Abstract
Crc protein is a global regulator involved in catabolite repression control of several pathways for the assimilation of carbon sources in pseudomonads when other preferred substrates are present. In Pseudomonas putida cells growing exponentially in a complete medium containing benzoate, Crc strongly inhibits the expression of the benzoate degradation genes. These genes are organized into several transcriptional units. We show that Crc directly inhibits the expression of the peripheral genes that transform benzoate into catechol (the ben genes) but that its effect on genes corresponding to further steps of the pathway (the cat and pca genes of the central catechol and β-ketoadipate pathways) is indirect, since these genes are not induced because the degradation intermediates, which act as inducers, are not produced. Crc inhibits the translation of target genes by binding to mRNA. The expression of the ben, cat, and pca genes requires the BenR, CatR, and PcaR transcriptional activators, respectively. Crc significantly reduced benABCD mRNA levels but did not affect those of benR. Crc bound to the 5′ end of benR mRNA but not to equivalent regions of catR and pcaR mRNAs. A translational fusion of the benR and lacZ genes was sensitive to Crc, but a transcriptional fusion was not. We propose that Crc acts by reducing the translation of benR mRNA, decreasing BenR levels below those required for the full expression of the benABCD genes. This strategy provides great metabolic flexibility, allowing the hierarchical assimilation of different structurally related compounds that share a common central pathway by selectively regulating the entry of each substrate into the central pathway.
Bacterial species that can thrive in very diverse habitats can usually assimilate many different compounds as carbon and energy sources. However, some compounds are frequently preferred over others. When the preferred compounds are present in sufficient concentrations, the assimilation of nonpreferred compounds is inhibited by a complex regulatory process termed catabolite repression control (6, 37, 40, 41). The regulatory networks involved interfere with transport and/or with the expression of the metabolic pathways for the nonpreferred compounds. The molecular mechanisms of catabolite repression can differ substantially among different bacterial species (41).
Pseudomonads are gram-negative saprophytic bacteria that can thrive in very diverse habitats and are important in the environment, in medicine, and in biotechnology (21, 31). Pseudomonas putida, in particular, can survive as a free-living organism in soils and water or can be associated with plant roots (23, 42). It has great metabolic versatility and can metabolize a wide range of aromatic compounds, many of which derive from the decomposition of plant material (28). The degradation of aromatic compounds takes place in two stages. Initial steps convert the individual substrates into a small number of structurally simple intermediates that can be fed into one of four chromosomally encoded central aromatic pathways, namely, the homogentisate, the catechol, the protocatechuate, and the phenylacetate pathways (3, 12, 17, 18). The catechol and protocatechuate pathways converge, leading to the β-ketoadipate pathway. It is within these central pathways that ring fission of the aromatic compound takes place. This strategy provides great metabolic versatility with a minimum number of enzymes.
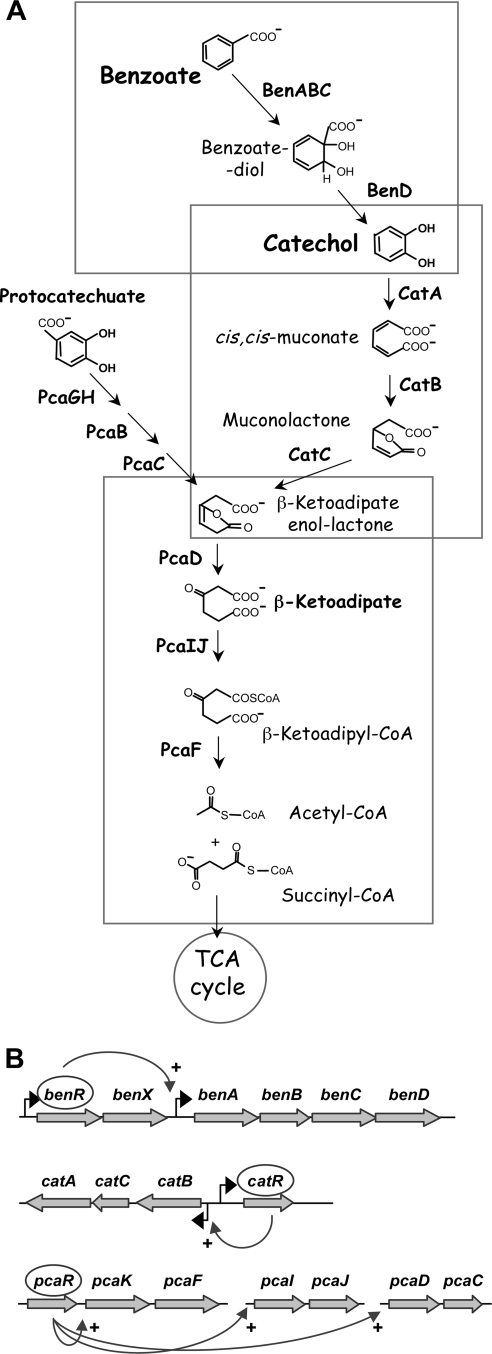
The assimilation of benzoate by P. putida involves the oxidation of benzoate into catechol in a two-step process catalyzed by benzoate dioxygenase, encoded by the benABC genes, and a cis-diol dehydrogenase, encoded by benD (12, 17), two enzymes that are peripheral in the catechol pathway (Fig. 1). The catechol aromatic ring is cleaved by catechol dioxygenase (CatA), producing cic,cis-muconate, which by the action of CatB and CatC is converted into β-ketoadipate-enol-lactone, which is transformed into acetyl coenzyme A (acetyl-CoA) and succinyl-CoA by enzymes of the β-ketoadipate pathway. The expression of the benABCD cluster requires the BenR transcriptional activator and benzoate as an effector (9), while the expression of the catBCA genes is activated by the CatR regulator, which needs cis,cis-muconate as an effector (30, 35) (Fig. 1). The pcaD, pcaIJ, and pcaF genes from the β-ketoadipate pathway are organized into separate transcriptional units (reviewed in reference 12), the expression of which is induced by β-ketoadipate and the PcaR transcriptional regulator (5, 12, 34). Therefore, although the ben genes need benzoate as an inducer, the cat and pca genes use metabolic intermediates (cis,cis-muconate and β-ketoadipate, respectively) as inducers.
FIG. 1.
The benzoate degradation pathway identified in P. putida KT2440. (A) The genes are organized into three units. The products of the ben genes transform benzoate into catechol. The enzymes encoded by the cat genes allow the conversion of catechol into β-ketoadipate-enol-lactone, while those encoded by the pca genes allow further assimilation of this compound into acetyl-CoA and succinyl-CoA, which are fed into the tricarboxylic acid (TCA) cycle. The point of entry of the products generated by the protocatechuate branch of the β-ketoadipate pathway is indicated. (B) Organization of the ben, cat, and pca genes. The expression of the benABCD genes requires the BenR transcriptional activator and benzoate as an effector, while that of the cat genes is regulated by the CatR transcriptional activator, which requires cis,cis-muconate as an effector (17). The expression of the pcaDC, pcaIJ, and pcaKF genes requires the PcaR transcriptional activator and β-ketoadipate as an effector (17).
When cells grow exponentially in a complete medium containing benzoate, the Crc regulator strongly inhibits the induction of the benABCD and catBCA genes (25). Crc influence on the expression of genes involved in further steps of the pathway, namely, pcaD, pcaIJ, and pcaF, has not been reported. While the Crc effect on benABCD may well be direct, that on catBCA may be direct or indirect, since the inhibition of benABCD expression impedes the transformation of benzoate into catechol, therefore hindering the activation of catBCA by CatR together with its effector cis,cis-muconate, which is not produced. This work analyzes which steps of the benzoate degradation pathway are controlled by Crc directly or indirectly and identifies the Crc target.
Crc is an important global regulator in pseudomonads. In addition to the above-mentioned pathways for aromatic compounds, it regulates the expression of genes involved in the assimilation of sugars, nitrogenated compounds, and several hydrocarbons (2, 8, 14, 15, 22, 45, 47). Crc represses gene expression posttranscriptionally (15, 27, 32, 47). However, its target and molecular mechanism have been established only for the P. putida OCT plasmid alkane degradation pathway. In this case, Crc inhibits the expression of the transcriptional activator of the pathway, the AlkS protein, by binding to the 5′ end of the corresponding mRNA and inhibiting translation (27). By keeping AlkS levels low, Crc modulates the induction of the alkane degradation genes when other preferred carbon sources are present in addition to alkanes. Here, we show that Crc inhibits the translation of the BenR transcriptional activator, responsible for the expression of the benABCD genes, suggesting that this process may be a general strategy for Crc action.
MATERIALS AND METHODS
Bacterial strains and culture media.
Strains were grown at 30°C in Luria-Bertani (LB) medium (38), supplemented where indicated with 5 mM benzoate or 5 mM catechol. Cell growth was monitored by measuring the turbidity at 600 nm. P. putida KT2442 is a rifampin-resistant derivative of P. putida KT2440 (11). P. putida KT2442-C1 derives from strain KT2442 by the inactivation of the crc gene (KT2442-C1 contains a crc::tet allele [36]). P. putida PBA1, also derived from KT2442, contains a benA::lacZ transcriptional fusion in its chromosome. The fusion includes a DNA segment spanning positions −267 to +17 relative to the transcription start site of the benA gene (−297 to −14 relative to the benA start codon). This DNA segment was PCR amplified with appropriate oligonucleotides containing restriction sites (underlined) for EcoRI (5′ end; 5′-CAGAGCGAATTCTTTTAACCTGG-3′) or BamHI (3′ end; 5′-GTCTCCGGATCCATTGTTTAGAC-3′), and the PCR product was cloned into the corresponding sites of plasmid pUJ8 (13), immediately upstream of a promoterless lacZ gene, to obtain plasmid pUJBA1. The sequence of the cloned DNA fragment was determined to ensure the absence of undesired mutations. The benA::lacZ fusion was excised from pUJBA1 as a NotI DNA fragment and inserted into the same site of the Mini-Tn5 suicide delivery plasmid pUT-Mini-Tn5Km (13), yielding pUTBA1. This plasmid was used to deliver the benA::lacZ fusion to the chromosome of P. putida KT2442 by means of triparental matings using pRK600 as a donor of transfer functions, as described previously (10). P. putida strain PBA1-C derives from KT2442-C1 through the insertion of the PbenA::lacZ transcriptional fusion into the chromosome of strain KT2442-C1 by the same procedure described for strain PBA1. Strains PBA1 and PBA1-C are representative of several independent transconjugants in which the benA::lacZ transcriptional fusion had similar expression patterns.
To generate a transcriptional fusion of benR to lacZ, a DNA segment spanning positions −568 to −14 relative to the translation start site (just upstream of the Shine-Dalgarno [SD] sequence) was PCR amplified using primers 5′-TTTTCGCGAATTCGGAGCTAAGCATT-3′ and 5′-CAAGGCGGATCCTATCGTTATTGTTC-3′ (the restriction sites are underlined). The PCR product was digested with EcoRI and BamHI and cloned between the corresponding sites of plasmid pUJ8, which contains a promoterless lacZ gene (13). After sequencing of the amplified region, the benR::lacZ fusion was excised from the resulting plasmid (pUJBR1) with NotI and cloned into the NotI site of suicide delivery plasmid pUT-Mini-Tn5Km (13), yielding pUTBR1. A similar strategy was followed to generate a translational fusion of the benR and lacZ genes, except that the oligonucleotides used were 5′-TTTTCGGAATTCGGAGCTAAGCATT-3′ and 5′-GGGTCGGGATCCTGAAACACGCTACT-3′ (the restriction sites are underlined), which generate a PCR product that spans positions −568 to +42 relative to the translation start site. After digestion with EcoRI and BamHI, the amplified DNA was cloned into the corresponding sites of plasmid pUJ9, designed to obtain translational fusion to lacZ (13), generating plasmid pUJBR2. After verification of the DNA sequence, the translational benR′-′lacZ fusion was excised from pUJBR2 with NotI and inserted into the corresponding site of pUTMini-Tn5Km, producing pUTBR2. The transcriptional and translational fusions contained in plasmids pUTBR1 and pUTBR2 were delivered to the chromosome of P. putida KT2442 or KT2442-C1 (a crc::tet derivative of KT2442) (36) by triparental matings using pRK600 as a donor of transfer functions. Several transconjugants were analyzed; representative ones were selected and named KTBR1 (containing a transcriptional fusion and a wild-type crc gene), KTBR1C (containing a transcriptional fusion and crc::tet), KTBR2 (containing a translational fusion and wild-type crc), and KTBR2C (containing a transcriptional fusion and crc::tet).
RNA purification and real-time reverse transcription-PCR (RT-PCR).
Cells were grown at 30°C in LB medium in aerated flasks. At mid-exponential phase (A600 = 0.7), 25-ml samples were collected and combined with a cold mixture containing 5% (vol/vol) phenol and 95% (vol/vol) ethanol in a volume equal to one-fifth of the culture volume to stabilize bacterial RNA. Cells were harvested by centrifugation, and the pellets were frozen at −70°C. At least three independent cultures of each strain were grown. RNA was purified from each pellet with the RNeasy RNA purification kit by following the instructions of the manufacturer (Qiagen). Purified RNA was treated with RNase-free DNase I (TURBO DNA free) as specified by the supplier (Ambion). RNA integrity was analyzed by agarose gel electrophoresis. The absence of DNA was checked by PCR using primers for rpoN, as described previously (26).
RNA samples (10 μg) were transformed into cDNA by using the high-capacity cDNA archive kit (Applied Biosystems). Real-time PCR was performed as described previously (26). The primer pairs used for each gene were 5′-TGCCTGTACCCCAACGTGTA-3′ and 5′-GGGCAACGCGGATCTG-3′ for benA, 5′-CGAGGTACGCGTCGATGTT-3′ and 5′-CCTGGCGTATGGCTACGAAA-3′ for catB, 5′-GCAAGGAAACCCGTGAGATC-3′ and 5′-GCGTGCAGCGGCATTT-3′ for pcaI, 5′-CCTGCTGGCCATGGTTTG-3′ and 5′-CCTGGGCAGCGAATCG-3′ for benR, 5′-CGGCGCGGGTGAAGT-3′ and 5′-TGCGCCACCTGCGTTACT-3′ for catR, and 5′-CAACGATGACGACGAATGG-3′ and 5′-ATCAGGGTCACGGCAATC-3′ for rpoN. PCR products were between 100 and 245 bp in length. Results were normalized relative to those obtained for the rpoN gene, since its expression in P. putida under several conditions is known to remain relatively constant throughout the growth phase (7, 16, 19, 46).
β-Galactosidase assays.
An overnight culture was diluted to a final turbidity (A600) of 0.04 in fresh LB medium. When the turbidity reached 0.08, benzoate was added to give a final concentration of 5 mM, where indicated, to induce the expression of the benA promoter (PbenA). Cultures were grown at 30°C. At different time points, aliquots were taken and β-galactosidase activity was measured as described by Miller (24) by using o-nitrophenyl-β-d-galactoside (ONPG) as the substrate. Where indicated, β-galactosidase activity was measured as well by chemiluminescence using Galacton-Plus (Tropix) as the substrate and Emerald-II (Tropix) as an enhancer. Reactions were performed in 96-well plates as described previously (43). Results were analyzed on a luminometer (TECAN Infinite 200); values shown (expressed in arbitrary units) correspond to the relative luminescence units displayed by the luminometer. The same amounts of bacterial cell samples were used in all assays. Four independent experiments were performed.
Protein purification.
Protein Crc-His6 was overproduced in Escherichia coli BL21(DE3)(pLysS) containing plasmid pCRCH and purified using a Ni-nitrilotriacetic acid column as described previously (27). The protein obtained was >95% pure.
RNA band shift assays.
Reaction mixtures contained, in 20 μl, 10 mM HEPES-KOH (pH 7.9), 35 mM KCl, 2 mM MgCl2, 0.1 nM radioactively labeled RNA, 1 μg of yeast tRNA, and where indicated, purified Crc-His6. After 1 h of incubation at room temperature, 4 μl of loading buffer (60% [vol/vol] glycerol, 0.025% [wt/vol] xylene cyanol) was added and samples were loaded onto a nondenaturing 4% (wt/vol) polyacrylamide gel containing TBM buffer (45 mM Tris-HCl [pH 8.3], 43 mM boric acid, 2 mM MgCl2, 5% [vol/vol] glycerol). Electrophoresis was performed at 4°C with TBM as the running buffer.
The radioactively labeled RNA fragments used as the substrates were obtained by in vitro transcription using plasmid pbenR51, pcatR46, or ppcaR47 as the template, which allowed the production of the desired RNA transcripts from the T7 promoter of the vector after linearization with PstI. Transcription was performed for 1 h at 37°C; reaction mixtures contained 40 mM Tris-HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 10 mM NaCl, 1 μg of DNA template (linearized with PstI), 10 mM dithiothreitol, 0.5 mM (each) ATP, CTP, GTP, and [α32P]UTP (3,000 Ci/mmol), and 20 U of T7 RNA polymerase. The reaction mixture was loaded onto a denaturing 6% (wt/vol) urea-polyacrylamide gel, and the RNA fragment corresponding to the benR, catR, or pcaR mRNA 5′ end was excised from the gel and purified. To generate plasmids pbenR51, pcatR46, and ppcaR47, DNA fragments of appropriate lengths were PCR amplified with oligonucleotides providing targets for ApaI (5′ end) and PstI (3′ end). These fragments were as follows (ApaI and PstI targets are underlined): 5′-GCGGTAGGGGGGCCCGACCAGA-3′ and 5′-TACTGCCGTCCTGCAGCAGGCG-3′ for benR, 5′-CAATATCGGGCCCATCTCCCACC-3′ and 5′-AAGACTTTGCTGCAGCGCAGGTG-3′ for catR, and 5′-TTTGTTCGAGGGCCCCACGAACC-3′ and 5′-GAATCGTCTGCAGGGGTTTCGT-3′ for pcaR. The DNA fragments obtained were digested with ApaI and PstI and cloned between the corresponding sites of pGEM-T Easy (Promega).
RESULTS AND DISCUSSION
Effect of Crc on the mRNA levels of key genes of the benzoate degradation pathway.
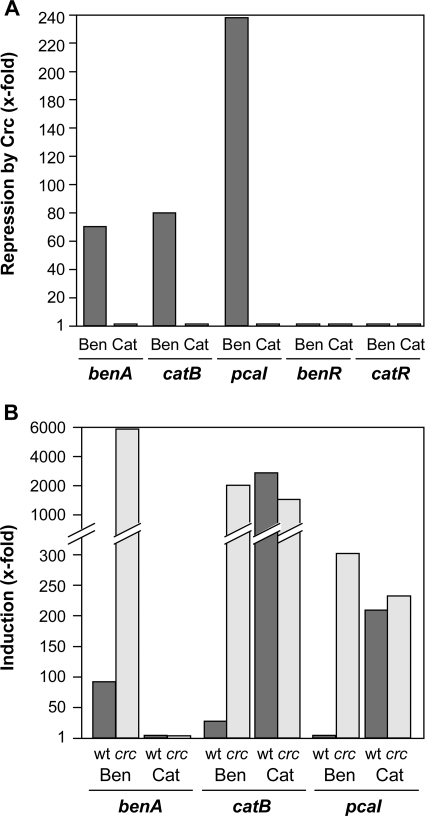
In P. putida, the inhibitory effect of Crc on gene expression is particularly strong in cells growing exponentially in a complete medium such as LB, and it fades away as soon as cells enter into stationary phase (47). Accordingly, to differentiate between direct and indirect effects of Crc on the expression of the genes involved in benzoate degradation, strain KT2442 and its isogenic Crc-deficient derivative KT2442-C1 were grown in complete LB medium in the absence or presence of either benzoate or catechol. At mid-exponential phase, total RNA was purified and the mRNA levels of the benA, catB, and pcaI genes were analyzed by real-time RT-PCR. The expression of benR and catR, which encode the transcriptional regulators of the ben and cat genes, respectively, was also analyzed. As shown in Fig. 2A, for cells growing exponentially in the presence of benzoate, the levels of benA, catB, and pcaI mRNAs in the strain lacking Crc were about two orders of magnitude higher than those in the wild-type strain; Crc repressed expression of benA about 70-fold, that of catB about 80-fold, and that of pcaI about 240-fold. The mRNA levels of benR and catR were not affected by the absence or presence of Crc. When cells were grown in the presence of catechol, rather than benzoate, the mRNA levels of benA, catB, pcaI, benR, and catR were essentially the same in the wild-type strain and in the strain lacking the Crc regulator (Fig. 2A). This result suggests that Crc affects the expression of the benA gene directly but that the effect on the catB and pcaI genes is indirect, due to the poor conversion of benzoate into catechol because of the weak expression of benzoate dioxygenase, which in turn would prevent the induction of the cat and pca genes due to the low levels of cis,cis-muconate and β-ketoadipate (effectors of CatR and PcaR, respectively) generated from benzoate (Fig. 1).
FIG. 2.
Effect of Crc on expression of the benzoate degradation pathway. The expression of the benA, catB, and pcaI genes was evaluated by real-time RT-PCR analysis of cells growing exponentially in LB medium in the absence or presence of either benzoate (Ben) or catechol (Cat). (A) The Crc repression factor was calculated by comparing mRNA levels (see Materials and Methods) for each gene in strain KT2442-C (which contains an inactivated crc allele) to those observed in the parental strain KT2442 (which contains wild-type crc). (B) Values of induction by benzoate or catechol corresponded to the mRNA levels observed for the indicated genes in cells of strain KT2442 (wild type [wt]) and KT2442-C (crc) growing in the presence of benzoate or catechol relative to those observed in these strains in the absence of benzoate or catechol.
It is worth noting that, even in the presence of Crc, the addition of benzoate to the growth medium led to significant induction of benA and catB genes (up to 85-fold for benA and 25-fold for catB) (Fig. 2B). For pcaI, the degree of induction was low. This finding indicates that, although Crc can be a strong modulator of gene expression, it does not necessarily switch off the affected pathways completely. Catechol did not induce benA, as could be predicted, but led to significant induction of catB and pcaI that was independent of Crc (Fig. 2B).
Effect of Crc on expression of a benA::lacZ transcriptional fusion.
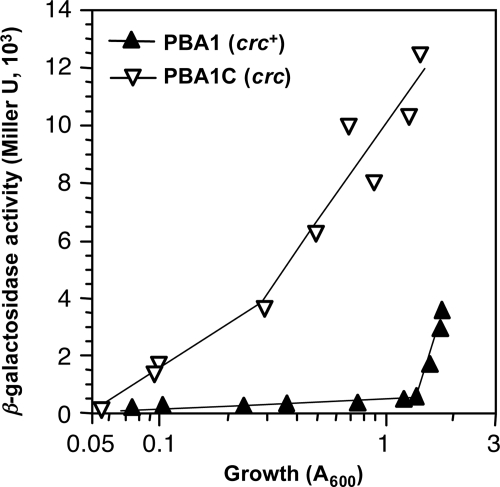
As an additional way to analyze the effect of Crc on the expression of the benABCD genes, a transcriptional fusion containing the PbenA promoter upstream of the lacZ reporter gene was constructed. The fusion was delivered into the chromosome of P. putida KT2442 by means of a Mini-Tn5 transposon, generating strain PBA1. The activity of PbenA in this strain was monitored throughout growth in cells grown in complete LB medium in the absence or presence of benzoate and was compared to that observed in strain PBA1-C, which harbors both the benA::lacZ fusion and an inactivated crc allele (crc::tet). The levels of β-galactosidase in the two strains in cultures lacking benzoate were very low throughout all the growth phase (data not shown). In the presence of benzoate, β-galactosidase levels in the strain containing a functional crc gene were very low during the exponential phase of growth and increased when the growth rate started to decline as cells entered into stationary phase, which occurred at turbidity values above 1.5 (Fig. 3). In contrast, in the case of the strain containing an inactivated crc::tet allele (strain PBA1-C), the activity of PbenA increased steadily right from the start of the exponential phase. This result further supports the idea that Crc exerts a potent inhibitory effect on the activity of the promoter for the benABCD genes during exponential growth in a complete medium.
FIG. 3.
Effect of Crc on PbenA activity in cells growing in LB medium. P. putida strains PBA1 and PBA1-C, both of which contain a benA::lacZ transcriptional fusion and either a wild-type crc gene (strain PBA1) or an inactive crc::tet alelle (strain PBA1-C), were grown in LB medium in the absence or presence of 5 mM benzoate. Aliquots were taken at different times, and β-galactosidase activity was measured. The plot shows the values obtained as a function of cell growth (as indicated by the turbidity at 600 nm). Values obtained for the cultures lacking benzoate were very low and are not represented.
Crc target in the benzoate degradation pathway.
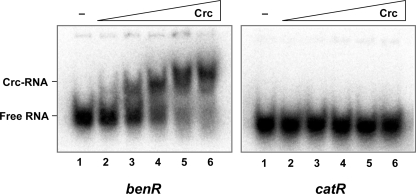
As explained above, although Crc has been proposed to act posttranscriptionally in several cases, the only direct target known to date is the AlkS transcriptional regulator of the P. putida OCT plasmid alkane degradation pathway. A detailed analysis showed that Crc inhibits the expression of the alkS gene by binding to the 5′ end of its mRNA and inhibiting translation (27). This inhibition in turn ensures that the amounts of AlkS protein produced are not large enough to achieve full induction of the alkane degradation genes. The results described above indicate that when cells grow exponentially in a complete medium containing benzoate, Crc strongly downregulates the mRNA levels of benA but not those of benR, the regulator that activates the expression of the benABCD operon in the presence of benzoate. This finding suggests that Crc may inhibit the translation of benR mRNA, decreasing the levels of the BenR transcriptional activator below those required to fully induce the promoter for the benABCD genes. In other words, Crc would affect the levels of benA mRNA indirectly. To test this hypothesis, the ability of purified Crc to bind to the 5′ end of benR mRNA was analyzed in vitro by band shift assays. The labeled RNA fragment used as the substrate, obtained by in vitro transcription, included positions −35 to +16 relative to the benR translation start site. Binding reactions were performed in the presence of a large excess of unlabeled tRNA, conditions under which Crc binds only to specific targets (27). As shown in Fig. 4, the purified Crc protein generated a clear retardation band when added at concentrations comparable to those that have been described to generate a Crc-RNA complex with the alkS mRNA (27). Under the same conditions, Crc did not bind to an RNA fragment including positions −26 to +20 of the catR activator mRNA (Fig. 4). Similarly, no binding to an RNA fragment including positions −31 to +16 of the pcaR activator mRNA was observed (data not shown). This result supports the idea that Crc inhibits the translation of the benR mRNA by binding to its 5′ end, as it does for the alkS mRNA.
FIG. 4.
Binding of the Crc protein to benR or catR mRNA. Band shift assay mixtures contained a radioactively labeled mRNA fragment including the 5′ end of benR or catR mRNA, 1 μg of yeast tRNA, and 30, 60, 125, 250, or 500 ng (lanes 2 to 6) of purified Crc. In both panels, lane 1 shows the migration of the RNA in the absence (−) of Crc. Protein-RNA complexes were resolved on a nondenaturing 4% (wt/vol) polyacrylamide gel. The position of the free-RNA substrate of the Crc-RNA complex is indicated.
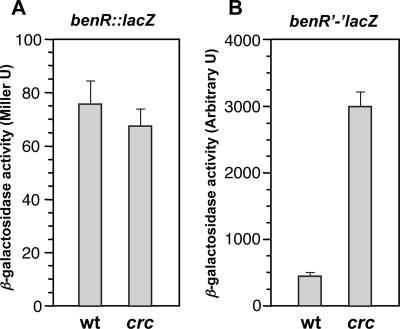
To further analyze this possibility, a transcriptional fusion and a translational fusion of benR to the lacZ reporter gene were generated. The transcriptional fusion included the 5′ end of benR mRNA but excluded the benR SD and downstream sequences, so that lacZ used its own SD sequence. The translational fusion included the benR sequence up to the 14th codon and, therefore, used the benR SD sequence. Each fusion was delivered into the chromosome of P. putida KT2442 (containing wild-type crc) or that of KT2442-C1 (containing an inactivated crc::tet allele) by means of a Mini-Tn5Km transposon. As shown in Fig. 5A, in cells growing exponentially in LB medium, the transcriptional benR::lacZ fusion generated similar β-galactosidase levels in the presence of Crc (strain KTBR1) and in its absence (strain KTBR1C). Under similar conditions, the level of expression of the translational benR′-′lacZ fusion in the presence of Crc (strain KTBR2) was fourfold lower than that in a Crc-deficient background (strain KTBR2C). However, the levels of β-galactosidase in the strains harboring the translational benR′-′lacZ fusion were very low (means ± standard deviations of 2 ± 0.7 Miller U for strain KTBR2 and 8 ± 1.5 Miller U for strain KTBR2C). Although these expression levels were reproducible, the assays were performed as well using the chemiluminescent substrate Galacton-Plus, which is much more sensitive than ONPG. In this case, the expression of the benR′-′lacZ translational fusion was sevenfold more efficient in the absence of Crc than in its presence (Fig. 5B). For both the transcriptional and translational fusions, the results from four independent transconjugants were similar. Therefore, the results of RNA band shift assays and the behavior of the transcriptional and translational fusions of benR to lacZ indicate that Crc inhibits the translation of benR mRNA. In this way, Crc would reduce the levels of the BenR transcriptional regulator below those required to achieve the efficient activation of promoter PbenA in the presence of benzoate. The final effect would thus be a reduction in the levels of the benABCD mRNA, which would impair the transformation of benzoate into catechol. This impairment would in turn prevent the production of cic,cis-muconate and β-ketoadipate, the effectors of the CatR and PcaR activators, which would keep the cat and pca genes uninduced. Our observation that Crc cannot bind to the 5′ ends of the catR and pcaR mRNAs, suggesting that it cannot regulate the translation of these mRNAs, is consistent with the idea that the Crc effect on the expression of the cat and pca genes is indirect.
FIG. 5.
Effect of Crc on the expression of a benR::lacZ transcriptional fusion (A) and of a translational fusion of the benR and lacZ genes (benR′-′lacZ) (B). P. putida strains KTBR1 (wild type [wt] containing benR::lacZ and a wild-type crc gene), KTBR1C (crc mutant [crc] containing benR::lacZ and an inactivated crc::tet allele), KTBR2 (wild type containing the translational benR′-′lacZ fusion and a wild-type crc gene), and KTBR2C (crc mutant containing the translational benR′-′lacZ fusion and an inactivated crc::tet allele) were grown in LB medium. At mid-exponential phase (turbidity of 0.5 at 600 nm), aliquots were taken and β-galactosidase activity was measured using either ONPG (A) or Galacton-Plus (B) as the substrate. Values correspond to the averages of results from four independent assays; the standard deviations are indicated.
The precise sequence or RNA structure recognized by Crc at the 5′ ends of the mRNAs it regulates is at present unknown. A comparison of the 5′ ends of alkS and benA mRNAs, to which Crc binds, did not show any clear sequence similarity. The prediction of the secondary structures that these two mRNAs may adopt by using the Mfold algorithm (www.bioinfo.rpi.edu; 48) did not show a clear common motif to which Crc could bind. The determinants for RNA recognition by proteins are normally subtle, frequently consisting of a relatively small number of nucleotides with a precise spatial distribution that relies on a particular RNA secondary structure (4, 20, 39, 44). Identification of the RNA determinants recognized by Crc will probably require the comparison of many more Crc targets and an extensive mutagenesis approach.
Conclusions.
The work presented here shows that Crc regulates the expression of the benzoate degradation pathway by acting at the first step, catalyzed by the products of the benABCD genes, while genes corresponding to further steps of the pathway (the cat and pca genes) are not induced because the appropriate degradation intermediates, which act as inducers, are not produced. The results presented herein also identify the Crc target as the 5′ end of the benR mRNA, indicating that the downregulation of benABCD expression is achieved by decreasing the levels of the BenR transcriptional activator below those required to fully induce PbenA. The effect of Crc on the expression of the cat and pca genes is, therefore, indirect. Thus, it is the induction of the peripheral genes that is regulated, not the central catechol and β-ketoadipate pathways. This strategy allows for the individual regulation of genes for the degradation of different carbon sources that share a common central pathway, facilitating the hierarchical assimilation of these sources. The benzoate, catechol, and β-ketoadipate pathways are widespread in bacteria and metabolize compounds that are abundantly produced by plants (17, 18). There is scattered evidence indicating that, when confronted by mixtures of aromatic compounds in sufficient concentrations, some soil bacteria metabolize certain aromatics preferentially over others. For example, root extracts from several plants, which contain several aromatic compounds, can inhibit the assimilation of phenanthrene by a strain of P. putida (33). Similarly, benzoate inhibits the catabolism of phenol in Ralstonia eutropha (1) and represses 4-hydroxybenzoate degradation genes in P. putida (29). This modulation can best be achieved if global regulation is targeted to the peripheral genes, rather than to the central pathways for aromatic compounds, and is likely to provide enhanced flexibility to select the preferred carbon source under a given circumstance.
Acknowledgments
We are grateful to L. Yuste for excellent technical assistance.
This work was supported by grant BFU2006-00767/BMC from the Spanish Ministry of Education and Science.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Ampe, F., D. Leonard, and N. D. Lindley. 1998. Repression of phenol catabolism by organic acids in Ralstonia eutropha. Appl. Environ. Microbiol. 641-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranda-Olmedo, I., J. L. Ramos, and S. Marqués. 2005. Integration of signals through Crc and PtsN in catabolite repression of Pseudomonas putida TOL plasmid pWW0. Appl. Environ. Microbiol. 714191-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arias-Barrau, E., E. R. Olivera, J. M. Luengo, C. Fernández, B. Galan, J. L. García, E. Díaz, and B. Miñambres. 2004. The homogentisate pathway: a central catabolic pathway involved in the degradation of l-phenylalanine, l-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 1865062-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auweter, S. D., F. C. Oberstrass, and F. H. Allain. 2006. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 344943-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertani, I., M. Kojic, and V. Venturi. 2001. Regulation of the p-hydroxybenzoic acid hydroxylase gene (pobA) in plant-growth-promoting Pseudomonas putida WCS358. Microbiology 1471611-1620. [DOI] [PubMed] [Google Scholar]
- 6.Cases, I., and V. de Lorenzo. 2005. Promoters in the environment: transcriptional regulation in its natural context. Nat. Rev. Microbiol. 3105-118. [DOI] [PubMed] [Google Scholar]
- 7.Cases, I., V. de Lorenzo, and J. Pérez-Martín. 1996. Involvement of sigma 54 in exponential silencing of the Pseudomonas putida TOL plasmid Pu promoter. Mol. Microbiol. 197-17. [DOI] [PubMed] [Google Scholar]
- 8.Collier, D. N., P. W. Hager, and P. V. Phibbs, Jr. 1996. Catabolite repression control in Pseudomonads. Res. Microbiol. 147551-561. [DOI] [PubMed] [Google Scholar]
- 9.Cowles, C. E., N. N. Nichols, and C. S. Harwood. 2000. BenR, a XylS homologue, regulates three different pathways of aromatic acid degradation in Pseudomonas putida. J. Bacteriol. 1826339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235386-405. [DOI] [PubMed] [Google Scholar]
- 11.Franklin, F. C., M. Bagdasarian, M. M. Bagdasarian, and K. N. Timmis. 1981. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl. Acad. Sci. USA 787458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harwood, C. S., and R. E. Parales. 1996. The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50553-590. [DOI] [PubMed] [Google Scholar]
- 13.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1726557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hester, K. L., J. Lehman, F. Najar, L. Song, B. A. Roe, C. H. MacGregor, P. W. Hager, P. V. Phibbs, Jr., and J. R. Sokatch. 2000. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 1821144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hester, K. L., K. T. Madhusudhan, and J. R. Sokatch. 2000. Catabolite repression control by crc in 2xYT medium is mediated by posttranscriptional regulation of bkdR expression in Pseudomonas putida. J. Bacteriol. 1821150-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishihama, A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54499-518. [DOI] [PubMed] [Google Scholar]
- 17.Jiménez, J. I., B. Miñambres, J. L. García, and E. Díaz. 2002. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4824-841. [DOI] [PubMed] [Google Scholar]
- 18.Jiménez, J. I., B. Miñambres, J. L. García, and E. Díaz. 2004. Genomic insights in the metabolism of aromatic compounds in Pseudomonas, p. 425-462. In J. L. Ramos (ed.), Pseudomonas, vol. 3. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 19.Jishage, M., A. Iwata, S. Ueda, and A. Ishihama. 1996. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 1785447-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapouge, K., E. Sineva, M. Lindell, K. Starke, C. S. Baker, P. Babitzke, and D. Haas. 2007. Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol. Microbiol. 66341-356. [DOI] [PubMed] [Google Scholar]
- 21.Lugtenberg, B. J., and L. C. Dekkers. 1999. What makes Pseudomonas bacteria rhizosphere competent? Environ. Microbiol. 19-13. [DOI] [PubMed] [Google Scholar]
- 22.MacGregor, C. H., S. K. Arora, P. W. Hager, M. B. Dail, and P. V. Phibbs, Jr. 1996. The nucleotide sequence of the Pseudomonas aeruginosa pyrE-crc-rph region and the purification of the crc gene product. J. Bacteriol. 1785627-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martins dos Santos, V. A., S. Heim, E. R. Moore, M. Stratz, and K. N. Timmis. 2004. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 61264-1286. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 25.Morales, G., J. F. Linares, A. Beloso, J. P. Albar, J. L. Martínez, and F. Rojo. 2004. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 1861337-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morales, G., A. Ugidos, and F. Rojo. 2006. Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidases. Environ. Microbiol. 81764-1774. [DOI] [PubMed] [Google Scholar]
- 27.Moreno, R., A. Ruiz-Manzano, L. Yuste, and F. Rojo. 2007. The Pseudomonas putida Crc global regulator is an RNA binding protein that inhibits translation of the AlkS transcriptional regulator. Mol. Microbiol. 64665-675. [DOI] [PubMed] [Google Scholar]
- 28.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. Chris Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. Eisen, K. N. Timmis, A. Dusterhoft, B. Tümmler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4799-808. [DOI] [PubMed] [Google Scholar]
- 29.Nichols, N. N., and C. S. Harwood. 1995. Repression of 4-hydroxybenzoate transport and degradation by benzoate: a new layer of regulatory control in the Pseudomonas putida beta-ketoadipate pathway. J. Bacteriol. 1777033-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsek, M. R., D. L. Shinabarger, R. K. Rothmel, and A. M. Chakrabarty. 1992. Roles of CatR and cis,cis-muconate in activation of the catBC operon, which is involved in benzoate degradation in Pseudomonas putida. J. Bacteriol. 1747798-7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pirnay, J. P., S. Matthijs, H. Colak, P. Chablain, F. Bilocq, J. Van Eldere, D. De Vos, M. Zizi, L. Triest, and P. Cornelis. 2005. Global Pseudomonas aeruginosa biodiversity as reflected in a Belgian river. Environ. Microbiol. 7969-980. [DOI] [PubMed] [Google Scholar]
- 32.Putrins, M., A. Tover, R. Tegova, U. Saks, and M. Kivisaar. 2007. Study of factors which negatively affect expression of the phenol degradation operon pheBA in Pseudomonas putida. Microbiology 1531860-1871. [DOI] [PubMed] [Google Scholar]
- 33.Rentz, J. A., P. J. Alvarez, and J. L. Schnoor. 2004. Repression of Pseudomonas putida phenanthrene-degrading activity by plant root extracts and exudates. Environ. Microbiol. 6574-583. [DOI] [PubMed] [Google Scholar]
- 34.Romero-Steiner, S., R. E. Parales, C. S. Harwood, and J. E. Houghton. 1994. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J. Bacteriol. 1765771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothmel, R. K., T. L. Aldrich, J. E. Houghton, W. M. Coco, L. N. Ornston, and A. M. Chakrabarty. 1990. Nucleotide sequencing and characterization of Pseudomonas putida catR: a positive regulator of the catBC operon is a member of the LysR family. J. Bacteriol. 172922-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruiz-Manzano, A., L. Yuste, and F. Rojo. 2005. Levels and activity of the Pseudomonas putida global regulatory protein Crc vary according to growth conditions. J. Bacteriol. 1873678-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saier, M. H., Jr., T. M. Ramseier, and J. Reizer. 1996. Regulation of carbon utilization, p. 1325-1342. In F. C. Neidhart, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, DC.
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Schubert, M., K. Lapouge, O. Duss, F. C. Oberstrass, I. Jelesarov, D. Haas, and F. H. Allain. 2007. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat. Struct. Mol. Biol. 14807-813. [DOI] [PubMed] [Google Scholar]
- 40.Shingler, V. 2003. Integrated regulation in response to aromatic compounds: from signal sensing to attractive behaviour. Environ. Microbiol. 51226-1241. [DOI] [PubMed] [Google Scholar]
- 41.Stülke, J., and W. Hillen. 1999. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2195-201. [DOI] [PubMed] [Google Scholar]
- 42.Timmis, K. N. 2002. Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ. Microbiol. 4779-781. [DOI] [PubMed] [Google Scholar]
- 43.Valls, M., M. Buckle, and V. de Lorenzo. 2002. In vivo UV laser footprinting of the Pseudomonas putida sigma 54-Pu promoter reveals that integration host factor couples transcriptional activity to growth phase. J. Biol. Chem. 2772169-2175. [DOI] [PubMed] [Google Scholar]
- 44.Williamson, J. R. 2000. Induced fit in RNA-protein recognition. Nat. Struct. Biol. 7834-837. [DOI] [PubMed] [Google Scholar]
- 45.Wolff, J. A., C. H. MacGregor, R. C. Eisenberg, and P. V. Phibbs, Jr. 1991. Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J. Bacteriol. 1734700-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuste, L., A. B. Hervás, I. Canosa, R. Tobes, J. I. Jiménez, J. Nogales, M. M. Pérez-Pérez, E. Santero, E. Díaz, J. L. Ramos, V. de Lorenzo, and F. Rojo. 2006. Growth phase-dependent expression of the Pseudomonas putida KT2440 transcriptional machinery analyzed with a genome-wide DNA microarray. Environ. Microbiol. 8165-177. [DOI] [PubMed] [Google Scholar]
- 47.Yuste, L., and F. Rojo. 2001. Role of the crc gene in catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 1836197-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]