Abstract
During infection, the phytopathogenic enterobacterium Erwinia chrysanthemi has to cope with iron-limiting conditions and the production of reactive oxygen species by plant cells. Previous studies have shown that a tight control of the bacterial intracellular iron content is necessary for full virulence. The E. chrysanthemi genome possesses two loci that could be devoted to iron storage: the bfr gene, encoding a heme-containing bacterioferritin, and the ftnA gene, coding for a paradigmatic ferritin. To assess the role of these proteins in the physiology of this pathogen, we constructed ferritin-deficient mutants by reverse genetics. Unlike the bfr mutant, the ftnA mutant had increased sensitivity to iron deficiency and to redox stress conditions. Interestingly, the bfr ftnA mutant displayed an intermediate phenotype for sensitivity to these stresses. Whole-cell analysis by Mössbauer spectroscopy showed that the main iron storage protein is FtnA and that there is an increase in the ferrous iron/ferric iron ratio in the ftnA and bfr ftnA mutants. We found that ftnA gene expression is positively controlled by iron and the transcriptional repressor Fur via the small antisense RNA RyhB. bfr gene expression is induced at the stationary phase of growth. The σS transcriptional factor is necessary for this control. Pathogenicity tests showed that FtnA and the Bfr contribute differentially to the virulence of E. chrysanthemi depending on the host, indicating the importance of a perfect control of iron homeostasis in this bacterial species during infection.
Erwinia chrysanthemi 3937, now called Dickeya dadantii, is a pathogenic enterobacterium that is able to cause soft rot diseases in a wide range of economically important crops (49, 55). The soft rot symptom produced by E. chrysanthemi consists of a progressive disorganization of parenchymatous tissues called maceration and results from several bacterial enzymatic activities, including pectinases and endoglucanases, which work in concert to degrade plant cell walls (43). The bacteria colonize the intercellular spaces of leaves and progress intercellularly to invade the other aerial parts of the plant (40). Under these conditions, E. chrysanthemi cells encounter an oxidative environment, as plant tissues produce active oxygen species in response to microbial attack (16). Active protective systems against oxidative damages, such as the methionine sulfoxide reductase MsrA and the manganese-containing superoxide dismutase, are essential for the outcome of the infection (13, 50). The production of the siderophores chrysobactin and achromobactin, two iron scavengers, is also essential for E. chrysanthemi cells to cause a systemic infection (14, 15, 19). Therefore, during the infectious process, a tight control of the intracellular iron content is important. Indeed, the redox properties of iron, which make this metal a valuable cofactor in a multitude of cellular process, can also lead to the production of harmful radicals. For instance, ferrous iron can exacerbate the oxidative stress through Fenton's reaction, which generates the highly toxic and reactive hydroxyl radical OH· (23, 30). We have demonstrated that the transcriptional repressor Fur, which controls the intracellular iron concentration, is involved in the pathogenicity of strain 3937 (18). The importance of the connection between iron metabolism and oxidative stress during the early steps of infection by E. chrysanthemi was emphasized with the discovery that the Suf machinery encoded by the sufABCDSE operon participates in the formation of Fe-S clusters under iron starvation and oxidative conditions and is necessary for full virulence (41, 42). The other mechanisms involved in the control of iron homeostasis during infection are currently not known.
In many bacteria, the removal of an excess of ferrous iron by its subsequent oxidation is achieved by ferritins or ferritin-like proteins. Ferritins constitute a broad superfamily of iron storage proteins, widespread in all domains of life, in aerobic or anaerobic organisms (3, 37). These proteins fall into three classes: ferritins that are heme free, found in pro- and eukaryotes, heme-containing bacterioferritins, found only in bacteria, and Dps proteins (DNA protein from starved cells), now called miniferritins, present only in prokaryotes (11, 52). They are composed of 24 identical subunits for ferritins and bacterioferritins and 12 identical subunits for the Dps proteins. These subunits assemble to make a spherical protein shell surrounding a central cavity able to hold up to 2,000 to 3,000 ferric iron atoms for ferritins and 500 atoms for miniferritins (3, 8). These iron storage proteins possess a binuclear di-iron center that constitutes the ferroxidase center involved in the oxidation of the ferrous iron (22, 27, 28, 53). Oxygen and hydrogen peroxide are the major cellular oxidants consumed during this reaction (7, 9, 60). Ferritins can concentrate and store iron as a mineral (hydrated ferric oxide) in their central cavity (10, 25, 32). This sequestered iron is nonreactive and can serve as a reserve when the exogenous availability of this metal becomes limited. Thus, besides their role in iron storage, maxi- and miniferritins are also involved in the detoxification of this metal, dioxygen, and hydrogen peroxide under certain conditions (24, 29, 31, 54, 56, 58, 59). This work was aimed at elucidating the role of the maxiferritins in the control of iron homeostasis in E. chrysanthemi 3937. The results obtained show that by participating in the control of iron homeostasis, these proteins also have a role in the virulence of this bacterium.
MATERIALS AND METHODS
Bacterial strains, phage, plasmids, and media.
The bacterial strains, bacteriophage, and plasmids used in this work are described in Table 1. The rich media used were L broth and L agar (48). To determine the effect of iron limitation on the growth of E. chrysanthemi cells, ethylenediamine-N,N′-bis-2-hydroxy-phenylacetic acid (EDDHA) (Sigma-Aldrich, Saint Quentin Fallavier, France) or 2,2′-dipyridyl (Sigma-Aldrich, Saint Quentin Fallavier, France) was added to L broth or L agar at a final concentration of 80 μM or 200 μM, respectively. Tris medium was used as the low-iron minimal medium (18). It was amended by adding EDDHA or 2,2′-dipyridyl at a final concentration of 40 μM or 100 μM, respectively, to achieve iron-poor conditions. For iron-rich conditions, it was supplemented with 20 μM FeCl3 or 20 μM FeSO4. Glucose (2 g/liter) was used as the carbon source. For genetic marker exchange by homologous recombination, minimal low-phosphate medium was used (18). Escherichia coli and E. chrysanthemi strains were grown at 37°C and 30°C, respectively. Antibacterial agents were added at the following concentrations: 50 μg/ml for ampicillin, 40 μg/ml for spectinomycin, 25 μg/ml for kanamycin, and 20 μg/ml for chloramphenicol. For oxidative growing conditions, cultures grown overnight were diluted 100-fold in L broth and grown under intensive shaking with the following compounds: 0.5 mM hydrogen peroxide (Acros Organics, Noisy le Grand, France), 6 μM paraquat (Sigma-Aldrich, Saint Quentin Fallavier, France), 2.5 μM streptonigrin (Sigma-Aldrich, Saint Quentin Fallavier, France), and 70 μM spermine NONOate (Acros Organics, Noisy le Grand, France).
TABLE 1.
Bacterial strains, bacteriophage, and plasmids used in this study
| Strain, phage, or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Erwinia chrysanthemi | ||
| 3937 | Wild type isolated from African violet | Our collection |
| L2 | Lac− derivative of 3937 | 21 |
| PPV40 | ftnAΩ-spec FtnA− Specr | This work |
| PPV41 | ftnA::uidA FtnA− Kmr | This work |
| PPV39 | furΩ-spec Fur− Specr | 18 |
| PPV42 | ftnA::uidA furΩ-spec FtnA− Fur− Kmr Specr | This work |
| PPV43 | bfrΩ-Km Bfr− Kmr | This work |
| PPV44 | bfr::uidA Bfr− Kmr | This work |
| A4109 | rpoS::Cm RpoS− Cmr | This work |
| PPV45 | bfr::uidA rpoS Bfr− RpoS− Kmr Cmr | This work |
| PPV46 | bfr::Ω-Km ftnAΩ-spec Bfr− FtnA− Kmr Specr | This work |
| PPV47 | ΔryhB::Ω-spec RyhB− Specr | This work |
| PPV48 | L2 fct::lacZ ftnAΩ-spec Fct− Cbs− FtnA− Kmr Specr | This work |
| Escherichia coli K-12 | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi relA1 | 48 |
| Phage | ||
| φEC2 | Generalized transducing phage from E. chrysanthemi strain 3690 | 47 |
| Plasmids | ||
| pGEM-T Easy | 3.015-kb vector, pGEM-5Zf derivative; Ampr | Promega |
| pBC | 3.4-kb vector, pUC19 derivative; Cmr | Stratagene |
| pUIDK1 | pNB4 vector carrying the uidA-Km cassette; Kmr Ampr | 5 |
| pHP45Ω | pBR322 derivative carrying the Ω interposon coding for spectinomycin resistance; Ampr Specr | 45 |
| pHP45Ω-Km | pHP45 derivative carrying the Ω interposon coding for kanamycin resistance; Ampr Kmr | 17 |
| pCKC15 | pUC19 derivative harboring a Cmr cassette from plasmid pSU9 flanked by EcoRI and SmaI restriction sites | Laboratory collection |
| pSR2488 | 650-bp amplified internal fragment from the rpoS gene cloned into pUC18; Ampr | This work |
| pTF40 | 1,500-bp amplified fragment of the ftnA gene region cloned into pGEM-T Easy; Ampr | This work |
| pTF41 | 1,500-bp amplified fragment of the ftnA gene region cloned at the EcoRI site of pBC; Cmr | This work |
| pTF42 | Interposon Ω-Spec cloned into the MunI site of the ftnA gene from pTF41; Cmr Specr | This work |
| pAB1 | uidA-Km cassette from pUIDK1 cloned into the MunI site of the ftnA gene from pTF41; Cmr Kmr | This work |
| pAB2 | 1,600-bp amplified fragment of the bfd bfr gene region cloned into pGEM-T Easy; Ampr | This work |
| pAB3 | 1,600-bp amplified fragment of the bfd bfr gene cloned into the ApaI and SpeI sites from pBC; Cmr | This work |
| pAB4 | uidA-Km cassette from pUIDK1 cloned into the blunted PstI site of the bfr gene from pAB3; Cmr Kmr | This work |
| pAB5 | Ω-Km cassette from pHP45Ω-Km cloned into the blunted PstI site of the bfr gene from pAB3; Cmr Kmr | This work |
| pAB6 | 5′ amplified region of the ryhB locus cloned into pGEM-T Easy; Ampr | This work |
| pAB7 | 3′ amplified region of the ryhB locus cloned into pGEM-T Easy | This work |
| pAB8 | 5′ and 3′ amplified regions of the ΔryhB locus cloned in pBC; Cmr | This work |
| pAB9 | Interposon Ω-Spec cloned into the BamHI site of the ΔryhB locus from pAB8; Cmr Specr | This work |
Construction of ferritin-deficient mutants.
Genomic fragments from the ftnA and bfd-bfr loci were amplified by PCR with the primers described in Table 2 and cloned into the pGEM-T Easy vector (Promega, Charbonnière, France). These fragments were subcloned into the pBC plasmid with appropriate restriction enzymes in order to gain unique restriction sites in the ftnA and bfr genes. The Ω interposon coding for spectinomycin resistance from pHP45Ω hydrolyzed with EcoRI was cloned into the MunI site of the ftnA gene, giving rise to plasmid pTF42. The Ω-Km interposon, coding for kanamycin resistance from pHP45Ω-Km hydrolyzed with SmaI, was cloned into the T4 polymerase-blunted PstI site of the bfr gene, giving rise to plasmid pAB5. The transcriptional ftnA::uidA and bfr::uidA fusions were constructed in vitro similarly. The rpoS and ΔryhB mutants were constructed as follows: a 650-bp genomic internal fragment of the rpoS gene was amplified by PCR with primers rpoS5 and rpoS6 (Table 2). These primers contained an extra BamHI site at the 5′ ends, and the resulting PCR fragment was cloned into the BamHI site of vector pUC18, giving rise to plasmid pSR2488. The chloramphenicol resistance cartridge from pCKC15 hydrolyzed with SmaI was then introduced into the unique HpaI site located in the PCR fragment internal to the rpoS gene. This insertion was then introduced into the E. chrysanthemi chromosome by marker exchange recombination between the chromosomal allele and the plasmid-borne mutated allele. The recombinants were selected after successive cultures in low-phosphate medium in the presence of chloramphenicol, conditions in which pBR322 derivatives are very unstable. Correct recombination was confirmed by PCR and by Western blotting using anti-RpoS antibodies (data not shown). The 5′ and 3′ parts of the ryhB locus were PCR amplified with two couples of primers containing one degenerated primer with an extra BamHI site and cloned into the pGEM-T Easy vector. These fragments were hydrolyzed with BamHI/EcoRI or BamHI/SpeI and cloned into the pBC vector cut with EcoRI and SpeI. The Ω interposon coding for spectinomycin resistance from pHP45Ω hydrolyzed with BamHI was cloned into the BamHI site of the ΔryhB gene, giving rise to plasmid pAB9. The wild-type strain of E. chrysanthemi was electroporated with the corresponding plasmids. Transformants were purified once on L agar plates containing the appropriate selection marker. Cultures grown with the corresponding antibiotics in L broth were 20-fold diluted in low-phosphate medium supplemented with iron in order to promote plasmid destabilization with exchange recombination of the disrupted DNA insert into the E. chrysanthemi chromosome. Recombinants were selected as described previously (18). The presences of disrupted ftnA or bfr genes as well a disrupted and deleted ΔryhB gene in these clones were confirmed by Southern blot hybridization experiments. Double or triple mutants were constructed by using the generalized transducing phage φEC2 (47).
TABLE 2.
Primers used in this work
| Primer | Sequence (5′-3′)a | Restriction site added |
|---|---|---|
| ftnA1s | GCTGCGTATGGTTATTTTCTG | |
| ftnA1r | GAAATAATCGGCGTGTATCC | |
| ftnA2s | CCATCAGTCACCTGCCAGTAA | |
| ftnA2r | GGATAATCAGCCGCCCAGCAA | |
| bfr1s | GGTCGTGTAGAGCGCGGCA | |
| bfr1r | CAGTACATAACCCCCATAT | |
| bfr2s | AAATGAGTTATCCGCATATGT | |
| bfr2r | GTGCTGACGAACGACCTGTCG | |
| ryhB1s | GTGGACCGCCTATACGCT | |
| ryhB1r | CGCGGATCCGATGAGTCACAAGGATGG | BamHI |
| ryhB2s | AGCGGATCCTGGTTTCCTATTTGTTTG | BamHI |
| ryhB2r | TCCAAAACCCTGTCCGCC | |
| rpoS5 | CGGGATCCGGATGATCGAGAGTAAC | BamHI |
| rpoS6 | CGGGATCCTTCAACCTGAATCTGGC | BamHI |
| rpoS1s | ATCACGGGTAGGAGCCACTTA | |
| rpoS1r | TTATTGTGCGAGTTATTCGCG |
Underlining indicates the restriction site.
Determination of siderophore production, iron accumulation, and total iron content.
Siderophore activities were detected as described previously (19). An iron accumulation assay was performed on 96-well culture plates. Cultures grown overnight were diluted 100-fold in wells containing L broth supplemented with 10 or 50 μM 55FeSO4 (40 mCi·mg−1 iron). After 18 h of growth, cells were collected with a cell harvester (Brandel) and washed with a solution of 50 mM Tris-5 mM EDTA (pH 7.5) on filters (Wallac, Gaithersburg, MD) before scintillation counting (Trilux MicroBeta; Wallac). Whole-cell iron content was analyzed by inductively coupled plasma atomic emission spectroscopy (ICP-AES) at the Service Commun d'Analyze from Centre National de la Recherche Scientifique (Vernaison, France).
59Fe labeling and preparation of whole-cell extracts.
Cultures of the wild-type strain and the ferritin-deficient mutants grown overnight were diluted 100-fold in L broth and grown to an optical density at 600 nm (OD600) of 0.4 when 1 μM of 59FeCl3 (20 mCi·mg−1 iron) was added to the cultures. Ninety minutes after the addition of iron, cells (OD600 of ∼1.5) were harvested by centrifugation. Cells were washed once with a solution containing 50 mM potassium phosphate (pH 7.8)-0.1 mM EDTA-10 mM MgCl2 and centrifuged. Pellets were resuspended in 400 μl of the same solution in which DNase I and lysozyme were added at final concentrations of 0.1 mg/ml and 0.2 mg/ml, respectively, and incubated 30 min at 4°C. Cell lysis was achieved by six freezing/thawing cycles, and samples were kept at −20°C. Twenty-five to thirty micrograms of proteins was loaded and run on a 10% polyacrylamide nondenaturing gel. Dried gels were autoradiographed at −80°C for 12 to 24 h using Kodak (Chalon sur Saône, France) X-ray film.
Mössbauer spectroscopy.
For each Mössbauer measurement, a 750-ml bacterial culture in 3-liter Erlenmeyer flasks was required in order to obtain approximately 750 μl of packed cells. Cultures of wild-type and bfr, ftnA, and bfr ftnA mutant strains were diluted 60-fold in Tris medium supplemented with glucose and grown to the early stationary growth phase. [57Fe(DHBA)3]3− was added to the cell suspensions at a final concentration of 5 μM 57Fe-100 μM DHBA. Cells were grown additionally for 30, 60, and 120 min, respectively. Cells were then cooled down to 4°C within 2 min, harvested, washed in cold Tris medium, and transferred into Delrin Mössbauer sample holders. Sample volumes were about 700 μl. Sample thickness did not exceed 9 mm. The containers were quickly frozen in liquid nitrogen and kept in a liquid nitrogen storage vessel until the measurement was performed. The Mössbauer spectra were recorded in horizontal transmission geometry using a constant acceleration spectrometer operated in conjunction with a 512-channel analyzer in the time scale mode. The source was at room temperature and consisted of 0.75 GBq [57Co] diffused in Rh foil (AEA, Braunschweig, Germany). The spectrometer was calibrated against a metallic α-iron foil at room temperature, yielding a standard line width of 0.24 mm/s. The Mössbauer cryostat was a helium bath cryostat (MD306; Oxford Instruments). A small field of 20 mT perpendicular to the γ-beam was applied to the tail of the bath cryostat using a permanent magnet. Isomer shift, δ, quadrupole splitting, ΔEQ, and the percentage of the total absorption area were obtained by least-square fits of Lorentzian lines to the experimental spectra. Experiments were performed two times, and data from one experiment are shown.
General DNA methods.
DNA manipulations (chromosomal DNA isolation, cloning, and electrophoresis) were described previously (18). Plasmids were extracted by using the QIAprep Spin Miniprep kit (Qiagen, Courtaboeuf, France). All cloning experiments were performed in the DH5α strain of E. coli. DNA/DNA hybridization analysis was performed by using Denhardt's method as described previously by Sambrook et al. (48). The primers used for PCR amplification of E. chrysanthemi genomic fragments are described in Table 2. PCR was performed using a DNA thermocycler (Hybaid PCR Express system) with a denaturation step at 94°C for 60 s, an annealing step at 52°C for 75 s, and an extension step at 72°C for 75 s, which was followed by an extension step at 72°C for 10 min. PCR products were cloned into the pGEM-T Easy plasmid according to the manufacturer's instructions. Nucleotide sequencing of PCR products was obtained from Genome Express (Meylan, France). For hybridization, DNA probes were prepared by using the Prime-a-Gene labeling system according to the manufacturer's recommendations (Promega, Charbonnière, France).
RNA isolation.
A culture grown overnight in L broth was diluted 60-fold in Tris medium-glucose or 100-fold in L broth. The culture was grown under shaking until an absorbance at 600 nm of 0.4 was reached, and iron (20 μM FeSO4) was added or not added. A total of 7.5 ml of culture was harvested by centrifugation for 10 min at 4°C (8,000 × g). The cell pellet was then resuspended in 600 μl of buffer A (20 mM sodium acetate [pH 5.5], 1 mM EDTA) at 4°C. After the addition of 33 μl of 10% sodium dodecyl sulfate (SDS) and 600 μl of hot acidic phenol (65°C) equilibrated with buffer A, the sample was vigorously mixed for 30 s and incubated for 10 min at 65°C. The aqueous phase was reextracted with phenol-chloroform (1:1) equilibrated with 10 mM Tris (pH 7). RNA was precipitated overnight with 30 μl of 3 M sodium acetate and 800 μl of ethanol. The RNA pellet was washed with 70% ethanol and resuspended in 35 μl of water treated with diethyl pyrocarbonate. Northern blot analysis was performed after electrophoresis: 3 μg of RNA were loaded and run on a 1% Tris-borate-EDTA agarose gel containing 7.2% formaldehyde. After electrophoresis, RNAs were transferred onto a positively charged nylon membrane (N+ Hybond; GE Healthcare), and hybridizations were carried out as described previously (12). Membranes were washed twice at 65°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% SDS and in 1× SSC-0.5% SDS. 16S RNA was used as a control. Membranes were placed against Kodak (Chalon sur Saône, France) X-ray film at −70°C for a few days.
Determination of β-glucuronidase activities in bacterial culture.
An inoculum of an L culture grown overnight was 60-fold diluted in Tris medium containing glucose or 100-fold in L broth with the appropriate antibiotics. The inoculated culture was divided into two subcultures supplemented or not with 20 μM FeSO4. Cultures were grown aerobically at 30°C. Samples were collected and immediately frozen. Enzymatic activities were assayed as reported previously (51). β-Glucuronidase activity is expressed in nanomoles of paranitrophenol liberated per minute per OD600 unit.
Pathogenicity assays.
Pathogenicity tests were performed on chicory leaves and on potted African violets (Saintpaulia ionantha) cv. Blue Rhapsody. Bacterial cells were plated onto L agar medium and incubated for 24 h at 30°C. Cells were suspended in an NaCl solution (9 g per liter) to give an OD600 of 0.4. The resulting suspension (4 μl) was used to inoculate chicory leaves, whereas 100 μl of inoculum was used for one leaf per African violet plant as described previously (51). Progression of the symptoms was scored during 4 days for the chicory test and 9 days for the Saintpaulia test.
RESULTS
Genetic organization of the loci encoding maxiferritins.
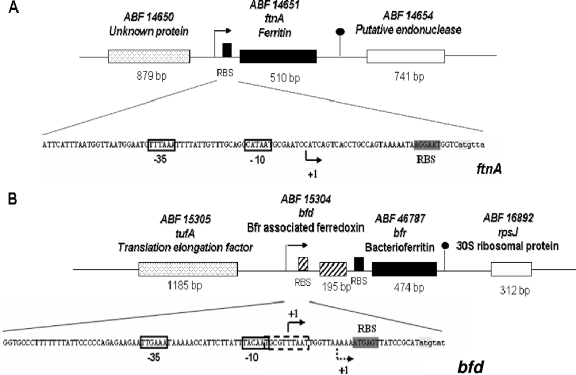
In the genomic sequence of strain 3937, two loci encode putative maxiferritin proteins involved in iron storage: the ftnA and bfr genes encode a ferritin and a bacterioferritin, respectively (https://asap.ahabs.wisc.edu/asap). The ftnA gene (coding sequence [CDS] ABF-0014651) is located at position 2293752 on the E. chrysanthemi genome. Sequence analysis of the ftnA locus revealed a putative monocistronic open reading frame of 510 nucleotides encoding a 169-amino-acid polypeptide (Fig. 1A). This 19.3-kDa protein is 78% and 64% identical to the FtnA ferritins from Yersinia pestis KIM and E. coli K-12. Amino acid residues E18, Y24, E50, H53, E94, and Q127, involved in the formation of the ferroxidase site, are conserved in those proteins (53). A putative σ70 promoter was identified by computational analysis. For confirmation, a primer extension experiment was performed. The extension reaction product showed that the transcription of the ftnA gene started at a C nucleotide situated 35 bases upstream of the ATG start codon, indicating that the anticipated promoter was functional (data not shown).
FIG. 1.
Genetic organization of the loci encoding the maxiferritins FtnA (A) and Bfr (B) of E. chrysanthemi 3937. The −35 and −10 promoter elements are boxed. Transcriptional starts identified by primer extension experiments are indicated by an arrow. The promoter element TGCGTTTAAT of the bfd-bfr operon, which is similar to the RpoS factor recognition consensus sequence, is boxed with a dotted line. Inverted repeats situated at the 3′ untranslated region of the bfr and ftnA genes that may form a hairpin are indicated by a small stem-loop. The bfr and ftnA genes are transcribed in opposite directions on the chromosome. RBS, ribosome binding site.
The bfr gene (CDS accession number ABF-0046787) appears to be clustered into an operon with the bfd gene (accession number ABF-0015304) (Fig. 1B). This putative polycistronic unit is located at position 4392896 on the reverse strand of the chromosome. The first gene, bfd, encodes a 64-amino-acid-long peptide showing 56% and 70% sequence identities with the bacterioferritin-associated [2Fe-2S] ferredoxins from E. coli K-12 and Y. pestis KIM, respectively (20). The translation start codon of the bfr gene is situated 53 nucleotides downstream of the end of the bfd gene (Fig. 1B). The bfr gene encodes a 157-amino-acid polypeptide that is 84% identical to the Bfr protein from E. coli K-12. Amino acid residues E18, Y25, E51, H54, E94, and H130, forming the ferroxidase center, are strictly conserved for both proteins (4, 53). Furthermore, methionine residue 52 of E. coli bacterioferritin, involved in the binding of heme, is also conserved in the Bfr protein from E. chrysanthemi (2). Primer extension experiments indicated that there are two transcriptional starts located 18 and 30 nucleotides upstream the of start codon of the bfd gene (Fig. 1B). Both promoters were predicted by computer analysis. The first one is a canonical σ70 promoter that can be associated with the transcription start situated ca. 30 nucleotides upstream from the ATG codon of the bfd gene. For the second transcriptional start, a −10 promoter element with the TGCGTTTAAT sequence could be identified. However, no −35 promoter element could be associated with this −10 region. The sequence of this −10 promoter element is reminiscent of the TGN0-2C(C/T)ATA(C/A)T consensus sequence identified for the rpoS-encoded alternative sigma factor σS in E. coli (26). At the 3′ untranslated region of the ftnA and bfr genes, inverted repeats may form a hairpin that could act as a transcription terminator.
Construction of the bfr and ftnA mutants.
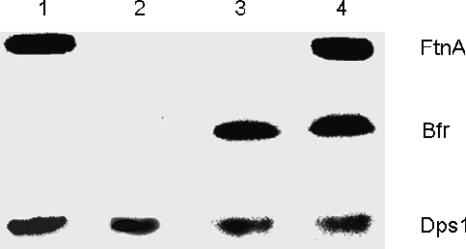
The bfd-bfr and ftnA loci were amplified by PCR and cloned into the pGEM-T Easy vector as described in Materials and Methods. The bfr and ftnA genes were inactivated by interposons or transcriptional uidA cassettes (see Materials and Methods). The disrupted ftnA and bfr genes were then introduced back into the chromosome of E. chrysanthemi by reverse genetics as described in Materials and Methods. The ftnA bfr double mutant was constructed by transducing the ftnA mutation into the bfr mutant with phage phiEC2. The presence of the disrupted genes in these mutants was confirmed by Southern experiments (data not shown). The absence of production of the FtnA and Bfr proteins in the corresponding mutants was checked by labeling exponentially growing cells with 59FeCl3. Crude cell extracts from the wild-type strain and the ftnA, bfr, and bfr ftnA mutants were analyzed by native polyacrylamide gel electrophoresis as described in Materials and Methods (Fig. 2). E. chrysanthemi wild-type extracts showed three major bands corresponding to iron-labeled protein species (Fig. 2). The upper band was missing in the ftnA mutant, whereas the central band was missing in the extracts of the bfr mutant. Both the upper and central iron-labeled bands were not detected in the cell extracts from the double mutant, indicating that the upper and central bands correspond to the FtnA and Bfr proteins, respectively.
FIG. 2.
Analysis of 59Fe-labeled E. chrysanthemi proteins from soluble cell extracts after electrophoresis on a native 8% polyacrylamide gel. Bacteria were grown in L medium to an OD600 of 0.4, and 59FeCl3 was added at a final concentration of 1 μM. Samples were collected 90 min after the addition of iron. Whole-cell extracts were prepared as described in Materials and Methods. The lower band probably corresponds to the Dps1 protein of E. chrysanthemi, as it is missing in a Dps1 gene mutant (data not shown). Lane 1, bfr mutant; lane 2, bfr ftnA mutant; lane 3, ftnA mutant; lane 4, wild-type strain.
Growth characteristics of the E. chrysanthemi bfr, bfr ftnA, and ftnA mutants.
We checked the growth properties of the ferritin-deficient mutants in L broth and in the low-iron minimal Tris medium. In L medium, the growth of the ftnA mutant was slower than that of the wild-type strain, giving rise to a final cellular density lower than that of the wild-type strain (data not shown). The decrease in the growth yield of the ftnA mutant was more important in Tris medium (Table 3). The addition of iron to Tris medium stimulated the growth of the ftnA mutant and the wild-type strain. However, the growth of the ftnA mutant still remained affected compared to that of the wild type (Table 3). The addition of the iron chelator EDDHA or 2,2′-dipyridyl to Tris medium severely reduced the growth capacity of the ftnA mutant in comparison to that of the wild-type strain. The intracellular ferrous iron chelator 2,2′-dipyridyl gave the highest inhibitory effect (Table 3). Under all growth conditions tested, the bfr mutant behaved like the wild-type strain, whereas the bfr ftnA double mutant displayed an intermediate growth phenotype (Table 3). The introduction of plasmid pTF41, containing the wild-type ftnA gene in the ftnA mutant, restored its growth capacity in the presence of the iron chelators (data not shown). The altered growth capacity of the ftnA mutant under conditions of iron starvation indicates that the absence of a functional ferritin alters the ability of the bacterium to overcome iron deprivation. On L agar plates, colony development was slower for the ftnA mutant than for the wild-type strain and other ferritin-deficient mutants. Under anaerobic conditions, only the ftnA mutant showed a reduced growth yield (data not shown).
TABLE 3.
Growth capacity of the E. chrysanthemi ferritin-deficient mutants compared to the wild-type strain in Tris medium with different iron avalaibilitiesa
| Mutant | Ratio of final cell density of mutant/final cell density of wild type ± SD
|
|||
|---|---|---|---|---|
| Tris medium | + Fe | + EDDHA | + Dipyridyl | |
| ftnA | 0.74 ± 0.03 | 0.80 ± 0.03 | 0.69 ± 0.03 | 0.63 ± 0.03 |
| bfr | 1 | 1 | 1 | 1 |
| bfr ftnA | 0.90 ± 0.02 | 0.91 ± 0.02 | 0.88 ± 0.03 | 0.87 ± 0.03 |
Ratios of the final cellular density of the mutants to that of the wild-type strain are given. Three experiments were performed, and standard deviations are shown.
Iron transport, whole-cell iron content, and Mössbauer spectroscopy.
The absence of functional ferritins can change the intracellular iron pool distribution and thus can lead to the metallation and activation of the Fur repressor. Therefore, we analyzed the production of siderophores, chrysobactin and achromobactin (44, 39), in the bfr, ftnA, and bfr ftnA mutants grown in Tris medium. Kinetics and levels of achromobactin and chrysobactin production were almost identical for the wild-type strain and the ferritin-deficient mutants (data not shown). We also found that the transcription of biosynthetic genes for achromobactin or chrysobactin was not reduced in these mutants (data not shown).
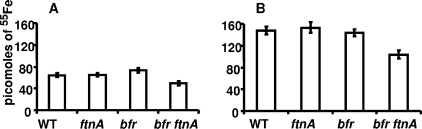
As the lack of functional ferritins could reduce the ability of bacteria to save iron when this metal becomes abundant, we checked the capacities of the different ferritin mutants to store iron by growing cells overnight in L medium supplemented with 1, 10, or 50 μM 55FeSO4. For 1 μM iron, the wild-type strain and the different ferritin mutants accumulated the same quantity of iron (data not shown). Amounts of 55Fe taken up and stored by the cells increased with increasing concentrations of this metal (Fig. 3A and B). For both iron concentrations, there was no significant difference in the amount of iron taken up by the wild-type strain and the bfr or the ftnA mutant (Fig. 3A and B). However, the bfr ftnA double mutant accumulated less iron than the wild-type strain (Fig. 3A and B). This defect in iron accumulation was more pronounced at the highest FeSO4 concentration tested. Indeed, the double mutant accumulated 20% less iron than the wild-type strain at an FeSO4 concentration of 10 μM and 33% less iron at a concentration of 50 μM (Fig. 3A and B). These data were corroborated by analyzing the total Fe content in the wild-type and ferritin-deficient mutant strains by ICP-AES. In L medium supplemented with 50 μM FeCl3, only the double mutant displayed a decrease of 30% in the total amount of intracellular iron in comparison to the iron content of the wild-type strain and the bfr and ftnA mutants (data not shown).
FIG. 3.
Iron accumulation in the wild-type strain and the ferritin-deficient mutants of E. chrysanthemi. Bacteria were grown overnight in L broth with 10 μM (A) or 50 μM (B) FeSO4. Cells were harvested on filters and washed as described in Materials and Methods. The amounts of 55Fe taken up by cells of the wild-type (WT) strain and the different ferritin-deficient mutants are indicated in picomoles of iron per OD600 unit. Experiments were performed in triplicate, and standard deviations are shown.
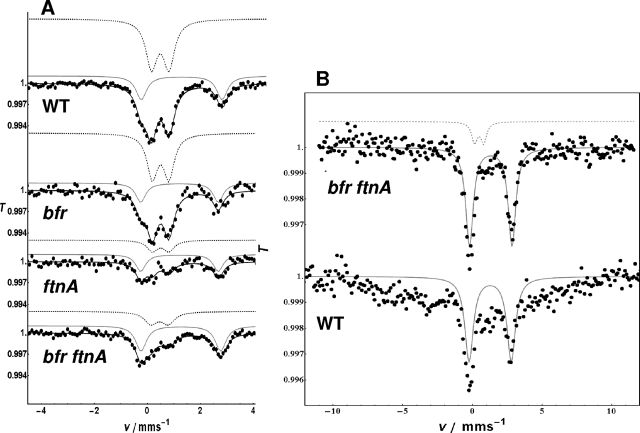
The natures of the intracellular iron pools of the wild-type strain and the mutants were analyzed by in situ Mössbauer spectroscopy. In principle, in situ Mössbauer spectroscopy enables the simultaneous identification of all main iron metabolites on a qualitative level as well as on a quantitative level without the destruction of cellular assembly (35, 36). Cells were grown in Tris medium to the beginning of the stationary phase in order to achieve a high bacterial yield. A ferric complex of 2,3-dihydroxybenzoic acid, [57Fe(DHBA)3]3−, was added to the culture, and 30, 60, and 120 min after the addition of this complex, cells were collected and prepared as described in Materials and Methods. Figure 4A displays Mössbauer spectra from cells of the E. chrysanthemi wild-type strain and its ferritin-deficient mutants after 120 min of incubation, which is the time when ferritin signals are easily observed (46). Mössbauer parameters of the spectra are listed in Table 4. The spectrum of the ferric DHBA complex (not shown here) displays a broad six-line pattern dominated by spin-spin relaxation, like other catecholate complexes at 77 K. Since such a spectrum is lacking in the cell spectra, it is safe to state that iron of this complex has been metabolized. The spectra exhibit two quadrupole doublets. One doublet corresponds to high-spin ferrous iron in an octahedral oxygen or nitrogen environment. Such a component has been observed in many bacterial systems. In E. coli, it was shown that this component is located in the cytoplasm and corresponds to an oligomeric ferrous sugar-phosphate complex (6). The second component represents a ferric high-spin species exhibiting Mössbauer parameters similar to those of a variety of bacterial ferritins. Figure 4B displays the Mössbauer spectra of the bfr ftnA mutant and the wild-type strain measured at 2 K. At this temperature, ferric iron of the wild-type strain displays magnetic broadening, and the doublet disappears. This feature was described previously (46) and is typical of bacterial ferritins. In contrast, the double mutant shows a ferric iron doublet even at 2 K, indicating the different natures of the two ferric iron components. The spectra of the wild-type strain and the bfr mutant exhibit very similar features (Fig. 4A). The majority of metabolized iron is found in FtnA (approximately 70%), and a smaller portion is ferrous iron (approximately 30%). In both strains, the overall iron incorporation is very similar, as estimated from the absolute areas. The most striking difference between the spectra of ftnA and bfr ftnA mutants compared to those of wild-type and bfr strains is the ferrous iron/ferric iron ratios, which are 1.65 and 1.98, respectively (Table 4). Both mutants are lacking a functional ftnA gene. The large contribution of Fe2+ reflects a lack of proper storage due to the absence of the ferritin FtnA. Moreover, Bfr is obviously not a replacement for FtnA. Therefore, the function of the bacterioferritin is not storage of bulk iron, indicating that Bfr is probably not involved in the regular iron storage transfer 2Fe2++ 1/2O2 + 2H2O + FtnA + (H2PO4)−→(Fe2O3H3 PO4) FtnA + 3H+. In addition, the observed excess of Fe2+ in spectra of the ftnA and bfr ftnA mutants indicates an impaired balance of general iron metabolism after [57Fe(DHBA)3]3− uptake (Fig. 4A). Due to the lack of bfr and ftnA genes in the double mutant, the ferric iron contribution cannot be attributed to the maxiferritin Bfr or FtnA. Rather, we suggest that this species represents ferric Dps. We further assume that the same is true for the ftnA mutant.
FIG. 4.
Mössbauer spectra of Erwinia chrysanthemi 3937 cells measured at 77 K (A) and 2 K (B). T represents the relative transmission, and v represents the energy scale measured as the velocity in mm/s. Genotypes of the different strains are indicated on the spectra. Cells were grown to an OD600 of 0.8 and incubated for 120 min with 5 μM 57Fe-100 μM DHBA. (A) Each spectrum is characterized by two quadrupole doublets, the parameters of which are listed in Table 4. The dashed gray line corresponds to the least-square fits of ferric high-spin iron to the experimental spectra, and full gray lines correspond to ferrous high spin. (B) Mössbauer spectra from cells of the bfr ftnA double mutant and the wild-type (WT) strain of E. chrysanthemi 3937 measured at 2 K. In both spectra, a ferrous high-spin component is observed (full gray lines). In the double mutant, the ferric ion doublet is still visible at 2 K (dashed gray line), whereas in the wild-type strain, this component broadens at temperatures below 4.3 K due to magnetic splitting, and the doublet disappears. The magnetic splitting is not resolved due to relaxation effects and was not fitted.
TABLE 4.
Mössbauer parameters of spectra of early-stationary-phase E. chrysanthemi cells after 120 min of incubation with [Fe(DHBA)3]3− determined by least-square fits of Lorentzian lines
| Strain | δ (mm s−1) | ΔEQ (mm s−1) | Γ (mm s−1) | Area (%) | Fe(II)/ Fe(III) ratio |
|---|---|---|---|---|---|
| Wild type (ferrous) | 1.30 | 3.05 | 0.48 | 32.6 | 0.48 |
| Wild type (ferric) | 0.49 | 0.66 | 0.50 | 67.4 | |
| bfr (ferrous) | 1.21 | 2.93 | 0.45 | 29.7 | 0.42 |
| bfr (ferric) | 0.48 | 0.63 | 0.48 | 70.3 | |
| ftnA (ferrous) | 1.27 | 2.93 | 0.35 | 62.3 | 1.65 |
| ftnA (ferric) | 0.51 | 0.78 | 0.31 | 37.7 | |
| bfr ftnA (ferrous) | 1.26 | 3.01 | 0.50 | 66.5 | 1.98 |
| bfr ftnA (ferric) | 0.48 | 0.65 | 0.50 | 33.5 |
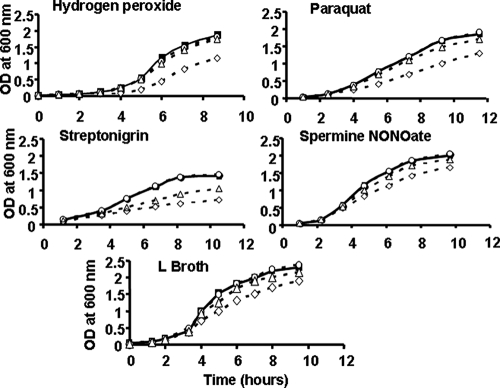
Sensitivity of the ferritin mutants to oxidative stress.
Since the intracellular iron pool could contribute to oxidative stress, we checked the sensitivities of ferritin mutants to hydrogen peroxide and to compounds generating the superoxide ion O2− (paraquat), radicals (streptonigrin), and nitric oxide (spermine NONOate). The addition of these compounds to L medium impaired the growth of the ftnA mutant (Fig. 5). The wild-type strain and the bfr mutant displayed the same sensitivities to all these compounds (Fig. 5). In addition, the bfr ftnA double mutant was sensitive to streptonigrin only. The introduction of plasmid pTF41, containing the wild-type ftnA gene, enabled the ftnA mutant to grow in the presence of these oxidative stress-generating compounds (data not shown). Thus, the increased sensitivity of the ftnA mutant to oxidative stress is caused by the lack of a functional ferritin.
FIG. 5.
Growth of the wild-type strain and the ferritin-deficient mutants of E. chrysanthemi under oxidative and nitrosative conditions. Cells were grown in L medium with intensive shaking under normal conditions or in the presence of 0.5 mM of H2O2, 6 μM paraquat, 2.5 μM streptonigrin, or 70 μM spermine NONOate, as indicated on the graphs. Filled squares, wild-type strain; open circles, bfr mutant; open diamonds, ftnA mutant; open triangles, bfr ftnA mutant. Experiments were performed five times, and the data reported are the means of three independent experiments, with standard deviations corresponding to less than 5%.
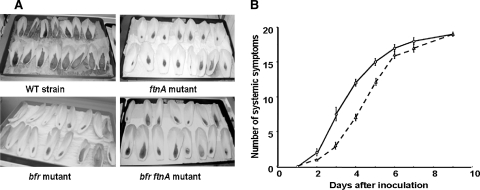
Pathogenicity of the ferritin mutants.
We examined the behaviors of the ferritin mutants of strain 3937 after inoculation onto chicory leaves. In comparison with the wild-type strain, the ftnA, bfr, and bfr ftnA mutants displayed a delay in the appearance of symptoms of maceration (Fig. 6A). However, once symptoms started, the progression of mutants and that of the wild-type strain were the same. For potted African violets, a lag in the appearance of maceration symptoms was observed only for the ftnA mutant (Fig. 6B). One week after inoculation, the numbers of systemic responses for the wild-type strain and those of its ftnA derivative mutant were identical (Fig. 6B). The bfr and bfr ftnA mutants were as virulent as the wild-type strain on African violets (data not shown). The delay in symptom appearance for the ftnA mutant is consistent with the fact that this mutant is sensitive to iron starvation and to oxidative stress, which are conditions encountered by bacterial cells during infection. Since symptom evolution for the wild-type strain was similar to that of the ftnA mutant, we analyzed the global production of pectate lyases in this mutant grown in Tris medium with polygalacturonic acid as a carbon source. This in vitro condition mimics the plant environment, where low iron availability triggers pectate lyase production (18, 51). The level of pectate lyase production was 2.5 higher for the ftnA mutant than for the wild-type strain (data not shown). This higher production of pectinases could enable the ftnA mutant to multiply in planta despite its lower growth capacity and its increased sensitivity to oxidative stress. Indeed, destruction of plant cells by pectate lyases increases nutrient and iron availability and limits the activation of the plant cell NADPH oxidase (16).
FIG. 6.
Pathogenicity of the wild-type strain and its ferritin-negative mutants. (A) Symptoms caused by Erwinia chrysanthemi strain 3937 and its ferritin-deficient mutants on chicory leaves 36 h postinoculation. (B) Pathogenicities of the wild-type (WT) strain (solid line) and the ftnA mutant (dashed line) on African violets. The progression of systemic symptoms (i.e., fully macerated leaf and petiole) was scored for 9 days as indicated. Experiments were performed in triplicate, and standard deviations are shown.
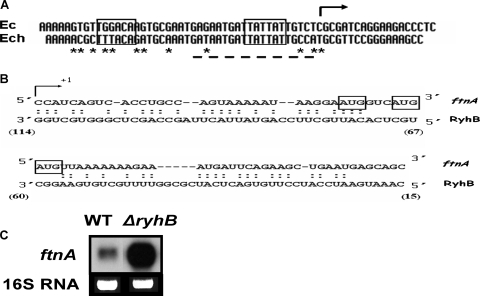
Regulation of the ftnA gene.
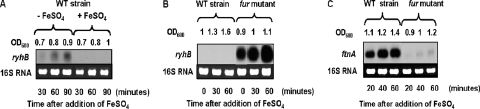
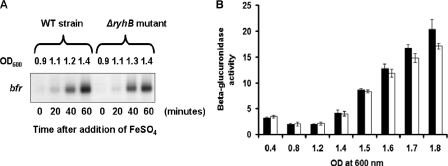
Since the ftnA gene is devoted to iron storage, we first examined its expression after the addition of iron. Cells harboring a transcriptional ftnA::uidA fusion were grown in L broth, and FeSO4 was added to a final concentration of 20 μM at an optical density of 0.5. Exogenous iron increased ftnA gene transcription fourfold (data not shown). In many enterobacteria, the iron control of the ftnA gene is mediated by the small antisense RNA RyhB (33). We identified a homologue of the E. coli ryhB gene in the E. chrysanthemi 3937 genome. The E. chrysanthemi ryhB gene (CDS accession number ABF-0061312) encodes a 120-nucleotide-long RNA, which may be untranslated. Potential −10 and −35 promoter elements were predicted by computational analysis. This promoter region is highly similar to that of the E. coli ryhB gene, with a putative Fur box matching the consensus sequence at 17 out of 19 positions which overlaps the −10 promoter element (Fig. 7A). Although the E. chrysanthemi RyhB RNA is longer than that of E. coli, the last 75 nucleotides are conserved in both RNAs (92% identity). We found sequence complementarity between the E. chrysanthemi RyhB RNA and the 5′ end of the ftnA gene that includes the 5′ leader sequence with the potential translational starts of the ftnA RNA (Fig. 7B). We constructed a ΔryhB mutant by reverse genetics (see Materials and Methods). Increased ftnA transcript levels were detected in total RNA isolated from the ΔryhB strain in comparison to RNAs isolated from the wild-type strain (Fig. 7C). RyhB RNA abundance was notably increased under conditions of iron starvation (Fig. 8A). Furthermore, the ryhB gene is highly expressed in a fur mutant regardless of the iron concentration (Fig. 8B), and this high level of transcription greatly decreased the amount of ftnA RNA, as shown in Fig. 8C. Thus, in E. chrysanthemi, the control of ftnA gene expression involves the transcriptional repressor Fur and a homolog of the small antisense RNA RyhB.
FIG. 7.
Occurrence of a ryhB gene in E. chrysanthemi. (A) Sequence alignment of the promoters of the ryhB genes of E. coli (Ec) and E. chrysanthemi 3937 (Ech). The −35 and −10 promoter elements are boxed, and the potential Fur box is underlined. The transcriptional start identified in E. coli is indicated by an arrow. Nonconserved nucleotides are indicated by asterisks. (B) Sequence complementarity between the E. chrysanthemi RyhB RNA and the 5′ part of the ftnA transcript. The identified transcriptional start of the ftnA gene is indicated by an arrow, and the potential translational starts are boxed. Numbers indicate nucleotide positions in RyhB RNA. (C) Northern blot analysis of E. chrysanthemi ftnA messenger accumulation in the wild-type (WT) strain and the ΔryhB mutant. Total RNAs were extracted from cells at the late exponential growth phase, and 3 μg of RNA was blotted as described in Materials and Methods. Northern blot analysis was performed as described in Materials and Methods.
FIG. 8.
Northern blot analyses of ryhB and ftnA gene expression in E. chrysanthemi. (A) The wild-type (WT) strain was grown in Tris medium until an OD600 of 0.7 was reached, and 20 μM FeSO4 was added. Samples were collected every 30 min. (B) RyhB RNA accumulation in the wild-type strain of E. chrysanthemi and its Fur− derivative. Cells were grown in L broth until an OD600 of 1 was reached, and 20 μM FeSO4 was added. Samples were collected every 30 min. (C) ftnA messenger accumulation in the wild-type strain and the fur mutant. Cells were grown in L broth until an OD600 of 1 was reached, and 20 μM FeSO4 was added. Samples were collected every 20 min. Northern experiments were performed as described in Materials and Methods.
Regulation of the bfd-bfr operon.
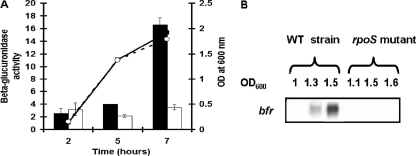
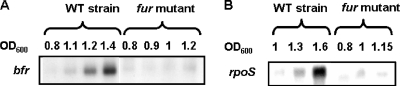
Since RyhB expression reduces bfr transcript abundance in E. coli (34), we first investigated whether the small RNA RyhB could pair to the E. chrysanthemi bfd and bfr genes. No sequence complementarity was detected between the polycistronic bfd-bfr transcript and RyhB RNA. Northern blot experiments corroborated this computational analysis by showing that the level of transcription of the bfd-bfr operon in the wild-type strain was identical to that in the ΔryhB mutant (Fig. 9A). Interestingly, in L broth, the expression of the bfr::uidA transcriptional fusion seemed to be dependent on cellular density since the highest expression occurred at the beginning of the stationary phase (Fig. 9B). Thus, we checked whether quorum sensing was implicated in this control. Transcription of the bfd-bfr operon in the wild-type strain was identical to that in the expR or canR mutant, which are altered in quorum-sensing regulation (data not shown). Since the σS transcription factor was shown to be involved in the regulation of the bfr gene in E. coli (26), we checked this possibility in E. chrysanthemi. We thus transduced the bfr::uidA fusion into an rpoS mutant of E. chrysanthemi 3937. In an rpoS genotype, the transcription of the bfd-bfr operon was reduced to a basal level, even at a high cellular density (Fig. 10A). Indeed, very low levels of bfr RNA were detected by Northern blot hybridization in total RNAs isolated from the E. chrysanthemi rpoS mutant (Fig. 10B). McHugh et al. (38) previously demonstrated that the Fur transcriptional repressor was involved in the regulation of the bfd-bfr operon in E. coli. We also examined the role of the Fur repressor in the expression of the bfd-bfr operon. In a fur genotype, there was only a very low level of transcription of the bfr gene (Fig. 11A). Since the E. chrysanthemi fur mutant is altered in its growth capacity and displays a lower final cellular density than that of the wild-type strain (18), we checked the levels of the rpoS gene transcripts in this mutant. As shown in Fig. 11B, there was a lower level of accumulation of rpoS transcripts in RNAs isolated from the fur mutant. In this mutant, the pattern of rpoS RNA accumulation correlated with the observed level of transcription of the bfr gene. Thus, in a fur mutant, the reduced level of expression of the bfr gene is likely caused by a decreased accumulation of the rpoS RNA.
FIG. 9.
Analysis of bfr gene expression in E. chrysanthemi. (A) bfr messenger accumulation in the wild-type (WT) strain and the ΔryhB mutant. Cells were grown in L broth until an OD600 of 0.9 was reached, and 20 μM FeSO4 was added. Samples were collected every 20 min, and Northern experiments were carried out as described in Materials and Methods. (B) Expression of a transcriptional bfr::uidA fusion during bacterial growth. Cells were grown in L medium. FeSO4 (20 μM) was added at an OD600 of 0.4. Samples were collected after the addition of iron as indicated. β-Glucuronidase activity was determined as described in Materials and Methods. White bars, with iron; black bars, no iron. Experiments were performed in triplicate, and standard deviations are shown.
FIG. 10.
Effect of an rpoS mutation on the expression of the E. chrysanthemi bfr gene. (A) Expression of a transcriptional bfr::uidA fusion in the wild-type strain and the rpoS mutant. Bacterial growth in L broth was assessed by measuring the OD600 (filled squares, bfr::uidA mutant; open circles, bfr::uidA rpoS mutant). β-Glucuronidase activity was determined as described in Materials and Methods. Black bars, bfr::uidA mutant; white bars, bfr::uidA rpoS mutant. Experiments were performed in triplicate, and standard deviations are shown. (B) bfd-bfr messenger accumulation in the wild-type (WT) strain and the rpoS mutant. Cells were grown in L medium, and samples were collected during growth. Northern blot analysis was performed as described in Materials and Methods.
FIG. 11.
Effect of a fur mutation on the expression of the bfr and rpoS genes of E. chrysanthemi. (A) bfr messenger accumulation in the wild-type (WT) strain and the fur mutant. (B) Expression of the rpoS gene in the wild-type strain and the fur mutant. Bacteria were grown in L medium and collected during growth as indicated by the OD600. bfr and rpoS transcripts were detected by Northern blot analysis as described in Materials and Methods.
DISCUSSION
In this work, we investigated the physiological roles of the E. chrysanthemi ferritin protein FtnA and bacterioferritin protein Bfr in iron storage. We show that the Bfr and FtnA proteins participate in different steps of iron storage in E. chrysanthemi. Relevantly, the different role of these proteins appears to be related to a differential regulation of expression of their genes.
In contrast to E. coli and Salmonella enterica serovar Typhimurium, strain 3937 of E. chrysanthemi harbors only two maxiferritins, since the ftnB locus is missing in its genome. The FtnB protein is a nonubiquitous ferritin-like protein that lacks the typical amino acids that form the ferroxidase center (1, 57). We constructed the ftnA, bfr, and bfr ftnA mutants by reverse genetics, and we characterized their phenotypes. No clear phenotype could be assigned to the bfr mutation. Indeed, this mutant behaved like the wild-type strain under conditions of iron deficiency or under oxidative stress conditions. Experiments measuring the total cellular iron content and Mössbauer analysis of the intracellular iron pool showed that Bfr plays no significant role in iron storage at stationary phase in E. chrysanthemi. This is in contrast to the role of the Bfr protein from Salmonella enterica serovar Typhimurium, which is an important iron reservoir and is implicated in resistance to H2O2 stress (57). Nevertheless, the E. chrysanthemi bfr mutant was less aggressive on chicory leaves, with a delay in the appearance of symptoms of maceration. Thus, the role of bacterioferritin in iron homeostasis in E. chrysanthemi remains to be elucidated.
As is the case in E. coli, the main iron storage protein in E. chrysanthemi 3937 is the ferritin FtnA (1). The absence of a functional FtnA leads to a pleiotropic phenotype. In comparison to the wild-type strain, growth of the ftnA mutant was impaired even in the presence of iron. This mutant is also more sensitive to iron deficiency than the wild-type strain. An increased sensitivity to compounds generating oxidative and nitrosative stress was also observed for the ftnA mutant. Interestingly, the ftnA mutant also grew less well under anaerobic conditions (data not shown). Thus, the reduced growth ability of this mutant not only is caused by the sensitivity to oxidative stress but is also a direct consequence of a lack of the FtnA-associated iron store. Increases in the ferrous iron/ferric iron ratio in the ftnA mutant, as determined by Mössbauer analysis, are probably the cause of its sensitivity to oxidative stress. Interestingly, inactivation by mutations of the high-affinity iron transport systems decreased the susceptibility of the ftnA mutant to oxidative stress (data not shown). Thus, by limiting the intracellular concentration of ferrous iron and reducing the cytotoxic effect of Fenton's chemistry, FtnA confers tolerance to oxygen metabolism in E. chrysanthemi. Surprisingly, the bfr ftnA double mutant is less susceptible to iron starvation than the ftnA mutant. In addition, the bfr ftnA mutant is sensitive only to oxidative stress caused by streptonigrin. However, its susceptibility is intermediate to those of the wild-type strain and the ftnA mutant. One explanation could be that there is a reduction in the total iron content of the double ferritin mutant as determined by the ICP-AES analysis. Although there is a higher Fe(II)/Fe(III) ratio, the decrease in the total iron content of this mutant could account for its intermediate phenotype. Analysis of Mössbauer spectra from the bfr ftnA mutant clearly demonstrated an Fe(III) signal that could be attributed to the ferric iron stored in Dps proteins. Indeed, we identified two genes encoding Dps proteins in the genome of E. chrysanthemi strain 3937. In the absence of maxiferritins, these proteins would limit the intracellular concentration of reactive iron by sequestering an excess of this metal. Another nonexclusive hypothesis might be that in an ftnA background, the absence of a functional Bfr results in changes in iron distribution in proteins or other components. The physiological consequences of the combination of the ftnA and bfr mutations on iron homeostasis remain to be determined.
The control of ftnA and bfr gene expression in E. chrysanthemi is similar to the regulation occurring in E. coli. Iron is a triggering signal for transcription to the ftnA gene. This control involves the Fur transcriptional repressor and a small RNA that is a functional homolog of the E. coli RyhB antisense RNA. E. chrysanthemi RNA is longer than that of E. coli (i.e., 120 versus 90 nucleotides). However, both RNAs show significant sequence identity in their last 80 nucleotides. This RNA possesses a long sequence complementarity with the 5′ end of the ftnA transcript, suggesting a base-pairing mechanism of regulation similar to that described previously for E. coli (33). Indeed, in a ΔryhB mutant, we observed an elevated level of the ftnA transcript. We also showed that the ryhB gene is negatively controlled by Fur. Thus, in a fur mutant, there was a very low level of the ftnA transcript in total RNA because of the overaccumulation of RyhB. No sequence complementarity between RyhB RNA and the bfd-bfr operon could be found. Our Northern experiments showed that the transcription of the bfd-bfr operon is not controlled by RyhB RNA. A high level of transcription of this operon was observed at the beginning of the stationary phase, and we found that quorum sensing is not involved in this regulatory process. The σS factor mediates this response, since transcription of the bfd-bfr operon was reduced in a rpoS mutant. This result is in agreement with the σS-regulated promoter that we identified for the bfd-bfr operon. Surprisingly, the expression of the bfd-bfr operon was also strongly decreased in a fur mutant. This regulatory response is probably indirect, since no potential Fur binding site was identified in the bfd-bfr operon sequence. However, we showed that there was a delay in the accumulation of the rpoS transcript in a fur mutant. This lower rpoS transcript level can account for the reduced expression of the bfd-bfr operon in the fur mutant. The mechanisms by which Fur modulates the transcription of rpoS or the stability of its RNA must be determined.
Mutations in the ftnA and bfr genes had different consequences on the virulence of E. chrysanthemi. All the ferritin-deficient mutants are less aggressive on chicory leaves, whereas only the ftnA mutant displayed a reduced virulence on African violets. These data are similar to those obtained for the E. chrysanthemi suf mutants, which are affected in the Suf machinery involved in the biogenesis/repair of the Fe-S centers under iron deficiency and oxidative conditions (41). Indeed, a sufA mutant lacking the [Fe-S] scaffold protein SufA is less virulent than the wild-type strain only on chicory leaves. A sufC mutant where the SufC ATPase component of the SufBCD complex is missing is altered in its pathogenicity to isolated organs and potted African violets. Our explanation may be that iron availability and oxidative stress conditions are different depending on the host tissues infected. Thus, under infection conditions where the environment is continually changing, a perfect control of iron homeostasis is important for E. chrysanthemi cells. In conclusion, although the E. chrysanthemi FtnA and Bfr proteins are highly similar to those of E. coli and S. enterica, their physiological roles seem to be different. When bacteria possess multiple maxiferritins, the respective functions of these proteins in the control of iron and dioxygen chemistry may depend on the iron metabolism machinery and the ecophysiology of the species.
Acknowledgments
We thank Jan Schorch for technical assistance. We are grateful to the reviewers for their suggestions to improve the manuscript.
This work was supported by the Institut National de la Recherche Agronomique (project SPE 0217-01) and the Procope program (grant 09636TF) from the Ministère des Affaires Etrangères. D. Expert is a researcher from the CNRS. A. Boughammoura was supported by a doctoral fellowship from the Ministère de l'Education Nationale, de l'Enseignement Supérieur, et de la Recherche.
Footnotes
Published ahead of print on 28 December 2007.
REFERENCES
- 1.Abdul-Tehrani, H., A. J. Hudson, Y. S. Chang, A. R. Timms, C. Hawkins, J. M. Williams, P. M. Harrison, J. R. Guest, and S. C. Andrews. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 1811415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C., N. E. Le Brun, V. Barynin, A. J. Thomson, G. R. Moore, J. R. Guest, and P. M. Harrison. 1995. Site-directed replacement of the coaxial heme ligands of bacterioferritin generates heme-free variants. J. Biol. Chem. 27023268-23274. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27215-237. [DOI] [PubMed] [Google Scholar]
- 4.Baaghil, S., A. Lewin, G. R. Moore, and N. E. Le Brun. 2003. Core formation in Escherichia coli bacterioferritin requires a functional ferroxidase center. Biochemistry 4214047-14056. [DOI] [PubMed] [Google Scholar]
- 5.Bardonnet, N., and C. Blanco. 1992. uidA-antibiotic resistance cassettes for insertion mutagenesis, gene fusions and genetic constructions. FEMS Microbiol. Lett. 93243-248. [DOI] [PubMed] [Google Scholar]
- 6.Böhnke, R., and B. F. Matzanke. 1995. The mobile ferrous iron pool in E. coli is bound to a phosphorylated sugar derivative. Biometals 8223-230. [DOI] [PubMed] [Google Scholar]
- 7.Bou-Abdallah, F., A. C. Lewin, N. E. Le Brun, G. R. Moore, and N. D. Chasteen. 2002. Iron detoxication properties of Escherichia coli bacterioferritin. Attenuation of oxyradical chemistry. J. Biol. Chem. 27737064-37069. [DOI] [PubMed] [Google Scholar]
- 8.Carrondo, M. A. 2003. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 221959-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceci, P., A. Ilari, E. Falvo, and E. Chiancone. 2003. The Dps protein of Agrobacterium tumefaciens does not bind to DNA but protects it toward oxidative cleavage: X-ray crystal structure, iron binding, and hydroxyl-radical scavenging properties. J. Biol. Chem. 27820319-20326. [DOI] [PubMed] [Google Scholar]
- 10.Chasteen, N. D., and P. M. Harrison. 1999. Mineralization in ferritin: an efficient means of iron storage. J. Struct. Biol. 126182-194. [DOI] [PubMed] [Google Scholar]
- 11.Chiancone, E., P. Ceci, A. Ilari, F. Ribacchi, and S. Stefanini. 2004. Iron and proteins for iron storage and detoxification. Biometals 17197-202. [DOI] [PubMed] [Google Scholar]
- 12.Dellagi, A., M. Rigault, D. Segond, C. Roux, Y. Kraepiel, F. Cellier, J. F. Briat, F. Gaymard, and D. Expert. 2005. Siderophore-mediated upregulation of Arabidopsis ferritin expression in response to Erwinia chrysanthemi infection. Plant J. 43262-272. [DOI] [PubMed] [Google Scholar]
- 13.El Hassouni, M., J. P. Chambost, D. Expert, F. Van Gijsegem, and F. Barras. 1999. The minimal gene set member msrA, encoding peptide methionine sulfoxide reductase, is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc. Natl. Acad. Sci. USA 96887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enard, C., A. Diolez, and D. Expert. 1988. Systemic virulence of Erwinia chrysanthemi 3937 requires a functional iron assimilation system. J. Bacteriol. 1702419-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Expert, D. 1999. Withholding and exchanging iron: interactions between Erwinia ssp. and their plant hosts. Annu. Rev. Phytopathol. 37307-334. [DOI] [PubMed] [Google Scholar]
- 16.Fagard, M., A. Dellagi, C. Roux, C. Périno, M. Rigault, V. Boucher, V. Shevchik, and D. Expert. 2007. Arabidopsis thaliana expresses multiple lines of defense to counter-attack Erwinia chrysanthemi. Mol. Plant-Microbe Interact. 20794-805. [DOI] [PubMed] [Google Scholar]
- 17.Fellay, R., J. Frey, and H. M. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52147-154. [DOI] [PubMed] [Google Scholar]
- 18.Franza, T., C. Sauvage, and D. Expert. 1999. Iron regulation and pathogenicity in Erwinia chrysanthemi strain 3937: role of the Fur repressor protein. Mol. Plant-Microbe Interact. 12119-129. [DOI] [PubMed] [Google Scholar]
- 19.Franza, T., B. Mahé, and D. Expert. 2005. Erwinia chrysanthemi requires a second iron transport route dependent of the siderophore achromobactin for extracellular growth and plant infection. Mol. Microbiol. 55261-275. [DOI] [PubMed] [Google Scholar]
- 20.Garg, R. P., C. J. Vargo, X. Cui, and D. M. Kurtz, Jr. 1996. A [2Fe-2S] protein encoded by an open reading frame upstream of the Escherichia coli bacterioferritin gene. Biochemistry 356297-6301. [DOI] [PubMed] [Google Scholar]
- 21.Hugouvieux-Cotte-Pattat, N., and J. Robert-Baudouy. 1985. Lactose metabolism in Erwinia chrysanthemi. J. Bacteriol. 162248-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilari, A., S. Stefanini, E. Chiancone, and D. Tsernoglou. 2000. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat. Struct. Biol. 738-43. [DOI] [PubMed] [Google Scholar]
- 23.Imlay, J. A. 2006. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 591073-1082. [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa, T., Y. Mizunoe, S. Kawabata, A. Takade, M. Harada, S. N. Wai, and S. Yoshida. 2003. The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J. Bacteriol. 1851010-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kauko, A., A. T. Pulliainen, S. Haataja, W. Meyer-Klaucke, J. Finne, and A. C. Papageorgiou. 2006. Iron incorporation in Streptococcus suis Dps-like peroxide resistance protein Dpr requires mobility in the ferroxidase center and leads to the formation of a ferrihydrite-like core. J. Mol. Biol. 36497-109. [DOI] [PubMed] [Google Scholar]
- 26.Lacour, S., and P. Landini. 2004. σS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of σS-dependent genes and identification of their promoter sequences. J. Bacteriol. 1867186-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Brun, N. E., S. C. Andrews, J. R. Guest, P. M. Harrison, G. R. Moore, and A. J. Thomson. 1995. Identification of the ferroxidase centre of Escherichia coli bacterioferritin. Biochem. J. 312385-392. [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, X., and E. C. Theil. 2004. Ferritin reactions: direct identification of the site for the diferric peroxide reaction intermediate. Proc. Natl. Acad. Sci. USA 1018557-8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, X., K. Kim, T. Leighton, and E. C. Theil. 2006. Paired Bacillus anthracis Dps (mini-ferritin) have different reactivities with peroxide. J. Biol. Chem. 28127827-27835. [DOI] [PubMed] [Google Scholar]
- 30.Luo, Y., Z. Han, S. M. Chin, and S. Linn. 1994. Three chemically distinct types of oxidants formed by iron-mediated Fenton reactions in the presence of DNA. Proc. Natl. Acad. Sci. USA 9112438-12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, J. F., U. A. Ochsner, M. G. Klotz, V. K. Nanayakkara, M. L. Howell, Z. Johnson, J. E. Posey, M. L. Vasil, J. J. Monaco, and D. J. Hasset. 1999. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J. Bacteriol. 1813730-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann, S., J. M. Williams, A. Treffry, and P. M. Harrison. 1987. Reconstituted and native iron-cores of bacterioferritin and ferritin. J. Mol. Biol. 198405-416. [DOI] [PubMed] [Google Scholar]
- 33.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 994620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massé, E., C. K. Vanderpool, and S. Gottesman. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 1876962-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matzanke, B. F. 1987. Mössbauer spectroscopy of microbial iron uptake and metabolism, p 251-284. In G. Winkelmann, D. van der Helm, and J. B. Neilands (ed.), Iron transport in microbes, plants and animals. Verlag Chemie, Weinheim, Germany.
- 36.Matzanke, B. F. 1991. Structures, coordination chemistry and functions of microbial iron chelates, p 15-60. In G. Winkelmann (ed.), Handbook of microbial iron chelates (siderophores). CRC Press, Boca Raton, FL.
- 37.Matzanke, B. F. 1997. Iron storage in microorganisms, p. 117-158. In G. Winkelmann, and C. J. Carrano, (ed.), Transition metals in microbial metabolism. Harwood Academic Publishers GmbH, Amsterdam, The Netherlands.
- 38.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 27829478-29486. [DOI] [PubMed] [Google Scholar]
- 39.Münzinger, M., H. Budzikiewicz, D. Expert, C. Enard, and J. M. Meyer. 2000. Achromobactin, a new citrate siderophore of Erwinia chrysanthemi. Z. Naturforsch. C 55328-332. [DOI] [PubMed] [Google Scholar]
- 40.Murdoch, L., J. C. Corbel, D. Reis, Y. Bertheau, and B. Vian. 1999. Differential cell wall degradation by Erwinia chrysanthemi in petiole of Saintpaulia ionantha. Protoplasma 21059-74. [Google Scholar]
- 41.Nachin, L., M. El Hassouni, L. Loiseau, D. Expert, and F. Barras. 2001. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol. Microbiol. 39960-972. [DOI] [PubMed] [Google Scholar]
- 42.Nachin, L., L. Loiseau, D. Expert, and F. Barras. 2003. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pérombelon, M. C. M. 2002. Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol. 511-12. [Google Scholar]
- 44.Persmark, M., D. Expert, and J. B. Neilands. 1989. Isolation, characterization and synthesis of chrysobactin, a compound with a siderophore activity from Erwinia chrysanthemi. J. Biol. Chem. 2643187-3193. [PubMed] [Google Scholar]
- 45.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29303-313. [DOI] [PubMed] [Google Scholar]
- 46.Rauscher, L., D. Expert, B. F. Matzanke, and A. X. Trautwein. 2002. Chrysobactin-dependent iron acquisition in Erwinia chrysanthemi: functional study of an homologue of the Escherichia coli ferric enterobactin esterase. J. Biol. Chem. 2772385-2395. [DOI] [PubMed] [Google Scholar]
- 47.Résibois, A., M. Colet, M. Faelen, E. Schoonejans, and A. Toussaint. 1984. Phi-EC2 a new generalized transducing phage of Erwinia chrysanthemi. Virology 137102-112. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Samson, R., J. B. Legendre, R. Christen, M. Fischer-Le Saux, W. Achouak, and L. Gardan. 2005. Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953, Brenner et al. 1973) and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov., and Dickeya zeae sp. nov. Int. J. Syst. Evol. Microbiol. 551415-1427. [DOI] [PubMed] [Google Scholar]
- 50.Santos, R., T. Franza, M. L. Laporte, C. Sauvage, D. Touati, and D. Expert. 2001. Essential role of superoxide dismutase on the pathogenicity of Erwinia chrysanthemi strain 3937. Mol. Plant-Microbe Interact. 14758-767. [DOI] [PubMed] [Google Scholar]
- 51.Sauvage, C., and D. Expert. 1994. Differential regulation by iron of Erwinia chrysanthemi pectate lyases: pathogenicity of iron transport regulatory cbr mutants. Mol. Plant-Microbe Interact. 771-77. [Google Scholar]
- 52.Smith, J. L. 2004. The physiological role of ferritin-like compounds in bacteria. Crit. Rev. Microbiol. 30173-185. [DOI] [PubMed] [Google Scholar]
- 53.Stillman, T. J., P. P. Connolly, C. L. Latimer, A. F. Morland, M. A. Quail, S. C. Andrews, A. J. Hudson, A. Treffry, J. R. Guest, and P. M. Harrison. 2003. Insights into the effects on metal binding of the systematic substitution of five key glutamate ligands in the ferritin of Escherichia coli. J. Biol. Chem. 27826275-26286. [DOI] [PubMed] [Google Scholar]
- 54.Theil, E. C., M. Matzapetakis, and X. Liu. 2006. Ferritins: iron/oxygen biominerals in protein nanocages. J. Biol. Inorg. Chem. 11803-810. [DOI] [PubMed] [Google Scholar]
- 55.Toth, I. K., K. S. Bell, M. C. Holeva, and P. R. J. Birch. 2003. Soft rot erwiniae: from genes to genomes. Mol. Plant Pathol. 417-30. [DOI] [PubMed] [Google Scholar]
- 56.Ueshima, J., M. Shoji, D. B. Ratnayake, K. Abe, S. Yoshida, K. Yamamoto, and K. Nakayama. 2003. Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect. Immun. 711170-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Velayudhan, J., M. Castor, A. Richardson, K. L. Main-Hester, and F. C. Fang. 2007. The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Mol. Microbiol. 631495-1507. [DOI] [PubMed] [Google Scholar]
- 58.Wang, G., Y. Hong, A. Olczak, S. E. Maier, and R. J. Maier. 2006. Dual roles of Helicobacter pylori NapA in inducing and combating oxidative stress. Infect. Immun. 746839-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamamoto, Y., M. Higuchi, L. B. Poole, and Y. Kamio. 2000. Role of the dpr product in oxygen tolerance in Streptococcus mutans. J. Bacteriol. 823740-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao, G., P. Ceci, A. Ilari, L. Giangiacomo, T. M. Laue, E. Chiancone, and N. D. Chasteen. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 27727689-27696. [DOI] [PubMed] [Google Scholar]