Abstract
The filamentous cyanobacterium Nostoc punctiforme forms symbioses with plants. Disruption of the catalytic domain of the N. punctiforme adenylate cyclase (CyaC) significantly increased symbiotic competence, whereas reduced infectivity was observed in a mutant with a disruption close to the N terminus of CyaC. The total cellular cyclic AMP levels were significantly reduced in both mutants.
The filamentous cyanobacterium Nostoc punctiforme is highly responsive to environmental changes and is able to regulate its cell type accordingly (29). The best-studied response is the patterned development of specialized dinitrogen-fixing cells, known as heterocysts (1, 43, 45). This cyanobacterium is also capable of differentiating motile filaments known as hormogonia (10, 30, 34), which act as the infective agents in symbiotic associations with plants. Potential host plants produce a hormogonium-inducing factor (3, 10, 13, 27, 34) and induce chemoattractive behavior in hormogonia (27, 31, 41).
Our knowledge of the molecular events involved in the differentiation of hormogonia and the factors affecting their behavior in response to plants is limited. In response to hormogonium-inducing factor, N. punctiforme mutants with disruptions in the hrmU and hrmA genes convert 80% of the filaments to hormogonia, compared with 30% of the filaments in the wild type (14). This increased capacity for hormogonium formation is associated with increased symbiotic competence in the host plant Anthoceros punctatus, a hornwort. Expression of hrmU and hrmA is induced by an aqueous extract of Anthoceros tissue, leading to the suggestion that a factor(s) in the extract prevents hormogonium formation, at least in part by its upregulation of hrmUA (13). Mutations in sigH and tprN (encoding an alternative group 2 sigma factor and a tetratricopeptide repeat protein, respectively) are also associated with increased symbiotic competence, although in these cases this is not associated with increased hormogonium differentiation, suggesting that the response of the hormogonia to plant-derived signals has been modified (9, 28). Hormogonia are clearly important in the establishment of symbiotic associations, but their formation alone does not guarantee infection. Campbell and Meeks (10) observed that a strain of Nostoc failed repeatedly to infect A. punctatus despite converting more than 90% of its vegetative filaments to hormogonia. Similarly, Johansson and Bergman (20) noted several noninfective cyanobacteria that, despite being able to differentiate motile hormogonia and being found in abundance on the stems and glands of the angiosperm Gunnera, failed to establish symbioses (16, 34). Campbell et al. (11) showed that 1,827 genes are differentially transcribed in hormogonia of Nostoc (estimated 24 h after their induction), a number almost five times higher than the number in akinete-forming or N2-fixing cultures. The majority of the 944 upregulated genes were found to be involved in signal transduction and transcriptional activation, and this indicates that the hormogonium is a rather complex, metabolically active motile filament that is highly adapted to sense and respond to its environment.
Transposon mutant H1, originally identified in our laboratory during a screen of nitrogen starvation-responsive N. punctiforme mutants (21), exhibits a prolonged hormogonial phase in the presence of the liverwort Blasia pusilla, another host plant, and increased symbiotic competence (which is 2.5-fold higher than that of the wild type). Analysis revealed that the transposon had inserted within the putative catalytic domain of cyaC encoding an adenylate cyclase (AC) (NpR0896 gene [http://genome.jgi-psf.org/finished_microbes/nospu/nospu.annotation.html]). ACs catalyze the formation of the intracellular messenger cyclic AMP (cAMP) (8, 12). Here we report reconstruction of the H1 cyaC mutant phenotype. In addition, we constructed a cyaC mutant in which the N terminus of CyaC was inactivated, which, by contrast, exhibited significantly reduced symbiotic competence. Total cAMP levels were dramatically reduced in both cyaC mutants, implying that reduced levels of cellular cAMP alone cannot explain the different levels of symbiotic competence observed in the two mutants.
Construction of cyaC mutants and phenotypic characterization of these mutants.
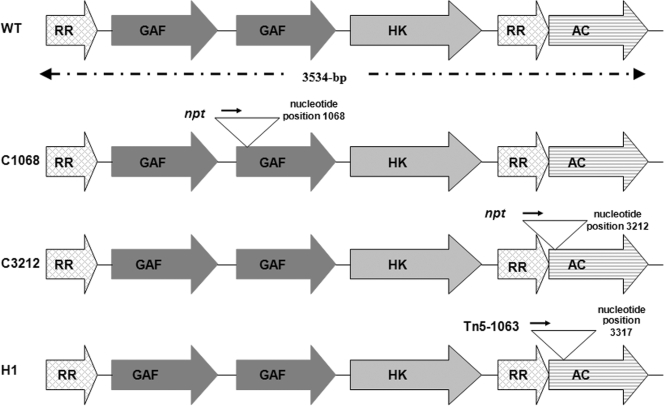
Attempts to reconstruct the transposon mutant H1 by introducing the recovered transposon and flanking Nostoc DNA into wild-type N. punctiforme using the strategy of Black and Wolk (7) were unsuccessful; therefore, an omega neomycin phosphotransferase gene (Ω-npt) from pSCR9 (13) was ligated to the 3′ region of cyaC (nucleotide position 3212, which is close to position 3317, the site of the transposon interruption in H1) (Fig. 1) to generate mutant C3212. The Ω-npt cassette was also introduced into the 5′ end of cyaC (nucleotide position 1068), generating mutant C1068 (Fig. 1). DNA sequences containing the cyaC gene and flanking DNA were PCR amplified from N. punctiforme genomic DNA, and constructs were introduced into wild-type N. punctiforme as described previously (14). The carboxy-terminal and most conserved region in cyanobacterial ACs consists of a catalytic domain (Fig. 1). The N-terminal region of ACs is characteristically variable, and the differences in this region are believed to account for the differences in the regulatory properties of the enzyme activity (24; see reference 12 for a review of the domain structure of cyanobacterial ACs).
FIG. 1.
Schematic illustration of cyaC in the wild type (WT) and cyaC mutants C1068, C3212, and H1. The positions and orientations of npt (neomycin phosphotransferase gene) and the transposon (Tn5-1063) are indicated by triangles. Domain abbreviations: RR, response regulator-like domain; GAF, GAF-like domain; HK, histidine kinase-like domain; AC, adenylate cyclase-like domain. The figure is not drawn to scale. GAF domains are part of a large superfamily of proteins that bind diverse ligands and regulate the biochemical output of proteins in which they are found (2, 4, 38, 40). GAF domains appear to represent sensor domains which receive a signal that is subsequently transmitted to the catalytic domain via the response regulator domain.
Upstream of the N. punctiforme CyaC catalytic domain are several distinct regulatory regions, which are also present in the CyaC proteins of the cyanobacterium Spirulina platensis (23) and Anabaena sp. strain PCC 7120 (24) and resemble the sensory kinase and response regulator proteins that comprise the bacterial two-component signal transduction systems (37, 47). The regulatory domains are arranged sequentially from the N terminus and comprise a response regulator-like domain, two GAF domains (GAF is an acronym derived from the proteins of initial identification, mammalian cGMP-phosphodiesterases, Anabaena adenylate cyclases, and Escherichia coli transcription factor FhlA [formate hydrogen lyase transcriptional activator] [2]), a histidine kinase-like domain, and a second response regulator-like domain (Fig. 1). The highly conserved histidine residue of histidine kinases and the aspartate residue characteristic of response regulator proteins are present in the Nostoc CyaC protein (His-549, Asp-59 and Asp-869) (data not shown) and align with the phosphorylation sites (His-524, Asp-59, and Asp-847) previously reported for the Anabaena sp. strain PCC 7120 CyaC protein (24). In two-component signaling systems the conserved histidine residue of the histidine kinase is able to transfer a phosphoryl group to conserved aspartate residues in the response regulator domains. It has been shown that autophosphorylation of His-572 in the transmitter domain and phosphotransfer to Asp-895 in the receiver (R2) domain of CyaC of S. platensis regulate the activity of AC. Autophosphorylation of CyaC is probably regulated by a specific signal transferred from a primary signal sensor (22).
Mutants C1068 and C3212 grew at rates comparable to those of the wild type in BG11 media (35) both with and without a source of combined nitrogen. Also, no obvious differences were observed in the frequency of proheterocysts and heterocysts expressed by mutant and wild-type cultures 24 h after transfer from nitrogen-replete to nitrogen-depleted conditions (data not shown). However, the mean number of cells per mutant filament was significantly higher than the mean number of cells per wild-type filament (P < 0.001). The following filament lengths (means ± standard errors) were calculated using cultures grown to mid-exponential phase in nitrogen-replete medium: for C3212, 116.5 ± 8.8 cells; for C1068, 136.7 ± 6.08 cells; and for wild-type N. punctiforme, 77.8 ± 3.52 cells.
Coculture experiments with the symbiotic partner B. pusilla were performed as described by Wong and Meeks (44). The mutant strains formed hormogonia approximately 12 h earlier than the wild-type strain (data not shown), and the maximum mean frequencies of hormogonia observed between 18 and 60 h of coculture were 70, 92, and 95% for the wild type, C1068, and C3212, respectively. The numbers of symbiotic colonies detected in the auricles (dome-shaped symbiotic cavities located on the ventral surface of the Blasia tissue) were estimated over a 28-day coculture period (Fig. 2). Wild-type cultures exhibited a steady increase in the frequency of infection from approximately 15% after 7 days to a maximum mean value of approximately 26% (observed after both 21 and 28 days of coculture). Initially, mutant C3212 infected the Blasia symbiotic tissues slowly, and the percentages of symbiotically associated colonies were just 4.16 and 19.5% after 7 and 14 days of coculture, respectively. However, after 21 and 28 days of coculture the mean percentage of infected colonies had increased dramatically to 40.36 and 67.12%, respectively (Fig. 2). This observation is in agreement with the results of the original study, in which the estimated percentage of infected H1 colonies was between 24 and 38% after 7 days of coculture and increased to 85 to 95% after 28 days (21). By contrast, mutant C1068 exhibited reduced symbiotic competence; the mean percentage of Blasia symbiotic cavities (auricles) occupied by the mutant filaments ranged from 2.27% (after 7 days) to 8.5% (after 28 days of coculture) (Fig. 2).
FIG. 2.
Mean frequencies of infection of B. pusilla tissue following coculture with N. punctiforme wild type (WT) and cyaC mutants C1068 and C3212. The frequency of infected symbiotic cavities (auricles) was estimated after 7, 14, 21, and 28 days of coculture with a host plant, the liverwort Blasia. The values are the mean number of infected auricles expressed as a percentage of the total number of auricles examined for each determination. At least eight cultures were examined for each strain, and at least 400 auricles were examined for each determination. The error bars indicate the standard errors of the means.
Analysis of cellular cAMP level.
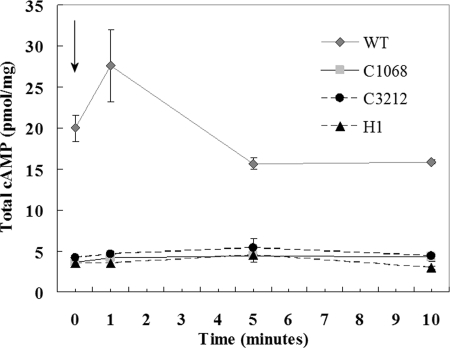
Since transfer of certain cyanobacteria from light to dark conditions results in an increase in cAMP levels (24, 32, 33, 36), we estimated cAMP levels at mid-exponential phase (optical density at 720 nm, 0.15 to 0.25) in the light and following transfer of the cells to dark conditions. Samples were collected after 1, 5, and 10 min of incubation in the dark (Fig. 3). The samples used for determination of cellular cAMP concentrations were prepared by the method of Terauchi and Ohmori (39), and the amount of cAMP in each lyophilized sample was determined using the cAMP Biotrak enzyme immunoassay system (Amersham Biosciences) according to the manufacturer's instructions. When wild-type cells were incubated in the dark, the cAMP levels rapidly increased approximately 1.4-fold in 1 min and then decreased within 5 min (Fig. 3). This is in agreement with previous observations and is consistent with the role of cAMP as a signaling molecule (25, 33, 36). By contrast, the cyaC mutants did not show any corresponding increase in cellular cAMP levels following transfer from light to dark growth conditions (Fig. 3). The total cAMP levels were three- to fourfold higher in the wild type than in any of the three cyaC mutants, suggesting that cyaC function in relation to cAMP production was effectively disrupted in all three mutants (since cAMP levels in the H1 mutant were not determined in the study of Joseph [21], they are included here). The results support previous proposals that cyaC is responsible for maintaining steady-state cAMP levels within certain cyanobacteria (24) and imply that CyaC is probably the major AC in N. punctiforme. Anabaena sp. strain PCC 7120 has at least six different AC genes (cyaA, cyaB1, cyaB2, cyaC, cyaD, and cyaE) (24; reference 12 and references therein), and searches of the N. punctiforme genome suggest that this organism also contains additional putative AC genes (including homologues of cyaA, cyaB1, cyaB2, cyaC, and cyaD), which may account for the small amounts of cAMP detected in all three of the mutant strains (Fig. 3). To our knowledge, this study is the first account of the determination of cAMP levels in N. punctiforme. The mean concentration of cAMP (20 pmol cAMP per mg chlorophyll a) estimated in wild-type cultures incubated in the light is almost fourfold higher than the concentration detected in Anabaena sp. strain PCC 7120 cells grown under similar conditions (25). By contrast, two other filamentous cyanobacteria, S. platensis and Anabaena cylindrica, grown in the light had more-than-twofold-higher intracellular concentrations of cAMP, 51 and 48 pmol cAMP per mg chlorophyll a, respectively (36). However, the values increased to 179 and 189 pmol cAMP per mg chlorophyll a in A. cylindrica and S. platensis, respectively, after incubation in the dark, leading to the suggestion that light suppresses cAMP synthesis or perhaps stimulates the degradation of cAMP in cyanobacteria (36).
FIG. 3.
Determination of cAMP levels (pmol cAMP mg chlorophyll a−1) for the N. punctiforme wild type (WT) and mutants C1068, C3212, and H1. Strains were grown to mid-exponential phase in nitrogen-replete medium. After the first sample was collected (time zero), the cells were immediately transferred to dark conditions (indicated by the arrow), and subsequent samples were collected after 1, 5, and 10 min. Four samples were analyzed for each strain, and the error bars indicate the standard errors of the means.
Mutant H1, containing the transposon Tn5 derivative Tn5-1063 (42) carrying the luxAB genes encoding the light-emitting protein complex luciferase, showed a higher level of luminescence under nitrogen-depleted conditions, and the luminescence was significantly higher in proheterocysts (21). Single filaments undergoing hormogonium formation under nitrogen-depleted conditions did not show any increase in luminescence (21).
Sequence analysis revealed that the transposon had inserted with luxAB oriented parallel to cyaC, implying that the increase in luminescence in response to nitrogen deprivation was driven by the cyaC promoter. Hood et al. (19) showed that nitrogen starvation increases intracellular cAMP levels fourfold in the filamentous cyanobacterium Anabaena variabilis. In Anabaena flos-aquae, transfer to nitrogen-depleted media causes an increase in cAMP levels (17). However, to a lesser degree, transfer to nitrogen-replete media also causes an increase in cAMP levels in this organism, raising doubts that nitrogen deprivation per se is a stimulus for cellular cAMP production (17). Similarly, mutational analysis of AC genes in another filamentous cyanobacterium, Anabaena sp. strain PCC 7120, did not reveal any nitrogen-dependent growth phenotypes (24), and cellular cAMP in Synechocystis sp. strain PCC 6803 appears to be unaffected by nitrogen deprivation (18).
It is not clear why the two cyaC mutants described here have very different phenotypes in terms of symbiotic competence, although distinct cyanobacterial phenotypes generated by mutations at different sites within the same gene (for example, taxAY1, which encodes a chemotaxis-related protein [6], and slr1443 encoding a protein kinase homologue required for motility in Synechocystis sp. strain PCC 6803 [26]), are not unusual. N. punctiforme CyaC is a large protein (1,177 amino acids) with multiple domains (Fig. 1). Mutant C1068 is inactivated in the second GAF domain, whereas C3212 is inactivated in the C terminus catalytic domain. It is not inconceivable that the different domains participate in different cellular activities and that the differences in symbiotic competence observed here may be a reflection of the different domain functions and the environmental signals that they respond to. Indeed, there is evidence that each domain (including a GAF domain, a kinase domain, and a receiver domain) of the tobacco plant ethylene receptor, NTHK1, may have specific roles in regulation of plant growth, the salt stress response, and gene expression (46). Therefore, the function with regard to the impact on symbiotic competence (if any) of the other domains within Nostoc CyaC requires further investigation.
Interestingly, mutants C1068 and C3212 (and H1) had low cAMP levels, implying that cAMP per se is not involved in symbiotic competence. There is evidence which suggests that pili expressed on the hormogonium surface are important in the establishment of Nostoc-Blasia symbioses (15). However, close examination, using shadow casting and transmission electron microscopy, revealed that the pili expressed on the hormogonium surface of cyaC mutants H1, C3212, and C1068 were, in terms of abundance, distribution, and structure, indistinguishable from the pili expressed on the surface of hormogonia differentiated by the wild type (data not shown), indicating that the explanation of the difference in the observed symbiotic competence phenotypes lies elsewhere. Recently, cAMP has been shown to be critical for the phototactic response of Synechocystis sp. strain PCC 6803 that is characterized by the formation of fingerlike projections from colonies on agar plates (5). The different symbiotic competence phenotypes reported here may imply that there are differences in the behavior of the mutant hormogonia in response to plant signals.
Acknowledgments
The financial support of BBSRC grant 24/C14515 is gratefully acknowledged.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Adams, D. G., and P. S. Duggan. 1999. Heterocyst and akinete differentiation in cyanobacteria. New Phytol. 1441-33. [Google Scholar]
- 2.Aravind, L., and C. P. Ponting. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22458-459. [DOI] [PubMed] [Google Scholar]
- 3.Bergman, B., A. Matveyev, and U. Rasmussen. 1996. Chemical signalling in cyanobacterial-plant symbioses. Trends Plant Sci. 1191-197. [Google Scholar]
- 4.Bhaya, D. 2004. Light matters: phototaxis and signal transduction in unicellular cyanobacteria. Mol. Microbiol. 53745-754. [DOI] [PubMed] [Google Scholar]
- 5.Bhaya, D., K. Nakasugi, F. Fazeli, and M. S. Burriesci. 2006. Phototaxis and impaired motility in adenylyl cyclase and cyclase receptor protein mutants of Synechocystis sp. strain PCC 6803. J. Bacteriol. 1887306-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaya, D., A. Takahashi, P. Shahi, and A. R. Grossman. 2001. Novel motility mutants of Synechocystis strain PCC 6803 generated by in vitro transposon mutagenesis. J. Bacteriol. 1836140-6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black, T. A., and C. P. Wolk. 1994. Analysis of a Het− mutation in Anabaena sp. strain PCC 7120 implicates a secondary metabolite in the regulation of heterocyst spacing. J. Bacteriol. 1762282-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, E. L., B. Brahamsha, and J. C. Meeks. 1998. Mutation of an alternative sigma factor in the cyanobacterium Nostoc punctiforme results in increased infection of its symbiotic plant partner, Anthoceros punctatus. J. Bacteriol. 1804938-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, E. L., and J. C. Meeks. 1989. Characteristics of hormogonia formation by symbiotic Nostoc spp. in response to the presence of Anthoceros punctatus or its extracellular products. Appl. Environ. Microbiol. 55125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell, E. L., M. L. Summers, H. Christman, M. E. Martin, and J. C. Meeks. 2007. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J. Bacteriol. 1895247-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cann, M. J. 2004. Signalling through cyclic nucleotide monophosphaqtes in cyanobacteria. New Phytol. 16123-34. [Google Scholar]
- 13.Cohen, M. F., and J. C. Meeks. 1997. A hormogonium regulating locus, hrmUA, of the cyanobacterium Nostoc punctiforme strain ATCC 29133 and its response to an extract of a symbiotic plant partner Anthoceros punctatus. Mol. Plant-Microbe Interact. 10280-289. [DOI] [PubMed] [Google Scholar]
- 14.Cohen, M. F., J. G. Wallis, E. L. Campbell, and J. C. Meeks. 1994. Transposon mutagenesis of Nostoc sp. strain ATCC 29133, a filamentous cyanobacterium with multiple differentiation alternatives. Microbiology 1403233-3240. [DOI] [PubMed] [Google Scholar]
- 15.Duggan, P. S., P. Gottardello, and D. G. Adams. 2007. Molecular analysis of genes involved in pilus biogenesis and plant infection in Nostoc punctiforme. J. Bacteriol. 1894547-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enderlin, C. S., and J. C. Meeks. 1983. Pure culture and reconstitution of the Anthoceros-Nostoc symbiotic association. Planta 158157-165. [DOI] [PubMed] [Google Scholar]
- 17.Francko, D. A., and R. G. Wetzel. 1981. Dynamics of cellular and extracellular cAMP in Anabaena flos-aquae (Cyanophyta): intrinsic culture variability and correlation with metabolic variables. J. Phycol. 17129-134. [Google Scholar]
- 18.Herdman, M., and K. Elmorjani. 1988. Cyclic nucleotides. Methods Enzymol. 167584-591. [Google Scholar]
- 19.Hood, E. E., S. Armour, J. D. Ownby, A. K. Handa, and R. A. Bressan. 1979. Effect of nitrogen starvation on the level of adenosine 3′,5′-monophosphate in Anabaena variabilis. Biochim. Biophys. Acta 588193-200. [DOI] [PubMed] [Google Scholar]
- 20.Johansson, C., and B. Bergman. 1994. Reconstitution of the Gunnera manicata Linde symbioses: cyanobacterial specificity. New Phytol. 126643-652. [Google Scholar]
- 21.Joseph, N. A. 2001. Use of luciferase as a reporter of gene expression in free-living and symbiotically associated cyanobacteria. Ph.D. thesis. The University of Leeds, Leeds, United Kingdom.
- 22.Kasahara, M., and M. Ohmori. 1999. Activation of a cyanobacterial adenylate cyclase, CyaC, by autophosphorylation and a subsequent phosphotransfer reaction. J. Biol. Chem. 27415167-15172. [DOI] [PubMed] [Google Scholar]
- 23.Kasahara, M., K. Yashiro, T. Sakamoto, and M. Ohmori. 1997. The Spirulina platensis adenylate cyclase gene, cyaC, encodes a novel signal transduction protein. Plant Cell Physiol. 38828-836. [DOI] [PubMed] [Google Scholar]
- 24.Katayama, M., and M. Ohmori. 1997. Isolation and characterization of multiple adenylate cyclase genes from the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 1793588-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katayama, M., and M. Ohmori. 1998. Expression of an adenylate cyclase gene of Anabaena cylindrica in the cyanobacterium Anabaena sp. strain PCC 7120. Plant Cell Physiol. 39786-789. [Google Scholar]
- 26.Kim, Y. H., Y. M Park, S.-J. Kim, Y.-I. Park, J.-S. Choi, and Y.-H. Chung. 2004. The role of Slr1443 in pilus biogenesis in Synechocystis sp. PCC 6803: involvement in post-translational modification of pilins. Biochem. Biophys. Res. Commun. 315179-186. [DOI] [PubMed] [Google Scholar]
- 27.Knight, C. D., and D. G. Adams. 1996. A method for studying chemotaxis in nitrogen fixing cyanobacterium-plant symbiosis. Physiol. Mol. Plant Pathol. 4973-77. [Google Scholar]
- 28.Meeks, J. C., E. C. Campbell., K. Hagen, T. Hanson, N. Hitzeman, and F. Wong. 1999. Developmental alternatives of symbiotic Nostoc punctiforme in response to its plant partner Anthoceros punctatus, p. 665-678. In G. A. Peschek, W. Löffelhardt, and G. Schmetterer (ed.), The phototrophic prokaryotes. Kluwer Academic/Plenum Publishers, New York, NY.
- 29.Meeks, J. C., E. C. Campbell, M. C. Summers, and F. C. Wong. 2002. Cellular differentiation in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. 178395-403. [DOI] [PubMed] [Google Scholar]
- 30.Meeks, J. C., and J. Elhai. 2002. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol. Mol. Biol. Rev. 6694-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsson, M., U. Rasmussen, and B. Bergman. 2006. Cyanobacterial chemotaxis to extracts of host and nonhost plants. FEMS Microbiol. Ecol. 55382-390. [DOI] [PubMed] [Google Scholar]
- 32.Ohmori, M. 1989. cAMP in Anabaena cylindrica: rapid changes in cellular levels in response to changes in extracellular environments. Plant Cell Physiol. 30911-914. [Google Scholar]
- 33.Ohmori, M., K. Ohomri, and K. Hasunuma. 1988. Rapid change in cyclic 3′,5′-AMP concentration triggered by a light-off or light-on signal in Anabaena cylindrica. Arch. Microbiol. 150203-204. [Google Scholar]
- 34.Rasmussen, U., C. Johansson, and B. Bergman. 1994. Early communication in the Gunnera-Nostoc symbiosis: plant-induced cell differentiation and protein synthesis in the cyanobacterium. Mol. Plant-Microbe Interact. 6696-702. [Google Scholar]
- 35.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Genetic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1111-61. [Google Scholar]
- 36.Sakamoto, T., N. Murata, and M. Ohmori. 1991. The concentration of cyclic AMP and adenylate cyclase activity in cyanobacteria. Plant Cell Physiol. 32581-584. [Google Scholar]
- 37.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 38.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terauchi, K., and M. Ohmori. 1999. An adenylate cyclase, Cya1, regulates cell motility in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 40248-251. [DOI] [PubMed] [Google Scholar]
- 40.Vreede, J., M. A. van der Horst, K. J. Hellingwerf, W. Crielaard, and D. M. van Aalten. 2003. PAS domains. Common structure and common flexibility. J. Biol. Chem. 27818434-18439. [DOI] [PubMed] [Google Scholar]
- 41.Watts, S. D., C. D. Knight, and D. G. Adams. 1999. Characterisation of plant exudates inducing chemotaxis in nitrogen-fixing cyanobacteria, p. 679-684. In G. A. Peschek, W. Löffenhardt, and G. Schmetter (ed.), The phototrophic prokaryotes. Kluwer Academic/Plenum Publishers, New York, NY.
- 42.Wolk, C. P., Y. Cai, and J. M. Panoff. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc. Natl. Acad. Sci. USA 885355-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 44.Wong, F. C., and J. C. Meeks. 2002. Establishment of a functional symbiosis between the cyanobacterium Nostoc punctiforme and the bryophyte Anthoceros punctatus requires genes involved in nitrogen control and initiation of heterocyst differentiation. Microbiology 148315-323. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, C.-C., S. Laurent, S. Sakr, L. Peng, and S. Bédu. 2006. Heterocyst differentiation and pattern formation in cyanobacteria: a chorus of signals. Mol. Microbiol. 59367-375. [DOI] [PubMed] [Google Scholar]
- 46.Zhou, H.-L., W.-H. Cao, Y.-R. Cau, J. Liu, Y.-J. Hao, J.-S. Zhang, and S.-Y. Chen. 2006. Roles of ethylene receptor NTHK1 domains in plant growth, stress response and protein phosphorylation. FEBS Lett. 5801239-1250. [DOI] [PubMed] [Google Scholar]
- 47.Zhulin, I. B., A. N. Nikolskaya, and M. Y. Galperin. 2003. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J. Bacteriol. 185285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]