Abstract
Members of a novel glycerate-2-kinase (GK-II) family were tentatively identified in a broad range of species, including eukaryotes and archaea and many bacteria that lack a canonical enzyme of the GarK (GK-I) family. The recently reported three-dimensional structure of GK-II from Thermotoga maritima (TM1585; PDB code 2b8n) revealed a new fold distinct from other known kinase families. Here, we verified the enzymatic activity of TM1585, assessed its kinetic characteristics, and used directed mutagenesis to confirm the essential role of the two active-site residues Lys-47 and Arg-325. The main objective of this study was to apply comparative genomics for the reconstruction of metabolic pathways associated with GK-II in all bacteria and, in particular, in T. maritima. Comparative analyses of ∼400 bacterial genomes revealed a remarkable variety of pathways that lead to GK-II-driven utilization of glycerate via a glycolysis/gluconeogenesis route. In the case of T. maritima, a three-step serine degradation pathway was inferred based on the tentative identification of two additional enzymes, serine-pyruvate aminotransferase and hydroxypyruvate reductase (TM1400 and TM1401, respectively), that convert serine to glycerate via hydroxypyruvate. Both enzymatic activities were experimentally verified, and the entire pathway was validated by its in vitro reconstitution.
Phosphorylation of glycerate by glycerate kinase (GK) is a key biochemical step connecting this important carbohydrate (a C3-sugar acid) with central carbon metabolism in a large variety of species. The diversity of the pathways supplying glycerate, including l-serine degradation, utilization of glyoxylate, glycolate, d-glucarate, and tartrate (see Fig. 1B), in different species is matched by the emerging diversity of GK enzymes. GK can convert glycerate to 3-phosphoglycerate (3PG; EC 2.7.1.31) or to 2-phosphoglycerate (2PG; EC 2.7.1.-). So far, at least three distinct GK families (GK-I, GK-II, and GK-III) with different phylogenetic distributions, which are classified based on their amino acid sequence and structure similarities, have been described (1, 2).
FIG. 1.
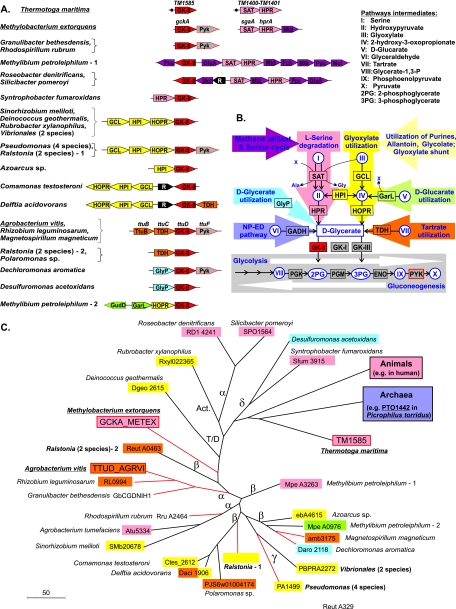
Genomic and functional context of glycerate kinase enzymes in bacteria. In all panels, matching colors are used to mark components of distinct pathways related to glycerate metabolism. (A) Examples of chromosomal clusters containing GK-II genes in bacteria. Genes (shown by arrows) from the same metabolic pathway are marked by matching colors. Transcriptional regulators are shown in black. The species with experimentally characterized pathways and enzymes are underlined. The presence of a common regulatory site upstream of the serine degradation genes in T. maritima is shown by black dots. (B) Reconstruction of metabolic pathways involving d-glycerate. Abbreviations for pathway intermediates (in circles and ovals) are given at the top. ENO, enolase. Different metabolic pathways and their respective enzymes are denoted by background color arrows and boxes marked by the same matching colors. (C) Phylogenetic tree of selected GK-II family proteins in bacteria whose biological role was tentatively identified using genome context analysis. Background colors reflect chromosomal clustering of GK-II genes with the genes from the respective metabolic pathways. Clustering of GK-II with the pyk genes is marked by red lines. The branching point of GK-II proteins from eukaryotes and archaea are indicated and colored according to their biological roles reported elsewhere. Abbreviations for taxonomic groups: α, β, γ, and δ denote alpha-, beta-, gamma-, and deltaproteobacteria; Act., Actinobacteria; T/D, Thermus/Deinococcus group.
Members of the historically first GK family (termed here GK-I) are represented by two paralogous genes, glxK and garK of Escherichia coli, and are involved in glyoxylate and glucarate/galactarate utilization pathways, respectively (5, 6, 18). GK-I family members that occur in many diverse bacteria are usually annotated as 3PG-forming enzymes, based on the original report of Doughty et al. (6). However, more-recent studies of GarK activity, using 1H nuclear magnetic resonance, revealed the formation of the 2PG rather than the 3PG product (12). Notwithstanding the importance of an accurate assignment of GK-I family reaction specificity, it does not affect the interpretation of its physiological role, as 2PG and 3PG are readily interconverted by a phosphoglycerate mutase (PGM). The three-dimensional (3D) structure of the GK-I enzyme from Neisseria meningitides was solved (PDB code 1to6) and classified as a separate structural family (2).
The first representative of a structurally distinct GK-II family was characterized in Hyphomicrobium methylovorum (33) as an enzyme involved in the serine cycle pathway coupled to formaldehyde assimilation. A gene, gckA, was later identified in another methylotrophic bacterium, Methylobacterium extorquens (3). GK-II orthologs are found in many bacterial, eukaryotic, and archaeal genomes. A GK-II enzyme was characterized and shown to play an important role in the serine degradation pathway in the mammalian liver (15). A deficiency of human GK-II activity leads to the hereditary disease d-glyceric aciduria (31), which was tentatively connected with alternative splicing of the GLYCTK gene (9). Recently, a GK-II family member from the archaeon Picrophilus torridus (encoded by the gck gene) was characterized as a key enzyme in the nonphosphorylating Entner-Doudoroff pathway, characteristic of thermoacidophilic archaea such as P. torridus and Thermoplasma acidophilum (24, 25). The first 3D structure recently reported for a GK-II family protein, TM1585, from Thermotoga maritima (PDB code 2b8n) revealed a novel fold and allowed tentative mapping of the active-site area (2, 29). Two characterized members of the GK-II family from M. extorquens (3) and P. torridus (24) were assigned 2PG-forming activity.
The third, structurally unrelated GK family (termed here GK-III and also known as GLYK) includes the recently characterized 3PG-forming enzyme from Arabidopsis thaliana (1) and its homologs in other plants, fungi, nearly all cyanobacteria, and a few other bacterial species. In plants, GK-III catalyzes the final reaction of the photorespiratory cycle, which converts glycolate-2P to 3PG (1).
Of the three GK families, GK-II, while less abundant in the current collection of bacterial genomes than GK-I, is the most universal GK family, spanning all three domains of life. However, until recently, functional assignments within this family remained somewhat controversial. Despite convincing genetic data first ascribing the GK function to the GckA protein in the methylotrophic bacterium M. extorquens (3), other representatives of the GK-II family in many bacterial genomes were tentatively assigned a seemingly unrelated function, hydroxypyruvate reductase, in various public archives such as NCBI and KEGG (see Results for our interpretation of this misannotation). The main objective of this study was to apply comparative genomics for the reconstruction of metabolic pathways associated with GK-II in a broad range of bacterial species with completely sequenced genomes integrated in the SEED platform (http://theseed.uchicago.edu [22]). This analysis, captured in the SEED subsystem “Glycerate metabolism” (http://theseed.uchicago.edu/FIG/subsys.cgi; see also Table S1 in the supplemental material), allowed us to infer a functional context (the metabolic pathways) of the GK-II family in fewer than 60 bacterial genomes.
In the case study of T. maritima, we performed the experimental verification and detailed characterization of GK activity of TM1585, the first GK-II family protein with a known 3D structure. We used directed mutagenesis and steady-state kinetic analysis to confirm the essential role of the two active-site residues, Lys-47 and Arg-325, which are highly conserved in the GK-II family. A novel variant of the three-step serine degradation pathway was inferred in T. maritima, based on the tentative identification of a regulon containing two additional enzymes, serine-pyruvate aminotransferase and hydroxypyruvate reductase (TM1400 and TM1401, respectively). Both enzymatic activities were experimentally verified, and the entire pathway, the conversion of serine to 2PG, was validated by its in vitro reconstitution.
MATERIALS AND METHODS
Genome resources and bioinformatics tools.
Metabolic reconstruction, genome context analysis (chromosomal clustering and phylogenetic profiling), and subsystem encoding, including annotations of relevant genes in a collection of ∼400 complete and almost complete genomes, was performed using SEED genomic software (http://theseed.uchicago.edu/FIG/index.cgi) as previously described (25, 29). The subsystems-based approach to genome analysis and the extensive use of a genome context allowed significant improvement of the accuracy of gene functional assignment and pathway reconstruction (20). The results of this analysis are captured in the novel subsystem, “Glycerate metabolism,” available at http://theseed.uchicago.edu/FIG/subsys.cgi?user=&ssa_name=Glycerate_metabolism&request=show_ssa. Genomes were uploaded from GenBank (http://www.ncbi.nlm.nih.gov/Genbank/). Preliminary genome sequence data for Thermotoga neapolitana was obtained from the Institute for Genomic Research (http://www.tigr.org). Tertiary structures were from PDB (http://www.rcsb.org/). ClustalX (30) and PHYLIP (7) were used to construct multiple protein alignments and phylogenetic trees by the maximum likelihood method (16). The GK box regulatory DNA motif was identified by an iterative signal detection procedure implemented in SignalX software (24); additional GK box regulatory sites were located in the Thermotoga genomes, using Genome Explorer software (17).
Bacterial strains, plasmids, and reagents.
E. coli strain DH5α (Invitrogen, Carlsbad, CA) was used for gene cloning, and BL21 and BL21(DE3) (Gibco-BRL, Rockville, MD) were used for protein overexpression. A pET-derived vector containing the T7 promoter, the His6 tag, and the tobacco etch virus-protease cleavage site (20) was used for protein expression. The resulting plasmid was transformed into E. coli BL21(DE3). Enzymes for PCR and DNA manipulations were from New England Biolabs, Inc. (Beverly, MA). Plasmid purification kits were from Promega (Madison, WI). PCR purification kits and nickel-nitrilotriacetic acid (Ni-NTA) resin were from Qiagen, Inc. (Valencia, CA). Oligonucleotides for PCR and sequencing were synthesized by Sigma-Genosys (Woodlands, TX). All other chemicals, including the assay components d-glycerate, 3-hydroxypyruvate, l-serine, pyruvate, glyoxylate, NADH, NADPH, NAD+, NADP+, ATP, phosphoenolpyruvate, enolase, lactate dehydrogenase, pyruvate kinase (PYK), 3PG kinase, and glyceraldehyde-3-phosphate dehydrogenase were purchased from Sigma-Aldrich (St. Louis, MO). The plasmids harboring the TM1585 (GK-II) or the TM1400 (serine-pyruvate aminotransferase [SAT]) gene with the arabinose promoter and the His6 tag (29) were a kind gift from S. Lesley at the Joint Center for Structural Genomics and were then transformed into E. coli BL21.
PCR amplification and cloning.
The TM1401 hydroxypyruvate reductase (HPR) gene from Thermotoga maritima MSB8 was amplified using the following primers: 5′-ggcgccATGGCGAGATACAGAGTGCACGTGAACGATCCTCTCGATAAAGAG and 3′-gaggtcgacTCAAATTCCCAGTTCTTTGAAAATCTTCTCCACGAGTTCTAT.
The introduced restriction sites (NcoI and PciI for the 5′ end and SalI for the 3′ end) are shown in boldface; mutations and added nucleotides are shown in lowercase. PCR amplification was performed using T. maritima MSB8 genomic DNA. PCR fragments were cloned into the pET-derived expression vector cleaved by NcoI and SalI. Selected clones were confirmed by DNA sequence analysis.
For site-directed mutagenesis of TM1585, the PCR primers 5′-cggaggacATGTTTGATCCTGAATCCTTGAAG and 3′-gacgcgtcgacCTAGACGATGAGGCCTATTATCAAG were used for the full-sized coding region; 5′-CTCGTTGCCGTTGGGgcAGCAGCGTGGCGAATGGC and 3′-GCCATTCGCCACGCTGCTgcCCCAACGGCAACGAG were used for the K47A mutant; 5′-CTCGTTGCCGTTGGGagAGCAGCGTGGCGAATGGC and 3′-GCCATTCGCCACGCTGCTcTCCCAACGGCAACGAG were used for the K47R mutant; 5′-GGAAACGGCATCGGAGGagcAAACCAAGAACTTGC and 3′-GCAAGTTCTTGGTTTgcTCCTCCGATGCCGTTTCC were used for the R325A mutant; and 5′-GGAAACGGCATCGGAGGAAagAACCAAGAACTTGC and 3′-GCAAGTTCTTGGTTctTTCCTCCGATGCCGTTTCC were used for the R325K mutant.
Site-directed mutagenesis.
The amino acid substitutions (K47A, K47R, R325A, and R325K) in TM1585 were generated using the sequential PCR steps technique. Briefly, mutated primers (sense and antisense) containing the separate base substitutions were synthesized. In the first step, two separate PCR runs were performed with two sets of primers, the 5′ end-flanking and antisense mutated primers as well as the 3′ end-flanking and sense mutated primers, and 10 ng of the expression plasmid encoding the full-sized TM1585 protein. Both of the amplified mutated fragments were purified, mixed in a 1:1 molar ratio, and used as the template for a second step of amplification with flanking primers under standard conditions to produce the full-length mutated fragment.
Protein overexpression and purification.
Recombinant proteins were overexpressed as N-terminal fusions with a His6 tag in E. coli strain BL21 or BL21(DE3). For the expression of TM1585 and TM1400, cells were grown on TB medium (24 g/liter yeast extract and 12 g/liter tryptone) containing 1% glycerol and 50 mM morpholinepropanesulfonic acid (pH 7.6), induced by 0.15% arabinose, and harvested after 3 h of shaking at 37°C. For the expression of the TM1401 and TM1585 mutants, cells were grown on LB medium to an optical density at 600 nm of 0.8 at 37°C, induced by 0.2 mM isopropyl-β-d-thiogalactopyranoside, and harvested after 12 h of shaking at 20°C. Protein purification was performed using a rapid Ni-NTA agarose minicolumn protocol as described previously (21). Briefly, harvested cells were resuspended in 20 mM HEPES buffer (pH 7) containing 100 mM NaCl, 0.03% Brij 35, and 2 mM β-mercaptoethanol supplemented with 2 mM phenylmethylsulfonyl fluoride and a protease inhibitor cocktail (Sigma-Aldrich). Lysozyme was added to a concentration of 1 mg/ml, and the cells were lysed by freezing-thawing, followed by sonication. After cells were centrifuged at 18,000 rpm, the Tris-HCl buffer (pH 8) was added to the supernatant (final concentration, 50 mM), and it was loaded onto a Ni-NTA agarose column (0.2 ml). After bound proteins were washed with the starting buffer containing 1 M NaCl and 0.3% Brij-35, they were eluted with 0.3 ml of the starting buffer containing 250 mM imidazole. For the purification of TM1400, 0.1 mM pyridoxal 5′-phosphate was added to the buffers, and the protein mixture was heated for 10 min at 65°C to weaken partial contamination by E. coli proteins because of the intrinsic thermostability of T. maritima proteins. The protein size, expression level, distribution between soluble and insoluble forms, and the extent of purification were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The soluble proteins for TM1401 and TM1585 were obtained with high yield (>1 mg from a 50-ml culture) and purified to >90% by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (see Fig. S2 in the supplemental material).
Enzyme assays.
In all coupled assays, the change in NADH or NADPH absorbance was monitored at 340 nm by using a Beckman DTX-880 multimode microplate reader. A NADH or NADPH extinction coefficient of 6.22 mM−1 cm−1 was used for rate calculation. Initial rate calculations were performed with Multimode Detection software (Beckman). Kinetic data for the wild-type and the mutant TM1585 were analyzed using XLfit4 software (IDBS). A standard Michaelis-Menten model was used to determine the apparent values of kcat and Km. Details of individual assays are provided below.
(i) Glycerate kinase activity.
Glycerate kinase activity was assayed by coupling the formation of ADP to the oxidation of NADH to NAD+ via PYK and lactate dehydrogenase and monitored at 340 nm. Briefly, 0.2 to 0.4 μg of purified glycerate kinase was added to 200 μl of reaction mixture containing 50 mM Tris buffer (pH 7.5), 10 mM MgSO4, 1.2 mM ATP, 1.2 mM phosphoenolpyruvate, 0.3 mM NADH, 1.2 U of PYK, 1.2 U of lactate dehydrogenase, and 1 mM d-glycerate at 37°C. No activity was detected in a control experiment in which an unrelated gene (TM1602) was expressed in the same vector and purified in parallel. For determination of the apparent kcat and the Km values of TM1585 and the mutants, the d-glycerate or the ATP concentration was varied in the range of 0.05 to 5 mM in the presence of a saturating concentration of the other substrate (ATP or d-glycerate, 2 mM). The temperature dependence of enzyme activity was measured between 37°C and 95°C, using a discontinuous assay. The assay mixture (200 μl) contained 50 mM Tris-HCl (pH 7.5; adjusted at the respective temperatures), 2 mM d-glycerate, 2 mM ATP, and 20 mM MgCl2, which ensured specific activities close to the Vmax value. After the mixture was preincubated, the reaction was started by the addition of glycerate kinase and incubation for 5 min and was stopped by the rapid addition of 20 μl of ice-cold 50% (vol/vol) perchloric acid. After it was rapidly vortexed, the mixture was kept on ice for 10 min and neutralized with 30% KOH, and the resulting precipitate was then removed by centrifugation. To 100 μl of the supernatant, 400 μl of 50 mM Tris-HCl (pH 7.5), containing 2 mM phosphoenolpyruvate and 0.25 mM NADH, was added. The amount of ADP formed by the glycerate kinase reaction was quantified by adding a mixture of PYK and lactate dehydrogenase and following the oxidation of NADH at 340 nm.
(ii) Phosphorylation specificity of glycerate kinase.
Phosphorylation specificity of glycerate kinase was performed using two assays (33). In assay I, the formation of 2PG was detected by coupling to the oxidation of NADH to NAD+ via consecutive enolase, PYK, and lactate dehydrogenase reactions and monitoring at 340 nm (Fig. 3A). The reaction mixture (200 μl) contained 50 mM Tris (pH 7.5), 20 mM MgCl2, 1 mM d-glycerate, 2 mM ATP, 0.3 mM NADH, 4.5 U of enolase, 7.2 U of PYK, 8.1 U of lactate dehydrogenase, and 6 μg of purified glycerate kinase. To test for the presence of the alternative 3PG product, phosphoglycerate mutase (10.0 U) was added to the same reaction mixture in a parallel sample. In assay II, the formation of 3PG was tested by adding 3PG kinase and glyceraldehyde-3-phosphate dehydrogenase and monitoring the oxidation of NADH to NAD+ at 340 nm (Fig. 3A). The reaction mixture (200 μl) contained 50 mM sodium phosphate buffer (pH 8.0), 20 mM MgCl2, 1 mM d-glycerate, 2 mM ATP, 0.3 mM NADH, 8.1 U of glyceraldehyde-3-phosphate dehydrogenase, 9.0 U of 3PG kinase, and 6 μg of purified glycerate kinase.
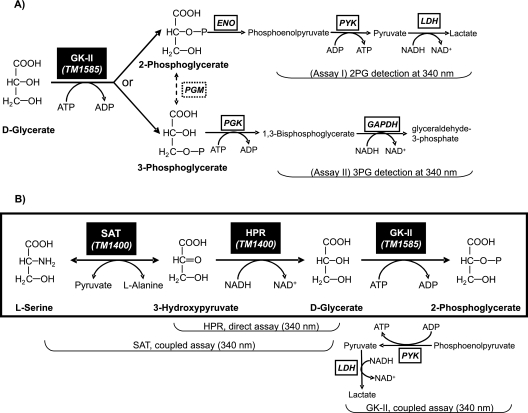
FIG. 3.
Enzymatic reactions and assays. (A) Coupled assays were used to establish the reaction specificity of d-glycerate phosphorylation by TM1585: (i) detection of 2PG was monitored at 340 nm by coupling to NADH oxidation via three consecutive reactions catalyzed by ENO, PYK, and lactate dehydrogenase (LDH); (ii) no indication of 3PG formation was obtained in the alternative assay with the addition of PGK and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (B) Biochemical transformations in the T. maritima serine degradation pathway and the assays used to assess the enzymatic activities of GK-II (TM1585) and of the two additional enzymes, SAT (TM1400) and HPR (TM1401).
(iii) Hydroxypyruvate reductase activity.
Hydroxypyruvate reductase activity was assayed by monitoring the decrease in absorbance at 340 nm due to the conversion of NADH to NAD+. Briefly, 5 to 10 μg of purified hydroxypyruvate reductase was added to 200 μl of reaction mixture containing 50 mM sodium phosphate buffer (pH 7.5), 0.3 mM NADH or NADPH, and 2 mM 3-hydroxypyruvate at 37°C. No activity was detected in a control experiment in which an unrelated gene (TM1602) was expressed in the same vector and purified in parallel. The reverse reaction of hydroxypyruvate reductase (i.e., glycerate dehydrogenase) was determined by measuring the reduction of NAD+ to NADH. The reaction mixture (200 μl) contained 50 mM Tris buffer (pH 8.0), 1 mM d-glycerate, 0.3 mM NAD+, and 10 μg of purified glycerate dehydrogenase.
(iv) PLP-dependent SAT activity.
Pyridoxal phosphate (PLP)-dependent SAT activity was assayed by coupling to the HPR reaction and monitoring the oxidation of NADH to NAD+ at 340 nm. The reaction mixture (200 μl) contained 50 mM sodium phosphate buffer (pH 7.5), 0.1 mM pyridoxal 5′-phosphate, 0.3 mM NADH, 2 mM pyruvate, 2 mM l-serine, and 10 to 20 μg of purified SAT and HPR. For testing glyoxylate as an alternative substrate, pyruvate was exchanged with 2 mM glyoxylate. No activity was detected in a control experiment in which an unrelated gene (TM1602) was expressed in the same vector and purified in parallel.
In vitro reconstitution of the serine degradation pathway.
The conversion of l-serine to d-glycerate by a mixture of purified SAT (TM1400) and HPR (TM1401) was determined by monitoring the decrease in absorbance at 340 nm due to the conversion of NADH to NAD+, which was described above as the SAT activity assay. In control samples, one enzyme was excluded from the mixture.
The conversion of hydroxypyruvate to 2PG by a mixture of purified HPR (TM1401) and GK (TM1585) was determined by monitoring the conversion of NADH to NAD+ and ATP to ADP, using high-performance liquid chromatography (HPLC). The reaction mixture contained 50 mM sodium phosphate buffer (pH 7.5), 20 mM MgCl2, 1 mM 3-hydroxypyruvate, 1 mM NADH, 1 mM ATP, and 5 to 20 μg of purified HPR and GK. In control samples, one enzyme was excluded from the mixture. After 2 h of incubation at 37°C, proteins were removed by microultrafiltration using Microcon YM-10 centrifugal filters (Amicon, Bedford, MA), and the filtrates were analyzed using a Shimadzu HPLC system with an LC-10AD solvent delivery system, an SIL-20A auto sampler, and an SPD-20AD UV/Vis detector. The separation of nucleotides was performed on a Supelco LC-18-T (15 cm by 4.6 mm; 3-μm particle size) chromatographic column equipped with Supelguard LC-18-T (2.0 cm by 4.0 mm; 5-μm particle size) guard column (Supelco, Bellefonte, PA) at room temperature, with detection at 254 nm. Elution was performed at 1 ml/min by a 1.5 to 30% gradient of methanol in 100 mM potassium phosphate (pH 6.0) with 8 mM tetrabutylammonium bromide.
RESULTS
Comparative genomics of the GK-II family and reconstruction of related metabolic pathways.
To assess the biological roles of TM1585 and other GK-II enzymes in a context of related metabolic pathways, we performed a comparative genomics survey of completely sequenced bacterial genomes. We used a subsystems-based approach implemented in the SEED genomic platform (http://theseed.uchicago.edu) containing a large collection of integrated genomes and tools for comparative analysis, annotation, and metabolic reconstruction (22). This approach was previously applied for the analysis of various metabolic subsystems and for gene and pathway discovery in a broad range of species (for example, see references 8, 27, and 32).
The results of this analysis are captured in the SEED subsystem “Glycerate metabolism,” available online (http://theseed.uchicago.edu/FIG/subsys.cgi). This subsystem includes all three types of glycerate kinases (GK-I, GK-II, and GK-III), as well as other key enzymes from the above-described metabolic pathways feeding glycerate to central carbon metabolism, including the utilization of serine, glyoxylate, d-glucarate, and tartrate (Fig. 1B). A condensed version of the subsystem spreadsheet is provided in Table S1 in the supplemental material.
A minimal serine degradation pathway (which is also a component of the so-called serine cycle in methylotrophs [3]) includes serine-glyoxylate (or serine-pyruvate) aminotransferase (or SAT) and HPR. The glyoxylate pathway is composed of three enzymes, glyoxylate carboligase (GCL), 2-hydroxy-3-oxopropionate reductase (HOPR), and hydroxypyruvate isomerase (HPI). The tartrate utilization pathway typically includes a bifunctional tartrate decarboxylase/dehydrogenase enzyme (TDH). The inferred novel pathway of exogenous d-glycerate utilization in various gammaproteobacteria (e.g., Alteromonadales, Pasteurellales, Pseudomonadales, Vibrionales, and Xanthomonadales) and Firmicutes (Clostridiales and Lactobacillales) involves GK-I and a candidate d-glycerate transporter (termed GlyP).
Genome context analysis techniques (implemented in SEED software, as well as in several other bioinformatics tools) play an important role in subsystems analysis, helping to reveal associated pathways, improve the accuracy of gene assignment, and infer functions for some previously uncharacterized genes (for a review, see reference 3). For example, the observed phylogenetic occurrence profile of tentatively assigned GK genes revealed an anticorrelation pattern (as defined in reference 19), with most species containing genes encoding only one of the three types, GK-I, GK-II, or GK-III (see Table S1 in the supplemental material). The GK-I family is the most widely distributed in bacteria, with 159 genes in 135 genomes, predominantly in gram-positive bacteria (the actinobacteria and the Bacillus/Clostridium groups) as well as in many betaproteobacteria and gammaproteobacteria (including E. coli). It is followed by the GK-II family, with 76 genes in 65 genomes, including the Thermus/Deinococcus groups and most alphaproteobacteria and deltaproteobacteria, as well as some betaproteobacteria and gammaproteobacteria. This family dominates in the superkingdoms Archaea and Eukaryota. Members of the GK-III family, characteristic of plants, were identified in 30 bacterial genomes, mostly in the Cyanobacteria phylum. Such a clear anticorrelation pattern points to the physiological equivalence of all three GK families despite their different reaction specificities (2PG forming versus 3PG forming) that are apparently compensated by the universal presence of PGM interconverting these two metabolites (Fig. 1B).
The identification of gene clustering on the chromosome, another powerful technique for genome context analysis, provides the most important evidence for their functional coupling (23). The analysis of conserved gene clusters in the genomic neighborhoods of GK-encoding genes revealed multiple cases of their colocalization with genes involved in the glycerate metabolism (see Table S1 in the supplemental material). Remarkably, either one of the nonorthologous GK-I or GK-II genes could be found embedded in the otherwise conserved chromosomal gene clusters, for example, in the most abundant glyoxylate utilization cluster (encoding orthologous GCL, HPI, and HOPR enzymes).
Several examples of chromosomal clusters observed for GK-II genes in bacteria are shown in Fig. 1A. TDH and the serine cycle (HPR and SAT) genes were found mostly in chromosomal clusters with GK-II genes. Among a few exceptions are the HPR-encoding genes in two Mycobacterium species and two gammaproteobacteria (Methylococcus capsulatus and Pseudoalteromonas tunicata) that occur in clusters with the GK-I and GK-III family genes, respectively. In addition to clustering with genes involved in the upstream pathways, some of the GK-I and GK-II family genes occurred in clusters with the PYK gene involved in the downstream glycolysis. In the three species of methylotrophic proteobacteria, GK-II genes were found within the extensive methane utilization gene clusters that include serine cycle genes (HPR and SAT), as well as other genes involved in tetrahydrofolate-linked C1 transfer.
Overall, ∼40% of the bacterial GK-II genes (30 out of 76) are functionally coupled with genes involved in various pathways of glycerate metabolism via clustering on the chromosome, the most frequent being the glyoxylate and tartrate utilization clusters. This information allowed us to confidently assert respective pathways in all these species and further project these pathways to other species where GK-II and glycerate metabolism genes occurred in separate chromosomal loci. The observed presence of one or another type of relevant pathway context in nearly all species containing GK-II family genes (clustered or not) provided stronger evidence in support of a uniform functional assignment of the entire family.
The phylogenetic tree was constructed for 30 representative bacterial GK-II family proteins with a well-defined genome context (Fig. 1C). Color coding that corresponds to various GK-II-related pathways helps to emphasize the remarkable diversity of physiologic specialization characteristic of this enzyme family, in addition to its taxonomic heterogeneity. Different metabolic pathways are often associated with GK-II enzymes from the same branch, whereas much more divergent members may participate in the same pathway. Some species contain pairs of GK-II paralogs from the distant branches of the tree (and often participating in distinct pathways), pointing to the possible role of horizontal gene transfer, as in the case of two distant GK-II paralogs in Ralstonia spp. that are coupled to glyoxylate and tartrate utilization pathways. One of the important observations from this analysis is that the sequence similarity or a relative position on the GK-II tree, as well as a taxonomic placement of species, may not be efficiently used for the accurate projection of physiological functions (or associated pathways) between individual members of the GK-II family. Such a projection requires a more-detailed analysis of genomic and functional context, e.g., as in the subsystems-based approach used in this study.
Case study of GK-II-related pathway in T. maritima.
The amino acid sequence of the GK-II enzyme TM1585 from T. maritima reveals the closest similarity with hyperthermophilic archaeal enzymes; however, in contrast with P. torridus (and other thermoacidophilic archaeal species), where the close homolog of TM1585 was characterized (24) as a part of the nonphosphorylating Entner-Doudoroff pathway, there is no genomic or physiological evidence for the presence of such pathways in T. maritima. Therefore, even though an evolutionary history of TM1585 could have included horizontal transfer from archaeal organisms, in T. maritima, this gene was adopted for operating in a different functional context. Although no candidate genes of glycerate metabolism could be found in the genomic neighborhood of the TM1585 gene, evidence for its functional coupling with a putative serine degradation operon (TM1400-TM1401) was obtained by the analysis of possible regulatory sites in their upstream DNA regions.
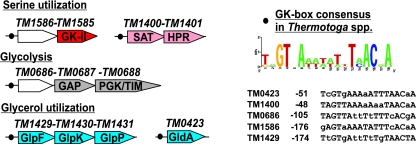
Computational identification of shared regulatory sites and putative regulons is another powerful genome context analysis technique that was successfully applied to reveal functional coupling of genes and operons in many metabolic subsystems and species (26). A conserved palindromic DNA motif (Fig. 2) was detected in the upstream region of TM1586-TM1585 and TM1400-TM1401, putative operons of T. maritima, and in the corresponding segments of T. neapolitana (the only closely related genome available at the time of this study). Whereas the function of TM1586 is unclear, TM1400 and TM1401, encoding distant homologs of SAT and HPR, respectively, appear to be the only candidate genes in the T. maritima genome with reliable homology to functional roles captured in the glycerate metabolism subsystem. Further scanning of the upstream regions in the Thermotoga spp. genomes with a derived DNA profile (Fig. 2, GK-box) revealed additional possible members of the same regulon, the gldA gene (TM0423) and the glpFKP (TM1429-1430-1431) genes, which are involved in glycerol utilization, and an operon containing the gap (TM0688) and pgk-tpiA (TM0689) genes, which are involved in glycolysis. While these two pathways do not involve GK-II enzymatic activity, they share a common 2PG intermediate with an inferred serine degradation pathway. It is tempting to speculate that the shared 2PG intermediary metabolite may constitute the actual effector for the entire GK-box regulon. Although a transcription factor for this putative regulon has yet to be established, our overall confidence in the main conclusion of this analysis is not affected: the GK-II enzyme TM1585 is involved in a version of a serine degradation pathway together with the SAT and HPR enzymes TM1400 and TM1401, respectively.
FIG. 2.
Candidate regulatory sites (GK-box) of GK-II and other members of a putative regulon in Thermotoga species. (Left) GK boxes are shown by black circles, and genes are shown by arrows. (Right) Sequence logo for the GK box consensus and individual GK boxes.
The proposed pathway illustrated in Fig. 3B, a three-step transformation of serine to 2PG via hydroxypyruvate, is similar to a part of the serine cycle previously described in methylotrophic bacteria (3). The only notable difference is due to the apparent absence of glyoxylate in T. maritima, as indicated by a draft genome-scale reconstruction of the entire metabolic network of this organism (I. Thiele, personal communication). Glyoxylate is usually considered a main cosubstrate in the first step of this pathway, transamination of serine to hydroxypyruvate by SAT. At the same time, some characterized enzymes of this family are known to accept alternative transamination cosubstrates such as pyruvate, which (in the absence of glyoxylate) appears to be the most likely component of the T. maritima version of the serine degradation pathway.
To provide experimental support for the bioinformatics analysis described above, we performed a detailed enzymatic characterization of the TM1585 protein, including its reaction specificity, and tested the predicted functional importance of the two conserved residues, Lys47 and Arg325. We also verified the inferred enzymatic activities of the TM1400 and TM1401 proteins and performed in vitro reconstitution of the proposed serine degradation pathway, as described in the following sections.
Enzymatic activity and active-site residues of TM1585, a glycerate 2-kinase from T. maritima.
We used the recombinant TM1585 protein, overexpressed in E. coli with the N-terminal His6 tag and purified using Ni-NTA affinity chromatography, to test for both types of reaction specificity known for GK enzymes, leading to the formation of 2PG and 3PG (Fig. 3A). The formation of the 2PG product was demonstrated by coupling assay I using enolase, PYK, and lactate dehydrogenase (Fig. 3A). PGM catalyzes the reversible 3PG to 2PG transformation, whereas assay I was irreversible because of the activities of PYK and lactate dehydrogenase. The addition of the PGM enzyme to the reaction mixture did not affect the overall response, suggesting that the reaction products did not contain any appreciable amounts of 3PG. This conclusion was additionally supported by assay II (Fig. 3A), which showed no indication of 3PG formation in the reaction mixture containing 3PG kinase and glyceraldehyde-3-phosphate dehydrogenase.
Therefore, TM1585 may be reliably classified as a highly specific 2PG-forming GK enzyme. Based on the data for other diverse members of the GK-II family (3, 24) and a high degree of sequence conservation within the tentative active-site area (as discussed below and in references 2 and 29), this functional assignment can be safely projected across the entire family (for at least 76 members in 65 sequenced bacterial genomes, as well as for 12 members in archaeal genomes).
To resolve another potential ambiguity related to contradictory annotations of the GK-II family mentioned above, we tested TM1585 for HPR activity. The spectroscopic assay was performed in both directions, from hydroxypyruvate in the presence of NADH or from glycerate in the presence of NAD+ (Fig. 3B), giving no indication of their respective activities. A detailed analysis of database records and publications suggests that the propagation of the apparently incorrect HPR annotation over many GK-II homologs originated from the erroneous functional assignment of TtuD involved in the tartrate utilization pathway of Agrobacterium vitis (4). In a more recent study, where this pathway was clarified (Fig. 1B), it was shown that a single enzyme, TDH, converts tartrate to d-glycerate in three steps with hydroxypyruvate as an intermediate (28). These results suggest that TtuD should have been assigned a GK function, consistent with its homology with other members of the GK-II family.
As is the case with many other T. maritima proteins, TM1585 is a thermostable enzyme with maximum enzymatic activity at 80 to 90°C (see Fig. S3 in the supplemental material) and shows high activity, even during ∼5 min of incubation at 100°C. However, in contrast to some other enzymes from hyperthermophiles (10, 11), TM1585 has an appreciable activity at 37°C. Therefore, we performed further comparative kinetic studies of wild-type and mutant TM1585 at 37°C, using a continuous coupled assay (Fig. 3B) which is convenient but not amenable to high temperature measurements.
The established steady-state kinetic parameters of TM1585 are shown in Table 1. The apparent Km values for d-glycerate (0.15 mM) and ATP (0.095 mM) are comparable to the respective values reported for the GK-II enzyme from Hyphomicrobium methylovorum (0.13 mM for both d-glycerate and ATP) (33).
TABLE 1.
Kinetic parameters of wild-type TM1585 and its derivatives obtained by site-directed mutagenesis
| Protein | Kinetic parameter ± SDa
|
|||
|---|---|---|---|---|
| kcat (s−1) | KmATP (mM) | kcat/KmATP (M−1s−1) | Km, d-glycerate (mM) | |
| Wild-type TM1585 | 14.4 ± 0.4 | 0.095 ± 0.009 | 1.5 × 105 ± 0.1 × 105 | 0.15 ± 0.03 |
| Mutants | ||||
| R325K | 0.84 ± 0.02 | 0.11 ± 0.01 | 7.7 × 103 ± 0.7 × 103 | 0.13 ± 0.02 |
| R325A | 0.90 ± 0.03 | 0.47 ± 0.05 | 1.9 × 103 ± 0.2 × 103 | 0.19 ± 0.05 |
| K47R | 0.08 ± 0.01 | 0.13 ± 0.02 | 6.7 × 102 ± 1.3 × 102 | 0.20 ± 0.04 |
| K47A | ND | ND | 0.15 ± 0.04 | ND |
kcat values for ATP are presented, and similar values were found for glycerate. ND, not determined. SD, standard deviation. All measurements were performed at 37°C. Values for Km, d-glycerate and KmATP were obtained by varying the substrate concentrations at saturating concentration of the second substrate (2 mM for both ATP and d-glycerate).
We used site-directed mutagenesis to assess the functional importance of the two residues, Lys47 and Arg325, highly conserved in the entire GK-II protein family (see Fig. S1 in the supplemental material) and located in the interdomain region of the TM1585 3D structure. This region is anticipated to comprise the GK-II active site, and although the catalytic mechanism for this family has yet to be established, conserved positively charged residues are expected to play a key role in the interactions with negatively charged substrates, glycerate, and, especially, ATP (2, 29). Two pairs of TM1585 mutations, K47A, K47R, R325A, and R325K, were obtained and kinetically characterized (Table 1). All these mutations led to a significant decrease in enzymatic activity, with most of the observed effects at the level of the apparent kcat values (180-fold for K47R, while a milder 17-fold effect resulted for R325A and R325K). The strongest effect was observed with the K47A mutant, where individual kinetic parameters could not be reliably measured due to the very low residual activity. Although these data did not allow us to assign a specific role (e.g., in substrate binding) to any of these residues, they confirmed their general importance for catalysis. A fivefold change of the apparent KmATP values in the R325A mutant compared to the unchanged KmATP value of the R325K and K47R mutants may, to some extent, reflect the importance of interactions with ATP substrate based on charge or side chain size. Overall, although these data did not provide insight into the GK-II mechanism, they support the tentative localization of the active-site area in the GK-II family, inferred from the sequence comparison and the TM1585 3D structure.
Experimental validation of the predicted serine degradation pathway in T. maritima.
Since both TM1400 and TM1401 belong to well-described enzyme families, we did not pursue their detailed characterization and limited our analysis to a qualitative verification of their inferred activities and in vitro validation of the entire pathway. This was performed using a panel of the coupled assays illustrated in Fig. 3B, combined with the HPLC-based detection of the pathway by-products ADP and NAD+ (see Fig. S4 in the supplemental material).
The HPR activity of the purified recombinant protein TM1401, an efficient conversion of hydroxypyruvate to d-glycerate, was confirmed by monitoring NADH-to-NAD+ conversion at 340 nm. This transformation is nearly irreversible as virtually no enzymatic transformation could be detected in the reverse assay setup, from d-glycerate to hydroxypyruvate in the presence of NAD+; this result agrees with reports for other distant members of the HPR family (14).
The PLP-dependent SAT activity of the purified recombinant protein TM1400, a conversion of l-serine to hydroxypyruvate using pyruvate as a cosubstrate, was confirmed by coupling this reaction to the consecutive HPR reaction in the presence of excess TM1401 enzyme and NADH (Fig. 3B). The presence of both proteins, TM1400 and TM1401, was necessary and sufficient for quantitative transformation of l-serine to d-glycerate (Table 2). This amounts to in vitro reconstitution and validation of the first two steps of the proposed pathway.
TABLE 2.
In vitro reconstitution of the serine degradation pathway by using recombinant purified T. maritima enzymes
| Substrate(s) added | Enzyme(s) added | Conversion of NADH to NAD+ and/or ATP to ADP |
|---|---|---|
| l-Serine and pyruvatea | SAT and HPR | + |
| SAT | − | |
| HPR | − | |
| 3-Hydroxypyruvateb | HPR and GK-II | + |
| HPR | − | |
| GK-II | − |
The conversion of l-serine to d-glycerate by a mixture of the enzymes SAT (TM1400) and HPR (TM1401) was determined by monitoring the decrease in absorbance at 340 nm due to the conversion of NADH to NAD+ (see the assay shown in Fig. 3B).
The conversion of hydroxypyruvate to 2-phospho-d-glycerate by a mixture of HPR (TM1400) and GK-II (TM1585) was detected by monitoring the conversion of NADH to NAD+ and ATP to ADP, using HPLC.
Similarly, in the last two steps, the transformation of hydroxypyruvate to 2PG was reconstituted in a mixture of two enzymes, TM1401 and TM1585, and monitored by the conversion of NADH to NAD+ and ATP to ADP, using HPLC (Table 2 and see Fig. S4 in the supplemental material). These results, taken together, provide the first confirmation of the physiological role of TM1585, a GK-II family enzyme in T. maritima, in the serine degradation pathway, inferred by comparative genomics and metabolic reconstruction.
DISCUSSION
We combined bioinformatics and experimental techniques to assess enzymatic properties and a possible physiological role for the TM1585 protein from T. maritima, the first glycerate 2-kinase of the GK-II family with a reported 3D structure (29). Members of the GK-II family are found in many diverse bacterial species, as well as in Archaea and Eukaryota, including mammals (Fig. 1A). GK-II is structurally distinct from the other two described families, GK-I (2), the most abundant family in bacteria, including E. coli, and GK-III, characteristic of plants (1) and cyanobacteria. Some representatives of the GK-II family, including GckA from M. extorquens (3), human GLYCK (9), and the enzyme from the archaeon P. torridus (24), have been characterized. Nevertheless, many GK-II family members with diverse bacterial genomes still have a misleading HPR annotation, a typical example of the error propagation in genomic databases originating from the incorrect functional assignment of the TtuD protein in A. vitis (4).
The detailed enzymatic analysis of the purified recombinant TM1585 presented here allowed us to rule out HPR activity and to confirm its GK activity with physiologically relevant steady-state kinetic parameters. A stringent reaction specificity observed for the TM1585 enzyme, the exclusive formation of 2PG (and not 3PG) product, together with previous reports for other GK-II family members, supports the unambiguous glycerate 2-kinase assignment for the entire family. Although two other families, GK-I and GK-III, were originally described as 3PG-forming enzymes, the recent data indicate that the GK-I family may, in fact, display a glycerate 2-kinase reaction specificity (12). The latter observation is consistent with the possibility of a common evolutionary origin of GK-I and GK-II families that reveal marginal sequence similarities within their respective Rossman-like domains (2).
Unlike many other carbohydrate kinases that belong to large and extensively studied enzyme families, GK-I and GK-II comprise distinct structural families with as-yet-unknown mechanisms of action. Crystallographic analysis of TM1585 allowed us to take the first steps toward structure-function understanding of the GK-II family by predicting the active-site area (2, 29), including two conserved, positively charged residues, Lys47 and Arg325. These residues were tentatively implicated in interactions with negatively charged substrates, ATP, and, possibly, glycerate (2, 29).
Our site-directed mutagenesis and steady-state kinetic data (Table 1) confirmed the functional importance of these residues. One of them, Lys47, appears to play a particularly important role in catalysis, as even the K47R mutant conserving the positive charge displays a 180-fold drop in kcat, without any appreciable change in the apparent Km value for either glycerate or ATP. The second and less-conserved Arg325 residue (replaced by proline in ∼25% of all compared bacterial enzymes) may be involved in charge or side chain interactions with ATP (but not glycerate), as suggested by a fivefold increase of the apparent KmATP value observed for the R325A, but not the R325K, mutant. Although an accurate mechanistic interpretation of these data awaits further analysis, an observed functional conservation of these residues provides additional support for the presumed conservation of the biochemical function within the entire GK-II family.
A remarkable homogeneity of substrate specificity across the GK-II family is in contrast with that of other large families of carbohydrate kinases, e.g., the FGGY protein family (Pfam accession number PF00294), which contains representatives with widely different substrate preferences toward a broad range of distinct sugars, including the xylulose kinase TM0116 (EC 2.7.1.17), the ribulokinase TM0284 (EC 2.7.1.16), the glycerol kinase TM1430 (EC 2.7.1.30), the gluconokinase TM0443 (EC 2.7.1.12), and the rhamnulokinase TM1073 (EC 2.7.1.5) in T. maritima (unpublished results). In addition to the experimental evidence accumulated for several divergent members of the GK-II family, its functional homogeneity is strongly supported by the metabolic reconstruction and genome context analysis performed in this study. A significant fraction (∼40%) of GK-II family genes in a collection of diverse bacterial genomes are clustered on the chromosome with genes involved in glycerate metabolism (Fig. 1), providing strong support for their functional assignment and for the assertion of respective metabolic pathways.
The comparative analysis of bacterial genomes included in the “glycerate metabolism” subsystem (see Table S1 in the supplemental material) revealed a remarkable diversity of pathways that involve GK-II enzymes (Fig. 1). Among the most frequent are the pathways of glyoxylate, serine, and tartrate utilization; whereas the utilization of d-glucarate and d-glycerate is relatively rare for GK-II, it is quite common for GK-I family enzymes. An observed mosaic phylogenetic distribution of genes corresponding to all three enzyme families, GK-I, GK-II, and GK-III, that are often involved in the same pathways and even in similarly organized chromosomal clusters confirms the equivalence of their functional roles in vivo.
An inventory analysis of genes associated with all functional roles included in the subsystem allowed us to expand pathway assertions toward many other genomes where GK-II genes are not involved in any suggestive chromosomal clusters. In the case of T. maritima, a remote operon encoding homologs of SAT and HPR enzymes (TM1400-TM1401) was deemed the only possible functional context for GK-II enzyme (TM1585). The inferred three-step serine degradation pathway was further supported by the identification of conserved putative regulatory sites in the upstream region of the respective genomic loci in T. maritima and T. neapolitana (Fig. 2).
Importantly, this analysis allowed us to suggest specific functional assignments for the TM1400 and TM1401 genes that belong to large enzyme families (PLP-dependent aminotransferase and d-isomer-specific 2-hydroxyacid dehydrogenase, respectively) with wide variations in substrate specificity between individual characterized representatives. Due to a divergent phylogenetic placement of T. maritima proteins, their precise substrate specificity could not be reliably predicted based solely on sequence similarity. Indeed, the current annotations of these proteins in most public archives are either imprecise (e.g., putative aminotransferase for TM1400) or incorrect (e.g., d-3-phosphoglycerate dehydrogenase).
Both inferred activities, SAT and HPR, were confirmed for the purified recombinant proteins TM1400 and TM1401, respectively, using specific assays (Fig. 3B). Although glyoxylate is considered a major physiological cosubstrate for other SAT enzymes, previously described as serine-glyoxylate aminotransferases (EC 2.6.1.45), it does not appear to be a relevant intermediate in the T. maritima metabolic network. At the same time, pyruvate, an important intermediary metabolite in T. maritima, was proven to be an efficient transamination cosubstrate of TM1400, which should be formally classified as SAT (EC 2.6.1.51). The challenge of distinguishing between biochemical and physiologically relevant activities is rather common for enzyme families with broad substrate specificities. Likewise, HPR enzymes (EC 1.1.1.81) from several species were shown to display an appreciable glyoxylate reductase (EC 1.1.1.26) activity. Although both of these activities are displayed in vitro by TM1401, only one of them, HPR, appears to be physiologically relevant for T. maritima.
Finally, the results obtained for two overlapping pairs of coupled reactions (TM1400 plus TM1401 and TM1401 plus TM1585) provided us with an experimental validation of the inferred three-step serine degradation pathway (Table 2). Despite the obvious shortcomings of in vitro data, in combination with bioinformatic analysis, these data constitute sufficient evidence for confident inclusion of this pathway in the reconstruction of the T. maritima metabolic network. Taking into account the organotrophic lifestyle of T. maritima (13), a possible physiological role for this pathway may be in the utilization of exogenous serine from its environment, enriched with amino acids and other carbon and energy sources.
In summary, the key results of this study obtained by applying a subsystems-based approach (22) to the analysis of bacterial metabolic pathways that involve members of GK-I, GK-II, and GK-III enzyme families are as follows: (i) >1,000 individual genes from ∼200 complete or nearly complete bacterial genomes and representing 12 distinct functional roles (mostly enzymes) were reliably and consistently annotated; (ii) a functional context of 76 members of the GK-II family identified in 65 diverse bacterial species was analyzed in detail, and specific pathways (or groups of pathways) were asserted in most of these species; (iii) in the T. maritima case study, a novel putative regulon was identified, covering an inferred serine degradation pathway and two other pathways, utilization of glycerol and a part of glycolysis, that share a common intermediary metabolite (and a likely effector) 2PG; and (iv) a proposed version of the serine degradation pathway in T. maritima implemented by the three enzymes, pyruvate-utilizing SAT (TM1400), HPR (TM1401), and GK-II (TM1585), was validated by in vitro reconstitution. A detailed experimental characterization of the glycerate 2-kinase TM1585, the main focus of this study, confirmed its substrate and reaction specificity and the functional importance of the two putative active-site residues, K47 and R237, largely conserved within the GK-II family.
Supplementary Material
Acknowledgments
We thank S. Lesley at the Joint Center of Structural Genomics for providing expression clones of T. maritima proteins and S. Krishna for help in the analysis of the TM1585 3D structure. We thank R. Overbeek and colleagues at the Fellowship for Interpretation of Genomes (FIG, IL) for help with the use of the SEED genomic resource, M. Gelfand and A. Mironov (IITP, Moscow, Russia) for their help with the analysis of regulons, and L. Sorci (BIMR, San Diego, CA) for help with the HPLC assay.
This work was partially supported by DOE grant DE-FG02-07ER64384, Integrated Genome-Based Studies of Shewanella Ecophysiology.
Footnotes
Published ahead of print on 21 December 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Boldt, R., C. Edner, U. Kolukisaoglu, M. Hagemann, W. Weckwerth, S. Wienkoop, K. Morgenthal, and H. Bauwe. 2005. d-GLYCERATE 3-KINASE, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. Plant Cell 172413-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheek, S., K. Ginalski, H. Zhang, and N. V. Grishin. 2005. A comprehensive update of the sequence and structure classification of kinases. BMC Struct. Biol. 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chistoserdova, L., and M. E. Lidstrom. 1997. Identification and mutation of a gene required for glycerate kinase activity from a facultative methylotroph, Methylobacterium extorquens AM1. J. Bacteriol. 1794946-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crouzet, P., and L. Otten. 1995. Sequence and mutational analysis of a tartrate utilization operon from Agrobacterium vitis. J. Bacteriol. 1776518-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cusa, E., N. Obradors, L. Baldoma, J. Badia, and J. Aguilar. 1999. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J. Bacteriol. 1817479-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doughty, C. C., J. A. Hayashi, and H. L. Guenther. 1966. Purification and properties of d-glycerate 3-kinase from Escherichia coli. J. Biol. Chem. 241568-572. [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17368-376. [DOI] [PubMed] [Google Scholar]
- 8.Gerdes, S. Y., O. V. Kurnasov, K. Shatalin, B. Polanuyer, R. Sloutsky, V. Vonstein, R. Overbeek, and A. L. Osterman. 2006. Comparative genomics of NAD biosynthesis in cyanobacteria. J. Bacteriol. 1883012-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, J. H., S. Hexige, L. Chen, G. J. Zhou, X. Wang, J. M. Jiang, Y. H. Kong, G. Q. Ji, C. Q. Wu, S. Y. Zhao, and L. Yu. 2006. Isolation and characterization of the human d-glyceric acidemia related glycerate kinase gene GLYCTK1 and its alternatively splicing variant GLYCTK2. DNA Seq. 171-7. [DOI] [PubMed] [Google Scholar]
- 10.Hansen, T., M. Musfeldt, and P. Schonheit. 2002. ATP-dependent 6-phosphofructokinase from the hyperthermophilic bacterium Thermotoga maritima: characterization of an extremely thermophilic, allosterically regulated enzyme. Arch. Microbiol. 177401-409. [DOI] [PubMed] [Google Scholar]
- 11.Hansen, T., and P. Schonheit. 2003. ATP-dependent glucokinase from the hyperthermophilic bacterium Thermotoga maritima represents an extremely thermophilic ROK glucokinase with high substrate specificity. FEMS Microbiol. Lett. 226405-411. [DOI] [PubMed] [Google Scholar]
- 12.Hubbard, B. K., M. Koch, D. R. Palmer, P. C. Babbitt, and J. A. Gerlt. 1998. Evolution of enzymatic activities in the enolase superfamily: characterization of the (d)-glucarate/galactarate catabolic pathway in Escherichia coli. Biochemistry 3714369-14375. [DOI] [PubMed] [Google Scholar]
- 13.Huser, B., B. K. C. Patel, R. M. Daniel, and H. W. Morgan. 1986. Isolation and characterization of a novel extremely thermophilic anaerobic chemo-organotrophic eubacterium. FEMS Microbiol. Lett. 37121-127. [Google Scholar]
- 14.Izumi, Y., T. Yoshida, H. Kanzaki, S. Toki, S. S. Miyazaki, and H. Yamada. 1990. Purification and characterization of hydroxypyruvate reductase from a serine-producing methylotroph, Hyphomicrobium methylovorum GM2. Eur. J. Biochem. 190279-284. [DOI] [PubMed] [Google Scholar]
- 15.Katayama, H., Y. Kitagawa, and E. Sugimoto. 1980. Purification of rat liver glycerate kinase and studies of its enzymatic and immunological properties. J. Biochem. (Tokyo) 88765-773. [DOI] [PubMed] [Google Scholar]
- 16.Matsuda, H. 1996. Protein phylogenetic inference using maximum likelihood with a genetic algorithm. Pac. Symp. Biocomput. Jan.:512-523. [PubMed]
- 17.Mironov, A. A., N. P. Vinokurova, and M. S. Gel'fand. 2000. Software for analyzing bacterial genomes. Mol. Biol. (Moskva) 34253-262. (In Russian.) [PubMed] [Google Scholar]
- 18.Monterrubio, R., L. Baldoma, N. Obradors, J. Aguilar, and J. Badia. 2000. A common regulator for the operons encoding the enzymes involved in d-galactarate, d-glucarate, and d-glycerate utilization in Escherichia coli. J. Bacteriol. 1822672-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morett, E., J. O. Korbel, E. Rajan, G. Saab-Rincon, L. Olvera, M. Olvera, S. Schmidt, B. Snel, and P. Bork. 2003. Systematic discovery of analogous enzymes in thiamin biosynthesis. Nat. Biotechnol. 21790-795. [DOI] [PubMed] [Google Scholar]
- 20.Osterman, A., N. V. Grishin, L. N. Kinch, and M. A. Phillips. 1994. Formation of functional cross-species heterodimers of ornithine decarboxylase. Biochemistry 3313662-13667. [DOI] [PubMed] [Google Scholar]
- 21.Osterman, A. L., D. V. Lueder, M. Quick, D. Myers, B. J. Canagarajah, and M. A. Phillips. 1995. Domain organization and a protease-sensitive loop in eukaryotic ornithine decarboxylase. Biochemistry 3413431-13436. [DOI] [PubMed] [Google Scholar]
- 22.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H. Y. Chuang, M. Cohoon, V. de Crecy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. D. Frank, S. Gerdes, E. M. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Ruckert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 335691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overbeek, R., N. Larsen, G. D. Pusch, M. D'Souza, E. Selkov, Jr., N. Kyrpides, M. Fonstein, N. Maltsev, and E. Selkov. 2000. WIT: integrated system for high-throughput genome sequence analysis and metabolic reconstruction. Nucleic Acids Res. 28123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reher, M., M. Bott, and P. Schonheit. 2006. Characterization of glycerate kinase (2-phosphoglycerate forming), a key enzyme of the nonphosphorylative Entner-Doudoroff pathway, from the thermoacidophilic euryarchaeon Picrophilus torridus. FEMS Microbiol. Lett. 259113-119. [DOI] [PubMed] [Google Scholar]
- 25.Reher, M., and P. Schonheit. 2006. Glyceraldehyde dehydrogenases from the thermoacidophilic euryarchaeota Picrophilus torridus and Thermoplasma acidophilum, key enzymes of the non-phosphorylative Entner-Doudoroff pathway, constitute a novel enzyme family within the aldehyde dehydrogenase superfamily. FEBS Lett. 5801198-1204. [DOI] [PubMed] [Google Scholar]
- 26.Rodionov, D. A. 2007. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 1073467-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodionov, D. A., O. V. Kurnasov, B. Stec, Y. Wang, M. F. Roberts, and A. L. Osterman. 2007. Genomic identification and in vitro reconstitution of a complete biosynthetic pathway for the osmolyte di-myo-inositol-phosphate. Proc. Natl. Acad. Sci. USA 1044279-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruszczycky, M. W., and V. E. Anderson. 2004. Tartrate dehydrogenase reductive decarboxylation: stereochemical generation of diastereotopically deuterated hydroxymethylenes. Bioorg. Chem. 3251-61. [DOI] [PubMed] [Google Scholar]
- 29.Schwarzenbacher, R., D. McMullan, S. S. Krishna, Q. Xu, M. D. Miller, J. M. Canaves, M. A. Elsliger, R. Floyd, S. K. Grzechnik, L. Jaroszewski, H. E. Klock, E. Koesema, J. S. Kovarik, A. Kreusch, P. Kuhn, T. M. McPhillips, A. T. Morse, K. Quijano, G. Spraggon, R. C. Stevens, H. van den Bedem, G. Wolf, K. O. Hodgson, J. Wooley, A. M. Deacon, A. Godzik, S. A. Lesley, and I. A. Wilson. 2006. Crystal structure of a glycerate kinase (TM1585) from Thermotoga maritima at 2.70 A resolution reveals a new fold. Proteins 65243-248. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Schaftingen, E. 1989. d-Glycerate kinase deficiency as a cause of d-glyceric aciduria. FEBS Lett. 243127-131. [DOI] [PubMed] [Google Scholar]
- 32.Yang, C., D. A. Rodionov, X. Li, O. N. Laikova, M. S. Gelfand, O. P. Zagnitko, M. F. Romine, A. Y. Obraztsova, K. H. Nealson, and A. L. Osterman. 2006. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J. Biol. Chem. 28129872-29885. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida, T., K. Fukuta, T. Mitsunaga, H. Yamada, and Y. Izumi. 1992. Purification and characterization of glycerate kinase from a serine-producing methylotroph, Hyphomicrobium methylovorum GM2. Eur. J. Biochem. 210849-854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.