Abstract
The anaerobic metabolism of catechol (1,2-dihydroxybenzene) was studied in the betaproteobacterium Thauera aromatica that was grown with CO2 as a cosubstrate and nitrate as an electron acceptor. Based on different lines of evidence and on our knowledge of enzymes and genes involved in the anaerobic metabolism of other aromatic substrates, the following pathway is proposed. Catechol is converted to catechylphosphate by phenylphosphate synthase, which is followed by carboxylation by phenylphosphate carboxylase at the para position to the phosphorylated phenolic hydroxyl group. The product, protocatechuate (3,4-dihydroxybenzoate), is converted to its coenzyme A (CoA) thioester by 3-hydroxybenzoate-CoA ligase. Protocatechuyl-CoA is reductively dehydroxylated to 3-hydroxybenzoyl-CoA, possibly by 4-hydroxybenzoyl-CoA reductase. 3-Hydroxybenzoyl-CoA is further metabolized by reduction of the aromatic ring catalyzed by an ATP-driven benzoyl-CoA reductase. Hence, the promiscuity of several enzymes and regulatory proteins may be sufficient to create the catechol pathway that is made up of elements of phenol, 3-hydroxybenzoate, 4-hydroxybenzoate, and benzoate metabolism.
Phenolic compounds contain a benzene ring to which one or more hydroxyl groups are attached. This class of compounds is widely distributed in nature. About 4,000 phenolic plant compounds are known, many of which serve as antioxidants, lignin precursors, and pigments, among other uses. Some of these compounds occur in food or are used in medicine.
The microbial metabolism of aromatic compounds under aerobic conditions has been well studied and documented (11). Catechol (pyrocatechol, 1,2-dihydroxybenzene) is one of the few central intermediates in aerobic aromatic metabolism. Catechol metabolism has also been observed under anoxic conditions, but pure-culture studies are rare. Growth on phenol and catechol requires CO2 (8, 16). Anaerobic catechol metabolism was observed previously in a methanogenic enrichment culture (12), in which phenol was considered the first intermediate. Extracts of the sulfate-reducing bacterium Desulfobacterium sp. strain Cat2 formed protocatechuyl coenzyme A (protocatechuyl-CoA) from catechol, bicarbonate, and CoA. This oxygen-sensitive reaction required high concentrations of both bicarbonate and protein, and only very low levels of enzyme activities were detected. In a second oxygen-sensitive step, protocatechuyl-CoA was reduced to 3-hydroxybenzoyl-CoA by reductive elimination of the p-hydroxyl group. Further dehydroxylation to benzoyl-CoA could not be detected (10). Anaerobic protocatechuate metabolism in Thauera aromatica strain AR-1 involves protocatechuate-CoA ligase and protocatechuyl-CoA reductase, leading to 3-hydroxybenzoyl-CoA (26).
According to this working hypothesis, the initial steps of catechol metabolism should be similar to those of phenol metabolism, i.e., phosphorylation and carboxylation to protocatechuate. Further degradation of protocatechuate may yield 3-hydroxybenzoyl-CoA (Fig. 1A). The genes coding for most enzymes involved in the anaerobic metabolism of aromatic compounds, such as phenol, 3-hydroxybenzoate, 4-hydroxybenzoate, and benzoate, are known to be present in T. aromatica, and the genome of the related organism “Aromatoleum aromaticum” strain EbN1 (proposed name for Azoarcus sp. strain EbN1), which is able to grow under denitrifying conditions on a variety of aromatic compounds, has been sequenced (28).
FIG. 1.
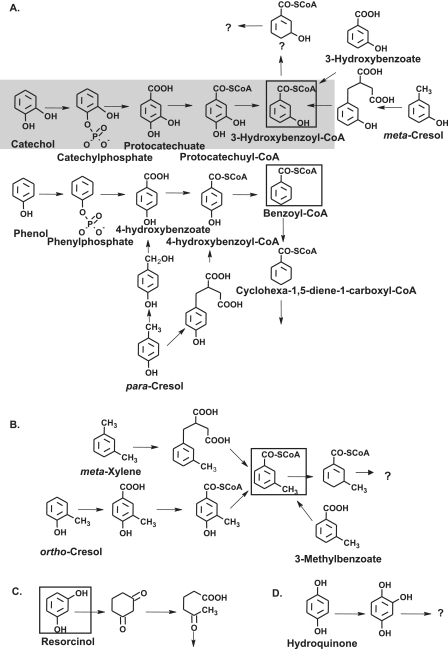
Anaerobic metabolism of representative aromatic compounds containing one or two phenolic hydroxyl groups. (A) Phenolic compounds metabolized via benzoyl-CoA or 3-hydroxybenzoyl-CoA. (B) Phenolic compounds and m-xylene metabolized via 3-methylbenzoyl-CoA. (C) Metabolism of resorcinol. (D) Metabolism of hydroquinone. The pathway of catechol was studied here, and the proposed scheme for the initial reactions is indicated by shading. The central intermediates that undergo aromatic ring reduction are enclosed in boxes. Note that several isomeric cresols are metabolized via different routes, depending on the organism and the redox potential of the terminal electron acceptor.
In this work we studied anaerobic catechol metabolism in T. aromatica type strain K172 that was grown on catechol with CO2 and nitrate. It appears that no enzyme of the pathway was specific for catechol. Rather, the promiscuity of several enzymes and regulatory proteins involved in phenol, 3-hydroxybenzoate, 4-hydroxybenzoate, and benzoate metabolism may be sufficient to create the catechol pathway. This provides an example of the great flexibility of bacterial metabolism that enables an organism to utilize a variety of different but chemically related substrates by using a limited set of genes and enzymes, which are even under proper transcriptional control, without creating dead end metabolites.
MATERIALS AND METHODS
Materials and equipment.
T. aromatica type strain K172 (= DSMZ 6984) (1) and Azoarcus sp. strain EbN1 (tentatively named Aromatoleum aromaticum DSMZ 9506) (28) have been deposited in the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). Chemicals were obtained from Sigma-Aldrich (Deisenhofen, Germany), Merck (Darmstadt, Germany), Roth (Karlsruhe, Germany), and Fluka (Neu-Ulm, Germany). Biochemicals were obtained from Roche (Mannheim, Germany) or Gerbu (Craiberg, Germany). [carboxyl-14C]protocatechuate, NaH14CO3, [U-14C]phenol, and [U-14C]catechol were obtained from American Radiolabeled Chemicals Inc./Biotrend Chemikalien GmbH (Cologne, Germany). Nitrogen, helium, and an N2-H2 gas mixture (95/5, vol/vol) were purchased from Sauerstoffwerke Friedrichshafen (Friedrichshafen, Germany). High-pressure liquid chromatography (HPLC) equipment (model L-6200A HPLC plus an L-4000A UV detector) was purchased from Merck (Darmstadt, Germany), Grom (Herrenberg-Kayh, Germany), and Raytest (Straubenhardt, Germany). Silica Gel 60 F254 plates (20 by 20 cm) for thin-layer chromatography analysis were purchased from Merck. The hybridization membrane used for immunoblotting was purchased from Schleicher & Schuell (Dassel, Germany). The antisera used were raised in rabbits by BioScience (Göttingen, Germany). Materials and equipment for protein purification were obtained from Amersham Biosciences (Freiburg, Germany) and Millipore (Eschborn, Germany).
Bacterial growth, cell harvest, and storage.
T. aromatica was grown anaerobically at 30°C on a mineral salts medium (30) with 1 mM catechol and 10 mM NaHCO3 as carbon sources and 10 mM NaNO3 as the electron acceptor. Azoarcus sp. strain EbN1 was grown anaerobically at 30°C on a mineral salt medium (28) with 5 mM benzoate as the carbon and energy source and 15 mM nitrate as the electron acceptor. Cells were harvested before each culture reached stationary phase. For large-scale fermentation in a 200-liter fed-batch fermentor 1 mM catechol and 3.6 mM nitrate were added initially. After growth had started, the substrates were added continuously from a stock solution containing 1 M catechol (or phenol) and 3.6 M nitrate by using a computer-controlled pump (3) until the optical density at 578 nm (OD578) (1-cm light path) reached 2. An A578 of 1 corresponded to 0.37 g (dry weight) per liter. T. aromatica was also grown at a small scale (2 liters) on other monocyclic aromatic acids (5 mM benzoate, 5 mM 3-hydroxybenzoate, or 5 mM 4-hydroxybenzoate) with 15 mM nitrate and with 1 mM protocatechuate and 3 mM nitrate to study simultaneous adaptation and protein expression and to isolate DNA. Cells were harvested by centrifugation at 35,000 × g at 4°C for 10 min, and the cell pellets were stored at −20°C.
Plasmid, primers, and Escherichia coli strain.
Plasmid pEX5-CT/TOPO was obtained from Invitrogen (Karlsruhe, Germany). Plasmid pET16b, E. coli BL31(DE3) [E. coli B F− ompT hsdS(rB− mB−) gal dcm], and E. coli BL31(DE3)/plysS [E. coli B F− ompT hsdS(rB− mB−) gal dcm/plysS Cmr] were obtained from Novagen (Bad Soden, Germany). E. coli cells were grown aerobically at 37°C on Luria-Bertani medium with 34 μg ml−1 chloramphenicol or 100 μg ml−1 ampicillin. Primers were synthesized by biomers.net GmbH (Ulm, Germany).
Preparation of chromosomal DNA.
Chromosomal DNA was prepared from 50 mg of frozen cells (T. aromatica or “A. aromaticum”) (21). It was suspended in water and stored at 4°C.
Simultaneous adaptation experiments with dense cell suspensions.
Simultaneous adaptation experiments with dense cell suspensions were carried out in anaerobic tubes. Fresh benzoate-, protocatechuate-, phenol-, or catechol-grown cells were precipitated by centrifugation and washed three times with mineral salts medium without organic substrates. The washed cells were resuspended in 10 ml of mineral salts medium containing 10 mM NaNO3 and 10 mM NaHCO3, resulting in a final OD578 of 7 (2.5 mg [dry weight] per ml). The tubes were incubated at 30°C with 1 mM substrates, and after 0.5, 5, 10, 15, 20, 30, and 60 min, 1-ml samples were removed anaerobically and immediately cooled on ice. The cells were precipitated by centrifugation (4°C, 35,000 × g, 10 min), and the concentrations of the substrates in the supernatants were determined by spectroscopic analysis (16, 19) using overlay spectra. The UV spectra for samples obtained from dense cell suspensions without a substrate were subtracted. The concentration of protocatechuate was determined at 300 nm using samples whose pH was adjusted to 11 immediately prior to measurement (ɛ300 = 8,100 M−1 cm−1).
Cell extracts.
Extracts were prepared under anaerobic conditions by suspending 1 part of frozen cells in 1 part of 10 mM Tris-HCl buffer (pH 7.8) containing 20% glycerol, 2 mM MgCl2, 2 mM dithioerythritol, and traces of DNase I. Each suspension was passed through a French pressure cell (137 MPa), the lysate was ultracentrifuged (100,000 × g, 4°C, 1 h), and the supernatant was stored at −20°C. The protein content was determined by the Bradford method (4) with bovine serum albumin as the standard.
SDS-PAGE and immunoblot experiments.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (13.5% polyacrylamide) was performed by the Laemmli method (18). Proteins were visualized by Coomassie blue staining (36). The following proteins were used as molecular mass standards: rabbit phosphorylase b (97 kDa), bovine serum albumin (67 kDa), egg ovalbumin (45 kDa), lactate dehydrogenase (34 kDa), carbonic anhydrase (29 kDa), and lysozyme (14 kDa). Cell extracts were separated by SDS-PAGE, and proteins were blotted onto a nitrocellulose membrane. Proteins that hybridized with antibodies were immunologically detected by luminescence using the ECL system from Amersham Biosciences (Freiburg, Germany).
Preparation of antibodies from antiserum.
The specific antibodies used were purified from serum by using the corresponding antigens bound to nitrocellulose. The method used was described by Smith and Fisher (34).
Enzyme assays.
All enzyme assays were performed at 30°C.
(i) Phenylphosphate/catechylphosphate formation and phenylphosphate carboxylation.
Phenylphosphate synthase (22) is composed of three subunits and is rather labile in cell extracts. Therefore, activities were determined in dense suspensions of whole cells (17). In brief, concentrated cell suspensions (optical density, 30) were incubated with [14C]phenol or [14C]catechol, and the formation of [14C]phenylphosphate or [14C]catechylphosphate was analyzed by thin-layer chromatography and phosphorimaging. Phenylphosphate carboxylase activity in cell extracts was measured as described previously (32). Phenol-grown cells were used as a positive control.
(ii) ATP-dependent protocatechuate formation from catechol and CO2.
The 500-μl assay mixture used routinely contained 100 mM Tris-HCl (pH 7.5), 20 mM MgCl2, 1 mM MnCl2, 10 mM ATP, 40 mM mercaptoethanol, 0.25 mM [U-14C]catechol (1.3 kBq/nmol), and cell extract (1.5 to 2 mg protein). The assay was performed under anaerobic conditions. NaHCO3 (20 mM) and KCl (2 mM) were added to start the carboxylase reaction. The reaction was stopped after 15 min by adding 50 μl of 3 M perchloric acid. After centrifugation of denatured protein, the radiolabeled substrate and products in the supernatant were separated by thin-layer chromatography and detected by phosphorimaging.
(iii) Aromatic acid-CoA ligases.
Benzoate-CoA ligase, 3-hydroxybenzoate-CoA ligase, and 4-hydroxybenzoate-CoA ligase activities were determined using a coupled spectrophotometric assay (19). The assay mixture (0.5 ml) for the protocatechuate-CoA ligase assay contained 100 mM Tris-HCl (pH 8.5), 2.5 mM MgCl2, 2 mM dithioerythritol, 2 mM ATP, 0.4 mM CoA, 0.5 mM aromatic acid, and 0.1 mg to 10 μg protein. The reaction was stopped by addition of 50 μl of 3 M perchloric acid, followed by centrifugation of the denatured protein, and CoA thioesters in the supernatant were analyzed by HPLC. In a direct spectrophotometric assay, the formation of protocatechuyl-CoA was followed at 365 nm. The molar absorption coefficient (ɛ365) was determined to be 5,790 M−1 cm−1. The Km and Vmax values of 3-hydroxybenzoate-CoA ligase were determined at saturating concentrations of the other (co)substrates (4 mM ATP, 4 mM MgCl2, 2 mM CoA, and 2 mM aromatic acid).
(iv) Reduction of 4-hydroxybenzoyl-CoA and protocatechuyl-CoA.
Each strictly anaerobic assay mixture contained 150 mM morpholinepropanesulfonic acid (MOPS)-KOH (pH 7), 2.5 mM MgCl2, 1 mM ATP, 0.2 mM CoA, 1 mM dithionite, 0.8 mM 4-hydroxybenzoyl-CoA or protocatechuyl-CoA, and cell extract (0.3 to 1.5 mg protein). Production of 3-hydroxybenzoyl-CoA and production of benzoyl-CoA were monitored by HPLC. The pH of the enzyme assay mixture was critical, since at pH values less than 6.5 protocatechuyl-CoA reduction was not observed. At pH 7.5 or higher protocatechuyl-CoA was completely hydrolyzed within a few minutes by cell extract, which obviously contained high thioesterase activity.
Synthesis and purification of CoA thioesters.
Protocatechuyl-CoA was synthesized with purified recombinant 3-hydroxybenzoate-CoA ligase using a 100-ml assay mixture. The reaction was stopped, and protein was precipitated with perchloric acid (0.3 M). The supernatant was applied to a 6-ml Strata X column (Phenomenex, Aschaffenburg, Germany), which was equilibrated with 1% methanol in 20 mM ammonium formate (pH 4) at 4°C. The column was washed with 18 ml of 1% methanol in 20 mM ammonium formate (pH 4), and protocatechuyl-CoA was eluted with 30 ml of 80% methanol. The level of recovery of protocatechuyl-CoA was 70%. 4-Hydroxybenzoyl-CoA was synthesized similarly, and the yield was 80%. The purified CoA thioesters were lyophilized and stored at −20°C.
HPLC analysis.
An RP-C18 column (Grom-Sil 120 ODS-4 HE; 5 μm; 120 by 4 mm; Grom) was used with a flow rate of 0.8 ml min−1. Aromatic compounds were detected by UV and/or radioactivity monitoring. Protocatechuate, 3-hydroxybenzoate, 4-hydroxybenzoate, and benzoate were separated with 15% methanol in 20 mM formic acid. The retention times were as follows: protocatechuate, 6.4 min; 4-hydroxybenzoate, 12 min; 3-hydroxybenzoate, 16 min; and benzoate, 20 min. CoA esters were analyzed using a 30-min linear 2 to 15% acetonitrile gradient in 50 mM potassium phosphate buffer (pH 6.8), followed by 5 min of elution with 15% acetonitrile in 50 mM potassium phosphate. The retention times were as follows: protocatechuyl-CoA, 22 min; 4-hydroxybenzoyl-CoA, 25 min; 3-hydroxybenzoyl-CoA, 27 min; and benzoyl-CoA, 32 min.
Thin-layer chromatography analysis.
Catechol and phenol and their phosphorylated products were loaded onto Silica Gel 60 F254 plates, and the plates were developed for between 3 and 3.5 h at room temperature with ethanol-dichloromethane-water (8:4:2, vol/vol/vol) as the solvent. The Rf of catechol was 0.9 and was similar to that of phenol; the Rf of phenylphosphate was 0.8. Protocatechuate and catechol were separated with the lower phase of dichloromethane-acetic acid-water (2:1:1, vol/vol/vol) as the solvent for 1 to 1.5 h at room temperature. The Rf of protocatechuate was 0.17, and the Rf of catechol was 0.68.
Purification of protocatechuate-CoA ligase from catechol-grown cells.
Protocatechuate-CoA ligase, which also acted on 3-hydroxybenzoate and was identical to 3-hydroxybenzoate-CoA ligase, was purified from 30 g of frozen cells under aerobic conditions as described by Laempe et al. (19), using ammonium sulfate precipitation and dialysis, DEAE-Sepharose chromatography, hydroxyapatite chromatography, and reactive green chromatography. The purified protein was analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry by Wolfgang Haehnel (Institut Biologie II, Universität Freiburg, Freiburg, Germany).
Expression of the 3-hydroxybenzoate-CoA ligase gene in E. coli BL21(DE3)/plysS.
The gene encoding 3-hydroxybenzoate-CoA ligase was amplified by PCR using chromosomal DNA of T. aromatica as the template. The sequence of the forward primer (5′ to 3′) was ATGTCTGAACAACTGCAGCCG, and the sequence of the reverse primer (5′ to 3′) was GAGGGTCGCCTGCAGCAT. The PCR conditions were as follows: 33 cycles of 0.5 min of denaturation at 95°C, 0.5 min of annealing at 65°C, and 1.5 min of synthesis at 72°C, followed by 1 min of annealing at 65°C and 5 min of synthesis at 72°C. The sample was loaded onto a 1% agarose gel, and the PCR product was purified with a gel elution kit from Qiagen (Hilden, Germany). The 3-hydroxybenzoate-CoA ligase gene was cloned into the vector pEX5-CT/TOPO with a TOPO cloning kit from Invitrogen (Karlsruhe, Germany). The ligation mixture (1 μl of vector [10 ng of DNA], 1 μl of 6× ligation buffer, 4 μl of purified PCR product [50 ng of DNA]) was incubated at room temperature for 20 min. One microliter of the TOPO ligation reaction mixture was added to 200 μl of competent cells (7) that were kept on ice for 10 min. This transformation mixture was heated at 42°C for 1 min and cooled on ice immediately for 5 min. Then 800 μl of LB medium was added to the mixture, and the transformed culture was incubated for 1 h at 37°C with shaking at 200 rpm. C-terminally His6-tagged 3-hydroxybenzoate-CoA ligase was produced after induction of the gene with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. Details of the procedure are described in the protocol provided by Invitrogen (Karlsruhe, Germany).
Expression of the 4-hydroxybenzoate-CoA ligase gene from Azoarcus sp. strain EbN1 in E. coli BL21(DE3).
The gene encoding 4-hydroxybenzoate-CoA ligase in Azoarcus sp. strain EbN1 was identified by a BLAST search of the National Center for Biotechnology Information (NCBI) database using the N-terminal protein sequence of 4-hydroxybenzoate-CoA ligase purified from T. aromatica (MNAA[E]LLXAGXA[D]AIXLI) (2). It was amplified by PCR using chromosomal DNA as the template. The sequence of the forward primer (5′ to 3′) was GAGGAGAAAATCCATATGAATGCTGCC, and the sequence of the reverse primer (5′ to 3′) was GCCTTGGCAGAATTCTCACAATG. The PCR conditions were as follows: 31 cycles of 45 s of denaturation at 95°C, 45 s of annealing at 55°C, and 200 s of synthesis at 72°C, followed by 5 min of synthesis at 72°C. The PCR product was purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany). Subsequently, the purified product and the vector pET16b (Novagen, Bad Soden, Germany) were restricted with the endonucleases NdeI and EcoRI (Fermentas, Leon-Rot, Germany). Samples were loaded onto a 1% agarose gel, and the restricted DNA was purified with a QIAquick gel extraction kit (Qiagen). Ligation of the fragments (220 ng of vector DNA and 360 ng of PCR product) was performed overnight at 16°C. The complete ligation reaction mixture was used for transformation of 200 μl of competent E. coli DH5α cells, which were plated on LB agar. Resulting clones were picked and grown overnight at 37°C in LB media, and the plasmids were isolated with a QIAprep spin miniprep kit (Qiagen). The constructs were verified by control restriction with PvuI endonuclease. Verified plasmid pET16bhbcL1 was subsequently used for transformation of E. coli BL21(DE3) cells that were cultured in LB medium containing 100 μg ml−1 ampicillin. N-terminally His10-tagged 4-hydroxybenzoate-CoA ligase was produced after induction of gene expression by 0.5 mM IPTG overnight at room temperature.
Purification of recombinant 3-hydroxybenzoate-CoA ligase.
Recombinant 3-hydroxybenzoate-CoA ligase protein was prepared from 30 g of E. coli cells by using DEAE-Sepharose chromatography (19) and nickel chelating Sepharose Fast Flow chromatography (Amersham Biosciences).
Purification of recombinant 4-hydroxybenzoate-CoA ligase.
Recombinant 4-hydroxybenzoate-CoA ligase protein was prepared from 2.3 g of fresh E. coli cells by using a HisTrap FF column (Amersham Biosciences). The binding buffer contained 250 mM KCl, 100 mM imidazole, and 20 mM Tris-HCl (pH 7.8). The elution buffer contained in addition 500 mM imidazole. Active fractions (6 ml) were combined, concentrated to 0.5 ml (Amicon concentrator cell), and stored with 50% glycerol at −20°C.
DNA sequencing and computer analysis.
The DNA sequence of the purified plasmid was determined by G. L. Igloi (Institut Biologie III, Universität Freiburg, Freiburg, Germany). DNA and amino acid sequences were analyzed using the BLAST network service at the NCBI (Bethesda, MD). A protein sequence alignment and a similarity tree of protein sequences were constructed using the multiple-alignment program (http://prodes.toulouse.inra.fr/multalin/multalin.html).
Nucleotide sequence accession number.
The sequence data for the 3-hydroxybenzoate-CoA ligase gene and other five open reading frames (bp 1 to 7072) have been deposited in the GenBank database under accession number AJ278289.
RESULTS
Anaerobic growth on catechol and protocatechuate.
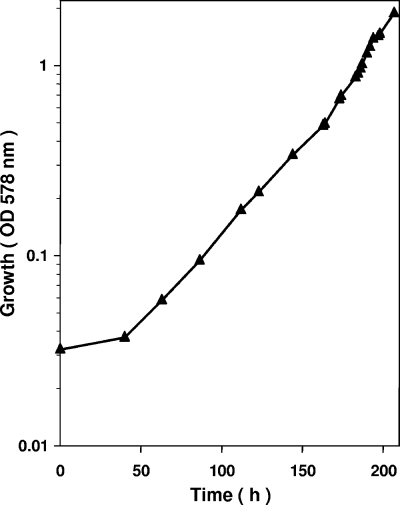
T. aromatica was grown anaerobically on catechol and CO2 with nitrate as the electron acceptor in a fed-batch fermentor (200 liter) to which nitrate and catechol were continuously added from a concentrated stock solution containing 3.6 M NaNO3 and 1 M catechol. This ratio of NaNO3 to catechol corresponded approximately to the ratio of consumption of nitrate and catechol and was chosen to avoid accumulation of toxic catechol and nitrite. The supply of substrate was growth limiting, and the pumping rate was increased exponentially so that a preset generation time of approximately 20 h was obtained. The growth curve obtained is shown in Fig. 2. At the end of the experiment (OD578, 1.9) 5.4 mol catechol and 19 mol nitrate had been consumed and 369 g (wet weight) of cells was obtained. The dry weight content was 0.37 g per liter at an OD578 of 1, resulting in a molar growth yield of approximately 25 g (dry weight) of cells formed per mol of catechol consumed. A similar value was obtained if it was assumed that 369 g of fresh, centrifuged cells contained approximately 30% dry cell mass. Under the experimental conditions, the calculated specific substrate consumption rate of the cells was approximately 22 nmol min−1 mg cell protein−1. The generation time was less than the optimum, which was 15 h; for comparison, T. aromatica grew on protocatechuate and nitrate with a 14-h generation time.
FIG. 2.
Anaerobic growth of T. aromatica on catechol with NaNO3 as the electron acceptor in a 200-liter continuous fed-batch fermentor. The ratio of the catechol concentration supplied to the nitrate concentration supplied was 1:3.6.
Simultaneous adaptation experiments with dense cell suspensions.
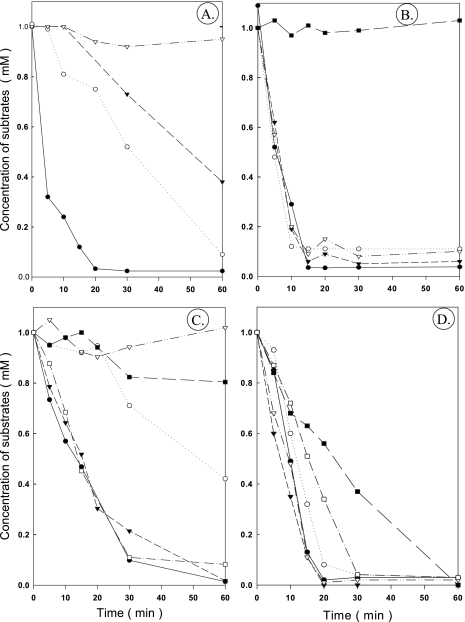
The ability of adapted cells to simultaneously metabolize benzoate, 3-hydroxybenzoate, 4-hydroxybenzoate, protocatechuate, catechol, and phenol was studied using dense suspensions of cells that were grown anaerobically on different substrates (Fig. 3). This kind of experiment does not take into account the possible selectivity of possible substrate transporters. Benzoate-grown cells degraded benzoate immediately (Fig. 3A). After a 10- to 15-min lag phase, 3-hydroxybenzoate and 4-hydroxybenzoate began to be consumed, whereas metabolism of protocatechuate did not start for >1 h. Protocatechuate-grown cells immediately consumed protocatechuate, benzoate, 3-hydroxybenzoate, and 4-hydroxybenzoate (Fig. 3B), whereas catechol was not metabolized in the first 1 h. Phenol-grown cells were not adapted to use catechol, protocatechuate, and 3-hydroxybenzoate, but they immediately consumed phenol, 4-hydroxybenzoate, and benzoate (Fig. 3C). Catechol-grown cells immediately consumed all substrates, but phenol and catechol were used more slowly than the other substrates (Fig. 3D).
FIG. 3.
Simultaneous adaptation experiments: substrate consumption by dense suspensions of cells (OD578, 7) that were grown anaerobically with nitrate on different aromatic substrates. (A) Cells grown on benzoate. (B) Cells grown on protocatechuate. (C) Cells grown on phenol. (D) Cells grown on catechol. •, benzoate; ○, 3-hydroxybenzoate; ▾, 4-hydroxybenzoate; ▿, protocatechuate; ▪, catechol; □, phenol.
Hence, phenol did not induce all of the genes required for catechol, protocatechuate, and 3-hydroxybenzoate metabolism, whereas catechol did induce these genes. These results are consistent with the hypothesis that catechol is metabolized via protocatechuate and 3-hydroxybenzoate (or CoA esters). Simultaneous adaptation of catechol-grown cells to phenol consumption would be expected if the initial steps in catechol metabolism were the same as those in phenol metabolism. However, further metabolism of catechol appears to require additional steps that are not involved in phenol metabolism.
Protein patterns of extracts of cells grown on different aromatic substrates and immunological detection of catabolic enzymes (benzoyl-CoA reductase and 3-hydroxybenzoate-CoA ligase).
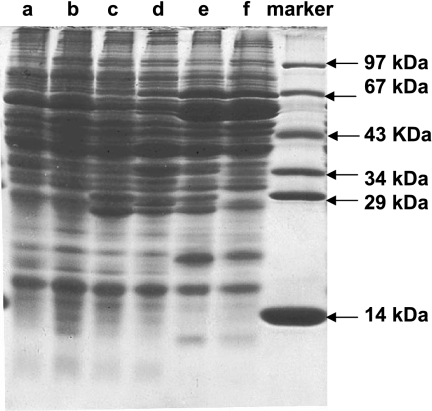
The soluble protein fractions of cells that were grown on different aromatic substrates were separated by SDS-PAGE (Fig. 4). The patterns observed might provide additional information about the induced proteins required for specific metabolic pathways. Extracts of phenol-grown cells (Fig. 4, lane f) and catechol-grown cells (lane e) contained large amounts of a 67-kDa polypeptide, which most likely represented protein 1 of the three-component enzyme phenylphosphate synthase (22, 30). In addition 53-, 54-, 18-, and 10-kDa polypeptides, presumably representing the four subunits of phenylphosphate carboxylase, were induced (32). 3-Hydroxybenzoate-induced 29- and 27-kDa polypeptides (lane c) were also observed in extracts of protocatechuate-grown cells (lane d) and catechol-grown cells (lane e), but not in extracts of phenol-grown cells (lane f). It appears that catechol-grown cells produced all protein bands that were induced in phenol-grown cells (compared with 4-hydroxybenzoate-grown cells [lane b]). In addition to these bands, protein bands induced in protocatechuate- and 3-hydroxybenzoate-grown cells (compared with benzoate-grown cells [lane a]) were seen.
FIG. 4.
SDS-PAGE (13.5%) of the soluble protein fraction (20 to 30 μg protein) of cells grown on different aromatic substrates. Lane a, benzoate-grown cells; lane b, 4-hydroxybenzoate-grown cells; lane c, 3-hydroxybenzoate-grown cells; lane d, protocatechuate-grown cells; lane e, catechol-grown cells; lane f, phenol-grown cells.
This comparison of protein patterns suggests that phenol metabolism and catechol metabolism may have common initial steps. Further catechol metabolism requires additional proteins that are characteristic of protocatechuate metabolism as well as 3-hydroxybenzoate metabolism. If catechol was initially carboxylated to protocatechuate, as phenol is carboxylated to 4-hydroxybenzoate, the further metabolism may proceed via 3-hydroxybenzoate (or the CoA ester). 3-Hydroxybenzoate metabolism requires benzoyl-CoA reductase or a similar enzyme for ring reduction, as well as some other proteins that are not involved in 4-hydroxybenzoate or benzoate metabolism (19).
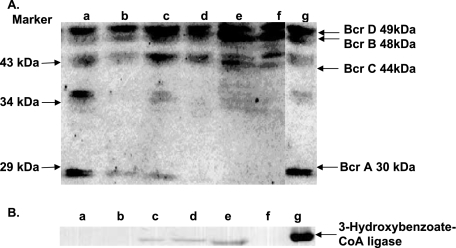
If this concept is correct, catechol-grown cells should contain benzoyl-CoA reductase (four subunits with molecular masses of 49, 48, 43, and 30 kDa) and possibly 3-hydroxybenzoate-CoA ligase (57 kDa), the initial enzyme of the 3-hydroxybenzoate pathway. This was tested by performing immunological hybridization experiments using polyclonal antibodies raised against these two enzymes (Fig. 5). In Western immunoblots of all cell extracts, the subunits of benzoyl-CoA reductase were stained (Fig. 5A); it is noteworthy, however, that some protein bands must have stained nonspecifically. 3-Hydroxybenzoate-CoA ligase was detected not only in 3-hydroxybenzoate-grown cells but also in catechol- and protocatechuate-grown cells, whereas this enzyme was not detected in phenol-, 4-hydroxybenzoate-, and benzoate-grown cells (Fig. 5B). These results are in line with the expectation that catechol is metabolized via protocatechuate and 3-hydroxybenzoate (or the CoA ester).
FIG. 5.
Detection of (A) benzoyl-CoA reductase and (B) 3-hydroxybenzoate-CoA ligase by Western blotting, using polyclonal antibodies raised against the purified enzymes. Extracts (20 to 30 μg protein) of cells grown on different substrates were separated by SDS-PAGE and then hybridized. Lane a, benzoate-grown cells; lane b, 4-hydroxybenzoate-grown cells; lane c, 3-hydroxybenzoate-grown cells; lane d, protocatechuate-grown cells; lane e, catechol-grown cells; lane f, phenol-grown cells; lane g, purified enzyme (2 μg) used as a positive control for (A) benzoyl-CoA reductase with four subunits (Bcr A, Bcr B, Bcr C, and Bcr D) and for (B) 3-hydroxybenzoate-CoA ligase.
Phenylphosphate carboxylase activity in extracts of cells grown on different aromatic substrates.
Carboxylation of phenol requires phenylphosphate synthase (forming phenylphosphate) and phenylphosphate carboxylase (forming 4-hydroxybenzoate). Carboxylation of catechol would similarly require catechylphosphate synthase activity (forming catechylphosphate) and catechylphosphate carboxylase (forming 3,4-dihydroxybenzoate [protocatechuate]). The presence of the carboxylase was examined following the carboxylation of phenylphosphate, as well as the exchange of 14CO2 into the carboxyl group of 4-hydroxybenzoate and protocatechuate (32). Catechol- and phenol-grown cells contained similar carboxylase activities. In catechol- and phenol-grown cells the phenylphosphate carboxylase activities were 139 and 97 nmol min−1 mg−1, respectively. The 14CO2 exchange activities with 4-hydroxybenzoate were 668 and 633 nmol min−1 mg−1, respectively, and the 14CO2 exchange activities with protocatechuate were 335 and 390 nmol min−1 mg−1, respectively. These enzyme activities were not present in any other cell batches.
Further experiments demonstrating catechol conversion to catechylphosphate and protocatechuate.
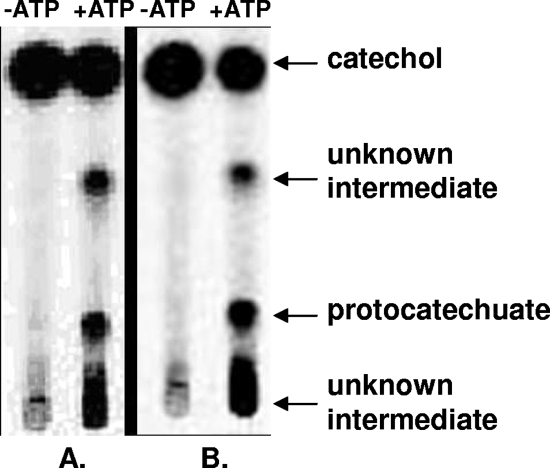
To demonstrate the formation of catechylphosphate from catechol, a dense suspension of catechol-grown cells was incubated with 1 mM [U-14C]catechol. The product formation was analyzed by thin-layer chromatography and phosphorimaging. After 30 s the formation of a labeled product was observed, which migrated similar to phenylphosphate (an authentic catechylphosphate standard was not available). The cells also rapidly converted [U-14C]phenol to a labeled product that comigrated with phenylphosphate. After 3 min of incubation the products accounted for the main labeled spots (not shown). The conversion of 1 mM [U-14C]catechol plus CO2 by extracts of phenol- or catechol-grown cells was studied both in the absence and in the presence of ATP (Fig. 6). In both cases [14C]protocatechuate was formed, provided that ATP was present. These results suggest that the two initial enzymes of phenol metabolism also convert catechol plus CO2 to protocatechuate with ATP via catechylphosphate.
FIG. 6.
Transformation of [U-14C]catechol to [14C]protocatechuate and other unknown products in 15 min by extracts of cells grown on (A) catechol and (B) phenol. The assays were conducted in the absence or in the presence of Mg-ATP. An autoradiogram for samples separated by thin-layer chromatography is shown. For details see Materials and Methods.
Protocatechuate-CoA ligase activity in extracts of cells grown on different aromatic substrates and enzyme assay.
Aromatic acid-CoA ligase activity was tested at pH 8.5 using extracts of cells grown on catechol, 4-hydroxybenzoate, 3-hydroxybenzoate, and benzoate. The activation of benzoate, 4-hydroxybenzoate, 3-hydroxybenzoate, and protocatechuate (aromatic acid + Mg-ATP + CoA → acyl-CoA + Mg-AMP + pyrophosphate) was studied by performing a HPLC analysis of CoA ester formation (Table 1). Similar results were obtained using spectrophotometric methods (see Materials and Methods). The products were identified by comparing the retention times and UV spectra with the properties of authentic synthesized compounds (19, 35). Benzoate-CoA ligase was present in cells grown on all four substrates. 4-Hydroxybenzoate-CoA ligase activity was present in all but the benzoate-grown cells. Protocatechuate and 3-hydroxybenzoate-CoA ligase activity were found in catechol- and 3-hydroxybenzoate-grown cells and, surprisingly, also in 4-hydroxybenzoate-grown cells (for an explanation, see below). The protocatechuate-CoA ligase activity was highest at pH 8.5, and at pH 8.0 and 9.0 the activity was 25% lower. The observed pattern of enzyme activities in the different cell batches can be explained by the substrate specificities of benzoate-CoA ligase (33), 3-hydroxybenzoate-CoA ligase (this study [see below]), and 4-hydroxybenzoate-CoA ligase (2; this study [see below]) and by differences in their induction during growth on different aromatic acids.
TABLE 1.
Various aromatic acid-CoA ligase activities in extracts of T. aromatica after anaerobic growth with nitrate on different aromatic substrates
| Enzyme | Activities (nmol min−1 mg−1) after growth on the following substratesa:
|
|||
|---|---|---|---|---|
| Benzoate | 4-Hydroxybenzoate | 3-Hydroxybenzoate | Catechol | |
| Benzoate-CoA ligase | 166 | 101 | 85 | 91 |
| 4-Hydroxybenzoate-CoA ligase | <5 | 284 | 168 | 120 |
| 3-Hydroxybenzoate-CoA ligase | <5 | <5 | 175 | 150 |
| Protocatechuate-CoA ligase | <5 | 110 | 214 | 282 |
The values are means of two individual cultures, and the deviations are less than ±15%. For the specific activities of the purified 3-hydroxybenzoate- and 4-hydroxybenzoate-CoA ligases, see the text. The benzoate-CoA ligase activities were as follows (33): with benzoate, 16.5 μmol min−1 mg−1; with 3-hydroxybenzoate, <1 μmol min−1 mg−1; with 4-hydroxybenzoate, <1 μmol min−1 mg−1; and with protocatechuate, <1 μmol min−1 mg−1.
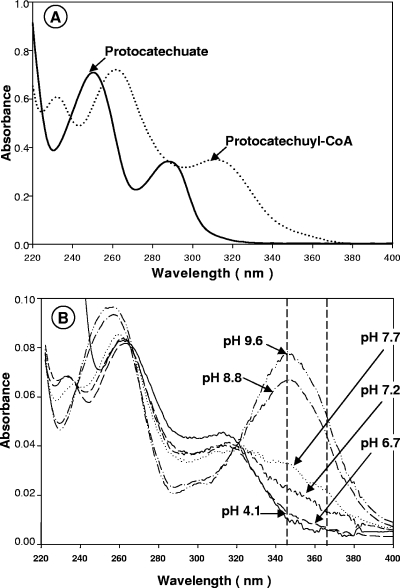
Due to the characteristic UV absorption of protocatechuyl-CoA (Fig. 7), the enzyme activity could also be followed using a direct spectrophotometric assay. At neutral pH the CoA ester formation with protocatechuate resulted in a shift of the absorption maximum at 290 nm to an absorption maximum at around 320 nm. At alkaline pH the dissociation of the phenolic hydroxyl group resulted in a strong increase in absorption with a maximum at 350 nm. Hence, protocatechuyl-CoA formation could be followed directly spectrophotometrically at pH 8.5 (routinely with a filter photometer at 365 nm).
FIG. 7.
UV spectra of (A) 75 μM protocatechuate and 33 μM protocatechuyl-CoA in 50 mM potassium phosphate buffer (pH 6.8) and (B) 4 μM protocatechuyl-CoA at different pH values. The maximum absorption of protocatechuyl-CoA at an alkaline pH in the near-UV region is at 345 nm. The enzyme activity was measured photometrically at pH 8.5 and 365 nm using a filter photometer with a strong 365-nm light source (Hg spectral line).
Purification of protocatechuate-CoA ligase and mass spectrometric analysis.
Protocatechuate-CoA ligase was purified in four steps, and the yield from 30 g fresh cells that were grown on catechol and nitrate was 34%. This protocol (19) resulted in an enzyme preparation that was not pure and had a specific activity of 13.8 μmol min−1 mg−1 (not shown). The active fraction, which eluted from a reactive green column with 20 mM protocatechuate, produced two major bands at approximately 60 and 40 kDa, in addition to some minor bands. The 60-kDa band was analyzed by MALDI-TOF mass spectrometry since all aromatic acid-CoA ligases have subunits of this size. The molecular mass determined was 57.6 kDa, and MALDI-TOF mass spectrometric analysis data were used to screen the sequence database at the NCBI (Bethesda, MD). The best fit was 3-hydroxybenzoate-CoA ligase from T. aromatica, with 74% coverage of 523 amino acids. The second best fit was benzoate-CoA ligase from T. aromatica, with 50% coverage of 532 amino acids.
Cloning of the 3-hydroxybenzoate-CoA ligase gene and purification and analysis of the recombinant enzyme.
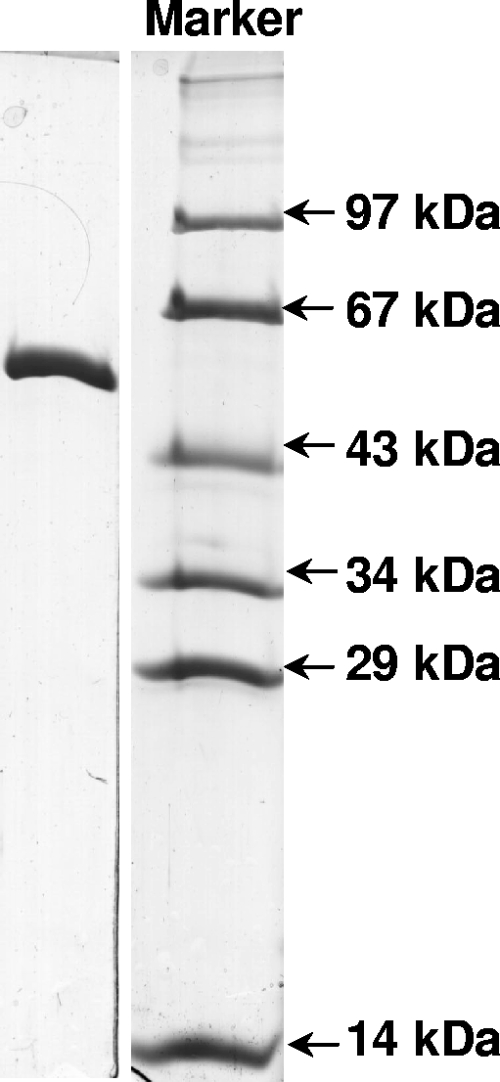
The gene for 3-hydroxybenzoate-CoA ligase was amplified by PCR and cloned, and a C-terminal His6-tagged protein was expressed in E. coli BL21(DE3)/plysS. This recombinant enzyme was purified in two steps from 30 g cells, which yielded 36 mg protein (Fig. 8), and it was also used to raise antibodies (see above). The expected molecular mass was 57.5 kDa. The enzyme acted not only on 3-hydroxybenzoate (specific activity, 14.3 μmol min−1 mg−1; Km, 0.1 ± 0.04 mM; Kcat, 15 s−1; Kcat/Km, 150 s−1 mM−1) but also on protocatechuate (22.7 μmol min−1 mg−1; Km, 0.34 ± 0.1 mM; Kcat, 24 s−1; Kcat/Km, 70 s−1 mM−1), 4-hydroxybenzoate (10.8 μmol min−1 mg−1; Km, 0.1 ± 0.03 mM; Kcat, 11.3 s−1; Kcat/Km, 113 s−1 mM−1), benzoate (7.6 μmol min−1 mg−1; Km, 0.5 ± 0.1 mM; Kcat, 8 s−1; Kcat/Km, 16 s−1 mM−1), and 3,4,5-trihydroxybenzoate (0.7 μmol min−1 mg−1; Km not determined; Kcat, 0.7 s−1). Neither 2-hydroxybenzoate nor 2,3-dihydroxybenzoate was used as a substrate. These results indicate that indeed 3-hydroxybenzoate-CoA ligase is promiscuous and can also act as protocatechuate-CoA ligase.
FIG. 8.
SDS-PAGE (13.5%) of purified 3-hydroxybenzoate-CoA ligase (3 μg, C-terminally His6-tagged protein). The protein band is at the expected molecular mass, 63 kDa. The purification protocol started with 30 g of E. coli BL21(DE3)/plysS/pET5C/TOPO-3OHBCL cells and yielded 36 mg of enzyme.
Cloning of the 4-hydroxybenzoate-CoA ligase gene and purification and analysis of the recombinant enzyme.
Surprisingly, 4-hydroxybenzoate-grown cells also contained protocatechuate-CoA ligase activity, although this ligase was reported not to transform protocatechuate. However, the coupled spectrophotometric assay used previously (2) cannot be used to assay protocatechuyl-CoA synthesis, because the absorption increase caused by the formation of protocatechuyl-CoA (which strongly absorbs at 365 nm) evens out the absorption decrease due to NADH oxidation. Therefore, the substrate specificity of this enzyme was reinvestigated. The gene encoding 4-hydroxybenzoate-CoA ligase in the closely related organism Azoarcus sp. strain EbN1, whose genome has been sequenced (27), was amplified by PCR and cloned, and the gene product was expressed as a C-terminal His10-tagged protein in E. coli BL21(DE3). The recombinant enzyme was purified in one step from 2.3 g cells, and the yield was 1.4 mg protein. The molecular mass was 54 kDa, as expected. The enzyme acted not only on 4-hydroxybenzoate (4.8 μmol min−1 mg−1, defined as 100% activity) but also on protocatechuate (20% activity), 3-hydroxybenzoate (44% activity), and benzoate (31% activity). Phenylacetate or acetate was not used as a substrate. We therefore assumed that the protocatechuate-CoA ligase activity in 4-hydroxybenzoate-grown cells was due to the presence of 4-hydroxybenzoate-CoA ligase that acted to some extent also on protocatechuate.
Reductive dehydroxylation of protocatechuyl-CoA and 4-hydroxybenzoyl-CoA by extracts of cells grown on different aromatic substrates.
The aromatic ring of protocatechuyl-CoA may be directly reduced to a nonaromatic product by a novel reductase, or one or both of the hydroxyl groups are reductively removed, leading either to 4-hydroxybenzoyl-CoA (which is further reduced to benzoyl-CoA) or to 3-hydroxybenzoyl-CoA. The only well-studied enzyme that removes the para-hydroxyl group is 4-hydroxybenzoyl-CoA reductase (4). Benzoyl-CoA and 3-hydroxybenzoyl-CoA are then reduced to a nonaromatic product by an ATP-dependent benzoyl-CoA reductase or a similar enzyme. Therefore, the conversion of protocatechuyl-CoA under reducing conditions with dithionite or Ti(III) was studied with cell extracts both in the presence and in the absence of Mg-ATP. The products were analyzed by HPLC (not shown). [Carboxyl-14C]protocatechuyl-CoA was transformed into labeled 3-hydroxybenzoyl-CoA and an unknown product, which had a retention time similar to that of benzoate but exhibited different spectroscopic properties. After prolonged incubation, other products were observed.
In all cases the ratio of the rate of 4-hydroxybenzoyl-CoA reduction to the rate of protocatechuyl-CoA reduction was approximately 5:1. Both enzyme activities were strongly induced in phenol-, catechol-, protocatechuate-, and 4-hydroxybenzoate-grown cells, induced less in 3-hydroxybenzoate-grown cells, and induced little in benzoate-grown cells. The specific activities for 4-hydroxybenzoyl-CoA reduction in these cells were 17, 17, 16, 21, 8, and 3 nmol min−1 mg−1, respectively.
DISCUSSION
Proposed catechol pathway.
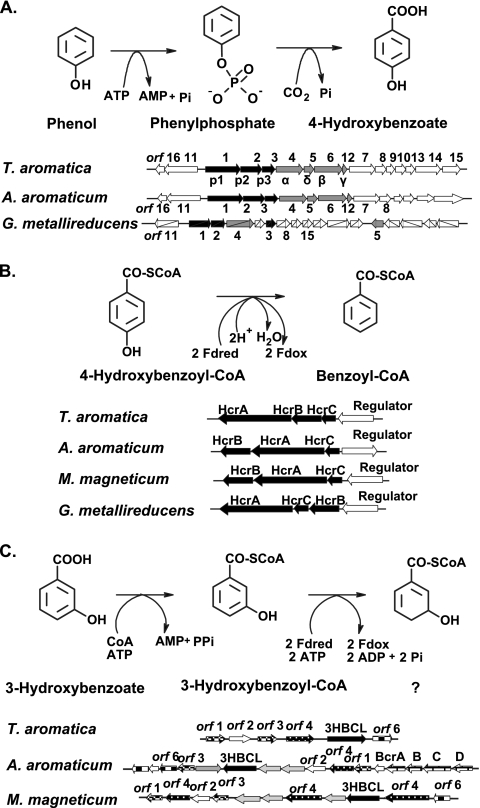
The proposed pathway by which catechol is anaerobically metabolized is composed of elements of phenol, 3-hydroxybenzoate, 4-hydroxybenzoate, and benzoate metabolism pathways. Within the limits of detection and separation, the gene products that were induced when cells were grown on catechol were the products postulated to be involved in these pathways. The gene clusters or operons containing genes coding for these pathways in T. aromatica are shown in Fig. 9.
FIG. 9.
Metabolic reactions and clusters of genes involved in (A) the catabolism of phenol to 4-hydroxybenzoate, (B) reductive dehydroxylation of 4-hydroxybenzoyl-CoA to benzoyl-CoA, and (C) metabolism of 3-hydroxybenzoate by T. aromatica, “A. aromaticum” (Azoarcus sp. strain EbN1), G. metallireducens, and M. magneticum. In panel A, orfs 1, 2, and 3 encode proteins 1, 2, and 3, the components of phenylphosphate synthase, which catalyzes the phosphorylation of phenol to phenylphosphate. orfs 4, 5, 6, and 12 encode subunits α, δ, β, and γ of phenylphosphate carboxylase, which catalyzes the carboxylation of phenylphosphate to 4-hydroxybenzoate and releases phosphate. In panel B, HcrA, HcrB, and HcrC are three subunits of 4-hydroxybenzoyl-CoA reductase, and the open arrows indicate a regulatory protein belonging to the MarR family. In panel C, the genes for 3-hydroxybenzoate-CoA ligase (3HBCL) and other open reading frames are shown. The annotation of 3HBCL in “A. aromaticum” and M. magneticum is tentative but is supported by the similar flanking genes and by the high e value compared with the Thauera enzyme. orfs 1 and 3 encode putative alcohol dehydrogenases. orf 4 is similar to the acyl-CoA dehydrogenase gene, and the orf 6 translated product is a putative hydrolase. The function of orf 2 is not known. The gray arrows indicate the open reading frames that were not found in the T. aromatica gene cluster.
Catechol metabolism is initiated by carboxylation to protocatechuate via catechylphosphate as an intermediate, a process catalyzed by the two enzymes required for phenol carboxylation to 4-hydroxybenzoate via phenylphosphate. The required enzymes are phenylphosphate synthase (composed of three proteins) and phenylphosphate carboxylase (composed of four proteins); the phosphorylated enzyme intermediate of phenylphosphate synthase is dephosphorylated by phenol and by catechol, indicating that both substrates become phosphorylated (23). The genes coding for these two enzymes are located in a large operon that contains other genes with unknown functions. Obviously, both phenol and catechol act as inducers of a putative regulatory protein encoded by a sequence next to the first gene of the operon. In vitro, the conversion of catechol to protocatechuate required Mg-ATP, which is consistent with intermediary phenylphosphate formation. “A. aromaticum” (Azoarcus sp. strain EbN1) harbors a similar phenol gene cluster; however, it is not known whether this strain can grow anaerobically with catechol (28).
Activation of the product protocatechuate requires 3-hydroxybenzoate-CoA ligase, which is encoded by another cluster of genes coding for 3-hydroxybenzoate-induced proteins (19). Nothing is known about the regulator protein. In contrast to our previous findings, 3-hydroxybenzoate-CoA ligase was found to activate protocatechuate effectively. The reason for the previous failure to detect this enzyme activity in a coupled spectrophotometric assay was the high absorption at pH 8 to 9 of protocatechuyl-CoA in the near-UV region. This absorption was approximately as high as the absorption decrease at 365 nm, which was due to the consumption of two NADH molecules in the coupled assay. The same caveat may apply to previous measurements of protocatechuate-CoA ligase activities by the coupled spectrophotometric assay (e.g., for purified 4-hydroxybenzoate-CoA ligase) (2). We postulate that both 3-hydroxybenzoate and protocatechuate act as inducers of the 3-hydroxybenzoate metabolic genes.
Protocatechuyl-CoA is reductively dehydroxylated, possibly by 4-hydroxybenzoyl-CoA reductase. This enzyme consists of three proteins, which are encoded by a gene cluster and may be controlled by a regulatory protein belonging to the MarR family. There is a similar gene cluster in “A. aromaticum” (Azoarcus sp. strain EbN1) (28), Magnetospirillum magneticum (NCBI accession number AP007255), and Geobacter metallireducens (NCBI accession number CP000148). We postulate that both protocatechuyl-CoA and 4-hydroxybenzoyl-CoA function as inducers.
The aromatic ring of 3-hydroxybenzoyl-CoA is thought to be reduced by benzoyl-CoA reductase in T. aromatica (19), a reaction that requires two ATP molecules. In the closely related organism “A. aromaticum” (Azoarcus sp. strain EbN1), a similar 3-hydroxybenzoate gene cluster contains a second copy of the genes coding for benzoyl-CoA reductase (13, 14, 28). Hence, at least in this organism a very similar isoenzyme of benzoyl-CoA reductase may act in ring reduction of 3-hydroxybenzoyl-CoA.
Comparison to anaerobic metabolism of other mono- and diphenolic compounds.
The anaerobic metabolism of some other diphenolic compounds has been studied (Fig. 1). Resorcinol (1,3-dihydroxybenzene) is reduced to 1,3-dioxocyclohexane in an ATP-independent reaction, followed by hydrolytic cleavage of the alicyclic ring (14) (Fig. 1C). Alternatively, direct hydrolytic cleavage of the resorcinol nucleus without an initial reduction has been discussed (10). In other bacteria resorcinol is first hydroxylated to hydroxyhydroquinone (1,2,4-trihydroxybenzene), which is further oxidized to 2-hydroxy-1,4-benzoquinone (24). Hydroxyhydroquinone can also be reduced to hydrohydroxyhydroquinone (31). Hydroquinone (1,4-dihydroxybenzene) is first oxidized to hydroxyhydroquinone, followed by reductive dearomatization (25). Alternatively, it could be degraded via gentisate by oxygen-sensitive carboxylation (9) (Fig. 1D). A carboxylation similar to that reported here for catechol may occur with o-cresol that is converted to 4-hydroxy-3-methylbenzoate. This intermediate is activated to its CoA thioester and reductively dehydroxylated to 3-methylbenzoyl-CoA (29) (Fig. 1B). This benzoyl-CoA analogue can also be formed from 3-methylbenzoate and m-xylene (1,3-dimethylbenzene) (15) (Fig. 1B). Further metabolism of this compound most likely involves ATP-driven ring reduction by benzoyl-CoA reductase, but the methyl substituent may require subsequent steps catalyzed by enzymes that are not required for benzoyl-CoA metabolism, notably a methylglutaryl-CoA dehydrogenase (J. Heider, Darmstadt, Germany, personal communication). m-Cresol, but not o-cresol, is oxidized at the methyl group very much like the toluene methyl group is oxidized to the carboxyl group by first adding fumarate (20) (Fig. 1A). p-Cresol oxidation may occur via methyl hydroxylase, yielding 4-hydroxybenzoate (29), as in the aerobic metabolism (6), or alternatively via fumarate addition and subsequent beta-oxidation, yielding 4-hydroxybenzoyl-CoA (20) (Fig. 1A). 2-Hydroxybenzoate appears to be anaerobically degraded via its CoA ester, followed by a reductive dehydroxylation reaction to enter the benzoyl-CoA degradation pathway (3), which is similar to the 4-hydroxybenzoate reaction (2, 5). Protocatechuate is one of the resorcylic acids (dihydroxybenzoic acids). Other compounds, including 2,3-dihydroxybenzoate (pyrocatechuate), 3,5-dihydroxybenzoate (α-resorcylate), 2,4-dihydroxybenzoate (β-resorcylate), 2,6-dihydroxybenzoate (γ-resorcylate), and gentisate (2,5-dihydroxybenzoate), are metabolized differently. 2,4-Dihydroxybenzoate and 2,6-dihydroxybenzoate are decarboxylated to resorcinol (14). 2,3-Dihydroxybenzoate is degraded via catechol by decarboxylation (23). 3,5-Dihydroxybenzoate is converted to hydroxyhydroquinone by oxidation and decarboxylation (7). 2,5-Dihydroxybenzoate is activated to its CoA ester and is reductively dehydroxylated to benzoyl-CoA (9).
Acknowledgments
This investigation was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG), by DFG-Graduiertenkolleg Biochemie der Enzyme, and by Fonds der Chemischen Industrie.
We thank Nasser Gad'on for invaluable help with growing the bacterium on a large scale and Wolfgang Haehnel, Freiburg, Germany, for performing MALDI-TOF mass spectrometry.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Anders, H. J., A. Kaetzke, P. Kampfer, W. Ludwig, and G. Fuchs. 1995. Taxonomic position of aromatic-degrading denitrifying Pseudomonas strains K 172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int. J. Syst. Bacteriol. 45327-333. [DOI] [PubMed] [Google Scholar]
- 2.Biegert, T., U. Altenschmidt, C. Eckerskorn, and G. Fuchs. 1993. Enzymes of anaerobic metabolism of phenolic compounds. 4-Hydroxybenzoate-CoA ligase from a denitrifying Pseudomonas species. Eur. J. Biochem. 213555-561. [DOI] [PubMed] [Google Scholar]
- 3.Bonting, C. F., and G. Fuchs. 1996. Anaerobic metabolism of 2-hydroxybenzoic acid (salicylic acid) by a denitrifying bacterium. Arch. Microbiol. 165402-408. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-252. [DOI] [PubMed] [Google Scholar]
- 5.Breese, K., and G. Fuchs. 1998. 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) from the denitrifying bacterium Thauera aromatica—prosthetic groups, electron donor and genes of a member of the molybdenum-flavin-iron-sulfur proteins. Eur. J. Biochem. 251916-923. [DOI] [PubMed] [Google Scholar]
- 6.Dagley, S., and M. D. Patel. 1957. Oxidation of p-cresol and related compounds by a Pseudomonas. J. Biochem. 66227-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallus, C., and B. Schink. 1998. Anaerobic degradation of α-resorcylate by Thauera aromatica strain AR-1 proceeds via oxidation and decarboxylation to hydroxyhydroquinone. Arch. Microbiol. 169333-338. [DOI] [PubMed] [Google Scholar]
- 8.Gorny, N., and B. Schink. 1994. Anaerobic degradation of catechol by Desulfobacterium sp. strain Cat2 proceeds via carboxylation to protocatechuate. Appl. Environ. Microbiol. 603396-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorny, N., and B. Schink. 1994. Hydroquinone degradation via reductive dehydroxylation of gentisyl-CoA by a strictly anaerobic fermenting bacterium. Arch. Microbiol. 16125-32. [Google Scholar]
- 10.Gorny, N., G. Wahl, B. Andreas, and B. Schink. 1992. A strictly anaerobic nitrate-reducing bacterium growing with resorcinol and other aromatic compounds. Arch. Microbiol. 15848-53. [DOI] [PubMed] [Google Scholar]
- 11.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50553-590. [DOI] [PubMed] [Google Scholar]
- 12.Healy, J. B. J., and L. Y. Young. 1978. Catechol and phenol degradation by a methanogenic population of bacteria. Appl. Environ. Microbiol. 35216-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heider, J., M. Boll, K. Breese, S. Breinig, C. Ebenau-Jehle, U. Feil, N. Gad'on, D. Laempe, B. Leuthner, M. E. Mohamed, S. Schneider, G. Burchhardt, and G. Fuchs. 1998. Differential induction of enzymes involved in anaerobic metabolism of aromatic compounds in the denitrifying bacterium Thauera aromatica. Arch. Microbiol. 170120-131. [DOI] [PubMed] [Google Scholar]
- 14.Kluge, C., A. Tschech, and G. Fuchs. 1990. Anaerobic metabolism of resorcyclic acids (m-dihydroxybenzoic acids) and resorcinol (1,3-benzenediol) in a fermenting and in a denitrifying bacterium. Arch. Microbiol. 15568-74. [Google Scholar]
- 15.Krieger, C. J., H. R. Beller, M. Reinhard, and A. M. Spormann. 1999. Initial reactions in anaerobic oxidation of m-xylene by the denitrifying bacterium Azoarcus sp. strain T. J. Bacteriol. 1816403-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lack, A., and G. Fuchs. 1992. Carboxylation of phenylphosphate by phenol carboxylase, an enzyme system of anaerobic phenol metabolism. J. Bacteriol. 1743629-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lack, A., and G. Fuchs. 1994. Evidence that phenol phosphorylation to phenylphosphate is the first step in anaerobic phenol metabolism in a denitrifying Pseudomonas sp. Arch. Microbiol. 161132-139. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 19.Laempe, D., M. Jahn, K. Breese, H. Schagger, and G. Fuchs. 2001. Anaerobic metabolism of 3-hydroxybenzoate by the denitrifying bacterium Thauera aromatica. J. Bacteriol. 183968-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller, J. A., A. S. Galushko, A. Kappler, and B. Schink. 2001. Initiation of anaerobic degradation of p-cresol by formation of 4-hydroxybenzylsuccinate in Desulfobacterium cetonicum. J. Bacteriol. 183752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray, M. G., and W. F. Thompson. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 84321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narmandakh, A., N. Gad'on, F. Drepper, B. Knapp, W. Haehnel, and G. Fuchs. 2006. Phosphorylation of phenol by phenylphosphate synthase: role of histidine phosphate in catalysis. J. Bacteriol. 1887815-7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostermann, A., C. Gallus, and B. Schink. 1997. Decarboxylation of 2,3-dihydroxybenzoate to catechol supports growth of fermenting bacteria. Curr. Microbiol. 35270-273. [Google Scholar]
- 24.Philipp, B., and B. Schink. 1998. Evidence of two oxidative reaction steps initiating anaerobic degradation of resorcinol (1,3-dihydroxybenzene) by the denitrifying bacterium Azoarcus anaerobius. J. Bacteriol. 1803644-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philipp, B., and B. Schink. 2000. Two distinct pathways for anaerobic degradation of aromatic compounds in the denitrifying bacterium Thauera aromatica strain AR-1. Arch. Microbiol. 17391-96. [DOI] [PubMed] [Google Scholar]
- 26.Philipp, B., D. Kemmler, J. Hellstern, N. Gorny, A. Caballero, and B. Schink. 2002. Anaerobic degradation of protocatechuate (3,4-dihydroxybenzoate) by Thauera aromatica strain AR-1. FEMS. Microbiol. Lett. 212139-143. [DOI] [PubMed] [Google Scholar]
- 27.Rabus, R., M. Kube, J. Heider, A. Beck, K. Heitmann, F. Widdel, and R. Reinhardt. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 18327-36. [DOI] [PubMed] [Google Scholar]
- 28.Rabus, R., and F. Widdel. 1995. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch. Microbiol. 16396-103. [DOI] [PubMed] [Google Scholar]
- 29.Rudolphi, A., A. Tschech, and G. Fuchs. 1991. Anaerobic degradation of cresols by denitrifying bacteria. Arch. Microbiol. 155238-248. [DOI] [PubMed] [Google Scholar]
- 30.Schmeling, S., A. Narmandakh, O. Schmitt, N. Gad'on, K. Schühle, and G. Fuchs. 2004. Phenylphosphate synthase: a new phosphotransferase catalyzing the first step in anaerobic phenol metabolism in Thauera aromatica. J. Bacteriol. 1868044-8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnell, S., B. Andreas, and B. Schink. 1991. Degradation of hydroxyhydroquinone by the strictly anaerobic fermenting bacterium Pelobacter massiliensis sp. nov. Arch. Microbiol. 55511-516. [Google Scholar]
- 32.Schühle, K., and G. Fuchs. 2004. Phenylphosphate carboxylase: a new C-C lyase involved in anaerobic phenol metabolism in Thauera aromatica. J. Bacteriol. 1864556-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schühle, K., J. Gescher, U. Feil, M. Paul, M. Jahn, H. Schägger, and G. Fuchs. 2003. Benzoate-coenzyme A ligase from Thauera aromatica: an enzyme acting in anaerobic and aerobic pathways. J. Bacteriol. 1854920-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, D. E., and P. A. Fisher. 1984. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J. Cell Biol. 9920-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webster, L. T., J. J. Mieyal, and U. A. Siddiqui. 1974. Benzoyl and hydroxybenzoyl esters of coenzyme A ultraviolet characterization and reaction mechanisms. J. Biol. Chem. 2492641-2645. [PubMed] [Google Scholar]
- 36.Zehr, B. D., T. J. Savin, and R. E. Hall. 1989. A one-step, low-background Coomassie staining procedure for polyacrylamide gels. Anal. Biochem. 182157-159. [DOI] [PubMed] [Google Scholar]