Abstract
The cip-cel cluster of genes plays an important role in the catabolism of the substrate cellulose by Clostridium cellulolyticum. It encodes several key components of the cellulosomes, including the scaffolding protein CipC and the major cellulase Cel48F. All the genes of this cluster display linked transcription, focusing attention on the promoter upstream from the first gene, cipC. We analyzed the regulation of the cipC promoter using a transcriptional fusion approach. A single promoter is located between nucleotides −671 and −643 with respect to the ATG start codon, and the large mRNA leader sequence is processed at position −194. A catabolite-responsive element (CRE) 414 nucleotides downstream from the transcriptional start site has been shown to be involved in regulating this operon by a carbon catabolite repression mechanism. This CRE is thought to bind a CcpA-like regulator complexed with a P-Ser-Crh-like protein. Sequences surrounding the promoter sequence may also be involved in direct (sequence-dependent DNA curvature) or indirect (unknown regulator binding) regulation.
Clostridium cellulolyticum, an anaerobic mesophilic bacterium, produces numerous enzymes that degrade plant cell wall polysaccharides. Most of the known enzymes are assembled into high-molecular-mass enzymatic complexes termed cellulosomes. These extracellular complexes efficiently degrade cellulose, the major polymer in plant cell walls (1, 36). Each complex contains up to eight enzymatic units bound to one scaffoldin (CipC). However, at least 30 different proteins are present in the complexes produced by cells cultured in cellulose-containing medium (2), indicating that there is considerable heterogeneity in cellulosome composition. The genes encoding the three major components (CipC, Cel48F, and Cel9E) are located in a large cluster spanning 26 kb (cip-cel cluster) (7, 31). cipC is the first gene of this cluster; it is followed by 11 genes (cel48F, cel8C, cel9G, cel9E, orfX, cel9H, cel9J, man5K, cel9M, rgl11Y, and cel5N), most of which encode cellulases (Cel proteins). Several genes encoding cellulosomal enzymes are located outside the cip-cel cluster (cel5A, cel5D, cel44O, man26A, cel9P, gal27A, gal59A, xyn10A, and cel9Q) (2, 36). Nevertheless, the cellulases encoded by the cluster have been shown to be essential for the formation of cellulosome complexes that efficiently degrade crystalline cellulose. Indeed, both the insertional mutant strain cipCMut1 and the cipC trans-complemented strain cipCMut1(pSOScipC), neither of which produces any of the enzymes encoded by the cip-cel cluster, have severely impaired cellulolytic activity (23). All the genes of the cip-cel cluster are transcribed together, as shown by the strong polar effect of the insertional cipC mutation, reverse transcription-PCR, and Northern blotting (22). Two large transcripts are detected in cells cultured in the presence of cellulose: a 14-kb mRNA carrying the cipC, cel48F, cel8C, cel9G, and cel9E coding sequences and a less abundant 12-kb mRNA carrying the coding sequences of the genes located in the 3′ part of the cluster. Several smaller transcripts have also been identified, which are assumed to be generated by posttranscriptional processing of a very large primary transcript corresponding to the entire cluster. The processing of this transcript is thought to lead to the production of several secondary transcripts with different stabilities. The differences in stability fine-tune the expression of individual genes.
A carbon catabolite repression (CCR) mechanism is thought to regulate the cellulolytic system of C. cellulovorans. This hypothesis is based on the relative abundance of transcripts, including several cellulase and hemicellulase genes, in various culture conditions (11). In Clostridium thermocellum, the expression of various cellulase genes and of the scaffoldin gene cipA increases with a decreasing growth rate (4-6). Zhang and Lynd (39) showed that the cellulase yields in continuous cultures were higher on Avicel cellulose than on cellobiose. These authors suggested that the cells may sense insoluble cellulose, triggering the synthesis of an intracellular regulatory molecule involved in cellulase gene induction. They also hypothesized that a CCR mechanism involving cellobiose might contribute to the regulation of cellulase genes (39).
Bacteria use several transcriptional and posttranscriptional mechanisms to regulate the differential production of proteins from polycistronic mRNAs. Primer extension analysis has identified two putative transcriptional start sites located 638 or 637 and 194 nucleotides upstream from the cipC translational start site, consistent with the presence of two promoters (22). We investigated the initiation of transcription of the cip-cel operon and the regulation of this process using fragments of the cipC promoter region and a catP gene fusion system in cells grown in cellulose- or cellobiose-containing media. We found only one functional promoter located upstream from the cipC open reading frame (ORF) and showed that a catabolite-responsive element (CRE) was involved in a CCR-based regulatory mechanism.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5α was used as the host strain for routine cloning. Luria-Bertani broth and agar were used for E. coli cultures. C. cellulolyticum strains were grown anaerobically at 32°C in basal medium or minimal medium supplemented with cellobiose (2 g liter−1; Sigma-Aldrich, Saint-Quentin Fallavier, France), MN300 cellulose (5 g liter−1; Serva, Heidelberg, Germany), or MN300 cellulose (5 g liter−1) plus one soluble sugar (cellobiose, glucose, arabinose, or xylose) at a concentration of 4 g liter−1. The minimal medium used was a modified form of the basal medium (9) in which 5 g liter−1 yeast extract was replaced by oligoelements and vitamin solutions (10). Colonies of C. cellulolyticum were isolated on solid medium (basal medium supplemented with 8 g liter−1 agar). pGEM-T Easy (Promega France, Charbonnières, France) was used as the PCR cloning vector in E. coli. Shuttle vector pPSV (24), which contains a promoterless catP gene, was used for the detection and quantification of promoter activity in C. cellulolyticum. Competent cells of C. cellulolyticum were prepared and electrotransformed as previously described (14, 37). The concentrations of antibiotics used for selection were as follows: ampicillin, 100 μg ml−1 for E. coli; and erythromycin, 300 μg ml−1 for E. coli and 10 μg ml−1 for C. cellulolyticum.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | F−endA1 hsdR17(rK− mK+) supE44 thi-1 λ−gyrA96 relA1 Δ(lacZYA argF)U169 (Φ80 lacZΔM15) recA1 | Roche |
| Clostridium cellulolyticum ATCC 35319 | Wild type | 30 |
| Plasmids | ||
| pPSV | pJIR418 derivative carrying the promoterless CAT gene (catP); Emr | 24 |
| pGEM-T Easy | Cloning vector; Apr | Promega |
| pSOS952 | E. coli-Clostridium shuttle vector (ColE1 and pIM13 replicons); Apr Emr; carries the adc-ctfA-ctfB operon transcribed from a modified thl promoter (two lac operators) | 29 |
| pPSVthl | pPSVΩ (SmaI Sos-dir/SosE 259-bp PCR fragment carrying thiolase promoter) | This study |
| pPSV1 | pPSVΩ (SphI/SacI 1,030-bp fragment of cipC promoter cloned in pGEM-T Easy) | This study |
| pPSV1ΔCRE | pPSVΩ (SphI/SacI 1,016-bp fragment of cipC ΔCRE promoter cloned in pGEM-T Easy) | This study |
| pPSV2 | pPSVΩ (SphI/SacI 155-bp fragment of cipC promoter cloned in pGEM-T Easy) | This study |
| pPSV3 | pPSVΩ (SphI/SacI 701-bp fragment of cipC promoter cloned in pGEM-T Easy) | This study |
| pPSV4 | pPSVΩ (SphI/SacI 574-bp fragment of cipC promoter cloned in pGEM-T Easy) | This study |
| pPSV5 | pPSVΩ (SphI/SacI 138-bp fragment of cipC promoter cloned in pGEM-T Easy) | This study |
| pPSV6 | pPSVΩ (SphI/SacI 250-bp fragment of cipC promoter cloned in pGEM-T Easy) | This study |
| pPSV7 | pPSVΩ (SphI/SacI 428-bp fragment of cipC promoter cloned in pGEM-T Easy) | This study |
| pPSV8 | pPSVΩ (SphI/SacI 452-bp fragment of cipC promoter cloned in pGEM-T Easy) | This study |
| pPSV9 | pPSVΩ (SphI/SacI 540-bp fragment of cipC promoter cloned in pGEM-T Easy) | This study |
DNA techniques.
Standard recombinant DNA techniques were used, as described elsewhere (22). DNA was amplified by PCR using synthetic oligonucleotide primers and Expand high-fidelity polymerase (Roche). The primers used for PCR amplification are listed in Table 2. The PCR products were purified with a Nucleospin extract purification kit (Macherey Nagel, Düren, Germany) and were inserted into the pGEM-T Easy vector (Promega) for sequence analysis. DNA was sequenced by Genome Express (Grenoble, France).
TABLE 2.
Primers
| Primer | Sequence (5′ to 3′)a | Position relative to cipC start codon or gene amplified |
|---|---|---|
| P1dir | GTCAAGAAGTAATTACAAGTCC | −967 |
| P1rev | TTTACATGAAAACCATTTGG | −22 |
| P2dir | TATTTTGAAAAATTTTATAACAGTAG | −243 |
| P2rev | ATTAGCTAAGTGGTTTTTCATAAG | −172 |
| P3dir | TTGAGGCATTAATAGTTATA | −638 |
| P4dir | AACATATTCATTTCAAATATCTGTAAG | −511 |
| P5dir | TTATTAGCGGCCGCGGGAATTCGATTAAGTACTTGTCATTTAAT | −677 |
| P5rev | TTACATGCGGCCGCGAATTCACTAGTGATTCTATTAATGCCTCAAA | −624 |
| P6dir | AAGTACTTGTCATTTAAT | −677 |
| P6rev | TTAAGCCGTTTACCTCAGGTGC | −512 |
| P7rev | CTATTAATGCCTCAAA | −624 |
| P8rev | TTTTTTTCATATAATTTCATATAAC | −600 |
| P1cre-dir | TAAGTTTAGCTTTGTTTTGAAAAATTTTATAACAGTAG | −269 |
| P1cre-rev | AAAATTTTTCAAAACAAAGCTAAACTTATCACATTTG | −228 |
| Sos-dir | ACTATTGGTTGGAATGGCGTG | thl promoter |
| SosE | TAAATTCTGGATCCTACGGGGTAACAGA | thl promoter |
| Qermdir | GCCATGCGTCTGACATCTAT | erm probe synthesis |
| Qermrev | AGACTTGAGTGTGCAAGAG | erm probe synthesis |
| Qcel48Fdir | TTGGTGGACAGTACGGATTC | cel48F probe synthesis |
| Qcel48Frev | CAGCCCAGTAAGTTGCTTGA | cel48F probe synthesis |
| QorfXdir | CTGCATCTGCTTCGGTATC | orfX probe synthesis |
| QorfXrev | CCTGTAGCTCCGCTTACTT | orfX probe synthesis |
Bold type indicates 5′ extensions.
Construction of cipC-catP transcriptional fusions and transformation of C. cellulolyticum.
Various regions of the cipC promoter region were amplified by PCR from C. cellulolyticum genomic DNA. Primer pairs P1dir/P1rev, P2dir/P2rev, P3dir/P1rev, P4dir/P1rev, P5dir/P5rev, P6dir/P6rev, P1dir/P7rev, P1dir/P8rev, and P1dir/P6rev (Table 2) were used to amplify cipC promoter regions P1, P2, P3, P4, P5, P6, P7, P8, and P9, respectively. Most of the PCR fragments were inserted directly into the pGEM-T Easy vector; the only exception was the P5 PCR fragment, which was first digested with the restriction enzyme NotI (Promega) and then inserted between the two NotI sites of the pGEM-T Easy vector. The oligonucleotides restored the pGEM-T Easy polylinker sequence. The P1Δcre promoter region was obtained by splice overlap extension-PCR. Two separate PCR fragments were amplified with primer pairs P1dir/P1cre-rev and P1cre-dir/P1rev. The two purified PCR fragments were then mixed, and a third PCR was performed using primers P1dir and P1rev. The P1 sequence with a 14-bp deletion was then ligated, like the other fragments, into pGEM-T Easy. The pPSV1, pPSV1ΔCRE, pPSV2, pPSV3, pPSV4, pPSV5, pPSV6, pPSV7 pPSV8, and pPSV9 vectors containing putative transcriptional fusions were constructed by ligating the SphI-SacI fragments from recombinant pGEM-T Easy vectors with the Clostridium perfringens promoter probe shuttle vector pPSV digested with the same enzymes. The pPSV1 to pPSV9 constructs contained the same 49-bp pGEM-T Easy cloning site/SacI polylinker sequence inserted between the cloned promoter PCR fragment and the SacI pPSV cloning site located 30 bp upstream from the catP Shine-Dalgarno sequence. Similarly, pPSVthl was constructed by inserting a 259-bp Clostridium acetobutylicum thiolase promoter-containing PCR product synthesized from pSOS952 with oligonucleotides Sos-dir and SosE (Table 2) into the SmaI site of pPSV. The pPSV derivatives were transferred to C. cellulolyticum by electrotransformation. Transformants were isolated on selective solid medium containing 10 μg ml−1 erythromycin.
Growth measurements.
The growth of the bacteria was followed on cellobiose-supplemented medium by monitoring changes in the optical density at 600 nm over time. For bacteria cultured on cellulose (5 g liter−1), growth measurements were based on bacterial protein evaluation, as described by Giallo et al. (9).
CAT activity assays.
Chloramphenicol acetyltransferase (CAT) activity was assayed by culturing C. cellulolyticum in minimal medium with the indicated carbohydrates. Cells in mid-logarithmic growth phase (optical density at 600 nm, 0.3 to 0.4) were harvested by centrifugation at 7,000 × g and 4°C for 10 min. For cultures on cellulose, cells were first centrifuged at 30 × g for 5 min to separate the cells from the cellulose. The cells were washed once with 40 mM KH2PO4 (pH 6.8) buffer, resuspended in 5 ml of the same buffer, and disrupted with a French press at 1,200 t/m2, where T is tons force per square meter. The disrupted cells were then subjected to two successive centrifugations at 20,000 × g for 30 min each, and the supernatants were assayed for CAT activity as described by Shaw (32). The protein contents of cell extracts were determined by the method of Lowry et al. (20). CAT activity was expressed in nanomoles per minute per milligram of protein.
Determination of the pPSV copy number.
Total DNA was purified from two cultures of C. cellulolyticum ATCC 35319(pPSV1). Cells were cultured in minimal medium supplemented with cellobiose and were harvested in mid-exponential growth phase. Oligonucleotides complementary to the erm gene of pPSV1 and to the chromosomal cel48F and orfX genes were used to determine the plasmid/chromosome ratio by the real-time quantitative PCR (QPCR) technique (19). The LightCycler FastStart DNA MasterPLUS SYBR green I (Roche, Mannheim, Germany) QPCR mixture was used with oligonucleotide pairs (1 μM) Qermdir/Qermrev, Qcel48Fdir/Qcel48Frev, and QorfXdir/QorfXrev and with 10 or 40 ng of total DNA. QPCR was performed with a LightCycler 1 apparatus. Absolute quantification of the target genes was achieved by relating the cycle threshold to standard curves.
RESULTS
Use of catP gene reporter transcriptional fusions in C. cellulolyticum.
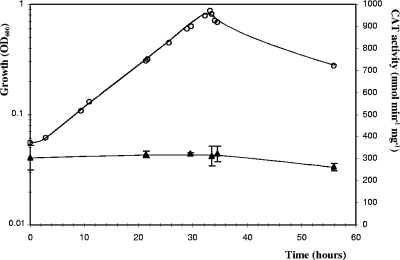
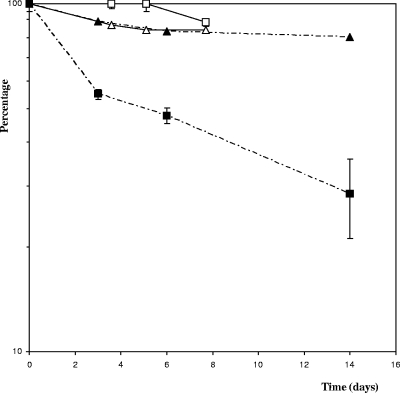
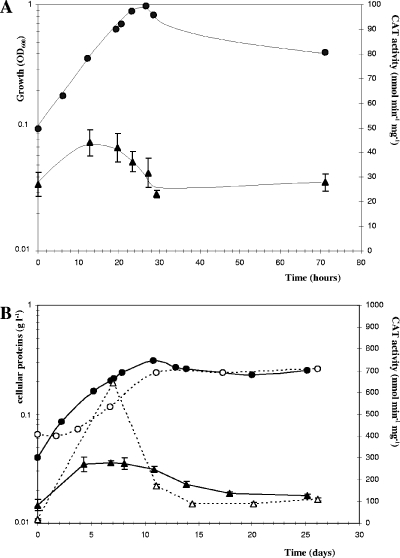
We tested a gene expression reporter system developed for C. perfringens as a possible means of studying promoter activities in C. cellulolyticum. The C. perfringens-E. coli pPSV shuttle vector (Table 1) harbors a promoterless C. perfringens CAT gene (catP) downstream from a multiple-cloning-site sequence (24). C. cellulolyticum lacks endogenous CAT activity (data not shown). This reporter gene is thus appropriate for evaluating promoter activity in cellulolytic Clostridium. This reporter system was tested and validated using the promoter of the thiolase gene of C. acetobutylicum (34) inserted into pPSV. This promoter is known to induce high levels of expression of the man5K gene, encoding mannanase, in C. cellulolyticum (28). The thl-catP transcriptional fusion was constructed by inserting a 259-bp fragment containing the thl promoter upstream from the catP gene. CAT activity was measured in strain ATCC 35319(pSPVthl) in basal medium with cellobiose as the substrate. An activity of 307 ± 23 nmol min−1 mg−1 (Fig. 1) was obtained during all growth phases, indicating that the thl promoter is not regulated in C. cellulolyticum. Furthermore, a very low level of CAT activity (<10 nmol min−1 mg−1) was detected in cell extracts of reference strain ATCC 35319(pPSV) containing the promoterless vector (see Fig. 5). Thus, the CAT reporter system is sensitive enough for evaluation of even weak promoters in C. cellulolyticum. The CAT protein has been reported to be very stable in C. perfringens (3). We estimated its stability in C. cellulolyticum by adding tetracycline (20 μg ml−1) to cultures of the ATCC 35319(pPSVthl) strain to stop translation. The cultures were then incubated at 32°C for several days. As expected, the total protein level ceased to increase following the addition of tetracycline to an exponentially growing culture in cellobiose-supplemented basal medium (Fig. 2). In these conditions, the CAT enzyme was found to be highly stable in vivo, with 88% of its activity remaining after 7.5 days of incubation. We studied CAT stability in stationary phase using cells from a culture grown on crystalline cellulose, as a lysis phase was observed in cells cultured on cellobiose (Fig. 1). The tetracycline-treated cells displayed a 52% decrease in the specific activity of the CAT after 6 days of incubation due to its degradation. CAT proteins therefore appear to be less stable during the stationary phase than during the exponential growth phase.
FIG. 1.
Expression of catP during the growth of C. cellulolyticum ATCC 35319(pPSVthl). The optical density at 600 nm (OD600) (○) was used to monitor the growth in basal medium supplemented with 2 g liter−1 cellobiose. CAT activities (▴) in soluble extracts were measured as described in Materials and Methods. The symbols indicate the means of three experiments, and the error bars indicate the standard deviations.
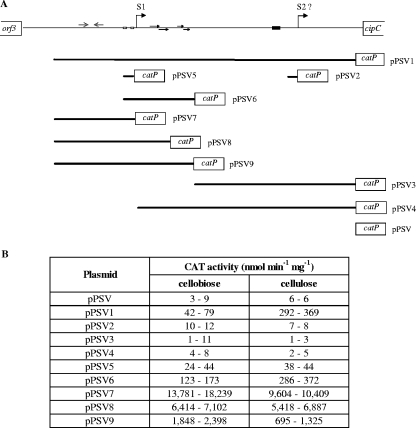
FIG. 5.
Analysis of transcription of the cip-cel operon based on deletions of the promoter region. (A) Various transcriptional fusions corresponding to different subregions of the cipC promoter were constructed by fusing promoter fragments (thick lines) to the SacI site located 30 bases upstream from the catP Shine-Dalgarno site. The putative transcriptional start sites, S1 and S2, are indicated by bent arrows, the −35 and −10 regions are indicated by open boxes, inverted and direct repeats are indicated by arrows, and the CRE element is indicated by a filled box. (B) CAT activity measured during the mid-exponential growth phase of C. cellulolyticum harboring the constructs. Cells were collected from 100-ml cultures containing 2 g liter−1 cellobiose (at optical densities at 600 nm between 0.35 and 0.5) or 5 g liter−1 cellulose (at cellular protein concentrations between 0.12 and 0.25 g liter−1). Values obtained in two independent experiments are shown.
FIG. 2.
Stability of CAT as a function of the growth phase. Exponential-phase cells (open symbols) and stationary-phase cells [ATCC 35319(pPSVthl)] (filled symbols) were incubated at 32°C in a culture medium supplemented with tetracycline (20 mg liter−1). CAT activity (squares) and total cell protein concentrations (triangles) were determined at specific time intervals. The symbols indicate the means of three experiments, and the error bars indicate the standard deviations.
Activity of the entire cipC promoter region.
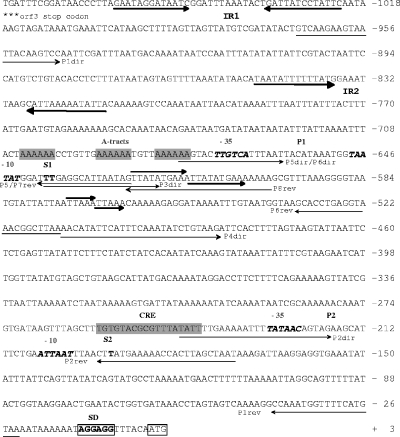
We evaluated cipC promoter activity during culture on cellobiose or cellulose by constructing a cipC promoter-catP fusion. A 1,076-base intergenic region was identified upstream from the cipC start codon (Fig. 3). A putative mRNA secondary structure was predicted using the Zuker algorithm (40) (ΔG = −20.4 kcal mol−1) in the region following the stop codon of the preceding ORF (orf3), suggesting the presence of a transcriptional terminator (IR1 in Fig. 3). We identified one σA-type promoter sequence (P1) in this intergenic region, which was preceded by three phased A tracts [poly(A) sequence repeated in phase with the DNA helical repeat and predicted to confer an intrinsic bend (15)], one inverted repeat upstream from P1 (from position −850 to position −813/ATG [IR2 in Fig. 3]), and two direct repeats located downstream from P1 (from position −623 to position −603 and from position −574 to position −562/ATG). The WWTGNAARCGNWWWCAWW (where W, R, and N indicate A or T, A or G, and any base, respectively) CRE consensus sequence from Bacillus subtilis (25) was used to search for putative regulatory sequences in the cipC promoter region. A sequence containing 15 of the 18 bases of the B. subtilis consensus sequence (TGTGTACGCGTTTATATT; italics indicate bases not corresponding to the consensus) was identified between positions −257 and −240 (Fig. 3). Primer extension analyses revealed two 5′ extremities for the cipC-carrying mRNA (22). The first 5′ end may correspond to a distal start of transcription (S1) immediately downstream from the putative P1 promoter; S1 is located 637 or 638 bases upstream from the ATG codon. The second 5′ end may correspond to a promoter-driven transcriptional start site (S2, position −194) or may be derived from a processing event. A cipC promoter-catP fusion was constructed by amplifying the 944-bp intergenic region between the orf3 transcriptional terminator and the cipC ribosome binding site using the P1dir and P1rev primers (Fig. 3) and inserting it into pPSV to construct the transcriptional fusion (pPSV1). Plasmid copy number may influence the results obtained, particularly due to possible titration effects of putative regulators. The copy number of pPSV1 in C. cellulolyticum was determined by the absolute QPCR method described by Lee et al. (19). The single-copy erm gene of pPSV and the single-copy cel48F or orfX gene of C. cellulolyticum chromosomal DNA were evaluated for the same total DNA samples. There was a mean of two copies of the plasmid per chromosome (data not shown). This very low copy number indicates that it is highly unlikely that there is a regulator titration effect. CAT activity was subsequently measured during growth in minimal medium with two different substrates: cellobiose, an easily assimilated disaccharide (Fig. 4A) and crystalline cellulose (Fig. 4B). With both substrates, the reporter activity was maximal in mid-exponential growth phase, remained steady until late exponential phase, and decreased in the late exponential to early stationary phase. This suggests that the promoter activity reaches a maximum in the second part of the exponential phase. Promoter activity may decrease in the late exponential growth phase, leading to a decrease in the specific activity of the CAT enzyme due to a dilution effect. CAT activity remained steady during the late stationary phase, although CAT proteins were shown to be quite unstable during this phase. Thus, the promoter was still active.
FIG. 3.
DNA sequence of the intergenic region between cipC and the preceding orf3 gene. Two putative transcriptional start sites, S1 and S2, are indicated by bold roman type. The −35 and −10 regions of the putative P1 and P2 promoters are indicated by bold italic type. Direct and inverted repeats (IR1 and IR2) are indicated by large arrows. The Shine-Dalgarno sequence (SD) is enclosed in a box. A tracts and the CRE element are indicated by shading. Oligonucleotides used for PCR amplification of parts of the cipC promoter region are indicated by small arrows. (For the sake of simplicity, 5′ extensions of primers [Table 2] are not shown.)
FIG. 4.
cipC promoter activity during the growth of C. cellulolyticum(pPSV1) on cellobiose (A) and cellulose (B). Transcriptional activity was monitored by measuring CAT activity for the P1-catP transcriptional fusion (▴) during growth in 1.3 liters of minimal medium supplemented with 2 g/liter cellobiose (A) or 5 g/liter MN300 cellulose (B). Cell growth (•) was monitored by measuring the optical density at 600 nm (OD600) with cellobiose and by determining the amount of cellular protein produced on cellulose. Medium containing cellobiose was inoculated with a culture of bacteria grown on cellobiose (A). Medium containing cellulose was inoculated with cultures grown on cellulose (filled symbols) or cellobiose (open symbols) (B). For CAT activities, the symbols indicate the means of three experiments, and the error bars indicate the standard deviations. Each culture was grown at least twice, and the growth kinetics of one representative culture are shown.
In cultures on cellulose, the CAT activity reached 272 nmol min−1 mg−1, a value sixfold higher than that observed in cultures on cellobiose (44 nmol min−1 mg−1). Moreover, inoculation of medium containing cellulose with bacteria cultured on cellobiose resulted in promoter activity (643 nmol min−1 mg−1) which was twice the activity obtained if the medium was inoculated with bacteria cultured on cellulose (Fig. 4B). Promoter activity was hypothesized to be stimulated during the acceleration phase. The decrease in CAT activity during the deceleration phase of growth (3.7-fold decrease in 4 days) may be explained by the combination of two phenomena: (i) CAT decay, which could be as great as 50% in 4 days, and (ii) the approximately 2.1-fold increase in biomass during this growth phase. The specific activity of the CAT enzyme therefore decreased due to a dilution effect.
Detailed characterization of the cipC promoter.
We broke down the 944-nucleotide sequence into several subregions to assess the efficiency of the S2 transcriptional start and to identify the sequences involved in regulating cipC promoter activity. These sequences, each of which included one of the presumed transcription promoters alone or the presumed transcription promoter plus adjacent DNA motifs suspected to play a role in the regulation, were used to generate transcriptional fusions with the catP reporter gene (Fig. 5A). CAT activity was determined for bacteria cultured on cellobiose- or cellulose-containing minimal medium and harvested at mid-exponential growth phase. With the entire promoter region (944-nucleotide sequence), the CAT activity was four- to ninefold higher in cultures on cellulose than in cultures on cellobiose (Fig. 5B). These data are consistent with the results of kinetic studies (Fig. 4). In strains containing pPSV2, pPSV3, and pPSV4, which carry the P2 putative promoter alone or the P2 putative promoter and the surrounding sequences, the CAT activity was found to be less than 12 nmol min−1 mg−1, which is within the range of values for the reference strain carrying the promoterless vector pPSV. Thus, the proximal region spanning from position −638 to the ATG codon and including the S2 region does not contain any functional promoter. The CAT activity of the strain containing the P1 promoter transcriptional fusion (pPSV5) was around 40 nmol min−1 mg−1 with both soluble and crystalline substrates. This activity suggests that the 53-base region located between positions −678 and −626/ATG contains the sequence responsible for the initiation of transcription solely at S1. This short unregulated promoter region contains only the −35 and −10 boxes. The presence of the downstream region carrying two direct repeats (pPSV6) in the transcriptional fusion increased promoter activity. The measured CAT activity was four times higher on cellobiose and eight times higher on cellulose than the activity with the minimal promoter in pPSV5. This suggests that the direct repeat region is involved in promoter activation on both cellobiose and cellulose. Addition of the upstream region which contained a putative inverted repeat (pPSV7) drastically increased the expression of the reporter gene. Very high levels of CAT activity were recorded (about 16,000 nmol min−1 mg−1 on cellobiose and 10,000 nmol min−1 mg−1 on cellulose). These much higher levels of CAT activity strongly suggest that the upstream region is also involved in the activation of the promoter. Nevertheless, addition of the first direct repeat (pPSV8) and addition of both direct repeats (pPSV9) located downstream from promoter P1 resulted in CAT activities that were 1.5- to 2- and 6- to 10-fold lower than the activity obtained with pPSV7, respectively.
Catabolic repression study.
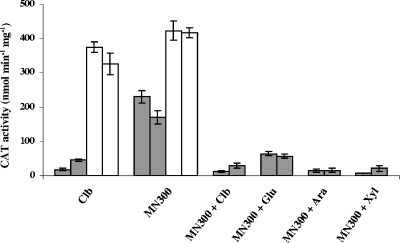
The CCR mechanism described for B. subtilis is known to operate in other low-G+C-content gram-positive bacteria (12, 18, 21, 26, 35). In such mechanisms, the use of one available carbon source may be prevented by the presence of a preferred substrate. Catabolite-repressing genes are negatively regulated by binding of the complex catabolite control protein (CcpA)-allosteric effector (P-Ser-HPr or P-Ser-Crh) with cis-acting CRE (8, 38). A putative CRE has been identified upstream of the cipC ORF (Fig. 3). We explored the possible negative regulation of the transcription of the cip-cel operon by a CCR mechanism by measuring the CAT activity driven by the pPSV1 transcriptional fusion in cells grown in cellobiose-containing minimal medium before and after transfer to medium containing cellulose alone and medium containing cellulose supplemented with cellobiose, glucose, arabinose, or xylose (Fig. 6). The expression levels quadrupled in medium containing only cellulose, whereas they remained low when the medium also contained a soluble sugar. These results strongly suggest that the operon is regulated by CCR. We therefore attempted to verify the role of the putative CRE site by comparing the activities of the P1-catP and P1ΔCRE-catP fusions in cellobiose and cellulose cultures. Deletion of the target sequence completely abolished the catabolite repression of catP (Fig. 6). These findings confirm that the CRE-like sequence located approximately 400 bases downstream from position 1 with respect to the transcriptional start site is involved in a CCR mechanism.
FIG. 6.
Regulation of the expression of the cip-cel operon by the carbon source and effect of the cis-acting CRE deletion on this regulation. Strains ATCC 35319(pPSV1) and ATCC 35319(pPSV1ΔCRE) grown in cellobiose (2 g liter−1)-containing minimal medium were used to inoculate various 100-ml cultures. Carbon sources were added (as indicated at the bottom) at final concentrations of 5 g liter−1 (MN300 cellulose) and 4 g liter−1 (cellobiose [Clb], glucose [Glu], arabinose [Ara], and xylose [Xyl]). The CAT activities determined by using cells collected in the mid-exponential growth phase reflect the expression driven by the P1 (filled bars) and P1ΔCRE (open bars) promoters. Experiments were performed twice. For CAT activities, the bars indicate the means of three experiments, and the error bars indicate the standard deviations.
DISCUSSION
We analyzed the contributions of various parts of the large cipC upstream region to the initiation and regulation of transcription by using the E. coli-Clostridium shuttle vector pPSV promoter probe developed by Matsushita et al. (24). The copy number of the reporter plasmid was shown to be low in C. cellulolyticum, ensuring a lack of bias that might affect the regulation study.
The orf3-cipC intergenic region (P1) was used to study promoter activity during growth. The activity was found to be maximal in mid-exponential growth phase on media containing cellobiose or cellulose. Cellulase activity reached a maximum at the end of the growth phase (9). This finding is consistent with the production and accumulation of a large amount of stable cellulosomes in the culture supernatant during the second part of the growth phase. In C. cellulovorans, Han et al. demonstrated the coordinate expression of several cellulosome genes (cbpA-exgS, engE, and xynA) whose mRNAs are most abundant in mid-exponential growth phase, regardless of the type of substrate (cellulose or cellobiose) (11).
Two primer extension endpoints were identified upstream from cipC in a previous study (22). Using reporter vectors carrying the regions considered (pPSV5 and pPSV2) (Fig. 5A), we showed that a single promoter located 643 or 671 bp upstream from the translation initiation codon is responsible for the initiation of transcription. The second extension endpoint clearly results from processing of the primary transcript. Such processing events have been reported previously for cip-cel transcripts (22). Larger quantities of the shortest transcript were also found to be present in primer extension experiments (data not shown), suggesting that the processing event may stabilize the 5′ end of the cipC-carrying transcripts.
Our results indicate that the operon is regulated by a CCR mechanism involving a CRE located 414 nucleotides downstream from the promoter. CCR is well documented in B. subtilis. It involves the CcpA transcriptional regulator, which is activated by interaction with the sugar phosphotransferase (PTS) transport HPr-Ser-phosphate protein or the Crh-Ser-phosphate non-PTS transport HPr-like protein. Little is known about CCR in cellulolytic clostridia, but analysis of the C. thermocellum genome identified putative CcpA, HPr-like protein, and HPr kinase/phosphatase orthologs. A BLASTP analysis of C. cellulolyticum partial genome data (NCBI accession no. GI 118665486) with the CcpA sequence of B. subtilis 168 identified a homologous protein in C. cellulolyticum (NCBI accession no. GI 118726636) displaying 32% identity (50% similarity) to CcpA (38). Furthermore, a Crh ortholog (accession no. GI 118663424; 36% identity and 65% similarity) and an HPr kinase ortholog (accession no. GI 118725184; 46% identity and 69% similarity) were also found. The B. subtilis Crh protein (catabolite repression HPr) displayed 45% sequence identity with HPr and contained the regulatory site serine (involved in CCR) but not the active-site histidine (involved in PTS transport). Consistent with these findings, Crh is inactive in the PTS but binds and activates CcpA when it is phosphorylated on serine 46 by the HPr kinase/phosphatase (38). The C. cellulolyticum Crh ortholog contains the serine residue at position 46, and, as in Crh of B. subtilis, no residue equivalent to His15 was found in the C. cellulolyticum Crh protein, suggesting that there is no involvement in any PTS transport system. This observation is consistent with the absence of a putative PTS enzyme EII-encoding gene in the C. cellulolyticum genome. It therefore seems highly likely that there is no transport system of this type in this Clostridium species. The CRE located 414 nucleotides downstream from the promoter should bind CcpA-like dimers complexed with its corepressor, Crh-Ser-phosphate (38). The complex would block either the initiation of transcription by interacting with the RNA polymerase, as described for the xyl operon of B. subtilis (17), or elongation (roadblock), as described for the ara operon of B. subtilis (13). A CCR mechanism has been evoked previously for the regulation of the cellulolytic system of the mesophilic species C. cellulovorans and the thermophilic species C. thermocellum. These hypotheses were based on quantification of transcripts for several components in various culture conditions (4-6, 11). Zhang and Lynd (39) reported higher cellulase yields in continuous cultures of C. thermocellum on cellulose than in continuous cultures of C. thermocellum on cellobiose. They suggested that a CCR mechanism involving cellobiose might be responsible for cellulase gene regulation, based on the strong negative correlation observed between cellobiose concentration and cellulase yield in continuous cultures. Nevertheless, unambiguous CRE sequences have yet to be identified in the promoter regions of cellulase genes.
We have shown that the sequences surrounding the cipC promoter also influence the promoter activity, but the underlying mechanisms remain unclear. Sequence-directed DNA curvature, also known as intrinsic DNA bending, occurs when special sequence motifs, such as A tracts, are repeated in phase with the DNA helical repeat. DNA curvature may play an active role in the formation of a transcriptionally competent complex (27). Three phased A tracts were identified immediately upstream from the cipC RNA polymerase binding site (Fig. 3). Similar sequences located upstream from the promoter of the phospholipase C gene of C. perfringens are assumed to facilitate the formation of the RNA polymerase-promoter complex by extending the contact region (16). The phased A tracts located upstream from the cipC promoter may stimulate promoter activity, as suggested by the high level of activity of the promoter regions present in the pPSV7, pPSV8, and pPSV9 constructs (Fig. 5). Further experiments are necessary to confirm this hypothesis. The possible role of regulatory proteins in this phenomenon also needs to be addressed. Indeed, A tracts and AT-rich sequences are known to be preferential binding sites for some DNA-binding proteins, such as the histone-like protein HU and integration host factor (33). The strong stimulation of the promoter activity by the upstream region was found to be modulated by the presence of downstream regions, as shown by the lower activities of the promoter regions present in pPSV8 and pPSV9 compared to the promoter region in pPSV7 (Fig. 5). These variations may be due to the sequence-dependent conformation of the region and/or to the binding of a combination of “regulators” upstream and downstream from the promoter site, resulting in a special conformation of the region that might modulate the promoter's activity. Furthermore, the promoter activity driven by the pPSV6 construction (which includes only the downstream direct repeats) was found to be higher in cellulose cultures than in cellobiose cultures, and this suggests that this region might be sensitive to substrate-dependent induction. The larger increase in P1 activity following transfer to cellulose-containing medium of cells grown in cellobiose-containing medium than following transfer to cellulose-containing medium of cells grown in cellulose-containing medium (Fig. 4B) might also be due to cellulose induction. Nevertheless, the possibility that a low level of CCR might contribute to the increase in the expression of the transcriptional fusion during the acceleration phase of growth cannot be excluded.
The present work can be considered a first step toward understanding the complex regulation of the cip-cel operon. The next step will be a study of the cis sequences and identification of the regulators involved. In this context, the possible role of the gene encoding a putative member of the LysR family, located immediately downstream from the cip-cel operon on the reverse strand, will be investigated.
Acknowledgments
We thank Odile Valette for expert technical assistance. We also thank Akinobu Okabe for providing pPSV and Anne Galinier and Vincent Méjean for fruitful discussions.
This research was supported by grants from the Centre Natíonal de la Recherche Scientifique and the Université de Provence. L.A. holds a fellowship from the French “Ministère de l'Enseignement Supérieur et de la Recherche”.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Bélaich, A., C. Tardif, H. P. Fierobe, S. Pagès, and J.-P. Bélaich. 2004. The clostridial cellulosome, p. 343-360. In M. M. Nakano and P. Zuber (ed.), Strict and facultative anaerobes: medical and environmental aspects. Horizon Bioscience, Norfolk, United Kingdom.
- 2.Blouzard, J. C., C. Bourgeois, P. de Philip, O. Valette, A. Belaich, C. Tardif, J. P. Belaich, and S. Pages. 2007. Enzyme diversity of the cellulolytic system produced by Clostridium cellulolyticum explored by two-dimensional analysis: identification of seven genes encoding new dockerin-containing proteins. J. Bacteriol. 1892300-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullifent, H. L., A. Moir, and R. W. Titball. 1995. The construction of a reporter system and use for the investigation of Clostridium perfringens gene expression. FEMS Microbiol. Lett. 13199-105. [DOI] [PubMed] [Google Scholar]
- 4.Dror, T. W., E. Morag, A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of the cellulosomal CelS (cel48A) gene of Clostridium thermocellum is growth rate dependent. J. Bacteriol. 1853042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dror, T. W., A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of expression of scaffoldin-related genes in Clostridium thermocellum. J. Bacteriol. 1855109-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dror, T. W., A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2005. Regulation of major cellulosomal endoglucanases of Clostridium thermocellum differs from that of a prominent cellulosomal xylanase. J. Bacteriol. 1872261-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gal, L., S. Pages, C. Gaudin, A. Belaich, C. Reverbel-Leroy, C. Tardif, and J. P. Belaich. 1997. Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl. Environ Microbiol. 63903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galinier, A., J. Deutscher, and I. Martin-Verstraete. 1999. Phosphorylation of either Crh and HPr mediates binding of CcpA to Bacillus subtilis xyn cre and catabolite repression of the xyn operon. J. Mol. Biol. 286307-314. [DOI] [PubMed] [Google Scholar]
- 9.Giallo, J., C. Gaudin, and J.-P. Bélaich. 1985. Metabolism and solubilization of cellulose by Clostridium cellulolyticum H10. Appl. Environ. Microbiol. 491216-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guedon, E., S. Payot, M. Desvaux, and H. Petitdemange. 1999. Carbon and electron flow in Clostridium cellulolyticum grown in chemostat culture on synthetic medium. J. Bacteriol. 1813262-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2003. Regulation of expression of cellulosomal cellulase and hemicellulase genes in Clostridium cellulovorans. J. Bacteriol. 1856067-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hueck, C. J., and W. Hillen. 1995. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol. Microbiol. 15395-401. [DOI] [PubMed] [Google Scholar]
- 13.Inacio, J. M., C. Costa, and I. de Sa-Nogueira. 2003. Distinct molecular mechanisms involved in carbon catabolite repression of the arabinose regulon in Bacillus subtilis. Microbiology 1492345-2355. [DOI] [PubMed] [Google Scholar]
- 14.Jennert, K. C., C. Tardif, D. I. Young, and M. Young. 2000. Gene transfer to Clostridium cellulolyticum ATCC 35319. Microbiology 1463071-3080. [DOI] [PubMed] [Google Scholar]
- 15.Katayama, S., O. Matsushita, C. M. Jung, J. Minami, and A. Okabe. 1999. Promoter upstream bent DNA activates the transcription of the Clostridium perfringens phospholipase C gene in a low temperature-dependent manner. EMBO J. 183442-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katayama, S., O. Matsushita, E. Tamai, S. Miyata, and A. Okabe. 2001. Phased A-tracts bind to the alpha subunit of RNA polymerase with increased affinity at low temperature. FEBS Lett. 509235-238. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J. H., Y. K. Yang, and G. H. Chambliss. 2005. Evidence that Bacillus catabolite control protein CcpA interacts with RNA polymerase to inhibit transcription. Mol. Microbiol. 56155-162. [DOI] [PubMed] [Google Scholar]
- 18.Leboeuf, C., L. Leblanc, Y. Auffray, and A. Hartke. 2000. Characterization of the ccpA gene of Enterococcus faecalis: identification of starvation-inducible proteins regulated by ccpA. J. Bacteriol. 1825799-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, C., J. Kim, S. G. Shin, and S. Hwang. 2006. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J. Biotechnol. 123273-280. [DOI] [PubMed] [Google Scholar]
- 20.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 21.Luesink, E. J., R. E. van Herpen, B. P. Grossiord, O. P. Kuipers, and W. M. de Vos. 1998. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol. Microbiol. 30:789-798. [DOI] [PubMed] [Google Scholar]
- 22.Maamar, H., L. Abdou, C. Boileau, O. Valette, and C. Tardif. 2006. Transcriptional analysis of the cip-cel gene cluster from Clostridium cellulolyticum. J. Bacteriol. 1882614-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maamar, H., O. Valette, H. P. Fierobe, A. Belaich, J. P. Belaich, and C. Tardif. 2004. Cellulolysis is severely affected in Clostridium cellulolyticum strain cipCMut1. Mol. Microbiol. 51589-598. [DOI] [PubMed] [Google Scholar]
- 24.Matsushita, C., O. Matsushita, M. Koyama, and A. Okabe. 1994. A Clostridium perfringens vector for the selection of promoters. Plasmid 31317-319. [DOI] [PubMed] [Google Scholar]
- 25.Miwa, Y., A. Nakata, A. Ogiwara, M. Yamamoto, and Y. Fujita. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 281206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monedero, V., M. J. Gosalbes, and G. Perez-Martinez. 1997. Catabolite repression in Lactobacillus casei ATCC 393 is mediated by CcpA. J. Bacteriol. 1796657-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez-Martin, J., F. Rojo, and V. de Lorenzo. 1994. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev. 58268-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perret, S., A. Belaich, H. P. Fierobe, J. P. Belaich, and C. Tardif. 2004. Towards designer cellulosomes in clostridia: mannanase enrichment of the cellulosomes produced by Clostridium cellulolyticum. J. Bacteriol. 1866544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perret, S., L. Casalot, H. P. Fierobe, C. Tardif, F. Sabathe, J. P. Belaich, and A. Belaich. 2004. Production of heterologous and chimeric scaffoldins by Clostridium acetobutylicum ATCC 824. J. Bacteriol. 186253-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petitdemange, E., F. Caillet, J. Giallo, and C. Gaudin. 1984. Clostridium cellulolyticum sp. nov., a cellulolytic mesophilic species from decayed grass. Int. J. Syst. Bacteriol. 34155-159. [Google Scholar]
- 31.Reverbel-Leroy, C., A. Belaich, A. Bernadac, C. Gaudin, J. P. Belaich, and C. Tardif. 1996. Molecular study and overexpression of the Clostridium cellulolyticum celF cellulase gene in Escherichia coli. Microbiology 1421013-1023. [DOI] [PubMed] [Google Scholar]
- 32.Shaw, W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43737-755. [DOI] [PubMed] [Google Scholar]
- 33.Shimizu, M., M. Miyake, F. Kanke, U. Matsumoto, and H. Shindo. 1995. Characterization of the binding of HU and IHF, homologous histone-like proteins of Escherichia coli, to curved and uncurved DNA. Biochim. Biophys. Acta 1264330-336. [DOI] [PubMed] [Google Scholar]
- 34.Stim-Herndon, K. P., D. J. Petersen, and G. N. Bennett. 1995. Characterization of an acetyl-CoA C-acetyltransferase (thiolase) gene from Clostridium acetobutylicum ATCC 824. Gene 15481-85. [DOI] [PubMed] [Google Scholar]
- 35.Tangney, M., A. Galinier, J. Deutscher, and W. J. Mitchell. 2003. Analysis of the elements of catabolite repression in Clostridium acetobutylicum ATCC 824. J. Mol. Microbiol. Biotechnol. 66-11. [DOI] [PubMed] [Google Scholar]
- 36.Tardif, C., A. Bélaich, H. P. Fierobe, S. Pagès, P. de Philip, and J. P. Bélaich. 2006. Clostridium cellulolyticum: cellulosomes and cellulolysis, p. 221-259. In I. Kataeva (ed.), Cellulosome. Nova Science Publishers Inc., Hauppauge, NY.
- 37.Tardif, C., H. Maamar, M. Balfin, and J. P. Belaich. 2001. Electrotransformation studies in Clostridium cellulolyticum. J. Ind. Microbiol. Biotechnol. 27271-274. [DOI] [PubMed] [Google Scholar]
- 38.Warner, J. B., and J. S. Lolkema. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, Y. H., and L. R. Lynd. 2005. Regulation of cellulase synthesis in batch and continuous cultures of Clostridium thermocellum. J. Bacteriol. 18799-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuker, M. 1989. Computer prediction of RNA structure. Methods Enzymol 180:262-288. [DOI] [PubMed] [Google Scholar]