Abstract
In this study, we determined the borders of the pathogenicity island in V. parahaemolyticus RIMD2210633 (Vp-PAI). Vp-PAI has features in common with Tn7 and other related elements at both terminal ends. Our findings indicate that the mobile element with a transposase which contains the DDE motif may have been involved in Vp-PAI formation.
Vibrio parahaemolyticus is a gram-negative bacterium which belongs to the Gammaproteobacteria (17, 22), and some V. parahaemolyticus strains cause food-borne gastroenteritis. The pathogenicity of this organism for humans has been shown by epidemiological studies to be strongly associated with the presence of the gene(s) encoding thermostable direct hemolysin (TDH) and/or TDH-related hemolysin (TRH) (11, 13, 19, 20). Whole-genome sequencing of tdh+ V. parahaemolyticus strain RIMD2210633 led to the proposal that there is a pathogenicity island (PAI) consisting of ca. 80 kb of DNA on the small chromosome (ChrII) (1.9 Mb). This region contains the genes for TDH and type III secretion system 2, which are involved in virulence (17). The term PAI refers to a horizontally acquired genetic region that contains a virulence gene(s), is found only in pathogenic strains, and is not detected in the same or closely related avirulent species (7, 21). PAIs play an important role in pathogenesis and bacterial evolution (8, 28). The PAI in V. parahaemolyticus RIMD2210633 (Vp-PAI) has a G+C content that is lower than the average G+C content of ChrII (17). In addition, experimental findings indicate that the genes for type III secretion system 2 on Vp-PAI are absent in tdh-lacking strains (24). Since the precise region of Vp-PAI and its genetic features as a mobile element have not been thoroughly identified, we performed the study described here.
All V. parahaemolyticus strains used in this study were obtained from the Laboratory for Culture Collection, Research Institute for Microbial Diseases, Osaka University, Osaka, Japan. Bacterial strains used in this study are listed in Table 1. V. parahaemolyticus strains were cultured in Luria-Bertani broth supplemented with 3% NaCl at 37°C with shaking. Purification of genomic DNA with a DNeasy tissue kit (Qiagen, Valencia, CA) and extraction of DNA from agarose gels using a Qiaex II gel extraction kit (Qiagen) were carried out according to the manufacturer's instructions. The primers used in this study were primer-C (5′-GAATCAAAAGATATGATAAATGCCC-3′) and primer-D (5′-CGCGCTAACTTCCACAAGGTTGCC-3′). A conventional PCR was performed using Ex Taq polymerase (TaKaRa Bio, Shiga, Japan) according to the manufacturer's instructions. Nucleotide sequencing was performed with an ABI PRISM 3100 sequencer (Applied Biosystems, Foster City, CA) and a BigDye v3.1 cycle sequencing kit (Applied Biosystems). The sequencing conditions were changed according to the manufacturer's protocols depending on the templates used for cycle sequencing. Direct sequencing with the sequencing using genomic DNA as a template method was performed as previously described (15). For homology searches of both nucleotide and amino acid sequences, the BLAST service at the Genome Information Research Center (http://genome.naist.jp/bacteria/vpara/) and the NCBI (http://www.ncbi.nlm.nih.gov) were used. Sequence information was obtained from the NCBI.
TABLE 1.
V. parahaemolyticus strains used in this study
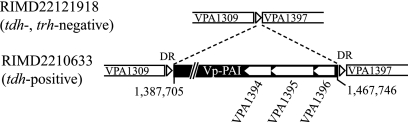
For determination of Vp-PAI boundaries, we compared RIMD2210633 and RIMD22121918, a tdh trh-lacking strain. The region corresponding to the outside of Vp-PAI in RIMD22121918 was sequenced by the sequencing using genomic DNA as a template method using genomic DNA as the template and PCR products (430 bp) amplified with primer-C and primer-D. Compared with RIMD2210633, the sequences showed loss of a large genetic segment (Fig. 1). A similar loss of a large Vp-PAI segment was also found in two other tdh trh-lacking strains, RIMD2212481 and RIMD2212470.
FIG. 1.
Schematic diagram of the exact region of Vp-PAI. The open triangles represent DRs in RIMD2210633, and the open pentagons represent VPA1394, VPA1395, and VPA1396 (see text).
The analysis showed that Vp-PAI ranged from 1,387,705 to 1,467,746 bp from oriCII (covering open reading frame [ORFs] VPA1310 to VPA1396) of RIMD2210633 and was flanked by 5-bp direct repeats (DRs) (5′-AACTC-3′). These DRs were presumably generated by target site duplication. Vp-PAI was inserted into an intergenic region between VPA1309 (an ORF encoding a hypothetical protein) and VPA1397 (an ORF encoding an acyl-coenzyme A thioester hydrolase-related protein). The size of Vp-PAI was 80,042 bp, and the G+C content was 39.2%, which is notably lower than the average G+C content of ChrII, which is 45.4%.
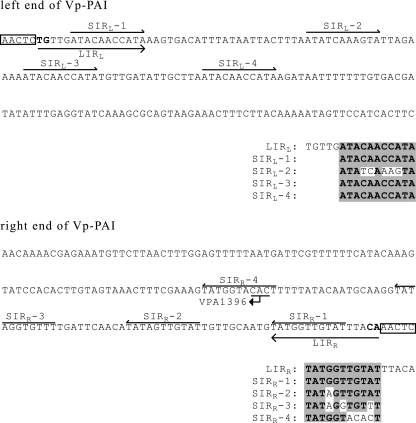
The relationship between a tRNA and an integrase has been the subject of reports on PAIs or genomic islands in a number of organisms, including V. parahaemolyticus (12, 18, 32). Vp-PAI, however, is not associated with an tRNA gene (the nearest tRNA gene, the tRNA-Gly gene, is more than 230 kb from Vp-PAI) and does not possess the gene for integrase. Moreover, we could not find any genes related to phages or conjugative transposons (recently renamed integrating conjugative elements) in Vp-PAI (3-5, 29). For these reasons, Vp-PAI may be atypical and distinct from other PAIs or genomic islands reported to date. We investigated the sequences at both ends of Vp-PAI since we thought that they could provide insight into the manner in which Vp-PAI was formed. We found that Vp-PAI possesses five inverted repeats (IRs) at both ends, consisting of one pair of long IRs (16 bp) and four pairs of short IRs (11 bp). The terminal ends are formed by the 5′-TG…CA-3′ dinucleotide (Fig. 2). These features are similar to those of Tn7, Mu, and other related transposable elements (10). For this reason, we searched for genes homologous to the genes present in such transposable elements and found that the products of VPA1394 and VPA1395 are similar to TnsC and TnsB (Tn7) or TniB and TniA (Tn5090), respectively (10, 26, 27). These proteins are responsible for transposition in Tn7 or binding to IRs in Tn5090 (1, 2, 6, 14, 30, 31). A central component of the catalytic reaction for TnsB and TniA, the DD(35)E motif, is conserved in VPA1395 (D226, D301, E347). The superfamily including Tn7 and Tn5090 is linked by the presence of the characteristic DD(35)E motif in the transposases and by the presence of the dinucleotide TG at the transposon 5′ ends (16). The structural features of Vp-PAI, therefore, indicate that Vp-PAI (or its precursor) may be related to this superfamily. It is difficult to imagine that an 80-kb stretch of DNA was transferred horizontally by the Tn7 superfamily because no such case has been reported. One assumption is that such an element was a founder that generated Vp-PAI, as previously described by Parks and Peters (25).
FIG. 2.
Left and right ends of Vp-PAI and DRs. The DRs are enclosed in boxes. LIR long IR; SIR, shorter. A subscript L indicates that the IR is embedded in the left end, and a subscript R indicates that the IR is embedded in the right end. For short IRs, the same nucleotides are shaded. The bent arrow indicates the initiation codon of VPA1396, GTG (complementary strand).
The TnsA homolog in Vp-PAI has not been identified using sequence information alone. The site of integration for Vp-PAI is different from that of Tn7. This seems to be consistent with the absence of TnsD and TnsE in Vp-PAI (9, 26).
We examined sequences that are at least 200 bp long around the target site in tdh+ and/or trh+ strains by direct genome sequencing (15). We aligned the sequences around the target site in tdh+ and/or trh+ strains with those in tdh trh-lacking strains. The results showed that unknown foreign elements become integrated into the same region of the Vp-PAI target site. Both ends of the elements were very similar to the ends of Vp-PAI (see the nucleotide sequences we deposited). Since this finding was obtained with the tdh+ and/or trh+ strains tested, it implied that elements with Vp-PAI-like end sequences are present not only in tdh+ strains but also in trh+ strains.
The Vp-PAI boundaries determined in this study are slightly different from those predicted by bioinformatics (12), but they are supported by experimental results obtained using three tdh trh-lacking strains (Fig. 1). The consolidated data should be helpful in future investigations of the pathogenicity and evolution of V. parahaemolyticus.
Nucleotide sequence accession numbers.
The nucleotide sequences described here have been deposited in the GenBank/DDBJ/EMBL database under accession numbers AB298911, AB298912, AB298913, AB298914, AB298915, AB298916, AB298917, AB298918, AB298919, AB298920, AB298921, and AB298922.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas and for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
We thank the staff of the Kanagawa Prefectural Institute of Public Health and Kansai International Airport Quarantine Station for supplying the V. parahaemolyticus strains.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Arciszewska, L. K., and N. L. Craig. 1991. Interaction of the Tn7-encoded transposition protein TnsB with the ends of the transposon. Nucleic Acids Res. 195021-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arciszewska, L. K., R. L. Mckown, and N. L. Craig. 1991. Purification of TnsB, a transposition protein that binds to the ends of Tn7. J. Biol. Chem. 26621736-21744. [PubMed] [Google Scholar]
- 3.Burrus, V., G. Pavlovic, B. Decaris, and G. Guédon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46601-610. [DOI] [PubMed] [Google Scholar]
- 4.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155376-386. [DOI] [PubMed] [Google Scholar]
- 5.Burrus, V., J. Marrero, and M. K. Waldor. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55173-183. [DOI] [PubMed] [Google Scholar]
- 6.Chaconas, G., B. D. Lavoie, and M. A. Watson. 1996. DNA transposition: jumping gene machine, some assembly required. Curr. Biol. 6817-820. [DOI] [PubMed] [Google Scholar]
- 7.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2414-424. [DOI] [PubMed] [Google Scholar]
- 8.Faruque, S. M., and J. J. Mekalanos. 2003. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 11505-510. [DOI] [PubMed] [Google Scholar]
- 9.Gringauz, E., K. A. Orle, C. S. Waddell, and N. L. Craig. 1988. Recognition of Escherichia coli attTn7 by transposon Tn7: lack of specific sequence requirements at the point of Tn7 insertion. J. Bacteriol. 1702832-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53245-281. [DOI] [PubMed] [Google Scholar]
- 11.Honda, T., Y. X. Ni, and T. Miwatani. 1988. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect. Immun. 56961-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurley, C. C., A. M. Quirke, F. J. Reen, and E. F. Boyd. 2006. Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates. BMC Genomics 7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iida, T., and K. Yamamoto. 1990. Cloning and expression of two genes encoding highly homologous hemolysins from a Kanagawa phenomenon-positive Vibrio parahaemolyticus T4750 strain. Gene 939-15. [DOI] [PubMed] [Google Scholar]
- 14.Kamali-Moghaddam, M., and L. Sundstrom. 2001. Arrayed transposase-binding sequences on the ends of transposon Tn5090/Tn402. Nucleic Acids Res. 291005-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasai, S., and T. Yamazaki. 2001. Identification of the cobalamin-dependent methionine synthase gene, metH, in Vibrio fischeri ATCC 7744 by sequencing using genomic DNA as a template. Gene 264281-288. [DOI] [PubMed] [Google Scholar]
- 16.Kholodii, G. Y., S. Z. Mindlin, I. A. Bass, O. V. Yurieva, S. V. Minakhina, and V. G. Nikiforov. 1995. Four genes, two ends, and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Mol. Microbiol. 171189-1200. [DOI] [PubMed] [Google Scholar]
- 17.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361743-749. [DOI] [PubMed] [Google Scholar]
- 18.Manson, J. M., and M. S. Gilmore. 2006. Pathogenicity island integrase cross-talk: a potential new tool for virulence modulation. Mol. Microbiol. 61555-559. [DOI] [PubMed] [Google Scholar]
- 19.Miyamoto, Y., T. Kato, Y. Obara, S. Akiyama, K. Takizawa, and S. Yamai. 1969. In vitro hemolytic characteristic of Vibrio parahaemolyticus: its close correlation with human pathogenicity. J. Bacteriol. 1001147-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishibuchi, M., M. Ishibashi, Y. Takeda, and J. B. Kaper. 1985. Detection of the thermostable direct hemolysin gene and related DNA sequences in Vibrio parahaemolyticus and other Vibrio species by the DNA colony hybridization test. Infect. Immun. 49481-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oelschlaeger, T. A., and J. Hacker. 2004. Impact of pathogenicity islands in bacterial diagnostics. APMIS 112930-936. [DOI] [PubMed] [Google Scholar]
- 22.Okada, K., T. Iida, K. Kita-Tsukamoto, and T. Honda. 2005. Vibrios commonly possess two chromosomes. J. Bacteriol. 187752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okura, M., R. Osawa, A. Iguchi, E. Arakawa, J. Terajima, and H. Watanabe. 2003. Genotypic analyses of Vibrio parahaemolyticus and development of a pandemic group-specific multiplex PCR assay. J. Clin. Microbiol. 414676-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, K. S., T. Ono, M. Rokuda, M. H. Jang, K. Okada, T. Iida, and T. Honda. 2004. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 726659-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parks, A. R., and J. E. Peters. 2007. Transposon Tn7 is widespread in diverse bacteria and forms genomic islands. J. Bacteriol. 1892170-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters, J. E., and N. L. Craig. 2001. Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol. 2806-814. [DOI] [PubMed] [Google Scholar]
- 27.Rádström, P., O. Sköld, G. Swedberg, J. Flensburg, P. H. Roy, and L. Sundström. 1994. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J. Bacteriol. 1763257-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raskin, D. M., R. Seshadri, S. U. Pukatzki, and J. J. Mekalanos. 2006. Bacterial genomics and pathogen evolution. Cell 124703-714. [DOI] [PubMed] [Google Scholar]
- 29.Seguritan, V., I. W. Feng, F. Rohwer, M. Swift, and A. M. Segall. 2003. Genome sequences of two closely related Vibrio parahaemolyticus phages, VP16T and VP16C. J. Bacteriol. 1856434-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skelding, Z., R. Sarnovsky, and N. L. Craig. 2002. Formation of a nucleoprotein complex containing Tn7 and its target DNA regulates transposition initiation. EMBO J. 213494-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stellwagen, A. E., and N. L. Craig. 1997. Avoiding self: two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J. 166823-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, H. Z., M. M. Wong, D. O'Toole, M. M. Mak, R. S. Wu, and R. Y. Kong. 2006. Identification of a DNA methyltransferase gene carried on a pathogenicity island-like element (VPAI) in Vibrio parahaemolyticus and its prevalence among clinical and environmental isolates. Appl. Environ. Microbiol. 724455-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]