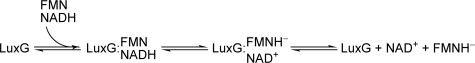Abstract
The luxG gene is part of the lux operon of marine luminous bacteria. luxG has been proposed to be a flavin reductase that supplies reduced flavin mononucleotide (FMN) for bacterial luminescence. However, this role has never been established because the gene product has not been successfully expressed and characterized. In this study, luxG from Photobacterium leiognathi TH1 was cloned and expressed in Escherichia coli in both native and C-terminal His6-tagged forms. Sequence analysis indicates that the protein consists of 237 amino acids, corresponding to a subunit molecular mass of 26.3 kDa. Both expressed forms of LuxG were purified to homogeneity, and their biochemical properties were characterized. Purified LuxG is homodimeric and has no bound prosthetic group. The enzyme can catalyze oxidation of NADH in the presence of free flavin, indicating that it can function as a flavin reductase in luminous bacteria. NADPH can also be used as a reducing substrate for the LuxG reaction, but with much less efficiency than NADH. With NADH and FMN as substrates, a Lineweaver-Burk plot revealed a series of convergent lines characteristic of a ternary-complex kinetic model. From steady-state kinetics data at 4°C pH 8.0, Km for NADH, Km for FMN, and kcat were calculated to be 15.1 μM, 2.7 μM, and 1.7 s−1, respectively. Coupled assays between LuxG and luciferases from P. leiognathi TH1 and Vibrio campbellii also showed that LuxG could supply FMNH− for light emission in vitro. A luxG gene knockout mutant of P. leiognathi TH1 exhibited a much dimmer luminescent phenotype compared to the native P. leiognathi TH1, implying that LuxG is the most significant source of FMNH− for the luminescence reaction in vivo.
Bacterial bioluminescence is a light-emitting phenomenon in bacteria resulting from a reaction catalyzed by an enzyme called luciferase. This enzyme catalyzes the oxidation of a long-chain aldehyde and reduced flavin mononucleotide (FMNH−) by molecular oxygen, resulting in a long-chain fatty acid, oxidized flavin mononucleotide (FMN), and water, with concomitant emission of blue-green light (13, 14). The overall reaction is as follows: FMNH− + H+ + RCHO + O2 → FMN + RCOOH + H2O + hν.
Previous studies have shown that tetradecanal (myristaldehyde) is a natural substrate for the luciferase reaction. A mutant of Vibrio harveyi with a dim phenotype could be restored to a luminescent phenotype when it was supplied with aldehydes and, of the aldehydes examined (C8, C10, C12, C14, and C16), myristaldehyde (C14) gave the highest quantum yield (45). It has also been shown that myristic acid, a product from the luciferase reaction, can be regenerated back to the aldehyde by myristic acid reductase from V. harveyi (46). The other substrate, FMNH−, which can be readily oxidized via an autocatalytic process in air-saturated solution, is supplied and transferred to the luciferase by NAD(P)H:FMN oxidoreductase (flavin reductase), which catalyzes the reduction of FMN by NAD(P)H (44).
Several flavin reductases from luminous bacteria have been identified and characterized, including two from Vibrio fischeri. Duane and Hastings first isolated FRaseI (8) from V. fischeri, a major FMN reductase in the cell that can utilize either NADH or NADPH as a reductant. FRaseI was cloned and overexpressed in Escherichia coli and found to contain one FMN per 26-kDa monomer (49). Another flavin reductase from V. fischeri, found in smaller quantities, has been cloned and expressed, and sequence analysis has shown that the enzyme is highly similar to Fre, a flavin reductase found in E. coli (33, 47). Both FRaseI and Fre-like flavin reductases can be used to supply FMNH− for the luciferase reaction (47, 49). Three flavin reductases have been identified in Vibrio harveyi (16). The most studied enzyme is FRP, a flavoprotein containing one FMN cofactor per subunit of 26.3 kDa. The enzyme was shown to be highly specific for NADPH as a reducing equivalent, in contrast to the FRaseI from V. fischeri that can utilize both NADPH and NADH. The gene encoding FRP has been cloned and overexpressed (24). Steady-state kinetics studies of FRP indicate a ping-pong type of reaction wherein the first half-reaction is the reduction of the enzyme-bound flavin by NADPH and the second half-reaction is the reduction and release of the FMN substrate from the enzyme (23, 40, 41, 44).
The enzymes involved in bacterial luminescence are encoded in a single operon (lux operon) composed of five common genes, luxCDABE, and these are found in all species of luminous bacteria (26). The luxAB genes encode the α and β subunits of the heterodimeric luciferase. The genes luxCDE encode three protein subunits of a long-chain fatty acid reductase that provides the long-chain fatty aldehyde for the luciferase reaction (25, 26). Marine luminous bacteria, such as Photobacterium phosphoreum, Photobacterium leiognathi, V. harveyi, and V. fischeri, have an additional gene, luxG, which follows luxE (22, 38, 39). The deduced amino acid sequences of luxG are similar to Fre, the flavin reductase found in E. coli (2, 15). Therefore, it has been postulated that the luxG gene product is a flavin reductase that provides the FMNH− substrate for the luciferase reaction (2, 48). However, the gene product of luxG had never been expressed, characterized, or shown to function as a flavin reductase. Only one report mentioned attempts to express luxG from V. fischeri. Neither additional flavin reductase activity nor an appreciable quantity of LuxG protein could be detected in cell extracts of E. coli expressing either the luxG gene or the entire lux operon genes (luxCDABEG) from V. fischeri. When a glutathione S-transferase (GST)-luxG fusion gene was expressed in E. coli, almost all of the GST-LuxG protein was recovered as inclusion bodies (47). Therefore, the role of luxG in bacterial luminescence has remained unproven.
In the present study, we report the cloning and overexpression in E. coli of luxG from P. leiognathi TH1. The gene products (LuxG) one without a tag and the other with a His6 tag at the C terminus were purified to homogeneity and characterized for their enzymatic properties. For the first time, LuxG was shown to be a flavin reductase that catalyzes reduction of flavins (FMN, FAD, and riboflavin) by NADH, and it could be coupled in vitro with the luciferases from P. leiognathi TH1 and V. campbellii to produce luminescence. A luxG knockout of P. leiognathi TH1 was constructed and shown to have a dim luminescence phenotype. The steady-state kinetics properties of the LuxG reaction indicate that the enzyme uses a ternary-complex type of mechanism.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
P. leiognathi TH1 was isolated from marine samples collected from the Southern part of Thailand and is the only P. leiognathi strain used in the present study (34). P. leiognathi TH1 genomic DNA was purified by using a DNA purification column (Qiagen). pGEM-T Easy vector, and IPTG (isopropyl-β-d-thiogalactopyranoside) were obtained from Promega. E. coli XL1-Blue was from Stratagene. pET-24b and pET-3a vectors and E. coli TUNER(DE3)/pLacI were from Novagen. Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. PCR used Taq or Pfu DNA polymerases from Fermentas. InsTAclone PCR cloning kits (pTZ57R) were purchased from Fermentas. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was purchased from GE Healthcare. All materials used for constructing the luxG knockout P. leiognathi strain were gifts from Skorn Mongkolsuk (Department of Biotechnology, Mahidol University). Oligonucleotide primers for PCR and DNA sequencing were synthesized by Sigma-Proligo. FAD and NADH were from Sigma. FMN was prepared by converting FAD to FMN with snake venom from Crotalus adamanteus (35). Bacterial luciferase (LuxAB) from V. campbellii was obtained according to protocols described previously (34). Chromatographic media were from GE Healthcare. All chemicals used were of analytical grade.
Construction of expression plasmids for LuxG and LuxG with a His6 tag at the C terminus.
Degenerate primers based on the conserved sequence of the lux operons of bioluminescent bacteria were used in amplifying the full-length gene of luxG from P. leiognathi TH1 (26). The sense strand primer was designed to be 5′-GA(A/G)(A/C/G)TIAA(C/T)ACITG(C/T)TT(C/T)TT(C/T)GA-3′, whereas the antisense strand was 5′-TC(A/G)TT(C/T)TCIC(G/T)(A/G)TC(C/T)TC(A/G)TC(A/G)TC-3′. The resulting PCR product was cloned into pGEM-T Easy to obtain the recombinant plasmids. Three independent positive recombinant plasmids were subjected to DNA sequencing, which indicated that the PCR product contained the entire luxG. To prevent carryover of errors from the previous PCR that might have been introduced by using Taq DNA polymerase with degenerate primers, specific primers that were based on the sequence of luxG of this P. leiognathi strain were designed to amplify the entire luxG directly from the genomic DNA using Pfu DNA polymerase. The sense strand primer was 5′-ATGGATGCTTCGTCTACCAGTT-3′, and the antisense strand primer was 5′-TGGCACCATCACCCATATATTA-3′. This new PCR product was inserted into pTZ57R to obtain the recombinant plasmid pTZ-tempG, which was used further as a template for constructing the expression vector for luxG.
Full-length luxG containing appropriate restriction sites for subcloning into an expression vector was amplified from pTZ-tempG by using the sense strand primer, 5′-GGGCTGACATATGATTTTTAATTGCAAGG-3′ (the NdeI site is underlined), and the antisense strand primer, 5′-ACAGTTGGATCCGCACTCATTATTAGCACCC-3′ (the BamHI site is underlined). The purified PCR product was cloned into pET-3a at the NdeI and BamHI sites to obtain the pET3G expression plasmid that was used for expressing the recombinant LuxG protein without any tag. To construct an expression vector for LuxG with a C-terminal His6 tag, a full-length luxG was amplified with the sense strand primer 5′-GGGCTGACATATGATTTTTAATTGCAAGG-3′ (the NdeI site is underlined) and the antisense strand primer 5′-ATAATGATTCTCGAGATAGTTAAATGC-3′ (the XhoI site is underlined). The purified PCR product was cloned into pET-24b at NdeI and XhoI sites to yield the pGhis expression plasmid. General cloning techniques were carried out according to protocols described by Sambrook et al. (31).
Construction of the expression plasmid for P. leiognathi LuxAB.
LuxAB genes were amplified from genomic DNA of P. leiognathi TH1 with degenerate primers; the sense strand primer was 5′-(A/G)TIGTI(C/T)TI(C/A)GIAA(C/T)TT(C/T)TA(C/T)CA-3′, and the antisense strand primer was 5′-GTIGG(A/G)AAIACIGG(A/T/G)AT(A/G)TC (A/G)TC(A/T/G)AT-3′. The PCR product was sequenced, and the resultant sequence was used for designing specific primers for amplifying the luxAB genes for expression. A full-length gene of luxAB was amplified from P. leiognathi TH1 genomic DNA by PCR using Pfu DNA polymerase with the sense strand primer 5′-GGAATAACATATGAAAATTAGTAA-3′ (the NdeI site is underlined) and the antisense strand primer 5′-TAGGATCCTCTATCTCTGTACTTA-3′ (the BamHI site is underlined). The resulting PCR product was cloned into pET-3a at the NdeI and BamHI sites, yielding the expression vector pPLAB.
Sequence analysis.
Protein similarity searches were performed by using the BLAST program via the National Center for Biotechnology Information (NCBI) server (http://www.ncbi.nlm.nih.gov/BLAST/). Protein sequence alignments were carried out with the CLUSTAL W program using default parameters. Molecular masses of the proteins were based on their amino acid sequences using the pI/MW tool in the Expasy proteomic server (http://www.expasy.org/tools/pi_tool.html).
Overexpression and purification of recombinant LuxG and His6-tagged LuxG.
E. coli TUNER(DE3)/pLacI cells harboring the pET3G expression vector (untagged) or the pGhis expression vector (His6 tagged) were cultured at 37°C in LB broth containing appropriate antibiotics (100 μg of ampicillin/ml or 30 μg of kanamycin/ml). The culture was cooled to 16°C, and protein expression was induced by adding IPTG (0.4 mM final conc.) when the optical density at 600 nm (OD600) of the cultures reached 1.0. After 7 h of growth at 16°C after the induction, cells were harvested by centrifugation and stored at −80°C until used.
Purification of LuxG was carried out at 4°C. E. coli frozen cell paste (from above) (∼30 g) was suspended in 20 mM sodium phosphate buffer (pH 7) containing 0.5 mM EDTA, 100 μM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), and 10% (vol/vol) glycerol, and the cells were disrupted by ultrasonication. Cell debris was removed by centrifugation at 35,000 × g for 60 min, which was followed by ultracentrifugation at 400,000 × g for 1.5 h. The clear supernatant was loaded onto a DEAE-Sepharose column (80 ml) pre-equilibrated with 20 mM sodium phosphate buffer (pH 7) containing 50 mM NaCl, 1 mM DTT, and 10% (vol/vol) glycerol. The column was washed with 10 column volumes of equilibration buffer, and the enzyme was eluted with a 2-liter gradient of 50 to 300 mM NaCl in the equilibration buffer. Active fractions were pooled, concentrated, and precipitated by salting out using ammonium sulfate (20 to 40% saturation) (32). The pellet was immediately dissolved in 50 mM Tris-Cl buffer (pH 8) containing 10% glycerol, 1 mM DTT, and 150 mM NaCl and then loaded onto a Sepharose S-200 gel filtration column (450 ml) pre-equilibrated with the same buffer. Fractions from the column were assayed for flavin reductase activity, and the active fractions were pooled, concentrated, and desalted by using a Sephadex G-25 gel filtration column previously equilibrated with 50 mM Tris-Cl buffer (pH 8) containing 10% glycerol and 1 mM DTT and stored at −80°C.
For His6-tagged LuxG purification, the E. coli cell paste (∼25 g) was thawed and resuspended in 50 mM Tris-Cl buffer (pH 8) containing 10% (vol/vol) glycerol and 100 μM PMSF. The cells were disrupted, and the cell debris was removed by the methods described above. The resulting supernatant was loaded onto a Ni-Sepharose column (20 ml) pre-equilibrated with 20 mM imidazole and 50 mM Tris-Cl buffer (pH 8) containing 250 mM NaCl and 10% (vol/vol) glycerol. The column (30 ml) was washed with 10 column volumes of 50 mM imidazole containing 50 mM Tris-Cl buffer (pH 8), 250 mM NaCl, and 10% (vol/vol) glycerol and then eluted by a 0.6-liter linear gradient of 50 to 250 mM imidazole in the same buffer. Active fractions were pooled and concentrated, and the buffer was exchanged by passing through a Sephadex G-25 column as described above.
Overexpression and purification of recombinant P. leiognathi LuxAB.
E. coli BL21(DE3) cells harboring the pPLAB plasmid were grown at 37°C in LB medium containing ampicillin (50 μg/ml). When the OD600 was ∼2.0, the temperature of the culture was adjusted to 16°C, and IPTG was added to make a final concentration of 1 mM. Cells were allowed to grow at this temperature until the OD600 reached ∼4.0 and harvested by centrifugation at 4°C.
Frozen cell paste (∼33 g) was thawed and suspended in buffer (50 mM sodium phosphate (pH 7.0) containing 100 μM PMSF and 0.3 mM EDTA), and the cells were disrupted by ultrasonication. Cell debris was removed by centrifugation. LuxAB was precipitated by using ammonium sulfate (35 to 60% saturation) (32). The pellet was resuspended in 50 mM sodium phosphate buffer (pH 7.0) and dialyzed overnight against 4 liters of 50 mM sodium phosphate buffer (pH 7.0). The dialyzed solution was loaded onto a DEAE-Sepharose column (80 ml) pre-equilibrated with 75 mM NaCl in 50 mM sodium phosphate buffer (pH 7). After a wash with the equilibration buffer, LuxAB was eluted with a 2-liter gradient of 75 to 350 mM NaCl in 50 mM sodium phosphate buffer (pH 7). Active fractions were loaded onto a Sephacryl S-200 column pre-equilibrated with 50 mM Tris-Cl (pH 8.0) and 1 mM DTT. The enzyme was eluted as a single peak from the column using the same equilibration buffer. The purified luciferase was concentrated and kept at −80 οC. The luciferase activity was assayed according to a previously described protocol (34).
Molecular mass determination.
The subunit molecular mass of LuxG was estimated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-15% PAGE) with low-range molecular weight markers from Bio-Rad. The estimate of native molecular mass was based on the elution volume of LuxG from a Superdex S-200 analytical gel filtration column operated by an AKTA FPLC system (GE Healthcare). The following proteins of known molecular mass were used as standard markers: cytochrome c, 12.4 kDa; carbonic anhydrase, 29 kDa; bovine serum albumin, 66 kDa; alcohol dehydrogenase, 150 kDa; β-amylase, 200 kDa; and blue dextran, 2,000 kDa. Buffer (50 mM Tris-Cl, 150 mM NaCl [pH 8.0]) was applied at a flow rate of 0.5 ml/min.
Protein identification.
The purified proteins were analyzed by LC/MS/MS (Thermo Electron) at the Proteomics Service Center of the Bioservice Unit, BIOTEC (Thailand). Proteins were digested by trypsin and then analyzed by liquid chromatography coupled with tandem mass spectrometry (MS/MS). Peptide ions were detected in a survey scan from 400 to 1,600 amu (atomic mass units) (one microscan), followed by one data-dependent MS/MS scan (two μscans each, isolation width of 2 amu, 30% CID (collision-induced dissociation) energy, dynamic exclusion of 180 s).
All MS/MS spectra were screened against the database by using the Sequest algorithm and the following criteria: enzyme, trypsin; static modification of cysteine, +57.05130 Da; differential modification of serine and threonine, +79.97990; and methionine +15.99940. The results of the search were filtered by Xcorr versus charge state (+1 ≥ 1.5, +2 ≥ 2.0, +3 ≥ 2.5) and protein probability (minimum, 1.00E-3).
Determination of protein concentration.
The molar absorption coefficient at 280 nm of the LuxG protein was calculated on the basis of the amino acid composition by using the Protparam program (http://ca.expasy.org/cgi-bin/protparam). The calculation yielded a value for LuxG at 280 nm of 34,755 M−1 cm−1 in denaturing conditions (6.0M guanidine hydrochloride, 0.02 M sodium phosphate buffer [pH 6.5]). Using this value as a guideline, the molar absorption coefficient of the native LuxG at 280 nm in 50 mM Tris-Cl buffer-10% (vol/vol) glycerol (pH 8.0) was determined to be 41,700 M−1 cm−1. This number was used throughout the present study to calculate the concentrations of LuxG and LuxG His6 tagged in 50 mM Tris-Cl buffer containing 10% (vol/vol) glycerol (pH 8.0). By the same protocol, the molar absorption coefficient at 280 nm of LuxAB was calculated to be 74,400 M−1 cm−1.
Enzyme assays.
Unless otherwise indicated, all assays were performed at 25°C in 50 mM Tris-Cl (pH 8) containing 10% (vol/vol) glycerol and 1 mM DTT. Flavin reductase activity was measured by monitoring the decrease of absorbance at 340 nm as NADH was depleted using FMN as the electron acceptor substrate. Because the assays were aerobic, the FMNH− produced was continuously reoxidized, so that the net reduction of FMN did not contribute measurably to the change in absorbance. Assay reactions were monitored by using Varian Cary 300 Bio or Shimadzu UV-2501PC spectrophotometers.
Steady-state kinetics.
Apparent kinetic parameters at 25°C were determined by fitting data to the Michaelis-Menten equation using a curve-fitting function in the KaleidaGraph program (for Windows; version 4.0). Apparent Km values for FMN, FAD, and riboflavin were determined from reactions containing 200 μM NADH and various concentrations of the flavin (1 to 40 μM). The apparent Km value for NADH was determined from the reactions containing 40 μM FMN and various concentrations of NADH (5 to 200 μM).
The steady-state kinetics at 4°C were investigated by using a stopped-flow spectrophotometer. Assay reactions in 50 mM Tris-Cl (pH 8) contained 10% (vol/vol) glycerol, 1 mM DTT, 100 nM concentrations of His6-tagged LuxG, and various concentrations of FMN (1 to 40 μM) and NADH (5 to 200 μM). Initial velocities (v) were calculated from slopes of absorbance decreases at 340 nm by using a molar absorption value of 6,220 M−1 cm−1 for NADH oxidation. Analyses were carried out according to established protocols (9).
The concentrations of the following compounds were determined by using known molar extinction coefficients: NADH, ɛ340 = 6.22 mM−1 cm−1; FAD, ɛ450 = 11.3 mM−1 cm−1; FMN, ɛ450 = 12.2 mM−1 cm−1; and riboflavin, ɛ445 = 11.3 mM−1 cm−1.
Coupled reaction of P. leiognathi LuxAB and LuxG.
Assay reactions of LuxAB from both P. leiognathi TH1 and V. campbellii were carried out in the presence of LuxG to determine whether LuxG can provide FMNH− for the luciferase reactions. V. campbellii luciferase was obtained by using the protocol described by Suadee et al. (34). The reaction mixtures were composed of 0.4 μM LuxAB, 0.4 μM LuxG, 100 μM dodecanal, 20 μM FMN, and 200 μM NADH in 50 mM Tris-Cl buffer (pH 8.0)-10% (vol/vol) glycerol-1 mM DTT. The same mixture without LuxG was used as a control. The light emitted was detected at room temperature by using a Shimadzu spectrofluorometer model RF-5301PC with the lamp off.
Construction of P. leiognathi TH1 with the luxG knockout.
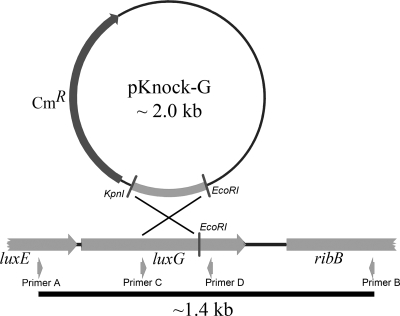
Part of luxG (311 bp, see the diagram in Fig. 1) was amplified by PCR using pTZ-tempG as a template and 5′-AGAAGGTACCATCGATGCACCACAC-3′ (Primer C, sense strand containing the KpnI site) and 5′-CGTCAATAACTGCGTCAATAACA-3′ (primer D, antisense strand). The purified PCR product was digested with the restriction enzymes KpnI and EcoRI, resulting in a 250-bp product after digestion. The 250-bp gene fragment was ligated into pKnock-CmR pretreated with KpnI and EcoRI to obtain the plasmid pKnock-G. “P. leiognathi-luxG” (P. leiognathi strain with the luxG gene inactivated) was constructed by conjugating the pKnock-G plasmid from E. coli strain BW20767 (oriT-RP4 terAR::Tn10 sacB Tets Sucr pir+) to the wild-type P. leiognathi TH1 in order to inactivate the luxG gene via homologous recombination. Positive recombinant plasmids of P. leiognathi-luxG were selected by screening for the chloramphenicol resistance phenotype and verified by PCR using the primers used previously in construction of pTZ-tempG (primer A, sense strand, 5′-ATGGATGCTTCGTCTACCAGTT-3′, and primer B, antisense strand, 5′-TGGCACCATCACCCATATATTA-3′). P. leiognathi-luxG and native P. leiognathi TH1 were compared for their native light-emitting phenotypes.
FIG. 1.
Map diagramming the construction of the P. leiognathi-luxG knockout.
RESULTS
Sequence analyses.
The complete sequence of luxG from P. leiognathi TH1 (accession number EU193661) encodes a protein of 237 amino acids, corresponding to a calculated molecular mass of ∼26.3 kDa. LuxAB (EU193662) from the same organism encodes a heterodimeric protein (α and β subunits) of 78.5 kDa. The deduced amino acid sequences of LuxG and LuxAB show greatest identity to the homologous proteins from P. leiognathi that has been reported very recently (3), 95% identity for LuxG (Fig. 2), and 97% identity for the α-subunit and 92% identity for the β-subunit of LuxAB. These results indicate that our isolated genes were indeed the luxG and luxAB from P. leiognathi TH1. The LuxG reported here is also homologous to the flavin reductase from E. coli (Fre) with 37% identity. Based on the structure of Fre, it is likely that the N-terminal part of LuxG is the flavin binding site and the C-terminal domain is the NAD(P)H binding site (15) (Fig. 2).
FIG. 2.
Pairwise alignment of the sequences of LuxG from P. leiognathi TH1 isolated in the present study (isolated_luxG) and LuxG previously reported (accession number AAA25621). The conserved residues are indicated by asterisk marks (*). Boldface letters represent the residues identical to those of E. coli Fre. Based on the structure of E. coli Fre (15), the flavin reductase can be divided into two domains; the N-terminal domain that binds flavin (indicated by “F”) and the C-terminal domain that probably binds NAD(P)H (indicated by “N”). Based on the structure of Fre (15), the secondary structure elements are shown above the text (β strands are indicated by arrows and α helices are indicated by solid blocks).
Expression and purification of recombinant LuxG and His6-tagged LuxG.
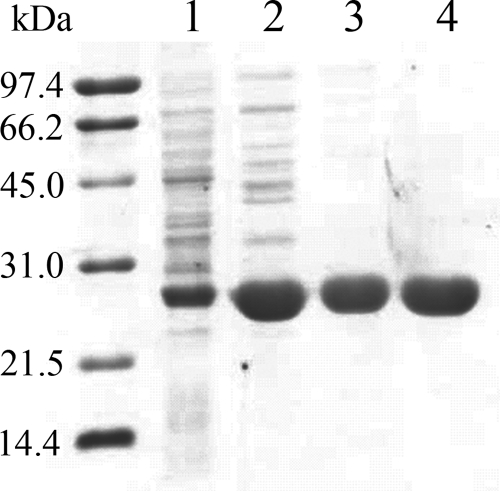
LuxG was expressed in E. coli TUNER(DE3)/pLacI using the pET expression system (Novagen) according to the protocols described in Materials and Methods. Soluble and particulate fractions of induced and noninduced cells were analyzed by SDS-PAGE. LuxG was expressed in the induced cells in both soluble and insoluble fractions in comparable quantities. Only the soluble fraction was further purified according to the protocols described in Materials and Methods. Recombinant LuxG was purified using ammonium sulfate fractionation, anion-exchange (DEAE-Sepharose), and gel filtration (Sephacryl-S200) chromatography. The enzyme had been highly purified as judged by SDS-PAGE (Fig. 3) and eluted as a single peak from the analytical gel filtration column. Recombinant His6-tagged LuxG was purified by using a single step of Ni-Sepharose chromatography. Most of the purification processes were done in the presence of 1 mM DTT, except for the Ni-Sepharose chromatography because DTT would reduce Ni2+. The flavin reductase activity was found to decrease over time in the absence of DTT. However, this inactivation could be reversed by adding DTT (final concentration, 1 mM) to the enzyme solution. In addition, it was found that glycerol in the buffers was necessary for preventing the enzyme at high concentrations from forming precipitation. The results of the protein purification are summarized in Table 1.
FIG. 3.
SDS-PAGE (15%) analysis of the purification of recombinant LuxG and His6-tagged LuxG. Lanes 1 to 3 (LuxG): 1, crude extract; 2, after purification by DEAE-Sepharose chromatography; 3, after purification by Sephacryl S-200 chromatography. Lane 4 shows the results for His6-tagged LuxG after purification by Ni+-Sepharose chromatography. The molecular size markers were phosphorylase b (97.4 kDa), bovine serum albumin (66.2 kDa), hen egg white ovalbumin (45 kDa), carbonic anhydrase (31 kDa), soybean trypsin inhibitor (21.5 kDa), and hen egg white lysozyme (14.4 kDa). The subunit molecular mass of LuxG was calculated to be ∼26 kDa.
TABLE 1.
Purification of recombinant LuxG and His6-tagged LuxG from E. coli TUNER(DE3)/pLacI
| Purification step | Total protein (mg) | Total activitya (U) | Sp act (U mg−1) | Yield (%) |
|---|---|---|---|---|
| Purification of recombinant His6-tagged LuxG | ||||
| Crude extract | 1,500 | 2,457 | 1.64 | 100 |
| Ni2+ column | 109 | 2,680 | 24.7 | 109 |
| Purification of recombinant LuxG | ||||
| Crude extract | 759 | 2,705 | 3.6 | 100 |
| DEAE-Sepharose | 190 | 2,732 | 14.3 | 101 |
| 20 to 40% (NH4)2SO4 precipitation | 83 | 1,567 | 18.9 | 58 |
| Sephacryl S200 | 38.4 | 803 | 20.9 | 30 |
One unit (U) is the amount of enzyme that catalyzes the oxidation of 1 μmol of NADH min−1.
Purified LuxG and His6-tagged LuxG were analyzed by liquid chromatography-mass spectrometry, which showed that the molecular masses of the purified LuxG and His6-tagged LuxG corresponded to the derived sequences from the cloned luxG, verifying that the purified proteins were indeed LuxG. Both forms of the purified enzyme were colorless, indicating that no prosthetic group was bound to the enzymes. SDS-PAGE (15%) revealed that the subunit molecular masses of LuxG and His6-tagged LuxG were about ∼26 kDa (Fig. 3), agreeing with the calculated values from the derived amino acid sequence (26.3 kDa). Analytical gel filtration (Superdex S-200) chromatography showed that the native enzyme eluted as a single peak and had a native molecular mass of 53 kDa, indicating that functional LuxG is a homodimer.
Expression and purification of recombinant P. leiognathi LuxAB.
The recombinant protein was largely expressed in a soluble form and purified to homogeneity as described in Materials and Methods. The results of the purification are summarized in Table 2. Typically, ∼ 480 mg of the purified enzyme can be obtained from ∼3.6 liters of E. coli culture. SDS-PAGE analysis revealed that purified LuxAB was a heterodimer composed of 40.5- and 38-kDa subunits (Fig. 4), a finding consistent with the calculated molecular masses from the amino acid sequences.
TABLE 2.
Purification of recombinant LuxAB from E. coli BL21(DE3)
| Purification step | Total protein (mg) | Total activitya (1018 photons) | Sp act (1014 photons mg−1) |
|---|---|---|---|
| Crude extract | 2,093 | 1.3 | 6.2 |
| 20 to 40% (NH4)2SO4 precipitation | 1,795 | 6.3 | 35.1 |
| DEAE-Sepharose | 1,080 | 4.2 | 38.9 |
| Sephacryl S200 | 480 | 2.5 | 52.1 |
The total photons from each assay were calculated by integrating the area under the luminescence trace and converting the value into quantum units by correlating them with the luminol reaction (34).
FIG. 4.
SDS-PAGE analysis of the purification of recombinant LuxAB. Lanes: 1, crude extract; 2, ammonium sulfate fraction (35 to 60%); 3, purified by DEAE-Sepharose chromatography; 4, extract purified by Sephacryl-200 chromatography. P. leiognathi luciferase is a heterodimeric protein with the α subunit (∼40.5 kDa) and β subunit (∼38 kDa).
NADH:flavin oxidoreductase activity of LuxG.
Based on the sequence similarity to Fre, researchers in previous studies have proposed that LuxG is the appropriate flavin reductase to provide FMNH− to the luciferase reaction (2, 47). We assayed the purified LuxG for flavin reductase activity by monitoring the decrease in absorbance at 340 nm in reaction mixtures containing 200 nM LuxG or His6-tagged LuxG, 200 μM NADH, 40 μM flavin (FMN, FAD, or riboflavin), 50 mM Tris-Cl, 10% (vol/vol) glycerol, and 1 mM DTT (pH 8.0). It was shown that LuxG could catalyze the oxidation of NADH in the presence of free flavins (FMN, FAD, or riboflavin), indicating that LuxG is indeed a flavin oxidoreductase.
The apparent kinetic parameters of LuxG and His6-tagged LuxG were measured and compared (Table 3). The two enzymes had similar kinetic parameters, implying that the His6 tag at the C terminus of LuxG did not interfere with the catalytic reaction. All three of the flavins tested were good substrates when NADH was used as an electron donor with similar apparent kcat and Km values (Table 3). With NADPH as an electron donor, there was no reaction with FAD, but the enzyme could utilize riboflavin and FMN. However, when NADPH was used, the Km values were very high (∼0.64 mM when using riboflavin and ∼0.51 mM when using FMN), suggesting that NADPH is not a physiological substrate for the LuxG reaction.
TABLE 3.
Apparent kinetic parameters of recombinant LuxG and His6-tagged LuxG from P. leiognathi TH1
| Substratea | Second substrate (concn [μM]) | Mean ± SDb
|
|
|---|---|---|---|
| Apparent Km (μM) | kcat (s−1) | ||
| FMN* | NADH (200) | 2.7 ± 0.3 (3.2 ± 0.3) | 10.6 ± 0.3 (11.3 ± 0.3) |
| FAD* | NADH (200) | 2.6 ± 0.4 (3.4 ± 0.4) | 7.1 ± 0.3 (8.0 ± 0.2) |
| Riboflavin* | NADH (200) | 1.2 ± 0.1 (1.2 ± 0.2) | 9.6 ± 0.2 (8.0 ± 0.3) |
| NADH† | FMN (40) | 20.5 ± 2.9 (19.6 ± 2.2) | 7.5 ± 0.3 (7.6 ± 0.2) |
Initial rates were measured by the flavin reductase assay described in Materials and Methods at 25°C and pH 8 with 0.2 μM concentrations of recombinant LuxG or His6-tagged LuxG, flavins (2 to 40 μM) at a fixed concentration of NADH (200 μM) or various concentrations of NADH (5 to 200 μM) at a fixed concentration of FMN (40 μM). Experiments were performed by using a stopped-flow spectrophotometer (*) or a standard spectrophotometer (†). Data were analyzed by using nonlinear regression analysis (Kaleidagraph software; Synergy).
The kinetic parameters of His6-tagged LuxG are shown in parentheses.
Steady-state kinetics of His6-tagged LuxG.
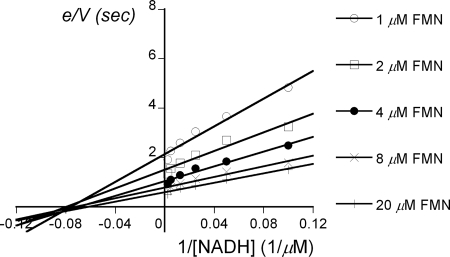
Based on the data in Table 3, FMN, FAD, and riboflavin are all good substrates, although the kcat/Km is ∼2-fold larger for riboflavin than for the other flavins. A more complete steady-state kinetic analysis was carried out at 4°C with NADH and FMN by using a stopped-flow apparatus as described in Materials and Methods. A primary plot of e/V versus 1/[NADH] revealed a series of convergent lines characteristic of a ternary-complex kinetics model (Fig. 5). The Dalziel equations (7, 9) were used for calculating Km for FMN (2.7 μM), Km for NADH (15.1 μM), and kcat (1.7 s−1) at pH 8.0.
FIG. 5.
Steady-state kinetics of the LuxG reaction. A primary double-reciprocal plot of initial rates of the His6-tagged LuxG reactions versus various concentrations of NADH and FMN. Each line represents e/V values at a fixed concentration of FMN. Assay reactions were performed in 50 mM Tris-Cl (pH 8), 10% (vol/vol) glycerol, 1 mM DTT, and 100 nM concentrations of His6-tagged LuxG with various concentrations of FMN as indicated (1 to 40 μM) and NADH (5 to 200 μM) using a stopped-flow apparatus at 4°C under aerobic conditions. The Dalziel parameters (7, 9) were determined: φ[NADH] = 8.97 μM s, φ[FMN] = 1.60 μM s, and φo = 0.59 s.
Coupled reactions of P. leiognathi His6-tagged LuxG and luciferase (LuxAB).
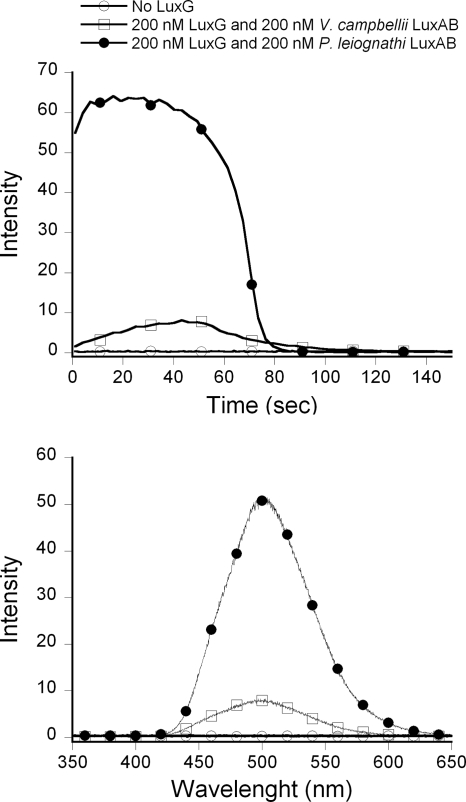
The ability of LuxG to supply FMNH− to support the luciferase reaction was investigated by the assay method described in Materials and Methods (34). Coupled reactions using luciferase emitted light with an λmax of ∼500 nm; no light was detected in a control reaction without LuxG added (Fig. 6). The intensity of light emission from P. leiognathi LuxAB was considerably higher than that of V. campbellii LuxAB, and this may be due to two factors. The first is that P. leiognathi LuxAB is a fast luciferase, whereas V. campbellii LuxAB is a slow luciferase (34). In these assays, the fast luciferase could catalyze multiple turnovers and consume a large fraction of the FMNH− before it was reoxidized in an uncoupled fashion. In the slow luciferase reaction, the enzyme was likely to turn over only once (or a few times) during the assay, whereas a large fraction of FMNH− was reoxidized in an uncoupled fashion (34). The second factor for the higher light yield of LuxAB from P. leiognathi may be due to the intrinsic properties of the enzyme. In the single-turnover reaction studied by the stopped-flow technique, LuxAB from P. leiognathi emits a larger integrated light yield than does LuxAB from V. campbellii (J. Wongratana, unpublished data). Nevertheless, these results clearly show that LuxG is functional in supplying luxAB with FMNH− in vitro.
FIG. 6.
In vitro luminescence kinetics of coupled reactions with LuxAB and His6-tagged LuxG. The reaction was composed of 400 nM P. leiognathi LuxAB (•) or V. campbellii LuxAB (□) (34), 100 μM dodecanal, 20 μM FMN, and 200 μM NADH in 50 mM Tris-Cl buffer (pH 8.0), 10% (vol/vol) glycerol, and 1 mM DTT in the presence or absence (○) of 400 nM His6-tagged LuxG under aerobic conditions. (Upper panel) Light emitted at 490 nm was observed with time in the spectrofluorometer with no excitation light. (Lower panel) Emission spectra of light emitted from the same reactions of the upper panel.
Phenotype of P. leiognathi-luxG.
pKnock-G was constructed by ligating part of luxG into the plasmid pKnock-CmR (1) according to the protocols described in Materials and Methods. To generate the P. leiognathi-luxG strain, pKnock-G was transferred into P. leiognathi TH1 by conjugation. Transconjugants were selected on LB agar plates containing 34 μg of chloramphenicol/ml. Interruption of luxG was validated by PCR. If pKnock-G (∼2 kb) were inserted into the luxG gene on the chromosome, PCR of the chromosome containing the interrupted luxG resulted in a product ∼2 kb larger than the product from the chromosome that did not contain pKnock-G (Fig. 1). P. leiognathi-luxG and the wild-type P. leiognathi TH1 were grown on the same LB plate, and their luminescence intensities were compared (Fig. 7). Clearly, P. leiognathi-luxG has some luminescence of much lower intensity compared to the wild type. Because P. leiognathi-luxG is still slightly luminescent, it is likely that LuxG is not the only flavin reductase that can supply FMNH− to LuxAB. However, the dim phenotype suggests that luxG is the most significant reductase supplying FMNH− to LuxAB reaction in vivo.
FIG. 7.
Comparison of the luminescence of P. leiognathi-luxG knockout and of native P. leiognathi TH1. Both strains were cultured on the same LB agar plate at 28°C for ∼16 h. The picture was taken in the dark by using a Canon Powershot A620 digital camera with an exposure time of 30 s and an aperture of 4.0. This result indicates that the luxG knockout strain was much less luminescent that the native strain.
DISCUSSION
Although LuxG (based on its sequence) has been proposed for more than a decade to be a flavin reductase functioning in marine luminous bacteria (2), the direct functional role has never been demonstrated. No studies have reported the expression of the protein, its enzymatic activity, or its role in bacterial luminescence (26). The work reported here is the first demonstration of the functional role of LuxG as a flavin reductase important for luminescence. We report herein the cloning, expression in E. coli, and biochemical characterization of LuxG from P. leiognathi TH1.
LuxG, LuxG with a His6 tag at the N terminus, and LuxG with a His6 tag at the C terminus were expressed, but only LuxG and LuxG His6 tagged at the C terminus could be expressed in soluble forms in E. coli TUNER(DE3)/pLacI. LuxG with a His6 tag at the N terminus was expressed in a particulate form (data not shown), indicating that the N-terminal part of LuxG is critical for the proper folding of LuxG during protein synthesis. Our observations suggest that lowering the temperature of the culture before adding IPTG and using E. coli TUNER(DE3)/pLacI (which allows the rate of protein expression to be adjusted by IPTG concentration) (28) are key factors for obtaining LuxG in a soluble form. Perhaps the rate of protein expression was slow under these conditions, allowing some expressed protein to correctly fold (28). It was also found that the enzyme (both LuxG and C-terminus His6-tagged LuxG) lost activity in the absence of DTT during the protein preparation, suggesting that some thiol groups of the enzyme are needed for the enzyme to function efficiently. It should be noted that LuxG has a total of seven cysteine residues in its 237-amino-acid sequence.
Neither form of purified P. leiognathi LuxG contains any flavin bound as a cofactor for mediating the electron transfer from the reduced pyridine nucleotide to free oxidized flavin. The lack of a bound redox cofactor suggests that LuxG is not a flavoprotein and is consistent with the steady-state kinetics data that suggest a ternary complex model in which the NADH and FMN substrates are both required to be bound to the enzyme active site simultaneously in order for hydride transfer to occur (Fig. 8). The enzyme also shows a broad tolerance toward substrate flavins (FMN, FAD, and riboflavin), while preferring NADH as the electron donor. With NADPH, LuxG showed some preference toward riboflavin and FMN, whereas FAD gave no activity. These properties are similar to those observed with E. coli Fre (10, 11), a homologue of LuxG. However, native LuxG was shown to form a homodimer in solution whereas Fre is a monomer. The molecular mass of the LuxG monomer (∼26 kDa) is similar to that of Fre (Fig. 3).
FIG. 8.
Proposed kinetic mechanism of the LuxG reaction.
Although Fre is a flavin reductase that does not use a bound flavin, structural data and mutagenesis studies show that it is closely related to flavoprotein enzymes such as ferredoxin oxidoreductases, which are in the FNR superfamily (15, 18, 27). We carried out sequence comparisons of LuxG, Fre, and other proteins in the FNR family to find likely substrate-binding domains and important catalytic amino acid residues in LuxG. Based on these analyses, we suggest that LuxG can be divided into an N-terminal domain that binds flavin and a C-terminal domain that binds NAD(P)H (15). Based on structural and mutagenesis studies of Fre and spinach FNR (4, 15, 18, 19), we suggest that Ser48 in LuxG (corresponding to Ser49 in Fre and Ser96 in spinach FNR) is important for catalysis and binding of the isoalloxazine moiety of the flavin. Ser116 and Tyr117 in LuxG, corresponding to Ser115 and Tyr116 in E. coli Fre, are also conserved. Based on structures of Fre, these residues are possibly important for forming the pocket that harbors the isoalloxazine ring of the flavin. The analogous loop between Fβ5 and Fα1 in Fre and LuxG (Fig. 2) serves as a binding site for the AMP moiety of FAD in FNR. However, this loop is shorter in Fre and in LuxG than in spinach FNR, possibly explaining why the flavins do not bind tightly to LuxG and Fre to form a prosthetic group. Residues important for binding NAD(P)H in Fre have not been experimentally located, but on the basis of the structure of FNR it was speculated that the conserved Gly201 may be involved (15).
Several different flavin reductases have been found in luminous bacteria, but LuxG is the only flavin reductase encoded in the lux operon. Prior to this work, no active LuxG preparation had been reported. In other two-component flavin-dependent oxygenases, such as p-hydroxyphenylacetate hydroxylases from A. baumannii (5, 35-37, 43), E. coli (12, 30, 33), and P. aeruginosa (6), the flavin reductases are found in the same operon as the partner oxygenases. Therefore, it is highly likely that LuxG is the flavin reductase that evolved to function in the luminescence reaction. However, when LuxAB and LuxG were mixed and analyzed by analytical gel filtration chromatography (data not shown), there was no evidence of the two proteins binding to each other. This suggests that no stable complex of the two proteins is necessary for efficient luciferase activity. Recently, we investigated how reduced flavin transfers from the reductase to the oxygenase of p-hydroxyphenylacetate hydroxylase. It was shown that no protein-protein interaction is necessary for efficiently transferring the reduced flavin to the oxygenase because the binding of reduced flavin to the oxygenase is much faster than the reaction of the reduced flavin with O2 (37). Our results here clearly indicate that LuxG can supply FMNH− for the luciferase reaction both in vitro (Fig. 6) and in vivo (Fig. 7) and, although it is not the only flavin reductase that can produce FMNH− for the luciferase in vivo, it appears to be the most effective reductase in the organism.
In conclusion, our results clearly show that LuxG from P. leiognathi functions as a nonflavoprotein flavin reductase in luminous bacteria, playing a significant role in bacterial bioluminescence. These studies serve as the basis for future more in-depth investigations on enzyme mechanism.
Acknowledgments
This study was supported by the Thailand Research Fund grants RMU4880028 and RTA4780006 and the Faculty of Science, Mahidol University (P.C.), and by a grant GM64711 from the National Institutes of Health (D.P.B.). S.N. and C.S. were recipients of scholarships under the Royal Golden Jubilee Ph.D. program of the Thailand Research Fund (PHD/0211/2546 and PHD/0032/2545). The Medical Scholars program at Mahidol University (S.N.) is also acknowledged. J.W. is a recipient of a scholarship of the institutional strengthening program offered by Faculty of Science, Mahidol University.
We thank Skorn Mongkolsuk for the generous gift pKnock-CmR and E. coli BW20767. We are grateful to Rattanon Suntivich and Kanlaya Prapainop for their experimental assistance. We thank Somchai Busarawit for suggestions about marine sample collection.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26824-826, 828. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, S. C., D. Shipley, J. N. Keen, J. B. Findlay, P. M. Harrison, and J. R. Guest. 1992. The hemoglobin-like protein (HMP) of Escherichia coli has ferrisiderophore reductase activity and its C-terminal domain shares homology with ferredoxin NADP+ reductases. FEBS Lett. 302247-252. [DOI] [PubMed] [Google Scholar]
- 3.Ast, J. C., H. Urbanczyk, and P. V. Dunlap. 2007. Natural merodiploidy of the lux-rib operon of Photobacterium leiognathi from coastal waters of Honshu, Japan. J. Bacteriol. 1896148-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruns, C. M., and P. A. Karplus. 1995. Refined crystal structure of spinach ferredoxin reductase at 1.7 Å resolution: oxidized, reduced and 2′-phospho-5′-AMP bound states. J. Mol. Biol. 247125-145. [DOI] [PubMed] [Google Scholar]
- 5.Chaiyen, P., C. Suadee, and P. Wilairat. 2001. A novel two-protein component flavoprotein hydroxylase. Eur. J. Biochem. 2685550-5561. [DOI] [PubMed] [Google Scholar]
- 6.Chakraborty, S., M. Ortiz-Maldonado, K. Eschenburg, B. Entsch, and D. P. Ballou. 2005. p-Hydroxyphenylacetate-3-hydroxylase from Pseudomonas aeruginosa, p. 161-166. In T. Nishino, R. Miura, M. Tanokura, and K. Fukui (ed.), Flavins and flavoproteins. ARchiTect, Inc., Tokyo, Japan.
- 7.Dalziel, K. 1957. Initial steady state velocities in the evaluation of enzyme-coenzyme-substrate reaction mechanisms. Acta Chem. Scand. 111706-1723. [Google Scholar]
- 8.Duane, W., and J. W. Hastings. 1975. Flavin mononucleotide reductase of luminous bacteria. Mol. Cell. Biochem. 653-64. [DOI] [PubMed] [Google Scholar]
- 9.Engel, P. C. 1981. Enzyme kinetics: the steady-state approach, 2nd ed. University Printing House, Cambridge, United Kingdom.
- 10.Fieschi, F., V. Niviere, C. Frier, J. Decout, and M. Fontecave. 1995. The mechanism and substrate specificity of the NADPH:flavin oxidoreductase from Escherichia coli. J. Biol. Chem. 27030392-30400. [DOI] [PubMed] [Google Scholar]
- 11.Fontecave, M., R. Eliasson, and P. Reichard. 1987. NAD(P)H:flavin oxidoreductase of Escherichia coli: a ferric iron reductase participating in the generation of the free radical of ribonucleotide reductase. J. Biol. Chem. 26212325-12331. [PubMed] [Google Scholar]
- 12.Galan, B., E. Diza, M. A. Prieto, and J. L. Garcia. 2000. Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: a prototype of a new flavin:NAD(P)H reductase subfamily. J. Bacteriol. 182627-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunsalus-Miguel, A., E. A. Meighen, M. Z. Nicoli, K. H. Nealson, and J. W. Hastings. 1972. Purification and properties of bacterial luciferase. J. Biol. Chem. 247398-404. [PubMed] [Google Scholar]
- 14.Hastings, J. W. 1996. Chemistries and colors of bioluminescent reactions: a review. Gene 1735-11. [DOI] [PubMed] [Google Scholar]
- 15.Ingelman, M., S. Ramaswamy, V. Niviere, M. Fontecave, and H. Eklund. 1999. Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli. Biochemistry 387040-7049. [DOI] [PubMed] [Google Scholar]
- 16.Jablonski, E., and M. DeLuca. 1977. Purification and properties of the NADH and NADPH specific FMN oxidoreductases from Beneckea harveyi. Biochemistry 162932-2936. [DOI] [PubMed] [Google Scholar]
- 17.Jeffers, C. E., and S. C. Tu. 2001. Differential transfers of reduced flavin cofactor and product by bacterial flavin reductase to luciferase. Biochemistry 401749-1754. [DOI] [PubMed] [Google Scholar]
- 18.Karplus, P. A., and C. M. Bruns. 1994. Structure-function relations for ferredoxin reductase. J. Bioenerg. Biomembr. 2689-99. [DOI] [PubMed] [Google Scholar]
- 19.Karplus, P. A., M. J. Daniels, and J. R. Herriott. 1991. Atomic structure of ferredoxin-NADP+ reductase: prototype for a structurally novel flavoenzyme family. Science 25160-66. [PubMed] [Google Scholar]
- 20.Kendrew, S. G., S. E. Harding, D. A. Hopwood, and E. N. G. Marsh. 1995. Identification of a flavin:NADH oxidoreductase involved in the biosynthesis of actinorhodin: purification and characterization of the recombinant enzyme. J. Biol. Chem. 27017339-17343. [DOI] [PubMed] [Google Scholar]
- 21.Lee, C. Y., and E. A. Meighen. 1992. The lux genes in Photobacterium leiognathi are closely linked with genes corresponding in sequence to riboflavin synthesis genes. Biochem. Biophys. Res. Commun. 186690-697. [DOI] [PubMed] [Google Scholar]
- 22.Lee, C. Y., R. B. Szittner, and E. A. Meighen. 1991. The lux genes of the luminous bacterial symbiont, Photobacterium leiognathi, of the ponyfish: nucleotide sequence, difference in gene organization, and high expression in mutant Escherichia coli. Eur. J. Biochem. 201161-167. [DOI] [PubMed] [Google Scholar]
- 23.Lei, B., and S. C. Tu. 1998. Mechanism of reduced flavin transfer from Vibrio harveyi NADPH-FMN oxidoreductase to Luciferase. Biochemistry 3714623-14629. [DOI] [PubMed] [Google Scholar]
- 24.Lei, B., M. Liu, S. Huang, and S. C. Tu. 1994. Vibrio harveyi NADPH-flavin oxidoreductase: cloning, sequencing and overexpression of the gene and purification and characterization of the cloned enzyme. J. Bacteriol. 1763552-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meighen, E. A. 1991. Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55123-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meighen, E. A. 1994. Genetics of bacterial bioluminescence. Annu. Rev. Genes. 28117-139. [DOI] [PubMed] [Google Scholar]
- 27.Niviere, V., F. Fieschi, J. L. Decout, and M. Fontecave. 1996. Is the NAD(P)H:flavin oxidoreductase from Escherichia coli a member of the ferredoxin-NADP+ reductase family? Evidence for the catalytic role of serine 49 residue. J. Biol. Chem. 27116656-16661. [DOI] [PubMed] [Google Scholar]
- 28.Novagen. 2006. Strain descriptions: features and application of competent cell strains, p. 103-105. In Novagen catalog 2006/2007. Novagen, La Jolla, CA.
- 29.Prieto, M. A., and J. L. Garcia. 1994. molecular characterization of 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli: a two-protein component system. J. Biol. Chem. 26922823-22829. [PubMed] [Google Scholar]
- 30.Prieto, M. A., A. Perez-Aranda, and J. L. Garcia. 1993. Characterization of an Escherichia coli aromatic hydroxylase with a broad substrate range. J. Bacteriol. 1752162-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring harbor Laboratory, Cold Spring Harbor, NY.
- 32.Scopes, R. K. 1994. Separation by precipitation: salting out at high salt concentration p. 76-85. In Protein purification principles and practice, 3rd ed. Narosa Publishing House, New Delhi, India.
- 33.Spyrou, G. E. Haggard-LjungQuist, M. Krook, H. Jornvall, E. Nilsson, and P. Reichard. 1991. Characterization of the flavin reductase gene (fre) of Escherichia coli and construction of a plasmid for overproduction of the enzyme. J. Bacteriol. 1733673-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suadee, C., S. Nijvipakul, J. Svasti, B. Entsch, D. P. Ballou, and P. Chaiyen. 2007. Luciferase from Vibrio campbellii is more thermostable and binds reduced FMN better than its homologues. J. Biochem. 142539-552. [DOI] [PubMed] [Google Scholar]
- 35.Sucharitakul, J., P. Chaiyen, B. Entsch, and D. P. Ballou. 2005. The reductase of p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii requires p-hydroxyphenylacetate for effective catalysis. Biochemistry 4410434-10442. [DOI] [PubMed] [Google Scholar]
- 36.Sucharitakul, J., P. Chaiyen, B. Entsch, and D. P. Ballou. 2006. Kinetic mechanisms of the oxygenase from a two-component enzyme, p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii. J. Biol. Chem. 28135104-35115. [DOI] [PubMed] [Google Scholar]
- 37.Sucharitakul, J., T. Phongsak, B. Entsch, J. Svasti, P. Chaiyen, and D. P. Ballou. 2007. Kinetics of a two-component p-hydroxyphenylacetate hydroxylase explain how reduced flavin is transferred from the reductase to the oxygenase. Biochemistry 468611-8623. [DOI] [PubMed] [Google Scholar]
- 38.Swartzman, E., C. Miyamoto, A. Graham, and E. A. Meighen. 1990. Delineation of the transcriptional boundaries of the lux operon of Vibrio harveyi demonstrates the presence of two new lux genes. J. Biol. Chem. 2653513-3517. [PubMed] [Google Scholar]
- 39.Swartzman, E., S. Kapoor, A. F. Graham, and E. A. Meighen. 1990. A new Vibrio fischeri lux gene precedes a bidirectional termination site for the lux operon. J. Bacteriol. 1726797-6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanner, J., B. Lei, M. Liu, S. C. Tu, and K. L. Krause. 1994. Crystallization and preliminary crystallographic analysis of NADPH:FMN oxidoreductase from Vibrio harveyi. J. Mol. Biol. 241283-287. [DOI] [PubMed] [Google Scholar]
- 41.Tanner, J. J., B. Lei, S. C. Tu, and K. L. Krause. 1996. Flavin reductase P: structure of a dimeric enzyme that reduces flavin. Biochemistry 3513531-13539. [DOI] [PubMed] [Google Scholar]
- 42.Thibaut, D., N. Tatet, D. Bisch, D. Faucher, L. Debussche, and F. Blanche. 1995. Purification of the two-enzyme system catalyzing the oxidation of the d-proline residue of pristinamycin IIB during the last step of pristinamycin IIA biosynthesis. J. Bacteriol. 1775199-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thotsaporn, K., J. Sucharitakul, J. Wongratana, C. Suadee, and P. Chaiyen. 2004. Cloning and expression of p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii: evidence of the divergence of enzymes in the class of two-protein component aromatic hydroxylase. Biochim. Biophys. Acta 168060-66. [DOI] [PubMed] [Google Scholar]
- 44.Tu, S. C. 2001. Reduced flavin: donor and acceptor enzymes and mechanisms of channeling. Antioxid. Redox Signal 3881-897. [DOI] [PubMed] [Google Scholar]
- 45.Ulitzur, S., and J. W. Hastings. 1979. Evidence for tetradecanal as the natural aldehyde in bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 76265-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulitzur, S., and J. W. Hastings. 1979. Control of aldehyde synthesis in the luminous bacterium Beneckea harveyi. J. Bacteriol. 137854-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zenno, H., and K. Saigo. 1994. Identification of the genes encoding NAD(P)H-flavin oxidoreductases that are similar in sequence to Escherichia coli Fre in four species of luminous bacteria: Photorhabdus luminescens, Vibrio fischeri, Vibrio harveyi, and Vibrio orientalis. J. Bacteriol. 1763544-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zenno, I., S. Inouye, and K. Saigo. 1992. Does the luxG gene in luminous bacteria code for an NAD(P)H-FMN oxidoreductase? Genetics (Life Sci. Adv.) 1185-91. [Google Scholar]
- 49.Zenno, H., K. Saigo, H. Kanoh, and S. Inouye. 1994. Identification of the gene encoding the major NAD(P)H-flavin oxidoreductase of the bioluminescent bacterium Vibrio fischeri ATCC 7744. J. Bacteriol. 1763536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]