed] [Google Scholar]
- 21.Huitema, E., S. Pritchard, D. Matteson, S. K. Radhakrishnan, and P. H. Viollier. 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 1241025-1037. [DOI] [PubMed] [Google Scholar]
- 22.Jenal, U., and L. Shapiro. 1996. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 152393-2406. [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, C. J., and R. M. Macnab. 1990. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J. Bacteriol. 1721327-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubori, T., S. Yamaguchi, and S. Aizawa. 1997. Assembly of the switch complex onto the MS ring complex of Salmonella typhimurium does not require any other flagellar proteins. J. Bacteriol. 179813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusumoto, A., K. Kamisaka, T. Yakushi, H. Terashima, A. Shinohara, and M. Homma. 2006. Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J. Biochem. (Tokyo) 139113-121. [DOI] [PubMed] [Google Scholar]
- 26.Lam, H., J. Y. Matroule, and C. Jacobs-Wagner. 2003. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev. Cell 5149-159. [DOI] [PubMed] [Google Scholar]
- 27.Lam, H., W. B. Schofield, and C. Jacobs-Wagner. 2006. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 1241011-1023. [DOI] [PubMed] [Google Scholar]
- 28.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 2902144-2148. [DOI] [PubMed] [Google Scholar]
- 29.Lupas, A. 1997. Predicting coiled-coil regions in proteins. Curr. Opin. Struct. Biol. 7388-393. [DOI] [PubMed] [Google Scholar]
- 30.Mackenzie, C., M. Choudhary, F. W. Larimer, P. F. Predki, S. Stilwagen, J. P. Armitage, R. D. Barber, T. J. Donohue, J. P. Hosler, J. E. Newman, J. P. Shapleigh, R. E. Sockett, J. Zeilstra-Ryalls, and S. Kaplan. 2001. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth. Res. 7019-41. [DOI] [PubMed] [Google Scholar]
- 31.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, I. Curtis, R., J. L. Ingraham, E. C. C. Lin, K. B. Low, and B. Magasanik (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 32.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 5777-100. [DOI] [PubMed] [Google Scholar]
- 33.Mason, J. M., and K. M. Arndt. 2004. Coiled coil domains: stability, specificity, and biological implications. Chembiochem 5170-176. [DOI] [PubMed] [Google Scholar]
- 34.Matroule, J. Y., H. Lam, D. T. Burnette, and C. Jacobs-Wagner. 2004. Cytokinesis monitoring during development: rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell 118579-590. [DOI] [PubMed] [Google Scholar]
- 35.McGrath, P. T., H. Lee, L. Zhang, A. A. Iniesta, A. K. Hottes, M. H. Tan, N. J. Hillson, P. Hu, L. Shapiro, and H. H. McAdams. 2007. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat. Biotechnol. 25584-592. [DOI] [PubMed] [Google Scholar]
- 36.Meisenzahl, A. C., L. Shapiro, and U. Jenal. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mullin, D. A., N. Ohta, A. H. Mullin, and A. Newton. 2001. Organization, expression, and function of Caulobacter crescentus genes needed for assembly and function of the flagellar hook. Mol. Genet. Genomics 265445-454. [DOI] [PubMed] [Google Scholar]
- 38.Murray, T. S., and B. I. Kazmierczak. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 1886995-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newton, A., N. Ohta, G. Ramakrishnan, D. Mullin, and G. Raymond. 1989. Genetic switching in the flagellar gene hierarchy of Caulobacter requires negative as well as positive regulation of transcription. Proc. Natl. Acad. Sci. USA 866651-6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nokhal, T. H., and F. Mayer. 1979. Structural analysis of four strains of Paracoccus denitrificans. Antonie van Leeuwenhoek 45185-197. [DOI] [PubMed] [Google Scholar]
- 41.Pandza, S., M. Baetens, C. H. Park, T. Au, M. Keyhan, and A. Matin. 2000. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol. Microbiol. 36414-423. [DOI] [PubMed] [Google Scholar]
- 42.Poggio, S., C. Abreu-Goodger, S. Fabela, A. Osorio, G. Dreyfus, P. Vinuesa, and L. Camarena. 2007. A complete set of flagellar genes acquired by horizontal transfer coexists with the endogenous flagellar system in Rhodobacter sphaeroides. J. Bacteriol. 1893208-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rath, A., A. R. Davidson, and C. M. Deber. 2005. The structure of “unstructured” regions in peptides and proteins: role of the polyproline II helix in protein folding and recognition. Biopolymers 80179-185. [DOI] [PubMed] [Google Scholar]
- 44.Roberts, R., C. Toochinda, M. Vedissian, R. Baldini, S. Gomes, and L. Shapiro. 1996. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J. Bacteriol. 1781829-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schäffer, A. A., L. Aravind, T. L. Madden, S. Shavirin, J. L. Spouge, Y. I. Wolf, E. V. Koonin, and S. F. Altschul. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 292994-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon, R., U. Prieffer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1784-790. [Google Scholar]
- 47.Sommer, J. M., and A. Newton. 1989. Turning off flagellum rotation requires the pleiotropic gene pleD: pleA, pleC, and pleD define two morphogenic pathways in Caulobacter crescentus. J. Bacteriol. 171392-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stepkowski, T., and A. B. Legocki. 2001. Reduction of bacterial genome size and expansion resulting from obligate intracellular lifestyle and adaptation to soil habitat. Acta Biochim. Pol. 48367-381. [PubMed] [Google Scholar]
- 49.Viollier, P. H., N. Sternheim, and L. Shapiro. 2002. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc. Natl. Acad. Sci. USA 9913831-13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walther-Mauruschat, A., M. Aragno, F. Mayer, and H. G. Schlegel. 1977. Micromorphology of gram-negative hydrogen bacteria. II. Cell envelope, membranes, and cytoplasmic inclusions. Arch. Microbiol. 114101-110. [DOI] [PubMed] [Google Scholar]
- 51.Wheeler, R. T., and L. Shapiro. 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol. Cell 4683-694. [DOI] [PubMed] [Google Scholar]
- 52.Wu, J., and A. Newton. 1997. Regulation of Caulobacter flagellar gene hierarchy; not just for motility. Mol. Microbiol. 24233-239. [DOI] [PubMed] [Google Scholar]
- 53.Yu, J., and L. Shapiro. 1992. Early Caulobacter crescentus genes fliL and fliM are required for flagellar gene expression and normal cell division. J. Bacteriol. 1743327-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.