Abstract
The lprG-Rv1410c operon is critical for the survival of Mycobacterium tuberculosis during infection, but very little is known about the functions of its proteins. LprG is a lipoprotein, and Rv1410c encodes the major facilitator superfamily small molecule transporter P55. P55 likely exports small molecules outside of the bacterial cell, but the function of LprG is unclear. A deletion of the homologous operon in Mycobacterium smegmatis is more susceptible to ethidium bromide, and drug resistance is restored by the intact operon from M. tuberculosis. The multidrug resistance pump inhibitor reserpine inhibits resistance to ethidium bromide in both wild-type M. smegmatis and the complemented mutant, suggesting that P55-mediated transport is responsible for drug resistance and that ethidium bromide is a novel substrate for P55. In addition to hypersensitivity to ethidium bromide, cells that lack the lprG-Rv1410c operon display abnormal colony morphology and are defective for sliding motility, properties that suggest an alteration of cell wall composition. Strikingly, both ethidium bromide transport and normal cell surface properties require functional P55 and LprG, as neither alone is sufficient to restore function to the deletion mutant. Thus, P55 requires the cell surface lipoprotein for normal function.
The two genes of the lprG-Rv1410c operon are crucial for the survival of Mycobacterium tuberculosis during infection. Transposon insertions in either gene are severely attenuating in mice, with no obvious defects during normal vegetative growth (13). A knockout of the operon is also profoundly attenuated in vivo (3).
Although lprG and Rv1410c are clearly required by the bacterium during infection, almost nothing is known about the biologic functions of the products of the operon. P55, the protein encoded by Rv1410c, is a small molecule transporter of the major facilitator superfamily (MFS) (14). The transporter belongs to the drug/H+ antiporter 14 transmembrane domain (DHA14) family, whose members are thought to export cationic small molecules by proton motive force (11). Characterized members of the DHA14 transporter family were identified based on their ability to confer drug resistance when heterologously expressed, and P55 from M. tuberculosis has been reported to confer resistance to tetracycline and aminoglycosides when expressed in Mycobacterium smegmatis (14). However, very few physiologic substrates are known for the DHA14 pumps, and none have been identified for P55. Much less information exists about the protein product encoded by lprG. Immediately upstream of Rv1410c is lprG, which codes for LprG, a lipoprotein with no conserved enzymatic or functional domains save for the lipid attachment site that defines lipoproteins (1). LprG is a secreted, surface-expressed lipoprotein and has been shown to modulate host immune function (7).
Whatever the individual proteins do, it is likely that their biologic roles relate to a common pathway. The genomic organization of lprG and Rv1410c in an operon (2) suggests that the protein functions are related. The lprG-Rv1410c operon is conserved across several species of mycobacteria, including M. bovis, M. leprae, and M. smegmatis, further supporting the notion of related functions. The severe attenuation of loss-of-function mutations in the lprG-Rv1410c operon in M. tuberculosis suggests that the products of the operon may be involved in response to host pathways. However, the conservation of the operon in nonpathogenic M. smegmatis suggests that at least part of the biologic role of the proteins is required during environmental growth.
The ability of P55 to transport substrate is likely crucial to its physiologic role in M. tuberculosis during infection. LprG, however, has no conserved functional or enzymatic domains. Within the M. tuberculosis genome, LprG shares significant homology to another lipoprotein, LppX, which is required for the translocation of the cell wall lipid phthiocerol dimycocerosate (PDIM) (15). LppX is functionally associated with the RND (resistance-nodulation-cell division) small molecule transporter Mmpl7, which exports PDIM across the cell membrane (5, 6). The residues in LppX that are shared by LprG constitute the pocket within which PDIM is thought to reside (15). Given the structural homology between LppX and LprG, we hypothesized that a functional relationship exists between LprG and P55 and that LprG could contribute to transport a substrate of P55.
Here we show that, in M. smegmatis, the lprG-Rv1410c operon is required for resistance to ethidium bromide and for maintenance of normal cell surface composition. We report that normal function requires functional P55 and the expression of both proteins and that these proteins act cooperatively in membrane transport.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains, plasmids, and primers used in this study are listed in Table 1. Escherichia coli cultures were maintained in LB supplemented with 100 μg/ml hygromycin B or 50 μg/ml kanamycin sulfate to maintain plasmids. M. smegmatis strains were maintained in Middlebrook 7H9 medium supplemented with albumin-dextrose-catalase and 0.05% Tween 80. Kanamycin sulfate was added at 50 μg/ml and 25 μg/ml to maintain the plasmids in E. coli and M. smegmatis, respectively. All cultures were routinely incubated at 37°C.
TABLE 1.
Bacterial strains and vectors used in this study
| Strain, plasmid, or oligonucleotide | Characteristic(s)a | Marker | Source |
|---|---|---|---|
| Strains | |||
| E. coli XL1Blue | Cloning strain | None | |
| M. smegmatis mc2155 | Wild type | None | |
| M. smegmatis d8.17 | lprG-Rv1410c operon deletion mutant in M. smegmatis mc2155 | None | This study |
| M. tuberculosis H37Rv | Wild type | None | |
| Plasmids | |||
| pJEB402 | Cloning vector | kan | D. Sherman |
| pJM1 | sacRB suicide vector | hyg chl | J. Murry |
| p402sacB | sacRB suicide vector | kan | This study |
| p503.505 | M. smegmatis lprG-Rv1410c operon knockout vector | kan | This study |
| pMB211 | Episomal E. coli-mycobacterial shuttle vector | kan | A. Jaeger |
| p555 | pMB211 with lprG-Rv1410c operon from M. tuberculosis | kan | This study |
| p563 | D22A point mutation in p555 | kan | This study |
| p564 | D22E point mutation in p555 | kan | This study |
| p548 | pMB211 with lprG from M. tuberculosis | kan | This study |
| p515.1a | pMB211 with Rv1410c from M. tuberculosis | kan | This study |
| p597.1 | pMB211 with C-terminal His8-tagged lprG from M. tuberculosis | kan | This study |
| Oligonucelotides | |||
| Apst1smeglprGf1.fd | GGGCTGCAGCTGCATCGTAGAGGCTCG | This study | |
| Axba1smeglprGf1.rv | GGGTCTAGAGGCGTACATCGGCAGCAG | This study | |
| Nsi1smeg14f2.fd | GGGATGCATGGGCGGGTCGGCGCGCATGG | This study | |
| Axba1pst1smeg14f2.rv | GGGTCTAGACTGCAGGCGGGGAACTCT | This study | |
| bgl2tblprGf | GGAGATCTATGCGGACCCCCAGACG | This study | |
| nhetb1410.rv | CCGCTAGCCTCGTTAGAGCGGCTCCAC | This study | |
| bamh1tb1410f | GGGGATCCATGCGAGCAGGACGTCGAG | This study | |
| nhetblprG.rv | CCGCTAGCTCAGCTCACCGGGGGCTTC | This study | |
| 1410D22A.fd | CTGCTGGGCGCCCTGGCCACCTATGTCGTGGTCACCb | This study | |
| 1410D22A.rv | GGTGACCACGACATAGGTGGCCAGGGCGCCCAGCAGb | This study | |
| 1410D22E.fd | CTGCTGGGCGCCCTGGAGACCTATGTCGTGGTCACCc | This study | |
| 1410D22E.rv | GGTGACCACGACATAGGTCTCCAGGGCGCCCAG CAGc | This study | |
| nhe1nde1tblprGHIS.rv | GGGGCTAGCCATATGTCAGTGGTGGTGGTGGTGGTGGTGGTGGCTCACCGGGGCCTTCG | This study | |
| KO.fd | CGCACCGACATCATGCTC | This study | |
| KO.rv | CACACTTCCGGTGCCGTC | This study |
The underlined text indicates a restriction site sequence.
The boldface text indicates the GAC-to-GCC codon change.
The boldface text indicates the GAC-to-GAG codon change.
Construction of knockout vector.
To make the suicide vector, pJEB402 was digested with NheI and XbaI to obtain a kanamycin resistance cassette and oriE. The sacRB locus was obtained from the plasmid pJM1, a chloramphenicol- and hygromycin-marked suicide vector. pJM1 was digested with SpeI and XbaI, and the two fragments were ligated after alkaline phosphatase treatment of the kan-oriE fragment. The resulting plasmid, p402sacB, was electroporated into XL1Blue and selected on kanamycin.
Generation of lprG-Rv1410c deletion mutant in M. smegmatis.
The M. smegmatis ΔlprG-Rv1410c deletion mutant was constructed by targeted mutagenesis. Two ∼900-bp DNA fragments flanking the operon were amplified by PCR with a 10:1 Taq-Pfu mixture using M. smegmatis mc2155 genomic DNA. The first flanking region (f1) was amplified using the primers Apst1smeglprGf1.fd and Axba1smeglprGf1.rv, and the second flanking region (f2) was amplified with the primers nsi1smeg14f2.fd and Axba1pst1smeg14f2.rv. After digestion with NsiI and XbaI, flanking region 2 (f2) was cloned into p402sacB digested with PstI and XbaI. The resulting plasmid was digested with PstI and XbaI, and f1 was cloned in to produce p503.505. The resulting vector was electroporated into XL1Blue and selected on kanamycin plates. Plasmid DNA was isolated from E. coli, the knockout construct was electroporated into M. smegmatis mc2155, and cells were plated on LB containing kanamycin. Kanamycin-resistant colonies (single crosses) were picked, grown overnight in 7H9 in the absence of kanamycin, and plated on LB containing 5% sucrose. Sucrose-resistant colonies were patched in duplicate onto LB plates containing kanamycin or 5% sucrose to identify kanamycin-sensitive, sucrose-resistant clones (putative double crosses). The mutant used for subsequent experiments was identified by PCR screening using primers KO.fd and KO.rv and confirmed by sequencing and Southern blotting.
Complementation of ΔlprG-Rv1410c strain.
To complement the ΔlprG-Rv1410c mutant, the operon from M. tuberculosis was amplified by PCR from genomic DNA using the primers bgl2tblprGf and nhe1tb1410.rv. After restriction digestion, the PCR product was cloned into the episomal plasmid pMB211 digested with BamHI and NheI. In the resulting construct, p555, the M. tuberculosis lprG-Rv1410c operon was under the control of the M. smegmatis hsp60 promoter. Similarly, lprG was amplified from M. tuberculosis genomic DNA using the primers bgl2tblprGf and nhe1tblprG.rv and cloned into pMB211 to make p548. Rv1410c from M. tuberculosis was amplified with primers bamh1tb1410f and nhe1tb1410.rv and cloned into pMB211 to make p515.1a. The M. smegmatis ΔlprG-Rv1410c mutant was transformed with these constructs and selected on kanamycin.
Site-directed mutagenesis of P55.
The conserved aspartic acid residue at position 22 in P55 was changed to alanine in the operon construct p555 by PCR mutagenesis. DNA was amplified with the completely overlapping primers 1410D22A.fd and 1410D22A.rv, using Pfu Turbo, for 16 cycles with an extension time in minutes equal to the size of the plasmid in kb. After DpnI digestion, the reaction mixture was transformed into XL1Blue and plated on LB kanamycin. The point mutant was identified by sequencing plasmid DNA from kanamycin-resistant clones. The conserved aspartic acid was similarly mutated to glutamic acid with the completely overlapping primers 1410D22E.fd and 1410D22E.rv.
Computer analysis.
Information on the M. tuberculosis lprG-Rv1410c operon was obtained from the TubercuList database (http://genolist.pasteur.fr/TubercuList/). Sequence databases were searched by using the program BLAST to search the genomes of Mycobacterium smegmatis, Mycobacterium leprae, and Mycobacterium bovis, which were made available by TIGR (http://www.tigr.org). Protein sequences were compared using ExPasy (http://ca.expasy.org/) and PROSITE (http://ca.expasy.org/prosite/).
Drug sensitivity assays.
M. smegmatis strains were grown to mid-log phase (optical density at 600 nm [OD600] between 0.4 and 0.7) and frozen in 2-ml aliquots at −80°C. Frozen aliquots were thawed and diluted in 7H9 medium to an approximate OD600 of 0.02. Bacterial growth was monitored by OD and compared to growth in the presence of ethidium bromide. For each experiment, ethidium bromide was titrated by twofold dilution to identify a concentration, 1.2 or 2.5 μg/ml as indicated, at which there was differential killing. Where indicated, reserpine (20 μg/ml) was added to cultures concurrent with ethidium bromide.
Generation of tagged LprG construct.
To obtain a C-terminal His8-tagged LprG construct, lprG from M. tuberculosis was amplified from genomic DNA with the forward primer bgl2tblprGf and reverse primer nhe1nde1tblprGHIS.rv. After digestion with NheI and BglII, the PCR product was cloned into pMB211 cut with BamHI and NheI to make p597.1.
Western blotting.
Constructs containing lprG from M. tuberculosis tagged with His8 were transformed into wild-type M. smegmatis and the lprG-Rv1410c deletion mutant. Bacteria were grown to mid-log phase (OD600 between 0.4 and 0.8), and cell lysates were prepared. Cells were centrifuged at 4,000 rpm for 10 min, washed once in phosphate-buffered saline (PBS), and resuspended in PBS plus protease inhibitor cocktail (1 tablet in 10 ml PBS). After resuspension, cells were lysed by bead beating and boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis buffer. All protein samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Western blots were performed using 1:8,000 rabbit anti-His primary antibody and 1:10,000 goat anti-rabbit conjugated to horseradish peroxidase and visualized by chemiluminescence using an Alpha Innotech Fluorchem SP charge-coupled device imager. To measure relative protein abundance on the blot, integrated intensity values of each band were calculated using AlphaEase FC software.
Congo red and motility assays.
For Congo red staining, bacteria were diluted in PBS and spread onto plates containing 7H9 salts, 1.5% Bacto agar, 100 μg/ml Congo red, 0.02% glucose, and 50 μg/ml kanamycin to maintain plasmid constructs. Pictures were taken after 1 week of incubation at 37°C. For motility assays, cells were spotted onto 7H9 salts plates containing 0.02% glucose, 0.4% agarose as a solidifying agent, and kanamycin to maintain plasmids.
RESULTS
Deletion of the lprG-Rv1410c operon in M. smegmatis results in increased susceptibility to ethidium bromide.
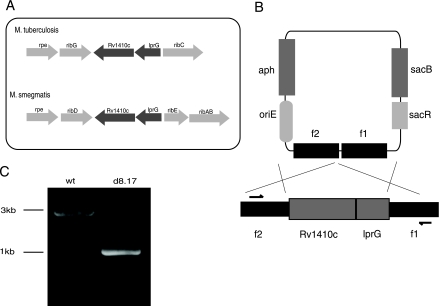
Two genes showing a high degree of similarity to the lprG-Rv1410c operon were identified in the M. smegmatis mc2155 strain (Fig. 1A). The corresponding amino acid sequences of the predicted M. smegmatis genes were ∼70% identical to M. tuberculosis lprG and Rv1410c. We generated a deletion of the homologous lprG-Rv1410c operon in M. smegmatis using a suicide vector containing sequences homologous to the flanking regions of the operon (Fig. 1B). Deletion of the operon was confirmed by PCR analysis (Fig. 1C), Southern blotting, and sequencing (data not shown).
FIG. 1.
Generation of lprG-Rv1410c deletion mutant in M. smegmatis. (A) Schematic diagram of the lprG-Rv1410c operon in M. tuberculosis and M. smegmatis. (B) Schematic diagram of the operon region on the chromosome of the wild-type M. smegmatis mc2155 strain and the suicide vector used to make the deletion strain. Arrows indicate the positions of primers used in PCR screening. (C) PCR analyses of wild-type (wt) M. smegmatis and the ΔlprG-Rv1410c mutant (d8.17).
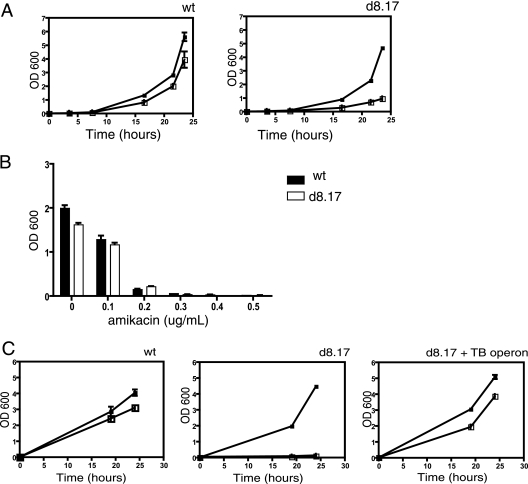
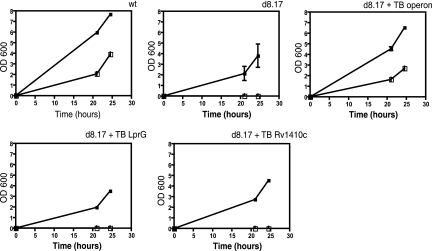
We searched for a synthetic substrate to allow us to measure P55 transport function. While published reports suggested that overexpression of M. tuberculosis P55 in M. smegmatis conferred resistance to streptomycin and aminoglycosides (14), we did not observe differential susceptibility to those drugs in the mutant (data not shown). Several MFS pumps are capable of transporting monovalent and divalent cationic molecules such as ethidium bromide. Therefore, we tested the ability of ethidium bromide to inhibit growth of wild-type and mutant M. smegmatis. Between 1.2 and 2.5 μg/ml, the M. smegmatis deletion mutant was more susceptible to ethidium bromide (Fig. 2A). This effect appears to be relatively specific for ethidium bromide. The deletion mutant does not appear to have a generalized increase in antibiotic susceptibility, as we observed no difference in susceptibility to amikacin (Fig. 2B), berberine, or DAPI (4′,6′-diamidino-2-phenylindole) (data not shown). Expression of the lprG-Rv1410c operon from M. tuberculosis restored wild-type levels of ethidium resistance to the M. smegmatis deletion strain (Fig. 2C).
FIG. 2.
Drug susceptibility of M. smegmatis and the ΔlprG-Rv1410c mutant. Bacterial strains grown to mid-log phase were diluted in 7H9 medium containing twofold dilutions of ethidium bromide. (A) Ethidium bromide susceptibility in M. smegmatis (wt) and the deletion mutant (d8.17) was determined by growth over time as indicated. Open squares, 2.5 μg/ml ethidium bromide; filled squares, no drug. (B) Growth of M. smegmatis strains in the presence of amikacin was measured after 24 h of incubation at 37°C. Black bars, wild-type M. smegmatis; gray bars, the ΔlprG-Rv1410c mutant (d8.17). (C) Complementation of the ΔlprG-Rv1410c mutant. Ethidium bromide resistance was measured for M. smegmatis mc2155 (wt), the deletion mutant (d8.17), or the mutant complemented with the M. tuberculosis operon (d8.17 + TB operon). Open squares, 1.2 μg/ml ethidium bromide; filled squares, no drug.
Transport is required to confer drug resistance.
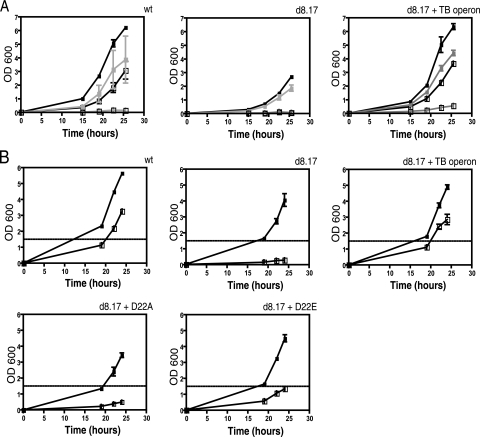
Because P55 is a member of a family of transporters, we believed it likely that its function is required to confer the ethidium bromide-resistant phenotype we observed. To determine if transport was the mechanism of drug resistance, we took two approaches. First, we assessed the ability of the multidrug resistance (MDR) pump inhibitor reserpine to reestablish sensitivity to ethidium bromide in both wild-type M. smegmatis and in the mutant complemented with the M. tuberculosis operon. We observed that, in the presence of reserpine, both wild-type M. smegmatis and the complemented deletion strain are sensitive to ethidium bromide (Fig. 3A).
FIG. 3.
Inhibition of M. smegmatis drug resistance. (A) Ethidium bromide resistance was measured in M. smegmatis (wt), the ΔlprG-Rv1410c mutant (d8.17), or the deletion mutant complemented with the M. tuberculosis operon (d8.17 + TB operon). Drug resistance assays were performed in the presence (light lines) or absence (dark lines) of 20 μg/ml reserpine (in dimethyl sulfoxide). Open squares, 1.2 μg/ml ethidium bromide; filled squares, no drug. (B) Ethidium bromide resistance of M. smegmatis (wt), the ΔlprG-Rv1410c mutant (d8.17), and the deletion mutant complemented with the indicated constructs. D22A, the lprG-Rv1410c operon from M. tuberculosis with a D22A mutant allele of P55; D22E, the lprG-Rv1410c operon from M. tuberculosis with a D22E mutant allele of P55. Open squares, 1.2 μg/ml ethidium bromide; filled squares, no drug.
Second, we took advantage of sequence conservation among the DHA14 family of proteins to probe P55 function genetically. The DHA14 MFS pumps have several conserved amino acid motifs that define the family. One of these, region D1, is strictly conserved among the DHA14 pumps (11). The presence of an aspartic acid at position 22, located within the first transmembrane domain of the protein, is characteristic of region D1. Given the conservation of this charged residue at this conserved location, we hypothesized that the aspartic acid residue would be necessary for transport, even if not directly involved in substrate translocation. Using the construct that contained an intact copy of the entire M. tuberculosis operon, we generated a mutant with a point mutation at position 22 of P55, D22A, which changed the aspartic acid to alanine. We reasoned that replacement of the charged residue with a nonpolar one would abolish P55's ability to transport substrate. The point mutant construct was transformed into the M. smegmatis deletion strain, and, when tested in ethidium bromide assays was found to be as sensitive to drug as the deletion mutant alone (Fig. 3B). To assess the requirement for a charged amino acid residue at position 22, we used the M. tuberculosis operon construct to make another point mutant, D22E, which replaced aspartic acid with the similarly charged residue glutamic acid. The D22E mutant, when expressed in the M. smegmatis deletion background strain, was also more susceptible to ethidium bromide than either wild-type M. smegmatis or the mutant complemented with a wild-type copy of the M. tuberculosis operon. However, the D22E mutant was more resistant to ethidium bromide than the D22A mutant, indicating that a charged amino acid at that position can provide some ability to transport (Fig. 3B).
Deletion of the lprG-Rv1410c operon results in morphological changes.
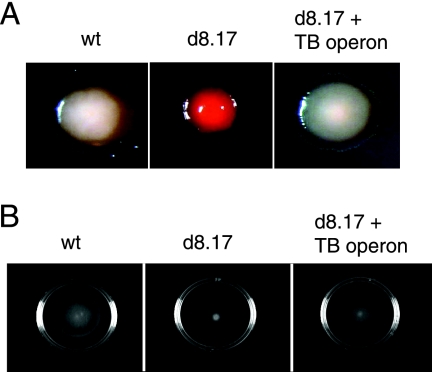
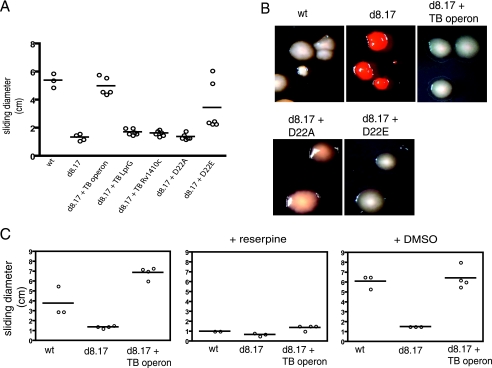
We noted that the mutant strain that lacks the lprG-Rv1410c operon displays altered colony morphology under conditions in which the carbon source is limiting. Colony morphology was assessed with plates containing Congo red, a dye that binds to lipids. Although wild-type M. smegmatis colonies are typically opaque, under carbon-limiting conditions M. smegmatis displays a translucent phenotype with a characteristic halo surrounding the colony (9) and has light staining on Congo red plates (Fig. 4A). Under these growth conditions, we find that the lprG-Rv1410c mutant remains opaque and stains dark red, with no evidence of a halo. Expression of the lprG-Rv1410c operon from M. tuberculosis restored wild-type-like characteristics to the deletion mutant.
FIG. 4.
Cell surface properties in M. smegmatis and the ΔlprG-Rv1410c mutant. (A) To assay colony morphology, bacteria were plated onto 7H9 plates containing 100 μg/ml Congo red and 0.02% glucose. (B) Sliding motility was measured in M. smegmatis strains grown to mid-log phase and spotted onto 7H9 salts-0.4% agarose plates containing 0.02% or 2% glucose. Sliding motility results in the formation of a transparent halo surrounding the point of inoculation. wt, wild-type M. smegmatis mc2155; d8.17, ΔlprG-Rv1410c mutant; d8.17 + TB operon, ΔlprG-Rv1410c mutant expressing the lprG-Rv1410c operon from M. tuberculosis.
Sliding motility has been described for several species of mycobacteria, including M. smegmatis (8) and is thought to depend on the hydrophobicity of the mycobacterial cell wall. We observed that the lprG-Rv1410c deletion mutant is defective for sliding motility under carbon-limiting conditions (Fig. 4B). Once again, motility was restored by expressing the operon from M. tuberculosis.
Function of the operon requires both proteins.
We found that the altered phenotypes of an M. smegmatis strain lacking both Rv1410c and lprG could be complemented by a plasmid that encodes both genes. Surprisingly, though, neither gene alone could restore activity. Strains expressing either Rv1410c or lprG alone remained hypersensitive to ethidium bromide (Fig. 5), displayed altered colony morphology and Congo red staining (data not shown), and were nonmotile when grown on plates with low glucose concentrations (Fig. 6A). The D22A mutant, when expressed in the knockout background, was nonmotile (Fig. 6A) and also displayed altered colony morphology (Fig. 6B). Both the motility and morphology of the D22E mutant more closely resembled wild-type M. smegmatis than the D22A mutant. In the presence of reserpine, both wild-type M. smegmatis and the complemented deletion mutant were nonmotile (Fig. 6C).
FIG. 5.
Contribution of LprG and P55 to ethidium bromide resistance. Constructs containing the lprG-Rv1410c operon, lprG alone, or the P55 gene alone from M. tuberculosis (TB) were expressed in the deletion mutant, and resistance to ethidium bromide was determined as before. wt, wild-type M. smegmatis mc2155; d8.17, ΔlprG-Rv1410c mutant. Open squares, 1.2 μg/ml ethidium bromide; filled squares, no ethidium bromide.
FIG. 6.
Contribution of LprG and P55 to M. smegmatis cell surface properties. (A) Sliding motility of the indicated M. smegmatis strains. (B) Colony morphology on Congo red plates was determined for M. smegmatis strains. (C) Effects of reserpine (20 μg/ml) on sliding motility in M. smegmatis. DMSO, dimethyl sulfoxide; TB operon, M. tuberculosis operon; wt, wild-type M. smegmatis mc2155; d8.17, ΔlprG-Rv1410c mutant. D22A, the lprG-Rv1410c operon from M. tuberculosis with a D22A mutant allele of P55; D22E, the lprG-Rv1410c operon from M. tuberculosis with a D22E mutant allele of P55.
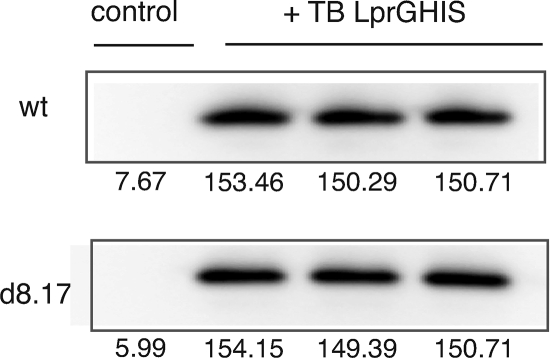
One possible explanation for this observation is that each protein is required to allow stable expression of the other. This is not the case for LprG. A modified LprG protein containing a polyhistidine tag incorporated into the M. tuberculosis allele was expressed equally well in the presence or absence of P55 (Fig. 7). Unfortunately, we were unable to express a P55 allele containing a C-terminal tag and were, therefore, unable to measure the abundance of this protein. However, it appears that P55 has no effect on the abundance of LprG.
FIG. 7.
Expression of LprG. Expression of LprG in M. smegmatis and the lprG-Rv1410c deletion mutant. Samples were normalized by Bradford assay and run in triplicate. Numbers indicate band intensity values. wt, wild-type M. smegmatis mc2155; d8.17, ΔlprG-Rv1410c mutant, TB, M. tuberculosis operon.
DISCUSSION
MDR pumps in bacteria confer resistance to a number of antibiotics and other drugs. While these proteins probably contribute to resistance to environmental toxins, they clearly have other functions. P55 is critical for bacterial survival during M. tuberculosis infection even in the absence of drug therapy and thus must have some other essential role. At the same time, P55 clearly functions as a transporter. It has strong sequence homology to other MFS pumps (11), and others have shown that heterologous expression of the MFS transporter P55 from M. tuberculosis and M. bovis in M. smegmatis confers resistance to drugs (14). Here we show that resistance to ethidium bromide is mediated by P55 in M. smegmatis. This requires transport, as resistance can be abolished by an MDR inhibitor and by mutation of a conserved amino acid likely to be critical for export function. Given that ethidium bromide is a substrate for other related pumps (10), it seems most likely that P55 directly exports this toxin. However, as mutants that lack P55 and LprG have numerous cell wall defects, lack of transport could result from other, indirect processes.
Surprisingly, however, P55 does not appear to act alone. We see no measurable resistance to ethidium bromide with expression of Rv1410c alone, strongly suggesting that LprG is required for transport mediated by P55. In addition, multiple other complex phenotypes that likely represent cell wall alterations (including colony morphology, Congo red staining, and motility) require both proteins. Major facilitator efflux pumps in other organisms, including the characterized members of the DHA14 family, are not thought to require accessory proteins or other factors (11). The vast majority of the DHA14 pumps are not associated with lipoproteins in other organisms, and the association of LprG with P55 has been largely unexplained. Although previous studies have found that heterologous expression of P55 alone was sufficient to confer drug resistance, those assays were performed in wild-type M. smegmatis cells containing an intact copy of the homologous operon (14). We presume that, in these experiments, endogenous levels of expression were adequate to allow full functionality.
Why does P55 need LprG? It seems unlikely that LprG is necessary for transport across the cell membrane, as DHA14 pumps in other organisms are sufficient to export substrates by proton motive force. It is possible that LprG is required for stable expression of P55, yet this also seems unlikely. While we were unable to produce epitope-tagged P55 to test this directly, there are no known examples of MDR pumps requiring accessory proteins for stability. While we cannot rule out an indirect role for LprG in the function of P55, we believe that LprG directly contributes to substrate transport mediated by the pump.
Translocation of substrates across the mycobacterial cell wall must be a complex process. In addition to the phospholipid bilayer, the membrane where P55 presumably resides, the cell wall is composed of several more covalently coupled layers, each of which produces a different type of diffusion barrier for small molecules (4). Thus, for many substrates, export across the inner membrane will result in trapping of the substrate within some cell wall layer. LprG might then play a role in moving substrates to the correct layer within the cell wall or out of the bacterium entirely. Thus, we propose a model in which LprG allows substrate translocation through the mycobacterial cell wall, perhaps acting as a chaperone or constituting a pore-forming channel.
This interpretation is supported by the sequence homology between LprG and LppX, a protein known to play a role in lipid transport (15). The LppX crystal structure indicates that it forms a half-beta barrel, containing a hydrophobic cavity within which lipid structures are thought to reside during translocation. The residues homologous to LppX in LprG constitute the portions of the protein that comprise the beta barrel cavity, and modeling of LprG indicates that it might form a similar structure (J. Sacchettini, personal communication). Given the structural relationship between LppX and LprG, it is easy to believe that LprG is similarly involved in translocation of a lipophilic substrate. LppX is functionally associated with a small molecule transporter, Mmpl7, which is thought to export the lipid PDIM across the cell membrane (5). We believe that P55 and LprG might have a similar functional (not necessarily physical) relationship.
What are the true physiologic substrates for these proteins? Because of the similarity between the two lipoproteins and the functional relationship between LprG and a small molecule transporter, it seems likely that LprG and P55 are required for the translocation of cell wall components or their precursors. We find that loss of the operon in M. smegmatis is associated with morphological differences. Wild-type M. smegmatis colonies are typically opaque, but in limiting carbon display a translucent phenotype (9). Under these conditions, we find that the lprG-Rv1410c mutant remains opaque, a change that has been shown to correlate with alterations in cell wall composition. We also find that the deletion mutant is defective for sliding motility and that this defect is most pronounced under carbon-limiting conditions. Cell motility in mycobacteria is thought to depend on the hydrophobicity of the mycobacterial cell wall (12), and, thus, the mutant's cell wall appears to be more hydrophilic. The morphological differences we report were observed under conditions in which the carbon source was limiting; in the presence of excess carbon source, the phenotypes are much less apparent. It may be that LprG and P55 contribute to cell wall composition only in carbon-limited environments and that this is crucial to the biology of the proteins. It is equally likely that the particular contribution to cell wall composition we observed in our assays is only detectable under such extreme conditions and that LprG and P55 functionally contribute to cell wall composition under normal carbon conditions as well. LprG and P55 may be responsible for the translocation of a class of related cell wall components or the scaffold from which many cell surface components are made. Thus, cell wall changes could be restricted to a small class of molecules or be more pleiotropic.
In M. smegmatis, both LprG and P55 are needed for ethidium transport and normal cell wall composition. Expression of the M. tuberculosis operon in an M. smegmatis strain carrying a deletion of both genes restores wild-type-like phenotypes in our in vitro assays. Thus, it seems likely that the functions are conserved in the pathogenic bacterium. The cell wall structure of M. tuberculosis is unique among prokaryotes and is a major determinant of virulence for the bacterium. Defects in the cell wall might fail to adequately protect the bacterium during infection and may contribute significantly to the attenuation of LprG and P55 mutants in vivo.
Acknowledgments
We thank David Sherman for plasmid pJEB402, Alejandra Jaeger for plasmid pMB211, and Jeff Murry for plasmid pJM1. We especially thank Jun-Rong Wei for advice and assistance with Southern blotting. We are also very grateful to Jeff Murry, Sarah Fortune, and Meera Unnikrishanan for review of the manuscript.
This work was supported by National Institutes of Health grants R01 AI48704 and P01 AI56296 (E.J.R.).
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Bigi, F., C. Espitia, A. Alito, M. Zumarraga, M. I. Romano, S. Cravero, and A. Cataldi. 1997. A novel 27 kDa lipoprotein antigen from Mycobacterium bovis. Microbiology 1433599-3605. [DOI] [PubMed] [Google Scholar]
- 2.Bigi, F., A. Alito, M. I. Romano, M. Zumarraga, K. Caimi, and A. Cataldi. 2000. The gene encoding P27 lipoprotein and a putative antibiotic-resistance gene form an operon in Mycobacterium tuberculosis and Mycobacterium bovis. Microbiology 1461011-1018. [DOI] [PubMed] [Google Scholar]
- 3.Bigi, F., A. Gioffre, L. Klepp, M. P. Santangelo, A. Alito, K. Caimi, V. Meikle, M. Zumarraga, O. Taboga, M. I. Romano, and A. Cataldi. 2004. The knockout of the lprG-Rv1410 operon produces strong attenuation of Mycobacterium tuberculosis. Microbes Infect. 6182-187. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 6429-63. [DOI] [PubMed] [Google Scholar]
- 5.Camacho, L. R., D. Ensergueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34257-267. [DOI] [PubMed] [Google Scholar]
- 6.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs, Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 40279-83. [DOI] [PubMed] [Google Scholar]
- 7.Gehring, A. J., K. M. Dobos, J. T. Belisle, C. V. Harding, and W. H. Boom. 2004. Mycobacterium tuberculosis LprG (Rv1411c): a novel TLR-2 ligand that inhibits human macrophage class II MHC antigen processing. J. Immunol. 1732660-2668. [DOI] [PubMed] [Google Scholar]
- 8.Martinez, A., S. Torello, and R. S. Kolter. 1999. Sliding motility in mycobacteria. J. Bacteriol. 1817331-7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ojha, A. K., S. Varma, and D. Chatterji. 2002. Synthesis of an unusual polar glycopeptidolipid in glucose-limited culture of Mycobacterium smegmatis. Microbiology 1483039-3048. [DOI] [PubMed] [Google Scholar]
- 10.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 621-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Recht, J., A. Martínez, S. Torello, and R. Kolter. 2000. Genetic analysis of sliding motility in Mycobacterium smegmatis. J. Bacteriol. 1824348-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 10012989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva, P. E. A., F. Bigi, M. de la Paz Santangelo, M. I. Romano, C. Martín, A. Cataldi, and J. A. Aínsa. 2001. Characterization of P55, a multidrug efflux pump in Mycobacterium bovis and Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 45800-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulzenbacher, G., S. Canaan, Y. Bordat, O. Neyrolles, G. Stadthagen, V. Roig-Zamboni, J. Rauzier, D. Maurin, F. Laval, M. Daffe, C. Cambillau, B. Gicquel, Y. Bourne, and M. Jackson. 2006. LppX is a lipoprotein required for the translocation of phthiocerol dimycocerosates to the surface of Mycobacterium tuberculosis. EMBO J. 251436-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]