Abstract
Microorganisms adapted to piezopsychrophilic growth dominate the majority of the biosphere that is at relatively constant low temperatures and high pressures, but the genetic bases for the adaptations are largely unknown. Here we report the use of transposon mutagenesis with the deep-sea bacterium Photobacterium profundum strain SS9 to isolate dozens of mutant strains whose growth is impaired at low temperature and/or whose growth is altered as a function of hydrostatic pressure. In many cases the gene mutation-growth phenotype relationship was verified by complementation analysis. The largest fraction of loci associated with temperature sensitivity were involved in the biosynthesis of the cell envelope, in particular the biosynthesis of extracellular polysaccharide. The largest fraction of loci associated with pressure sensitivity were involved in chromosomal structure and function. Genes for ribosome assembly and function were found to be important for both low-temperature and high-pressure growth. Likewise, both adaptation to temperature and adaptation to pressure were affected by mutations in a number of sensory and regulatory loci, suggesting the importance of signal transduction mechanisms in adaptation to either physical parameter. These analyses were the first global analyses of genes conditionally required for low-temperature or high-pressure growth in a deep-sea microorganism.
The bulk of deep-sea environments select for microorganisms which grow preferentially at low temperatures (psychrophiles) and high hydrostatic pressures (piezophiles) and which are capable of tolerating sporadic inputs of organic nutrients (9, 10).
Photobacterium profundum strain SS9 (69) is a psychrotolerant and moderately piezophilic bacterium. It was first isolated from an amphipod homogenate enrichment from the Sulu Sea (28). This microorganism, which is suitable for a variety of systems biology investigations, is capable of growth at temperatures of <2 to >20°C (optimal temperature, 15°C) and at pressures from 0.1 MPa to nearly 90 MPa (optimal pressure, 28 MPa). Analysis of the SS9 genome and transcriptome has suggested that two important aspects of the deep-sea adaptations of this organism are the nature of its pressure- and temperature-responsive genes and its high degree of metabolic diversity and redundancy (87).
Like the genes of other Vibrionaceae, the genes of P. profundum are partitioned onto two circular chromosomes (19, 87). The two chromosomes are thought to be functionally distinct, with most “established” and essential genes located on chromosome 1 and most strain-specific and horizontally acquired genes located on chromosome 2 (39, 87). In addition, P. profundum SS9 has a dispensable 80-kbp plasmid carrying mostly genes with unknown functions (19, 87). Intraspecific gene variation between strains of P. profundum that differ in their degrees of piezoadaptation has been examined, and sequences acquired by lateral gene transfer or which could be important in high-pressure growth have been identified (19).
Genetic approaches have also been used to elucidate mechanisms of deep-sea adaptation in P. profundum SS9. During the course of investigation of factors that influence the pressure regulation of outer membrane protein abundance, the membrane-localized transcription factor ToxR was identified as a pressure sensor (92), and the rseC and recD genes have been found to be important for low-temperature and high-pressure growth and for high-pressure growth, respectively (15, 23). More recently, site-directed insertional mutagenesis was employed to identify genes involved in fatty acid unsaturation important for high-pressure growth (4-6). None of the studies mentioned above included a global analysis of genes conditionally required for growth at high pressure or low temperature.
To complement and expand these studies, a collection of mini-Tn5 and mini-Tn10 transposon mutants was screened for high-pressure sensitivity and low-temperature sensitivity. In this paper we describe isolation and characterization of these mutants.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and mutant screening.
P. profundum strains were routinely cultured under aerobic conditions in 0.75× 2216 medium (28 g/liter; Difco Laboratories) at 15°C. Escherichia coli strains were grown aerobically at 37°C in Luria-Bertani medium.
Antibiotics were used at the following final concentrations: rifampin (Rif), 100 μg/ml; kanamycin (Km), 100 μg/ml for E. coli and 200 μg/ml for P. profundum; and streptomycin (Sm), 50 μg/ml for E. coli and 150 μg/ml for P. profundum. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was added to solid medium at a concentration of 40 μg/ml in N,N-dimethylformamide.
Bacterial strains and plasmids used in this study are listed in Table 1. Plasmids were introduced into P. profundum strain SS9 by triparental conjugation using an E. coli strain containing the helper plasmid pRK2073 or pRK2013 as previously described (24).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| P. profundum SS9R | Rifr SS9 derivative | 24 |
| E. coli strains | ||
| ED8654 | Used for pRK2073 maintenance | 67 |
| DH5α | recA strain used for cloning | 37 |
| XL1-Blue | recA strain used for cloning | Stratagene, La Jolla, CA |
| TOP10 | recA strain used for cloning | Invitrogen, Carlsbad, CA |
| Plasmids | ||
| pRK2073 | Carries tra genes for conjugal transfer | 14 |
| pRK2013 | Carries tra genes for conjugal transfer | 14 |
| pLOF | Mini-Tn10 transposon, Kmr | 40 |
| pRL27 | Mini-Tn5 transposon, Kmr | 52 |
| pFL122 | Broad-host-range vector, Smr | 54 |
| pFL400 | PBPRB0212 in pFL122 | This study |
| pFL401 | PBPRA0747 in pFL122 | This study |
| pFL402 | PBPRA3239 in pFL122 | This study |
| pFL403 | PBPRB1757 in pFL122 | This study |
| pFL404 | PBPRA2596 in pFL122 | This study |
Transposon mutagenesis was performed by mating E. coli S17-1 containing the mini-Tn10 donor plasmid pLOF (40) or E. coli BW20767 containing the mini-Tn5 donor plasmid pRL27 (52) with P. profundum strain SS9R (a rifampin-resistant derivative of SS9) and screening for exconjugants growing with double selection (kanamycin plus rifampin) on 2216 marine agar plates (Difco laboratories, BD, Franklin Lakes, NJ).
Screening for auxotrophic mutants was performed by replica plating onto morpholinepropanesulfonic acid (MOPS)-glucose minimal marine medium (11). Colonies unable to grow on this medium were considered auxotrophs. Screening for mutants whose growth at low temperatures was impaired was performed by replica plating transposon mutants onto 2216 marine agar plates and incubating the plates at 15 and 4°C. The clones that grew poorly compared to the wild type after 120 h of incubation at 4°C were saved. They were subsequently rescreened in liquid medium, and growth plots were determined in triplicate. A cold sensitivity ratio was computed by dividing the low-temperature growth rate of each mutant by the maximum growth rate at 15°C. When a mutant reproducibly displayed the cold-sensitive phenotype on plates but not in liquid, it was still recorded as cold sensitive.
High-pressure growth of P. profundum SS9 transposon mutants was performed at 17°C in 2216 marine medium supplemented with 20 mM glucose, 10 mM HEPES (pH 7.5), 200 μg/ml of kanamycin, and 50 mg/liter of phenol red as a color indicator for fermentative growth. Transposon mutants were individually inoculated into wells of a PCR plate with 96 elevated wells (Aygen Scientific, Union City, CA), and the wells were filled to the top with the high-pressure growth medium described above. The plate was sealed with a 96-well PCR Axymat (Axygen Scientific, Union City, CA), and we were careful not to trap any air bubbles. The microtiter plates were then pressurized using water and a hydraulic pump in a custom-designed stainless steel 2-liter pressure vessel (Autoclave Engineers, Erie, PA). Screening for pressure sensitivity was done by replica plating transposon mutants, incubating them at high pressure (45 MPa) and low pressure (0.1 MPa) at 15°C, and looking for wells that retained the red color after 48 h, which indicated that no fermentative growth had occurred,. The pressure-sensitive mutants were saved and rescreened by determining their growth plots in triplicate.
Pressure-dependent growth curves were obtained by using 1,000-fold dilutions of 48-h-old cultures of the mutants that were aliquoted in heat-sealable plastic bulbs and grown at 0.1 and 45 MPa. At each time point, one bulb was removed from the stock and transferred to a glass tube, and the turbidity of the culture at 600 nm was measured with a Spectronic 20 spectrophotometer (Milton Roy, New York). The pressure sensitivity ratio of strains was assessed as previously described (23). Briefly, when the low-pressure culture entered early logarithmic phase (optical density at 600 nm [OD600], 0.1 to 0.3), two values were recorded: the OD600 of the 45-MPa culture (a) and the OD600 of the 0.1-MPa culture (b). The pressure sensitivity ratio (a/b) was then computed. Under these conditions the pressure sensitivity ratio of the wild type was 1 to 1.1. Clones having a pressure sensitivity ratio of <0.5 were considered pressure sensitive. Clones having a pressure sensitivity ratio of >1.5 were considered pressure enhanced.
DNA extraction, purification, manipulation, and sequencing.
Genomic DNA was extracted from 3 ml of 48-h-old P. profundum SS9 cultures by using a Wizard genomic kit (Promega, Madison, WI). The DNA was then further purified by extracting it once with a phenol-chloroform mixture and once with chloroform alone as described elsewhere (80).
Plasmid DNA was extracted using Qiagen (Valencia, CA) Miniprep (high-copy-number plasmids) and Midiprep (low-copy-number plasmids) kits and following the manufacturer's instructions. All enzymatic reactions were prepared using standard protocols (80). Enzymes were purchased from Invitrogen (Carlsbad, CA) or from New England Biolabs (Beverly, MA). DNA sequences were determined by thermal cycle fluorescent dideoxy sequencing with a MegaBACE 1000 (Amersham Biosciences, Piscataway, NJ) automated sequencer used as instructed by the manufacturer.
Arbitrary PCR amplification and transposon cloning.
Rapid identification of the flanking sequences of the transposon insertions in P. profundum was accomplished by using a rapid arbitrary PCR method similar to the method used by Watnick and Kolter (90). This method involved two rounds of PCR. Briefly, during the first round genomic DNA from the mutant was PCR amplified with a primer unique to one end of mini-Tn10 (10extdx2 [5′-AGAGCATTACGCTGACTTG-3′] or 10extsx [5′-CACCCCTTGTATTACTGTTTATGT-3′]) or mini-Tn5 (pRL27Extdx [5′-CCAGAAAGTGAGGGAGCCA-3′] or pRL27Extsx [5′-GACAACAAGCCAGGGATG-3′]) in combination with a degenerate primer (SS9arb1 [5′-GACCACGAGACGCCACACTNNNNNNNNNNCATGC-3′], SS9arb2-[5′-GACCACGAGACGCCACACTNNNNNNNNNNACTAG-3′], or SS9arb8 [5′-GACCACGAGACGCCACACTNNNNNNNNNNGATAT-3′]) that was designed to hybridize to an arbitrary sequence on the chromosome and has a 5′ GC clamp. The conditions used for the first round of amplification were five cycles of 94°C for 30 s, 30°C for 30 s, and 72°C for 60 s, followed by 30 cycles of 94°C for 30 s, 45°C for 30 s, and 72°C for 60 s. Two microliters of the PCR product was subjected to a second round of amplification (30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s) using a nested primer unique to the mini-Tn10 end (10int [5′-GTATGAGTCAGCAACACCTTCTTC-3′]) or the mini-Tn5 end (pRL27Intdx1 [5′-GAGTCGACCTGCAGGCATGC-3′] or pRL27Intsx [5′-CGCACTGAGAAGCCCTTAGAGC-3′]) and a primer with sequence identity to the 5′ GC clamp of the arbitrary primer (arb3 [5′-GACCACGAGACGCCACACT-3′]). After the products of the second amplification were electrophoresed on a gel, the PCR products producing single bands were purified using an UltraClean PCR clean-up kit (MoBio Laboratories, Solana Beach, CA) and sequenced using a third transposon-specific nested primer.
The mini-Tn10 insertion points difficult to characterize by arbitrary PCR were determined by transposon cloning. Genomic DNA from the mutant was digested overnight with EcoRI, which did not cut inside the transposon. The fragments obtained were ligated into the EcoRI site of pUC18, transformed in E. coli TOP10 cells, and selected with kanamycin (100 μg/ml) and ampicillin (100 μg/ml). The transformants growing with this selection contained the pUC18 plasmid with the cloned kanamycin resistance gene from Tn10 together with flanking sequences. The flanking sequences were then determined by sequencing with primers specific to the multiple cloning site of pUC18.
Similarly, the mini-Tn5 mutants difficult to characterize by arbitrary PCR were analyzed by performing genomic DNA extraction, followed by complete digestion with BamHI, relegation and transformation into E. coli EC100D pir+ (Epicenter, Madison, WI), and plating onto Luria-Bertani agar plates containing kanamycin (50 μg/ml).
The sequence flanking the transposon insertion was searched with BLASTN (7) using the nucleotide sequence of the P. profundum SS9 genome, which allowed retrieval of whole open reading frame (ORF) sequences. Downstream genes were analyzed using the genome browser available at http://SS9.cribi.unipd.it. The translated ORF sequence was classified using the COG database (84), the cellular localization was analyzed using SubLoc 1.0 (42), and the presence of signal peptides was analyzed using SignalP (13).
Complementation of selected mutants.
Some of the mutants obtained were characterized further by reintroducing the wild-type copy of the disrupted gene. The ORF sequence, including the predicted promoter and additional upstream sequence, was PCR amplified using an Expand long-template PCR kit (Roche, Indiana) and ligated into the mobilizable broad-host-range vector pFL122 (54). The recombinant clones selected after blue-white screening were sequenced to determine the accuracy of the insert. These constructs were then conjugated into the appropriate mutants by triparental mating using previously described procedures (23). The cold sensitivity ratios and the pressure sensitivity ratios of the merodiploid mutants were then redetermined as described above and compared to those of the isogenic parental mutant strain containing the plasmid vector alone.
RESULTS AND DISCUSSION
Twenty thousand transposon mutant derivatives of P. profundum SS9R were obtained using either mini-Tn10 (6,000 mutants) or mini-Tn5 (14,000 mutants) transposable elements. During preliminary screening, Southern blot and arbitrary PCR analysis of 12 auxotrophic mutants obtained by mini-Tn10 mutagenesis revealed the presence of hotspots for the mini-Tn10 transposition (4/12 unique insertions, located in PBPRA0199, PBPRA0269, PBPRA0280, and PBPRA0289). Tn10 has previously been found to exhibit strong sequence insertion bias (47). The same analysis performed with mutants obtained by using mini-Tn5 did not show the same bias (data not shown). Curiously, despite these limitations, the Tn10 mutants evaluated in this study did not provide a disproportionately low number of the cold-sensitive or pressure-sensitive mutants (see below).
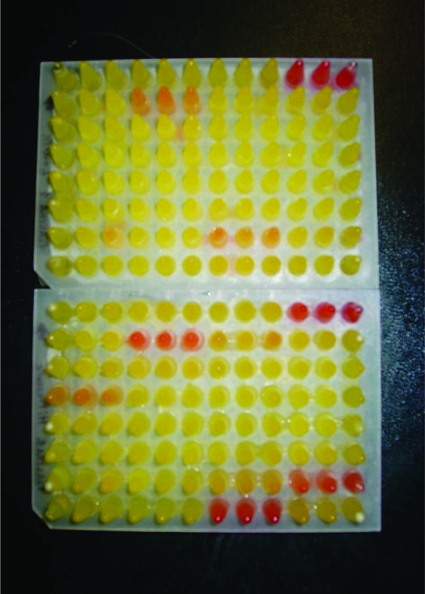
Mutants were examined for defects in growth at a low temperature (4°C) at atmospheric pressure and at a high pressure (28 MPa) at 15°C. In the latter case a phenol red fermentation color screening method in pressurizable microtiter plates was used (Fig. 1). Putative cold-sensitive and pressure-sensitive mutants were rescreened, and bona fide cold-sensitive and pressure-sensitive mutants were evaluated further by generating a detailed growth curve; 1.5% (11/720) of the originally isolated mutants tested were auxotrophs. In contrast, only 0.13% (27/20,000) of the mutants screened were cold sensitive, and only 0.04% (8/20000) of the mutants screened were pressure sensitive. In addition, two pressure-enhanced mutants were also recovered. These mutants were initially isolated as weak fermenters at high pressure, but subsequent analyses revealed that they actually had higher growth rates and the yields were higher at high pressure, indicating that high pressure partially compensated for their growth deficiencies. This is consistent with the view developed following transcriptome experiments, which indicates that P. profundum SS9 is under greater stress at atmospheric pressure than at elevated pressure, presumably reflecting its deep-sea origin (87).
FIG. 1.
Microtiter plate growth assay used in the first screening of the transposon mutants. Growth was detected by addition of phenol red to the growth medium, which changed color following the production of acid by the bacteria growing fermentatively. Yellow, growth; red, no growth.
When the cold-sensitive and pressure-affected mutants were considered together, 77% of the transposon insertions were located on chromosome 1, 23% were located on chromosome 2, and none were located on the plasmid. The gene disruptions and their locations are shown in Table 2 and Fig. 2. The finding that a larger percentage of loci required for growth at high pressure and/or low temperature were present on chromosome 1 was true even after normalization for the larger number of ORFs on this replicon. Transcriptome and codon adaptation index analyses indicated that the most actively transcribed genes are also located on chromosome 1 of P. profundum SS9 (87).
TABLE 2.
Mutants obtained in this study
| Mutant | Genea | Annotation | COG class(es) | Type of transposon insertions recovered in ORF | Phenotypeb | Notesc |
|---|---|---|---|---|---|---|
| FL1 | PBPRB2014 | Transcriptional regulator; LuxR family | T | a-Tn5, b-Tn10 | CS | |
| FL2 | PBPRB0212 | ATP-dependent DEAD box RNA helicase | L, K, J | Tn5 | CS | Complementation analysis |
| FL3 | PBPRA2678 | Hypothetical protein | M | a-Tn5, b-Tn5 | CS | Missing or divergent in P. profundum strains 3TCK and DSJ4 |
| FL4 | PBPRA3093 | rpoE regulatory protein RseB | T | Tn5 | CS, PS | Previously observed (23) suppressors; overexpressed at 28 MPa |
| FL5 | PBPRA2681 | Glycosyl transferase | M | a-Tn5, b-Tn5 | CS | |
| FL6 | PBPRA1774 | ATP-dependent protease; LA related | O | Tn10 | CS | Overexpressed at 28 MPa |
| FL7 | PBPRA2684 | Hypothetical protein involved in polysaccharide biosynthesis | R | Tn5 | CS | Missing or divergent in P. profundum strains 3TCK and DSJ4 |
| FL8 | PBPRA0396 | Hypothetical integral membrane protein | R | a-Tn5, b-Tn5 | CS | Pfam00892 DUF6 domain |
| FL9 | PBPRA2686 | Tyrosine protein kinase | M | Tn5 | CS | Missing or divergent in P. profundum strains 3TCK and DSJ4 |
| FL10 | PBPRA0428 | Pyruvate kinase I | G | a-Tn10, b-Tn10 | CS, PS | Suppressors; underexpressed at 28 MPa |
| FL11 | PBPRA1082 | DNA-binding protein H-NS | R | Tn5 | CS, PE | Enhanced biofilm mutant |
| FL12 | PBPRA0747 | Suppressor protein SuhB | G | Tn10 | CS, PS | Complementation analysis |
| FL13 | PBPRA0189 | Guanosine-3,5-bis(diphosphate) 3-pyrophosphohydrolase SpoT | T, K | Tn5 | CS, PS | |
| FL14 | PBPRA2407 | Periplasmic linker protein | M | Tn5 | CS | |
| FL15 | PBPRA3239 | Hypothetical protein | S | Tn5 | CS | Pfam06295; DUF1043 domain; upstream of degQ/degS orthologues; complementation analysis |
| FL16 | PBPRA2282 | Conserved hypothetical protein | G | Tn5 | CS | pfam04074 DUF386 domain |
| FL17 | PBPRB2009 | Phosphotransferase cellobiose-specific component IIc | G | Tn10 | CS | Possibly missing or divergent in P. profundum strain DSJ4 |
| FL18 | PBPRA0667 | Conserved hypothetical protein | S | Tn10 | CS | Missing or divergent in P. profundum strain 3TCK; pfam05943; DUF877 |
| FL19 | PBPRB1941 | Hypothetical protein | Tn10 | CS | Signal peptide | |
| FL20 | Unknown | Transposase | L | Tn10 | CS | Multiple copies of the sequence: 8 copies on chromosome 2 and 5 copies on chromosome 1 |
| FL21 | PBPRB1757 | Hypothetical protein | T | Tn10 | CS | Signal peptide; missing or divergent in strain DSJ4; complementation analysis |
| FL22 | PBPRB0828 | Beta-hexosaminidase | G | Tn10 | CS | |
| FL23 | PBPRA3229 | Phosphoheptose isomerase | G | Tn10 | CS, PS | Probably diaA (44) |
| FL24 | PBPRA0917 | Polar flagellar protein FlaJ | N, U, O | Tn10 | CS | Phenotype only on plates |
| FL25 | PBPRA2700 | Hypothetical protein | M | a-Tn5, b-Tn5 | CS | pfam04464; phenotype only on plates |
| FL26 | PBPRA0218 | O-antigen ligase | M | Tn5 | CS | Underexpressed at 4°C; phenotype only on plates |
| FL27 | PBPRA0674 | Conserved hypothetical protein | S | Tn5 | CS | Missing or divergent in P. profundum strain 3TCK; pfam05638.3 (hcp); phenotype only on plates |
| FL28 | PBPRA1039 | SeqA protein | L | Tn10 | PE | |
| FL29 | PBPRA2596 | l-Asparaginase | E, J | a-Tn5, b-Tn5 | PS | Complementation analysis; possibly overexpressed at 45 MPa |
| FL30 | PBPRA2658 | 3-Oxoacyl-(acyl carrier protein) synthase I | I, Q | Tn5 | PS | Previously observed (3) |
| FL31 | PBPRB0001 | Chromosome II replication protein RctB | Tn5 | PS | —d |
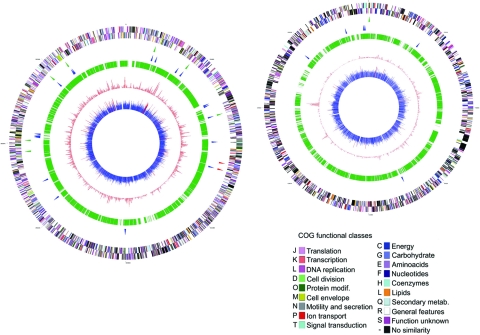
FIG. 2.
Genomic localization of the transposon insertions in the two chromosomes of P. profundum SS9 (not to scale). (Left panel) Chromosome 1. (Right panel) Chromosome 2. From the outside in, the first two circles show the predicted protein coding on the two strands, with the colors indicating the COG functional classes. The third circle shows the locations of the pressure-sensitive (green) and pressure-enhanced (red) genes. The fourth circle shows the locations of the cold-sensitive (blue) genes. The fifth circle shows the syntheny with the draft genome of P. profundum 3TCK (www.venterinstitute.org). The sixth circle shows the mean fluorescence intensities obtained in the microarray experiments at 28 MPa, and the sixth circle shows the codon adaptation index, with scores of >0.5 indicated by red (87).
Although temperature and pressure are distinct thermodynamic parameters, in many cases the effect of pressure is synergistic with the effect of low temperature (28). In the case of E. coli, which is mesophilic with respect to both temperature and pressure, decreased temperature and increased pressure both perturb membrane structure and function and DNA replication, transcription, and translation (9). The ability of 16% of the mutants obtained here to grow was altered both at low temperature and at high pressure.
Isolation of cold-sensitive mutants.
Mutants with mutations in 27 loci displayed a cold-sensitive phenotype (Fig. 3 and Table 2). Of these mutants, 21 were exclusively cold sensitive, 5 were also pressure sensitive, and one was also pressure enhanced. These mutants could be functionally divided into six categories based on the COG identity of the disrupted gene. Genes belonging to the poorly characterized COG functional classes (classes R and S) were clustered based on additional information provided by other analyses.
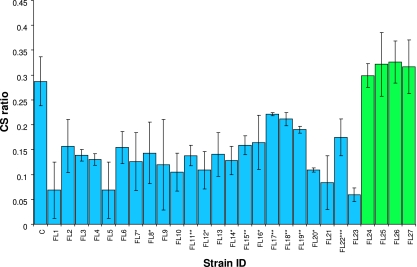
FIG. 3.
Cold sensitivity (CS) ratios for the mutants isolated in this study. The values were computed as described in Materials and Methods. The green bars indicate the ratios for the mutants that displayed a reproducible phenotype only on plates. C, control strain. For some strains, the lag phase was 40 to 59 h (indicated by one asterisk), 60 to 79 h (two asterisks), or >79 h (three asterisks) longer than the lag phase for the control strain. The error bars indicate one standard deviation.
(i) Signal transduction mechanisms (class T) (strains FL1, FL4, FL11, FL13, and FL21).
Microorganism sense and coordinate metabolic functions during cold stress (72). These responses occur through temperature-induced changes in DNA supercoiling (30, 49, 64), via effects on ribosomes (86), through the modulation of guanosine tetraphosphate (ppGpp) levels (46), and by gene regulation through the activity of alternative sigma factors (74, 94).
The expression of genes required for low-temperature growth in SS9 could be modulated by specific regulators, such as the PBPRB2014 protein (strain FL1), a transcriptional regulator of the LuxR family, and the PBPRB1757 protein (strain FL21), a hypothetical protein with a response regulator receiver domain (pfam00072) and a partial domain of a signal transduction histidine kinase (COG0642), suggesting its possible role as a hybrid two-component system. Related regulatory systems have been observed in Bacillus subtilis and some cyanobacteria (79). In these organisms, the expression of cold-inducible genes, in particular the genes responsible for membrane unsaturation, is controlled by specific two-component systems.
(ii) Cell envelope biogenesis and outer membrane (class M) (strains FL3, FL5, FL7, FL9, FL14, FL25, FL26, and FL27).
Workers have speculated about the involvement of capsular polysaccharide in adaptation to low temperature for some time (25, 60, 62).
FL26 contains a disruption in the gene encoding the PBPRA0218 protein, a predicted O-antigen ligase for lipopolysaccharide (LPS) synthesis whose expression is reduced at low temperature (19, 87). The mutations in four of the remaining mutants are mutations in part of a large gene cluster that is highly divergent in the members of the species P. profundum (19). This cluster is approximately 35 kb long and contains many genes that are involved in various aspects of LPS O-antigen biosynthesis, extracellular polysaccharide (EPS) biosynthesis, and flagellar filament glycosylation (36). Some of these genes are also differentially expressed under different temperature and pressure conditions (PBPRA2692, PBPRA2701, and PBPRA2710). Their functions are currently being characterized further (G. Ferguson et al., unpublished data). Strain FL7 (disruption of PBPRA2684, hypothetical protein, COG functional class R) is included in this category of mutants based solely on the fact that its insertion is located within the predicted polysaccharide biosynthesis cluster. Using similar reasoning, we included strain FL27 among the envelope mutants. The mutated gene in strain FL27, PBPRA0674, is homologous to the gene encoding hemolysin-coregulated protein (Hcp) of Vibrio species. While the function of Hcp has not been determined (95), Enos-Berlage et al. have shown that a mutation in hcp results in alterations in the formation of Vibrio parahaemolyticus biofilms, and thus Hcp could be associated with polysaccharide production (32).
Interestingly, strains FL25, FL26, and FL27 displayed the cold-sensitive phenotype when they were grown on agar plates but not when they were grown in liquid media (Fig. 3), suggesting that there are different EPS and LPS requirements for growth at low temperature under these two conditions. The basis for the distinction is not clear at this time. E. coli with a deletion of the rfa locus, which is responsible for assembly of the core oligosaccharide of LPS, is cold sensitive and nonmotile (70). The role of LPS in stabilizing the bacterial cell envelope is exerted both through its interaction with the EPS and through the fluidizing effects that its lipid components can have on the outer membrane (50).
Corsaro et al. (25), have suggested that the psychrophile Pseudoalteromonas haloplanktis TAC125 is unable to complete the biosynthesis of lipooligosaccharide at a suboptimal temperature as the phosphorylation of both lipooligosaccharide and EPS decreases with temperature. In these molecules, phosphate groups bind to divalent cations, such as Ca2+ and Mg2+, stabilizing the extracellular leaflet of the outer membrane (50, 76) and modulating its permeability. LPS integrity is essential for correct incorporation of many proteins, such as porins, in the outer membrane (70). It is possible that changes in LPS and EPS, which change the surface properties of bacteria, are used in processes affected by low temperature, such as membrane fluidity and substrate transport (32).
(iii) Carbohydrate transport and metabolism (class G) (strains FL10, FL12, FL16, FL17, FL22, and FL23).
Low-temperature growth was affected by mutations in a number of genes involved in the transport and the central metabolism of carbohydrates. These genes included the genes encoding the glycolytic enzyme pyruvate kinase I (PBPRA0428), a component of a group translocation transporter specific for cellobiose (PBPRB2009), and the conserved hypothetical protein PBPRA2282, which may participate in the metabolism of N-acetylneuraminic acid and its derivatives (48).
Two genes deserve special attention. PBPRA0747 is in this category because it is homologous to suhB, whose product was shown to possess inositol monophosphatase activity (68). However, the role of this product in the cell appears to involve modulating the processing activity of RNase III, and because of this, in E. coli a suhB mutation suppresses a wide variety of other mutations (43). An E. coli suhB mutant is cold sensitive (43).
Strain FL23 has an insertion in PBPRA3229, which was originally annotated as a phosphoheptose isomerase gene (87) but after closer inspection turned out to be orthologous to diaA, a gene required during the initiation of chromosome replication (44). Both PBPRA0747 and PBPRA3229 also affect the growth of P. profundum SS9 at high pressure.
(iv) Protein export (class U) (strains FL18, FL19, and FL24).
Strain FL19 has a mutation in a gene encoding a hypothetical protein with a well-defined signal peptide (13). While it is possible that the product of the PBPRB1941 gene is directly involved in some aspect of temperature adaptation, another hypothesis is that the insertion results in a blockage of the general secretory pathway (sec). This pathway is inherently cold sensitive in E. coli (73).
Other secretion routes could also be important for low-temperature growth. Strain FL18 has an insertion in PBPRA0667, a homolog of impC, a gene of Rhizobium leguminosarum (16) that is part of a locus important for temperature-dependent secretion and establishment of the symbiotic interaction with the host plant roots. Strain FL24 has a mutation in PBPRA0917, an orthologue of the flagellar chaperone gene fliS of E. coli. FliS functions as a substrate-specific chaperone facilitating the export of flagellin axial-filament subunits and preventing their polymerization in the cytosol (8). The cold-sensitive phenotype of strain FL24 is visible only on plates and might be a result of the deleterious accumulation of flagellin oligomers in the cytosol, especially at low temperatures, when the activity of proteases involved in recycling of nonfunctional peptides is lower.
(v) Protein synthesis and turnover (classes J and O) (strains FL2, FL6, and FL15).
Strain FL2 has a mutation in PBPRB0212. This gene was annotated as a member of a family of genes encoding ATP-dependent helicases known as DEAD box helicases because of their characteristic amino acid motif (Asp-Glu-Ala-Asp). RNA helicases are involved in unwinding duplex RNA and, because of their regulation and role in ribosome biogenesis and translation initiation, have been linked to cold stress in both cyanobacteria (20) and Archaea (56).
The insertion in strain FL6 is in PBPRA1774, which codes for an LA-related protease. Similar ATP-dependent proteases have been shown to degrade nonfunctional proteins in the cytoplasm of E. coli (78) and to be important for cold acclimation in the marine cyanobacterium Synechococcus (75). Strain FL15 has an interruption in the gene for the hypothetical protein PBPRA3239, which is upstream of and in the same transcriptional unit as the degQ and degS genes encoding the periplasmic serine proteases.
Cold shock can result in protein unfolding and aggregation (33). The aggregates are considered “dead ends,” and accumulation of them may cause severe damage to the organism (78). It is possible that the protease mutations described above lead to accumulation of protein aggregates that affect growth.
(vi) Unknown (classes L and R) (strains FL8 and FL20).
The mini-Tn10 insertion of mutant FL20 is in the region coding for a transposase. Transposable elements are very abundant in the genome of P. profundum SS9 (87). The basis for the cold sensitivity of this mutant is not clear at this time. The basis for the cold sensitivity of strain FL8 is also unknown. The interrupted gene, PBPRA0396, codes for a hypothetical integral membrane protein and is part of an operon with genes encoding two ribosomal proteins, L21 and L27.
Isolation of pressure-altered mutants.
Table 2 and Fig. 4 show the transposon mutants displaying a pressure-altered phenotype together with their pressure sensitivity ratios. The number of loci recovered in the pressure sensitivity screen was less than one-third the number of the cold-sensitive loci. Three separate hypotheses could account for this difference: the screen for pressure-sensitive mutants was less sensitive, adaptation to high hydrostatic pressure requires fewer genes, or adaptation to high hydrostatic pressure requires a higher proportion of essential genes.
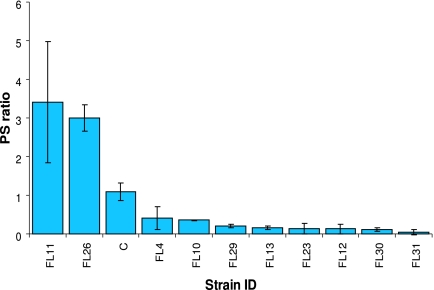
FIG. 4.
Pressure sensitivity (PS) ratios for the mutants isolated in this study. The values were computed as described in Materials and Methods. C, control strain. The error bars indicate one standard deviation.
Two lines of evidence support one or both of the latter two hypotheses: (i) previously identified pressure-sensitive mutants with site-directed mutations were identified using the screen, thus demonstrating its sensitivity and arguing against the first hypothesis, and (ii) with the exception of PBPRA2596 (l-asparaginase) and PBPRA2658 [3-oxoacyl-(acyl carrier protein) synthase I], all the genes conferring piezoadaptation have been shown to have pleiotropic effects in other bacteria because of their central role in the cell (12, 23, 41, 43, 58, 59, 65, 83), which is consistent with the last hypothesis. These mutants could be functionally divided into four categories based on prior research and the COG identities of the disrupted genes.
(i) Previously isolated genes (classes T and I) (strains FL4 and FL30).
PBPRA2658 (fabB) codes for the KASI enzyme (77) and has been linked to piezophily because of its role in the production of monounsaturated fatty acids which are essential for the proper membrane physical state at high pressure (3). Inactivation of rseB (PBPRA3093), a gene belonging to the rpoE cluster, results in a pressure- and cold-sensitive phenotype. Chi and Bartlett (23) have suggested that this is due to a polar effect on rseC. At the present time it is not clear if the observed phenotype is determined by RseC involvement in the regulation of the alternative sigma factor RpoE (63) or by its role in other cellular functions, such as thiamine synthesis, as observed in Salmonella enterica serovar Typhimurium (12).
(ii) Metabolism (classes E and G) (strains FL10 and FL29).
Strain FL29 contains an insertion in PBPRA2596 encoding l-asparaginase. This enzyme is essential under conditions in which asparagine is the only nitrogen source (27). It is possible that the l-asparaginase activity is a key route for nitrogen assimilation at high pressure under the glucose and peptide fermentation conditions used. The culture medium employed contained only 15 μM nitrate. The importance of an organic nitrogen source for metabolism is further reinforced by the upregulation at 45 MPa of PBPRA1174 (periplasmic l-asparaginase), PBPRA3391 (aspartate-ammonia lyase), and PBPRA2173 (histidine-ammonia lyase), all of which contribute to the catabolism of amino acids.
Another target of pressure resulting in global alteration of metabolism was observed in strain FL10, which has a mutation in pyruvate kinase I (PBPRA0428). Pyruvate kinase has previously been found to be a pressure-sensitive enzyme and to undergo adaptational changes in deep-sea animals (26, 57). The requirement for this isozyme could have been a reflection of the medium conditions employed (growth under glucose-fermenting conditions). Alternatively, because of the role of pyruvate kinase in glycolytic regulation (65), the mutant could be a regulatory mutant. Transcriptome studies indicated that five of the nine steps of glycolysis involve pressure- and temperature-regulated genes, one of which is PBPRA0428. The growth of this mutant is also impaired at low temperature.
(iii) Chromosome replication (classes L, R, and G) (strains FL11, FL23, FL28, and FL31).
Almost one-half of the pressure-altered mutants were associated with some aspect of chromosome structure and partitioning during cell division. Cell division and chromosome replication and segregation are among the most pressure-sensitive processes in a mesophilic bacterial cell (9, 96). Therefore, proteins underlying these cellular functions are expected to be under strong selective pressure to acquire functionality at depth. Bidle and Bartlett (15) reported impairment of growth at high pressure of a recD mutant of P. profundum. These workers were also able to show that high-pressure impairment of cell division in E. coli could be rescued by heterologous expression of the recD gene from P. profundum (15).
PBPRB0001 is a gene that is considered essential for the replication of chromosome II in the family Vibrionaceae (31). Egan and Waldor (31) were unable to obtain a null mutant with a mutation in VCA0002, the orthologue of PBPRB0001, in Vibrio cholerae. Because the insertion in strain FL31 is close to the 3′ end of the ORF, we hypothesized that it results in only a partial loss of function. This further implies that the C terminus of PBPRB0001 is required only under high-pressure conditions.
Two additional genes whose products are important for chromosome replication at high pressure were also discovered. The first gene, seqA (PBPRA1039), is a negative regulator of the cell cycle, and a strain having a mutation in PBPRA1039 is pressure enhanced. E. coli SeqA mutants have irregular growth caused by asynchronous patterns of replication (58). The stress imposed by growth at low pressure might exacerbate this phenotype in the case of SS9, but it is not clear if this is due to the role of SeqA as a cell cycle regulator (58), its importance for chromosome partitioning (55), or its effect on the structure of cell membranes (91).
While a seqA mutation resulted in a pressure-enhanced phenotype, pressure sensitivity and cold sensitivity were observed for strain FL23, which contains an insertion in diaA. This gene has recently been identified as a novel DNA-binding protein involved in ensuring the initiation of chromosomal replication at the right time (44). Interestingly, studies with E. coli have shown that both seqA and diaA can suppress temperature-sensitive phenotypes associated with mutations in dnaA, which codes for the initiator of chromosome replication (44, 88).
The nucleoid-associated protein H-NS also influences growth at elevated pressure. Ishii et al. (45) observed that a Δhns strain of E. coli is at least 1,000-fold more sensitive to high hydrostatic pressure than the isogenic wild-type strain. In contrast, mutant strain FL11, which contains an insertion in the hns orthologue PBPRA1082, grows better at high hydrostatic pressure than at atmospheric pressure. However, H-NS-deficient mutants of both E. coli (29) and P. profundum are cold sensitive, suggesting that the hns gene has different roles in the adaptation to temperature and in the adaptation to pressure and has evolved piezo-specific traits in P. profundum.
The cold sensitivity of FL11 could be caused by the role of H-NS in modulating the cold shock response (18, 45) or, alternatively, by an alteration in the EPS-LPS matrix. Enos-Berlage et al. (32) have reported that insertional inactivation of H-NS results in modifications of the capsular polysaccharide. FL11 produces a visibly thicker biofilm when it is grown in liquid media (data not shown).
(iv) Translation, ribosomal structure, and biogenesis (classes J, G, and T) (strains FL2, FL12, and FL13).
Mutations in PBPRA0747 (suhB), PBPRB0212 (DEAD box helicase), and PBPRA0189 (spoT) result in both cold and pressure sensitivity and are predicted to affect the structure or function of the ribosome.
Mesophilic ribosomes are extremely sensitive to high hydrostatic pressure (35, 51). P. profundum SS9 has the record number of ribosomal operons, and there is a large amount of intragenomic variation (53, 87). The intragenomic variation in the 23S rRNA of P. profundum SS9 is concentrated in helices 25 and 45. There is no evidence that these sequences are retained in the processed ribosome, and if the intervening sequences are removed, it is by the action of RNase III. Interestingly, RNase III activity is modulated by SuhB (43), the product of PBPRA0747.
Helix 25 is also the major site of interaction with ribosomal protein L13, a protein essential for assembly of the 50S subunit of the ribosome. In a correctly assembled ribosome, L13 is located within a few angstroms of loop 2475 (66). Two members of the DEAD box family of RNA helicases have been implicated in this step of ribosome biogenesis. The first enzyme, DbpA, was shown to interact with residues 2454 to 2606 of the 23S rRNA in vitro (85). The second enzyme, SrmB, was implicated directly in the assembly of L13 because an srmB deletion results in accumulation of incomplete large ribosomal subunits (40S) lacking L13 (22). Interestingly, such mutants are also cold sensitive (22). Therefore, if PBPRB0212 performs a similar function in the P. profundum cell, this possibly explains the cold-sensitive phenotype of the P. profundum mutant. The genome of P. profundum SS9 codes for at least nine DEAD box helicases (PBPRA0562, PBPRA1748, PBPRA3542, PBPRB0199, PBPRB0212, PBPRB0427, PBPRB1008, PBPRB1232, and PBPRB1761), almost twice as many as the genome of E. coli, in which some of the proteins have been shown to have unique but partially overlapping functions in the cell (21). A similar expansion of genes encoding the DEAD box family of helicases has also been observed in the psychrophilic gammaproteobacterial genomes of Colwellia psychrerythraea 34H (62) and P. haloplanktis TAC125 (61), suggesting that this phenomenon might be important for temperature adaptation.
In P. profundum, microarray analysis revealed differential expression of some of the DEAD box helicases under certain temperature and pressure conditions: the orthologue (PBPRB0427 protein) most closely related to E. coli DbpA (NCBI accession no. P0A9P6) (17) is underexpressed at 28 or 45 MPa and 4°C (19, 87), the PBPRB1232 protein is underexpressed at 45 Mpa, and the PBPRB1761 protein is underexpressed at 4°C. Notably, this pattern of expression, with the DEAD box helicases repressed at lower temperatures, is opposite the pattern that was observed in the Antarctic methanogen Methanococcoides burtonii (56).
Another aspect of the pressure and temperature ribosome connection appears to be modulation of the stringent response. This phenomenon results in dramatic downregulation of ribosomal components following a variety of stresses and was first observed in E. coli cells subjected to amino acid starvation (81). A cell not undergoing the stringent response is said to be in a relaxed state. The effector molecule of the stringent response is ppGpp, which is generated by the gene products of spoT and relA.
In E. coli a temperature downshift induces a relaxed state through a decrease in the levels of ppGpp (46). Similarly, one might predict that a pressure-induced decrease in ppGpp levels would result in the production of increased amounts of ribosomal proteins L7/L12, S6, the elongation factor EF-G, and cold shock proteins (46). Most of these markers can in fact be detected by proteomic analysis of E. coli subjected to a sudden pressure upshift (93) and microarray analysis of high-pressure-shocked Lactobacillus sanfranciscensis (71).
In P. profundum, a mutant with a mutation in the spoT orthologue PBPRA0189 is both cold and pressure sensitive. This phenotype might be caused by disruption of the delicate interplay between the stringent, cold, and pressure responses. SpoT is responsible for both the synthesis and the degradation of ppGpp, and spoT mutants have higher basal levels of ppGpp even under steady-state conditions (82).
An alternative hypothesis for the cold and pressure sensitivity of a P. profundum spoT mutant originated from the observation that in a V. cholerae mutant producing lower-than-normal levels of ppGpp toxR was transcriptionally repressed (38). ToxR functions as a piezosensor in SS9. It is therefore feasible that upregulation of toxR might occur in a spoT mutant background, and overexpression of toxR in P. profundum does indeed result in a pressure-sensitive phenotype (D. H. Bartlett, unpublished results). Moreover, because in E. coli RelA generates ppGpp in response to amino acid starvation while SpoT is responsible for sensing other stresses (59), it would be interesting to analyze the pressure and cold sensitivity of a P. profundum relA mutant and a spoT relA double mutant.
Complementation of selected mutants.
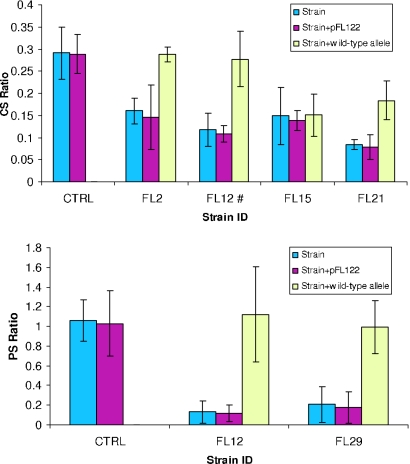
A subset of mutant strains (strains FL2, FL12, FL15, FL21, and FL29) was selected for complementation analysis. In most cases (strains FL2, FL12, and FL29) reintroduction of the wild-type copy of the gene resulted in a wild-type phenotype (Fig. 5), confirming the role of the disrupted ORF in growth at high hydrostatic pressure or low temperature. In one case (FL21, PBPRB1757), the cold-sensitive phenotype was intermediate, suggesting the possibility of a partially dominant negative mutant or that a downstream gene might also have a role in low-temperature growth. In another case (FL15, PBPRA3239) reintroduction of the wild-type copy did not complement the mutant phenotype. The insertion in PBPRA3239 could exert a polar effect on transcription of the downstream genes PBPRA3240 and PBPRA3241 coding for orthologues of the DegQ and DegS serine proteases involved in recycling of nonfunctional proteins in the periplasmic space (89). Curiously, degS has been shown to have a function in the regulation of rpoE activity by proteolytically degrading the periplasmic domain of RseA (34). A degS mutant of E. coli is unable to respond to extracytoplasmic stress (2). Alternatively, it is possible that PBPRA3239 functionally interacts with PBPRA3240 in the periplasm. In fact, it has been predicted that PBPRA3239 is localized in the periplasmic space (42), and the synteny conservation of the two ORFs in all the members of the family Vibrionaceae suggests that they are part of the same regulon.
FIG. 5.
Cold sensitivity (CS) and pressure sensitivity (PS) ratios for a selected subset of mutants after reintroduction of the wild-type copy of the allele on the plasmid vector pFL122. (Top panel) Complementation of cold-sensitive mutants. (Bottom panel) Complementation of pressure-sensitive mutants. The error bars indicate one standard deviation. Complementation of the mutation eliminated the increase in the lag phase observed in FL12 for the cold sensitivity ratio. CTRL, control.
Conclusions.
Previous to this work, only Abe and Iida (1) had described a comprehensive analysis of nonessential genes influencing the growth of any microorganism at high hydrostatic pressure. The mesophile Saccharomyces cerevisiae was examined by these workers. This study expanded the number of P. profundum cold-sensitive mutants from 2 (6, 23) to 28 and the number of pressure-sensitive mutants from 4 (3, 4, 15, 23) to 10.
During the screening some transposon insertions were in genes previously implicated in low-temperature or high-pressure growth (3, 23). However, not all the piezoadaptive genes obtained previously were recovered. This can be explained by the limited coverage of the transposon screen. Based upon Poisson statistics, we inferred that the minimal number of mutants that must be screened in order to have a 95% probability of hitting every ORF in the genome is ∼16,500. Taking into consideration the existence of hotspots for mini-Tn10 transposition, screening of 20,000 mutants was not likely to have been saturating. Indeed, of the 31 pressure-sensitive cold-sensitive loci discovered, only 7 were hit more than once. This study also highlights the fact that while there is some functional overlap between the adaptive responses to temperature and pressure, each condition affects microbial cells in a unique way. Most cold-sensitive mutants are not pressure sensitive, and most pressure-sensitive mutants are not cold sensitive. Almost all the cold-sensitive mutants discovered could be clustered into six COG functional classes (class T, signal transduction mechanisms; class M, cell envelope biogenesis and outer membrane; class G, carbohydrate transport and metabolism; class U, intracellular trafficking, secretion, and vesicular transport; class L, DNA replication, recombination, and repair; and class O, posttranslational modification, protein turnover, and chaperones). The fewer pressure-sensitive mutants were more diverse, although at least four of them could be associated with chromosome partitioning (classes L, R, and G) and three could be associated with ribosomal function (classes J, G, and T).
This is also the first study that provides direct genetic evidence for a crucial role of EPS genes in adaptation to low temperature (60). It has been hypothesized that one function of EPS at low temperature might be as a cryoprotectant under freezing conditions (60). This cannot be the only low-temperature function in the case of P. profundum as the cold-sensitive phenotype of the EPS mutants appears at temperatures well above the freezing point of water. Further studies are needed to evaluate the relationship between EPS structure and function and cold sensitivity.
Additionally, because of the wide variety of sensory and regulatory mutants with mutations affecting growth at high pressure and low temperature, it would be interesting to identify the genes under transcriptional control of each of the regulators. Keeping in mind that piezophilic bacteria are under stress at atmospheric pressure, it might also be interesting to embark upon a hunt for mutants with low-pressure sensitivity.
Acknowledgments
We are grateful to Yoko Philips for technical assistance, Bianca Brahamsha for help with transposon mutagenesis, and Gail Ferguson for insightful discussions.
This work was supported by NSF grants MCB02-37059, MCB04-009, and MCB05-44524 to D.H.B.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Abe, F., and H. Iida. 2003. Pressure-induced differential regulation of the two tryptophan permeases Tat1 and Tat2 by ubiquitin ligase Rsp5 and its binding proteins, Bul1 and Bul2. Mol. Cell. Biol. 237566-7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ades, S. E., L. E. Connolly, B. M. Alba, and C. A. Gross. 1999. The Escherichia coli σE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 132449-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, E. E. 2002. Piezophysiology of membrane-based adaptations in the deep-sea bacterium Photobacterium profundum strain SS9. University of California, San Diego, La Jolla.
- 4.Allen, E. E., and D. H. Bartlett. 2000. FabF is required for piezoregulation of cis-vaccenic acid levels and piezophilic growth of the deep-sea bacterium Photobacterium profundum strain SS9. J. Bacteriol. 1821264-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen, E. E., and D. H. Bartlett. 2002. Structure and regulation of the omega-3 polyunsaturated fatty acid synthase genes from the deep-sea bacterium Photobacterium profundum strain SS9. Microbiology 1481903-1913. [DOI] [PubMed] [Google Scholar]
- 6.Allen, E. E., D. Facciotti, and D. H. Bartlett. 1999. Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl. Environ. Microbiol. 651710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auvray, F., J. Thomas, G. M. Fraser, and C. Hughes. 2001. Flagellin polymerisation control by a cytosolic export chaperone. J. Mol. Biol. 308221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartlett, D. H. 2002. Pressure effects on in vivo microbial processes. Biochim. Biophys. Acta 1595367-381. [DOI] [PubMed] [Google Scholar]
- 10.Bartlett, D. H., F. M. Lauro, and E. A. Eloe. 2007. Microbial adaptation to high pressure, p. 333-351. In C. Gerday and N. Glansdorff (ed.), Physiology and biochemistry of extremophiles. American Society for Microbiology Press, Washington, DC.
- 11.Bartlett, D. H., and T. J. Welch. 1995. OmpH gene expression is regulated by multiple environmental cues in addition to high pressure in the deep-sea bacterium Photobacterium species strain SS9. J. Bacteriol. 1771008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beck, B. J., L. E. Connolly, A. De Las Penas, and D. M. Downs. 1997. Evidence that rseC, a gene in the rpoE cluster, has a role in thiamine synthesis in Salmonella typhimurium. J. Bacteriol. 1796504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 14.Better, M., and D. R. Helinski. 1983. Isolation and characterization of the recA gene of Rhizobium meliloti. J. Bacteriol. 155311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bidle, K. A., and D. H. Bartlett. 1999. RecD function is required for high-pressure growth of a deep-sea bacterium. J. Bacteriol. 1812330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bladergroen, M. R., K. Badelt, and H. P. Spaink. 2003. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. 1653-64. [DOI] [PubMed] [Google Scholar]
- 17.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 18.Brandi, A., R. Spurio, C. O. Gualerzi, and C. L. Pon. 1999. Massive presence of the Escherichia coli ‘major cold-shock protein’ CspA under non-stress conditions. EMBO J. 181653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campanaro, S., A. Vezzi, N. Vitulo, F. M. Lauro, M. D'Angelo, F. Simonato, A. Cestaro, G. Malacrida, G. Bertoloni, G. Valle, and D. H. Bartlett. 2005. Laterally transferred elements and high pressure adaptation in Photobacterium profundum strains. BMC Genomics 6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chamot, D., and G. W. Owttrim. 2000. Regulation of cold shock-induced RNA helicase gene expression in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 1821251-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charollais, J., M. Dreyfus, and I. Iost. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 322751-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charollais, J., D. Pflieger, J. Vinh, M. Dreyfus, and I. Iost. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 481253-1265. [DOI] [PubMed] [Google Scholar]
- 23.Chi, E., and D. H. Bartlett. 1995. An RpoE-like locus controls outer-membrane protein synthesis and growth at cold temperatures and high pressures in the deep-sea bacterium Photobacterium sp. strain SS9. Mol. Microbiol. 17713-726. [DOI] [PubMed] [Google Scholar]
- 24.Chi, E., and D. H. Bartlett. 1993. Use of a reporter gene to follow high-pressure signal transduction in the deep-sea bacterium Photobacterium sp. strain SS9. J. Bacteriol. 1757533-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corsaro, M. M., R. Lanzetta, E. Parrilli, M. Parrilli, M. L. Tutino, and S. Ummarino. 2004. Influence of growth temperature on lipid and phosphate contents of surface polysaccharides from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC 125. J. Bacteriol. 18629-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Felice, F. G., V. C. Soares, and S. T. Ferreira. 1999. Subunit dissociation and inactivation of pyruvate kinase by hydrostatic pressure oxidation of sulfhydryl groups and ligand effects on enzyme stability. Eur. J. Biochem. 266163-169. [DOI] [PubMed] [Google Scholar]
- 27.Del Casale, T., P. Sollitti, and R. H. Chesney. 1983. Cytoplasmic l-asparaginase: isolation of a defective strain and mapping of ansA. J. Bacteriol. 154513-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLong, E. F., D. G. Franks, and A. A. Yayanos. 1997. Evolutionary relationships of cultivated psychrophilic and barophilic deep-sea bacteria. Appl. Environ. Microbiol. 632105-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dersch, P., S. Kneip, and E. Bremer. 1994. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol. Gen. Genet. 245255-259. [DOI] [PubMed] [Google Scholar]
- 30.Drlica, K. 1992. Control of bacterial DNA supercoiling. Mol. Microbiol. 6425-433. [DOI] [PubMed] [Google Scholar]
- 31.Egan, E. S., and M. K. Waldor. 2003. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114521-530. [DOI] [PubMed] [Google Scholar]
- 32.Enos-Berlage, J. L., Z. T. Guvener, C. E. Keenan, and L. L. McCarter. 2005. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol. Microbiol. 551160-1182. [DOI] [PubMed] [Google Scholar]
- 33.Franks, F., R. H. Hatley, and H. L. Friedman. 1988. The thermodynamics of protein stability. Cold destabilization as a general phenomenon. Biophys. Chem. 31307-315. [DOI] [PubMed] [Google Scholar]
- 34.Grigorova, I. L., R. Chaba, H. J. Zhong, B. M. Alba, V. Rhodius, C. Herman, and C. A. Gross. 2004. Fine-tuning of the Escherichia coli σE envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev. 182686-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gross, M., and R. Jaenicke. 1990. Pressure-induced dissociation of tight couple ribosomes. FEBS Lett. 267239-241. [DOI] [PubMed] [Google Scholar]
- 36.Gryllos, I., J. G. Shaw, R. Gavin, S. Merino, and J. M. Tomas. 2001. Role of flm locus in mesophilic Aeromonas species adherence. Infect. Immun. 6965-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 38.Haralalka, S., S. Nandi, and R. K. Bhadra. 2003. Mutation in the relA gene of Vibrio cholerae affects in vitro and in vivo expression of virulence factors. J. Bacteriol. 1854672-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herrero, M., V. Delorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1726557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 4020-36. [DOI] [PubMed] [Google Scholar]
- 42.Hua, S., and Z. Sun. 2001. Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17721-728. [DOI] [PubMed] [Google Scholar]
- 43.Inada, T., and Y. Nakamura. 1995. Lethal double-stranded RNA processing activity of ribonuclease III in the absence of suhB protein of Escherichia coli. Biochimie 77294-302. [DOI] [PubMed] [Google Scholar]
- 44.Ishida, T., N. Akimitsu, T. Kashioka, M. Hatano, T. Kubota, Y. Ogata, K. Sekimizu, and T. Katayama. 2004. DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J. Biol. Chem. 27945546-45555. [DOI] [PubMed] [Google Scholar]
- 45.Ishii, A., T. Oshima, T. Sato, K. Nakasone, H. Mori, and C. Kato. 2005. Analysis of hydrostatic pressure effects on transcription in Escherichia coli by DNA microarray procedure. Extremophiles 965-73. [DOI] [PubMed] [Google Scholar]
- 46.Jones, P. G., M. Cashel, G. Glaser, and F. C. Neidhardt. 1992. Function of a relaxed-like state following temperature downshifts in Escherichia coli. J. Bacteriol. 1743903-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Junop, M. S., and D. B. Haniford. 1997. Factors responsible for target site selection in Tn10 transposition: a role for the DDE motif in target DNA capture. EMBO J. 162646-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolker, E., K. S. Makarova, S. Shabalina, A. F. Picone, S. Purvine, T. Holzman, T. Cherny, D. Armbruster, R. S. Munson, Jr., G. Kolesov, D. Frishman, and M. Y. Galperin. 2004. Identification and functional analysis of ‘hypothetical’ genes expressed in Haemophilus influenzae. Nucleic Acids Res. 322353-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krispin, O., and R. Allmansberger. 1995. Changes in DNA supertwist as a response of Bacillus subtilis towards different kinds of stress. FEMS Microbiol. Lett. 134129-135. [DOI] [PubMed] [Google Scholar]
- 50.Kumar, G. S., M. V. Jagannadham, and M. K. Ray. 2002. Low-temperature-induced changes in composition and fluidity of lipopolysaccharides in the Antarctic psychrotrophic bacterium Pseudomonas syringae. J. Bacteriol. 1846746-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landau, J. V. 1967. Induction transcription and translation in Escherichia coli—a hydrostatic pressure study. Biochim. Biophys. Acta 149506-512. [DOI] [PubMed] [Google Scholar]
- 52.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178193-201. [DOI] [PubMed] [Google Scholar]
- 53.Lauro, F. M., R. A. Chastain, L. E. Blankenship, A. A. Yayanos, and D. H. Bartlett. 2007. The unique 16S rRNA genes of piezophiles reflect both phylogeny and adaptation. Appl. Environ. Microbiol. 73838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lauro, F. M., E. A. Eloe, N. Liverani, G. Bertoloni, and D. H. Bartlett. 2005. Conjugal vectors for cloning, expression, and insertional mutagenesis in gram-negative bacteria. BioTechniques 38708-712. [DOI] [PubMed] [Google Scholar]
- 55.Lemon, K. P., I. Kurtser, and A. D. Grossman. 2001. Effects of replication termination mutants on chromosome partitioning in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim, J., T. Thomas, and R. Cavicchioli. 2000. Low temperature regulated DEAD-box RNA helicase from the Antarctic archaeon, Methanococcoides burtonii. J. Mol. Biol. 297553-567. [DOI] [PubMed] [Google Scholar]
- 57.Low, P. S., and G. N. Somero. 1976. Adaptation of muscle pyruvate kinases to environmental temperatures and pressures. J. Exp. Zool. 1981-11. [DOI] [PubMed] [Google Scholar]
- 58.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77413-426. [DOI] [PubMed] [Google Scholar]
- 59.Magnusson, L. U., A. Farewell, and T. Nystrom. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13236-242. [DOI] [PubMed] [Google Scholar]
- 60.Marx, J. C., T. Collins, S. D'Amico, G. Feller, and C. Gerday. 2007. Cold-adapted enzymes from marine Antarctic microorganisms. Mar. Biotechnol. 9293-304. [DOI] [PubMed] [Google Scholar]
- 61.Medigue, C., E. Krin, G. Pascal, V. Barbe, A. Bernsel, P. N. Bertin, F. Cheung, S. Cruveiller, S. D'Amico, A. Duilio, G. Fang, G. Feller, C. Ho, S. Mangenot, G. Marino, J. Nilsson, E. Parrilli, E. P. C. Rocha, Z. Rouy, A. Sekowska, M. L. Tutino, D. Vallenet, G. von Heijne, and A. Danchin. 2005. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 151325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Methe, B. A., K. E. Nelson, J. W. Deming, B. Momen, E. Melamud, X. J. Zhang, J. Moult, R. Madupu, W. C. Nelson, R. J. Dodson, L. M. Brinkac, S. C. Daugherty, A. S. Durkin, R. T. DeBoy, J. F. Kolonay, S. A. Sullivan, L. W. Zhou, T. M. Davidsen, M. Wu, A. L. Huston, M. Lewis, B. Weaver, J. F. Weidman, H. Khouri, T. R. Utterback, T. V. Feldblyum, and C. M. Fraser. 2005. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. USA 10210913-10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Missiakas, D., M. P. Mayer, M. Lemaire, C. Georgopoulos, and S. Raina. 1997. Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 24355-371. [DOI] [PubMed] [Google Scholar]
- 64.Mizushima, T., K. Kataoka, Y. Ogata, R. Inoue, and K. Sekimizu. 1997. Increase in negative supercoiling of plasmid DNA in Escherichia coli exposed to cold shock. Mol. Microbiol. 23381-386. [DOI] [PubMed] [Google Scholar]
- 65.Munoz, M. E., and E. Ponce. 2003. Pyruvate kinase: current status of regulatory and functional properties. Comp. Biochem. Physiol. B 135197-218. [DOI] [PubMed] [Google Scholar]
- 66.Muralikrishna, P., and B. S. Cooperman. 1995. Ribosomal components neighboring the 2475 loop in Escherichia coli 50S subunits. Biochemistry 34115-121. [DOI] [PubMed] [Google Scholar]
- 67.Murray, N. E., W. J. Brammar, and K. Murray. 1977. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol. Gen. Genet. 15053-61. [DOI] [PubMed] [Google Scholar]
- 68.Nigou, J., L. G. Dover, and G. S. Besra. 2002. Purification and biochemical characterization of Mycobacterium tuberculosis SuhB, an inositol monophosphatase involved in inositol biosynthesis. Biochemistry 414392-4398. [DOI] [PubMed] [Google Scholar]
- 69.Nogi, Y., N. Masui, and C. Kato. 1998. Photobacterium profundum sp. nov., a new, moderately barophilic bacterial species isolated from a deep-sea sediment. Extremophiles 21-7. [DOI] [PubMed] [Google Scholar]
- 70.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 1742525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pavlovic, M., S. Hormann, R. F. Vogel, and M. A. Ehrmann. 2005. Transcriptional response reveals translation machinery as target for high pressure in Lactobacillus sanfranciscensis. Arch. Microbiol. 18411-17. [DOI] [PubMed] [Google Scholar]
- 72.Phadtare, S. 2004. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6125-136. [PubMed] [Google Scholar]
- 73.Pogliano, K. J., and J. Beckwith. 1993. The Cs sec mutants of Escherichia coli reflect the cold sensitivity of protein export itself. Genetics 133763-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Polissi, A., W. De Laurentis, S. Zangrossi, F. Briani, V. Longhi, G. Pesole, and G. Deho. 2003. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 154573-580. [DOI] [PubMed] [Google Scholar]
- 75.Porankiewicz, J., J. Schelin, and A. K. Clarke. 1998. The ATP-dependent Clp protease is essential for acclimation to UV-B and low temperature in the cyanobacterium Synechococcus. Mol. Microbiol. 29275-283. [DOI] [PubMed] [Google Scholar]
- 76.Ray, M. K., G. S. Kumar, and S. Shivaji. 1994. Phosphorylation of lipopolysaccharides in the Antarctic psychrotroph Pseudomonas syringae: a possible role in temperature adaptation. J. Bacteriol. 1764243-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rock, C. O., and J. E. Cronan. 1996. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim. Biophys. Acta 13021-16. [DOI] [PubMed] [Google Scholar]
- 78.Rosen, R., D. Biran, E. Gur, D. Becher, M. Hecker, and E. Z. Ron. 2002. Protein aggregation in Escherichia coli: role of proteases. FEMS Microbiol. Lett. 2079-12. [DOI] [PubMed] [Google Scholar]
- 79.Sakamoto, T., and N. Murata. 2002. Regulation of the desaturation of fatty acids and its role in tolerance to cold and salt stress. Curr. Opin. Microbiol. 5208-210. [DOI] [PubMed] [Google Scholar]
- 80.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 81.Sands, M. K., and R. B. Roberts. 1952. The effects of a tryptophan-histidine deficiency in a mutant of Escherichia coli. J. Bacteriol. 63505-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sarubbi, E., K. E. Rudd, H. Xiao, K. Ikehara, M. Kalman, and M. Cashel. 1989. Characterization of the spoT gene of Escherichia coli. J. Biol. Chem. 26415074-15082. [PubMed] [Google Scholar]
- 83.Siddiquee, K. A., M. J. Arauzo-Bravo, and K. Shimizu. 2004. Effect of a pyruvate kinase (pykF-gene) knockout mutation on the control of gene expression and metabolic fluxes in Escherichia coli. FEMS Microbiol. Lett. 23525-33. [DOI] [PubMed] [Google Scholar]
- 84.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2833-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tsu, C. A., and O. C. Uhlenbeck. 1998. Kinetic analysis of the RNA-dependent adenosine triphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry 3716989-16996. [DOI] [PubMed] [Google Scholar]
- 86.VanBogelen, R. A., and F. C. Neidhardt. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA 875589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vezzi, A., S. Campanaro, M. D'Angelo, F. Simonato, N. Vitulo, F. M. Lauro, A. Cestaro, G. Malacrida, B. Simionati, N. Cannata, C. Romualdi, D. H. Bartlett, and G. Valle. 2005. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 3071459-1461. [DOI] [PubMed] [Google Scholar]
- 88.von Freiesleben, U., K. V. Rasmussen, and M. Schaechter. 1994. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol. Microbiol. 14763-772. [DOI] [PubMed] [Google Scholar]
- 89.Waller, P. R., and R. T. Sauer. 1996. Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J. Bacteriol. 1781146-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wegrzyn, A., B. Wrobel, and G. Wegrzyn. 1999. Altered biological properties of cell membranes in Escherichia coli dnaA and seqA mutants. Mol. Gen. Genet. 261762-769. [DOI] [PubMed] [Google Scholar]
- 92.Welch, T. J., and D. H. Bartlett. 1998. Identification of a regulatory protein required for pressure-responsive gene expression in the deep-sea bacterium Photobacterium species strain SS9. Mol. Microbiol. 27977-985. [DOI] [PubMed] [Google Scholar]
- 93.Welch, T. J., A. Farewell, F. C. Neidhardt, and D. H. Bartlett. 1993. Stress response of Escherichia coli to elevated hydrostatic pressure. J. Bacteriol. 1757170-7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wiegeshoff, F., C. L. Beckering, M. Debarbouille, and M. A. Marahiel. 2006. Sigma L is important for cold shock adaptation of Bacillus subtilis. J. Bacteriol. 1883130-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Williams, S. G., L. T. Varcoe, S. R. Attridge, and P. A. Manning. 1996. Vibrio cholerae Hcp, a secreted protein coregulated with HlyA. Infect. Immun. 64283-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yayanos, A. A., and E. C. Pollard. 1969. A study of effects of hydrostatic pressure on macromolecular synthesis in Escherichia coli. Biophys. J. 91464-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]