Abstract
Three clock proteins—KaiA, KaiB, and KaiC—have been identified as essential components of the circadian oscillator in cyanobacteria, and Kai-based chemical oscillation is thought to be the basic circadian timing mechanism in Synechococcus elongatus PCC 7942. Transcription and translation of kaiBC in cyanobacterial cells was quantitatively studied to elucidate how these processes are coupled to the chemical oscillator using a strain in which circadian oscillation is under the control of IPTG (isopropyl-β-d-thiogalactopyranoside). The kinetics of repression of kaiBC promoter triggered by IPTG allowed estimation of transient response at 10 h. This response time is suitable for cyanobacterial transcription and/or translation to match with the Kai-based oscillator. Interestingly, kaiBC promoter activity and KaiC phosphorylation showed robust circadian rhythms, whereas trc promoter-driven kaiBC mRNA levels and KaiC accumulation were almost arrhythmic. These results indicate that cyanobacterial circadian rhythms can be generated even if kaiBC expression is constitutive. Moreover, there was a positive correlation between activation of the kaiBC promoter and an increase in the KaiC phosphorylation ratio in three rhythmic conditions. Based on these observations, it is likely that the KaiC phosphorylation ratio is the main factor in the activation of kaiBC promoter. Finally, we quantitatively compared the threshold level of phosphorylated KaiC for the repression or derepression of kaiBC promoter and found that this parameter is an important factor in repressing the kaiBC promoter.
The circadian clock is an endogenous biological timing mechanism found in almost all organisms (18). Cyanobacteria are the simplest organisms known to exhibit circadian rhythms. KaiA, KaiB, and KaiC were identified as essential genetic components for circadian oscillation in the cyanobacterium Synechococcus elongatus PCC 7942 (6). Activities of the kaiBC promoter as assayed by a luciferase reporter showed robust circadian oscillation and were strongly repressed by overexpression of KaiC. This feedback regulation was first assumed to be the basic circadian timing mechanism of cyanobacteria, as has been proposed for many eukaryotic circadian oscillators (3).
KaiC interacts with KaiA and KaiB in a circadian fashion (8). KaiA enhances the autokinase activity of KaiC (7, 24) and/or inhibits its autophosphatase activity (26), whereas KaiB attenuates the activity of KaiA (9, 24, 26). In 2005, we obtained two important results that substantially altered the basic hypothesis for circadian rhythm generation. First, the circadian rhythm of KaiC phosphorylation persisted even in continuous darkness, under which conditions the transcription in Synechococcus was stopped completely (23). Second, we found that KaiC phosphorylation autonomously oscillates with a period of ∼24 h when the three recombinant Kai proteins are incubated in vitro in the presence of ATP (15). Since the period of this oscillation is refractory to changes in temperature and coincides with the in vivo circadian rhythm, Kai-based chemical oscillation is now thought to be the basic circadian timing mechanism in Synechococcus. However, in living cyanobacterial cells, even though transcription and translation of kai genes are not essential for the generation of the KaiC phosphorylation rhythm, feedback regulation of kaiBC transcription is thought to be crucial for maintaining robust circadian rhythms. In fact, under continuous light conditions (LL), KaiC exhibits circadian rhythms both in phosphorylation level and accumulation, and the kaiBC operon is transcribed in a robust circadian fashion that precedes the rhythm of KaiC accumulation by ∼6 h (5, 6, 7, 25). This type of time lag between mRNA and protein rhythms has also been observed for some negative regulators in eukaryotic clock systems and is thought to be important in causing feedback loops to oscillate (4). Alternatively, in cyanobacteria, the feedback regulation might be adjusted to facilitate coupling of the Kai-based oscillator with its output. Therefore, quantitative assessment of the kinetics of feedback regulation by KaiC is of interest to evaluate the intracellular dynamics of the cyanobacterial clock.
To date, the following examples of transcriptional regulation by KaiC have been demonstrated. (i) KaiC and KaiA proteins cooperatively regulate the kaiBC promoter (7). (ii) Overexpression of a mutant version of KaiC that cannot be phosphorylated transiently represses kaiBC promoter activity but fails to repress it over extended times (16). This suggests that both phosphorylation and accumulation of KaiC are involved in transcriptional feedback regulation. (iii) In contrast to eukaryotes, in which many clock gene products primarily regulate cis-acting elements of clock genes in a promoter-specific manner, KaiC coordinates genome-wide gene expression, as monitored by bioluminescence reporter (11). Moreover, the overexpression of KaiC strongly represses the rhythmic expression of both kaiBC and other genes (14). (iv) Recent studies suggest that temporal information from the Kai-based oscillator is transmitted by the SasA-RpaA two-component regulatory system and LabA to generate genome-wide circadian gene expression (20, 21).
These results have demonstrated qualitative aspects of gene regulation by KaiC, but little is known about quantitative aspects. We report here the basic kinetics of KaiC-mediated transcriptional regulation. To provide external control of kaiBC expression, we designed a strain in which endogenous kaiBC was inactivated, and these genes were exogenously expressed under the control of the Escherichia coli trc promoter. After the addition or removal of IPTG (isopropyl-β-d-thiogalactopyranoside), temporal profiles of kaiBC mRNA accumulation, KaiC protein accumulation and phosphorylation, and luxAB mRNA accumulation driven from the kaiBC promoter were quantitatively examined. The kinetics of the responses reveal the basic properties of cyanobacterial regulation of gene expression and indicate that both KaiC phosphorylation ratio and accumulation of phosphorylated KaiC regulate kaiBC promoter activity but in different fashions.
MATERIALS AND METHODS
Bacterial strains and culture.
S. elongatus PCC 7942 was used as the background strain for the reporter strains in the present study. NUC42 and NUC0203 (14) are wild-type and kaiBC deletion strains, respectively. Both strains carry the PkaiBC-luxAB reporter construct at neutral site I. A DNA fragment containing the kaiB and kaiC open reading frames was amplified by PCR using pCkaiABC as the template and was then introduced into the NcoI-BamHI site of pNS2KmTΔHincII-Ptrc to yield pNS2KmPtrc-kaiBC (see Fig. 1A). pNS2KmTΔHincII-Ptrc is a pNS2KmT (10)-based vector with a deleted HincII region and an insertion of a BglII fragment from pTrc99A at a BamHI site (Imai, unpublished). NUC0203 was transformed with pNS2KmPtrc-kaiBC to generate cells harboring an IPTG-inducible kaiBC cassette in neutral site II (NUC0220: PkaiBC-luxAB, ΔkaiBC, and Ptrc-kaiBC). Synechococcus cells were grown in modified BG-11 medium (1) under LL conditions (46 μmol m−2 s−1 from a white fluorescent lamp) at 30°C.
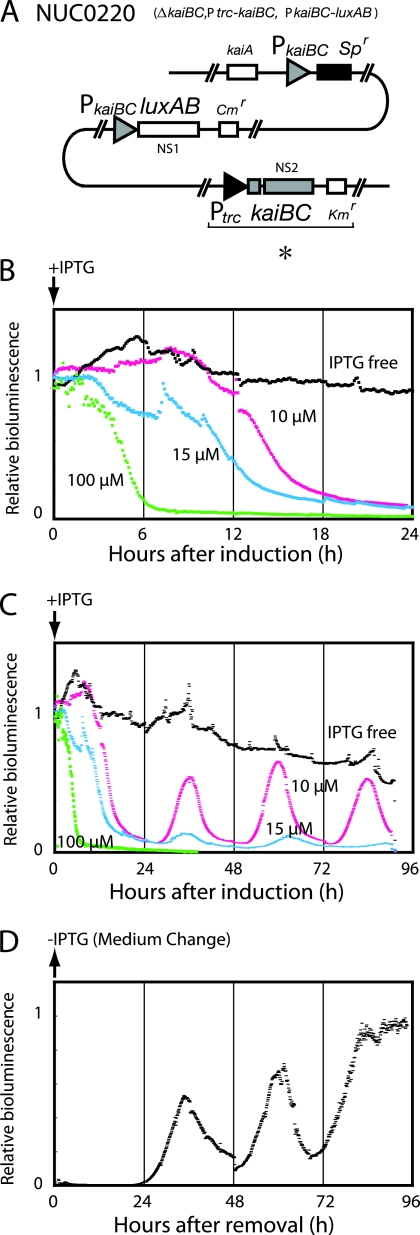
FIG. 1.
Transcriptional regulation by KaiC monitored by bioluminescence. (A) Genotype of the NUC0220 strain. In this strain, kaiBC is driven from the E. coli trc promoter (*) and can be induced by IPTG in a dose-dependent manner. (B and C) Initial responses (B) and subsequent profiles (C) of bioluminescence in NUC0220 after kaiBC induction at various concentrations of IPTG. Bioluminescence was analyzed in liquid culture under continuous light (LL) in the absence or presence of IPTG, administered at hour 0 (indicated by arrow) at the final concentrations as shown. The bioluminescence was plotted against the time after induction. The bioluminescence intensity before induction was normalized to 1. (D) Response of bioluminescence to the removal of IPTG in NUC0220. Cells were cultured in the presence of 100 μM IPTG for at least 36 h, washed, suspended in fresh medium lacking IPTG, and cultured under LL at 30°C. Bioluminescence was plotted against time after the removal of IPTG (indicated by the arrow). The maximum bioluminescence intensity was normalized to 1. All traces in this figure are representative of at least three replicates.
Bioluminescence assay.
Cells were cultured in a continuous culturing system to maintain an optical density (at 730 nm) of 0.25. The cells were exposed to two cycles of 12-h light and 12-h dark to synchronize their clocks and were then released into LL conditions. For kaiBC induction, IPTG was administered to the continuous culture at hour 12 in LL at final concentrations of 10, 15, or 100 μM. To remove IPTG from the culture, the cells were collected by centrifugation and washed twice with fresh BG-11. Cells were resuspended in fresh BG-11 medium to continue the culture. To monitor bioluminescence from the liquid culture, cells were introduced into silicon tubing exposed to 10% decanal. Bioluminescence was monitored in a flow cell using a photomultiplier (H7360, Hamamatsu) tube-based bioluminescence monitoring system.
Northern and Western blotting analyses.
Cells were harvested at various time points after exposure to LL, immediately frozen, and stored at −80°C. RNA was extracted from each sample as described previously (6), samples were subjected to electrophoresis on 1.0% agarose gels containing ethidium bromide (0.1 μg/ml, final concentration), blotted onto positively charged nylon membranes (Amersham), and hybridized with digoxigenin (DIG)-labeled kaiBC and luxAB probes in DIG-Easy-Hyb (Roche) (21). Transcripts were detected by enzyme-linked immunoassay using an anti-DIG antibody-alkaline phosphatase conjugate and the chemiluminescent substrate CSPD (Roche). Northern signal of kaiBC mRNA appeared as double band (see Fig. 2, 3, and 4). We assumed the double bands is caused by excess amount rRNA overlapping at the kaiBC mRNA position. We estimated kaiBC mRNA amount as the sum of both bands.
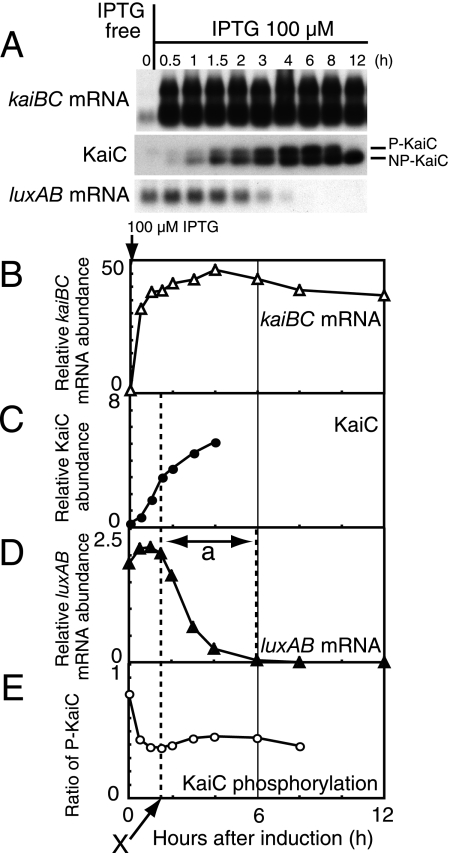
FIG. 2.
Transcriptional repression of the kaiBC promoter activity by kaiBC overexpression. (A) Temporal profiles of kaiBC mRNA accumulation (Northern blot analysis; upper panel), KaiC protein accumulation (Western blot analysis; middle panel), and luxAB mRNA accumulation (Northern blot analysis; lower panel) in NUC0220 after the addition of 100 μM IPTG. Cells for analysis were collected before induction (hour 0) and 0.5, 1, 1.5, 2, 3, 4, 6, 8, and 12 h after induction. Total protein (0.5 μg) and total RNA (1 μg) were used for the analyses. In the middle panel, the upper band represents the phosphorylated forms of KaiC (P-KaiC), and the lower band corresponds to the unphosphorylated form (NP-KaiC). The data shown are representative of at least five independent experiments. (B to E) Quantitative estimation of kaiBC mRNA (B), KaiC protein (C), luxAB mRNA (D), and the phosphorylation ratio of KaiC (E). The phosphorylation ratio of KaiC was calculated as the ratio of phosphorylated KaiC to total KaiC (from panel A). The signals from Western and Northern blot analyses were measured by densitometry. Relative accumulation levels were calculated (see Materials and Methods) and plotted against time after the addition of 100 μM IPTG. Due to the smearing of NP-KaiC signal to the P-KaiC position, data for hour 12 are not shown in Fig. 2E. Dashed lines and the arrow (“X”) indicate the time point at which luxAB mRNA levels (which reflected kaiBC promoter activity) began to decrease. “a” indicates the transition time for repression of PkaiBC to baseline level.
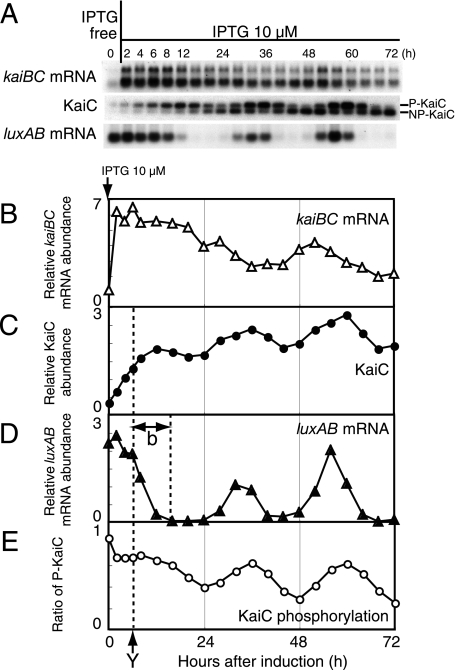
FIG. 3.
Circadian rhythm generation by moderate kaiBC induction. (A) Temporal profiles of kaiBC mRNA accumulation (upper panel), KaiC protein accumulation (middle panel), and luxAB mRNA accumulation (lower panel) in NUC0220 after the addition of 10 μM IPTG. Protein and mRNA levels were examined by Western and Northern blot analyses with cells collected at the indicated time points (every 2 h for the first 8 h and then every 4 h until hour 72) after induction. Total protein (1 μg) and total RNA (1 μg) were used for the analyses. The data shown are representative of at least four independent experiments. (B to E) Quantitative estimation of kaiBC mRNA (B), KaiC protein (C), luxAB mRNA (D), and the phosphorylation ratio of KaiC (E). The experimental procedures and the presentation of the data are the same as for Fig. 2. Dashed lines and the arrow (“Y”) indicate the time point at which luxAB mRNA levels began to decrease. “b” indicates the transition time for the repression of PkaiBC to the baseline.
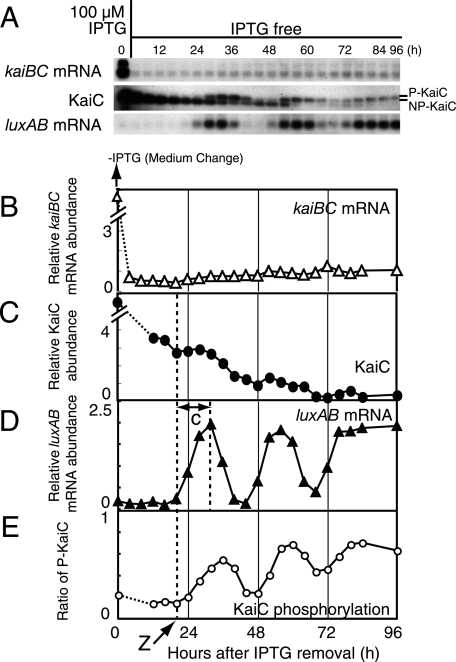
FIG. 4.
Circadian rhythm generation after recovery from strong repression of PkaiBC. (A) Temporal profiles of kaiBC mRNA accumulation (upper panel), KaiC protein accumulation (middle panel), and luxAB mRNA accumulation (lower panel) after the removal of 100 μM IPTG. NUC0220 cells were grown in the presence of 100 μM IPTG for 36 h and then transferred to fresh medium lacking IPTG. Removal of the IPTG was performed as in Fig. 1D. After the removal of IPTG, the cells were collected every 4 h for 84 h and at hour 96. Protein and mRNA levels were examined by Western and Northern blot analyses. Total protein (1 μg) and total RNA (1 μg) were used for the analyses. The data shown are representative of at least three independent experiments. (B to E) Quantitative analyses of kaiBC mRNA (B), KaiC protein (C), luxAB mRNA (D), and phosphorylation ratio of KaiC (E). The experimental procedures and the presentation of the data are the same as for Fig. 2. Dashed lines and arrow (“Z”) indicate the time point at which the initial increase in luxAB mRNA began. “c” indicates the transition time for the derepression of PkaiBC to the maximum level. The signals of mRNA (B) and KaiC (C) at time zero were saturated because of the presence of 100 μM IPTG. The signals at this time points could be estimated as 10, at least.
For protein assays, cells were disrupted by using the Multi-bead-shocker (Yasui Kikai) with zirconium beads (diameter, 0.1 mm; Yasui Kikai) for 10 cycles of 30 s of agitation and 30 s of rest at 4°C. After centrifugation, the supernatants were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and immunoblotted with anti-KaiC antibodies as described previously (16). KaiC protein was detected with an enhanced chemiluminescence detector (Amersham). The protein concentration was determined with the bicinchoninic acid method with bovine serum albumin as a standard.
Normalization of blots.
Densitometric analysis of blots was performed with a GS-800 calibrated densitometer and QuantityOne software (Bio-Rad). In the present study, estimations of accumulation in NUC0220 were always accompanied by wild-type samples loaded on the same gel. The levels of target (kaiBC mRNA, KaiC protein, and luxAB mRNA) in NUC0220 were normalized with the respective signals of wild type set to 1. The wild-type signal was obtained as an average of signals at six time points in one circadian cycle. KaiC phosphorylation ratio was calculated as the ratio of phosphorylated KaiC to total KaiC.
RESULTS
Transcriptional feedback regulation by KaiC monitored by bioluminescence.
The endogenous kaiBC in a PkaiBC-luxAB reporter strain (NUC42) was inactivated, and kaiBC was introduced into the genome under the control of the E. coli trc promoter (NUC0220 ΔkaiBC, Ptrc-kaiBC, PkaiBC-luxAB) (14) (Fig. 1A). In this strain, expression of kaiBC can be induced by the administration of IPTG in a dose-dependent manner, and the activity of the kaiBC promoter (PkaiBC) can be monitored by bioluminescence. To examine the initial response to IPTG induction, Synechococcus cells were grown in liquid medium to an optical density at 730 nm of 0.25 for the delivery of IPTG. In the absence of IPTG, bioluminescence driven from PkaiBC was higher due to low KaiC accumulation in the cell (Fig. 1B and C). The onset of bioluminescence decrease occurred at 4, 8, and 10 h after the addition of 100, 15, and 10 μM IPTG, respectively (Fig. 1B). This dose-dependent delay of bioluminescence decrease is likely due to a lag time for accumulation of KaiC to a level at which PkaiBC is repressed. After this initial response, bioluminescence was maintained at a low background level in the presence of 100 μM IPTG (Fig. 1C). In contrast, Ptrc-driven expression of kaiC generated rhythms of ca. 24 h, as reported previously (ΔkaiBC+PtrckaiCB strain [14], ΔkaiC+PtrckaiC [26]).
We also examined the derepression of KaiC by removal of 100 μM IPTG from the culture. In this case, bioluminescence remained at a baseline for a considerable period and then began to increase at 24 h after the removal of IPTG (Fig. 1D). From hour 24 to hour 84, the bioluminescence oscillated for two and a half cycles and was maintained thereafter at a higher level. In a separate experiment, we confirmed that the bioluminescence level stayed constant up to 120 h. Interestingly, the period of this fluctuation was again approximately 24 h.
Regulation of PkaiBC by KaiC monitored by luxAB mRNA accumulation.
Although monitoring gene expression using bioluminescence reporters is precise and sensitive, the time lag due to transcription, translation, and posttranslation processes may interfere with the accurate temporal estimation of the induction or repression of target genes. Thus, we examined the accumulation of luxAB mRNA. Since the half-life of luxAB mRNA is ∼5 min (see Fig. S1 in the supplemental material), luxAB mRNA accumulation should more closely reflect PkaiBC activity.
To understand the processes between kaiBC induction and transcriptional repression of PkaiBC by KaiC protein, we quantitatively analyzed the temporal profiles of Ptrc-driven kaiBC mRNA, KaiC accumulation, KaiC phosphorylation, and PkaiBC-driven luxAB mRNA after strong induction of kaiBC by 100 μM IPTG (Fig. 2A). Under these conditions, the level of Ptrc-driven kaiBC mRNA promptly increased by at least 30-fold within 0.5 h of induction (Fig. 2B). We confirmed that kaiBC mRNA levels were elevated considerably within 5 min after IPTG administration (see Fig. S1 in the supplemental material). The levels of KaiC protein elevated gradually to fivefold the level in wild-type cells by 4 h after induction (Fig. 2C).
luxAB mRNA driven by the PkaiBC began to drop at 1.5 h after induction (“X”), when KaiC levels were elevated to two- to threefold the level in the wild type. We assume that this KaiC level is necessary for the effective repression of PkaiBC activity. It took 4 to 5 h for luxAB mRNA to drop at the zero level (“a” in Fig. 2D). A transient increase of KaiC in the cell would also cause a quick reduction of KaiC phosphorylation ratio just after IPTG treatment because newly synthesized KaiC should be unphosphorylated (Fig. 2E). Note also that the decrease in bioluminescence began at 4 h after addition of IPTG (Fig. 1B), that is, the accumulation of luxAB mRNA preceded bioluminescence output by 2 to 4 h (12).
Kinetics of circadian rhythm generation by moderate kaiBC induction.
Next, we analyzed the kinetics of rhythm generation with 10 μM IPTG induction in NUC0220 cells. As in the case of addition of 100 μM IPTG, we analyzed the temporal profiles of kaiBC mRNA, KaiC, and luxAB mRNA (Fig. 3A). Figure 3B to E depict time courses of these parameters. The amount of kaiBC mRNA driven from the trc promoter increased within 2 h of addition of 10 μM IPTG, but the level (sixfold) was lower than that induced by 100 μM IPTG. KaiC accumulated from hour 0 to hour 12 to ∼2-fold the wild-type level. luxAB mRNA levels increased slightly at hour 2 (the second point of Fig. 3D) and stayed at a higher level for 4 h (hours 4 and 6 in Fig. 3D). The levels then abruptly decreased after hour 6 (“Y”), when the amount of KaiC was 1.3-fold the wild-type level. Finally, the luxAB mRNA level dropped to baseline by hour 16 of induction. Thus, it took 10 h (“b”) for luxAB mRNA levels to reach a minimum.
As observed with bioluminescence, this strain exhibited robust circadian oscillation of luxAB mRNA after hour 24 of induction. Although both kaiBC mRNA and KaiC accumulation showed circadian rhythms, the amplitudes of both rhythms were weaker than those of the wild type. Note that the periods of the rhythms in this strain were similar to those of the wild type, whereas these factors accumulated to two- to threefold more than those in the wild type. On the other hand, luxAB mRNA accumulation and phosphorylation of KaiC showed rhythms as robust as those in the wild type (Fig. 3E). The phase relationships of these rhythms were consistent with those in the wild type.
Kinetics of circadian rhythm recovery from strong repression of PkaiBC.
As shown in Fig. 1D, we found that a circadian rhythm was generated when strong induction of kaiBC was ended by the removal of IPTG. We examined the time course of kaiBC mRNA, KaiC, and luxAB mRNA after the removal of IPTG (Fig. 4A), and quantitative time profiles are shown in Fig. 4B to E. Upon the removal of IPTG, the amount of kaiBC mRNA expressed from the trc promoter decreased immediately to less than the wild-type levels and stayed at a low level without detectable fluctuation. During incubation with 100 μM IPTG, KaiC had accumulated in the cell to at least 10-fold more than the wild type. After removal of the IPTG, the level gradually decreased to two- to threefold the wild-type levels by 24 h. The KaiC levels then continued to slowly decrease with a weak circadian oscillation.
As expected, the decrease in KaiC levels released PkaiBC repression. The amount of luxAB mRNA began to increase at hour 20 (“Z”) when the amount of KaiC was approximately two- to threefold that of the wild type (Fig. 4D). The luxAB mRNA level increased from 20 and 32 h after induction. The transition to maximum levels (“c”) took ∼12 h. From 20 to 76 h, luxAB mRNA levels showed a circadian rhythm as robust as those of the wild type. The phosphorylation ratio of KaiC also oscillated with the luxAB mRNA accumulation rhythm with an ∼4-h delay. Note that both profiles were rhythmic (Fig. 4D and E), whereas the amount of kaiBC mRNA constitutively stayed at low levels (Fig. 4B). These results are compatible with the hypothesis that the feedback regulation of clock gene expression is not essential for circadian rhythm generation in LL conditions.
DISCUSSION
In eukaryotes, a transcription/translation feedback regulation of clock genes is thought to underlie the circadian oscillator, and the time required for steps in the feedback loop is thought to be the basis of the circadian period. Quantitative assessment of individual steps in the feedback loop in Neurospora (13) have shown that times in several steps of the feedback loop are consistent with the 24-h periodicity. However, in cyanobacteria, we reconstituted an in vitro oscillation by mixing three Kai proteins and ATP in a test tube (15). Our recent results in vitro have determined that the biochemical activities of KaiC, including kinase/phosphatase and ATPase activities, are basic timekeepers of circadian period length in cyanobacteria (17, 19, 22). Therefore, the transcription and translation processes in cyanobacterial cells need to resonate with the Kai-based oscillator for efficient temporal information transfer.
Using an inducible system that allows starting and stopping of circadian oscillation, we have quantitatively analyzed the accumulation and phosphorylation of KaiC, as well as PkaiBC activity (as luxAB mRNA level), to understand the kinetic properties of PkaiBC regulation in cyanobacteria. Repression of PkaiBC by KaiC showed dose-dependent kinetics. After stronger induction (100 μM IPTG), it took 4 to 5 h for the transition of luxAB mRNA levels from derepressed (“X”) to baseline levels (“a” in Fig. 2), whereas it took about 10 h after moderate (10 μM IPTG) induction (“b” in Fig. 3). Since the transition time is close to half of the circadian period, our estimation time for repression (“b”) is apparently compatible with PkaiBC to cycle in the circadian period. We also found that the time for the accumulation of luxAB mRNA from baseline (“Z”) to the first peak was also 12 h (“c” in Fig. 4). Although the transition of derepression process of PkaiBC was accompanied by elevation of the KaiC phosphorylation ratio (Fig. 4E), the response of kaiBC expression potentially took a similar range of time. Thus, temporal coincidence between the sums of transition times of PkaiBC for repression and derepression (“b” + “c”) and the circadian period supports precise matching between the pacemaker of the cyanobacterial clock and its output. Namely, feedback of kaiBC expression has been adjusted to cycle with a period of ca. 24 h to match the Kai-based chemical oscillator.
Our quantitative analyses with NUC0220 permitted the study of the functional relationship between parameters in the rhythmic transcription by PkaiBC, observed by inducing KaiC production by 10 μM IPTG (Fig. 3) or aborting it by removing 100 μM IPTG (Fig. 4). In both cases, PkaiBC activity (luxAB mRNA) and KaiC phosphorylation showed circadian rhythms as robust as those in wild-type cells (see Fig. S2 in the supplemental material). In contrast, Ptrc-driven kaiBC mRNA and the accumulation of KaiC displayed only weak rhythmicity (Fig. 3) or were almost arrhythmic (Fig. 4), whereas these parameters showed robust rhythms in wild-type cells. These observations could be ascribed to an open-loop regulatory structure in NUC0220 (Fig. 6). In this strain, Ptrc-driven kaiBC mRNA expression and accumulation of KaiC should be constitutive, as observed. The robust rhythms in PkaiBC activity and KaiC phosphorylation could be ascribed to regulation by autonomous KaiC phosphorylation oscillation as observed in vitro and in cells in darkness. Weak rhythms observed in KaiC levels could be ascribed to minor and indirect effects of global regulation of gene expression in cyanobacteria by KaiC (14).
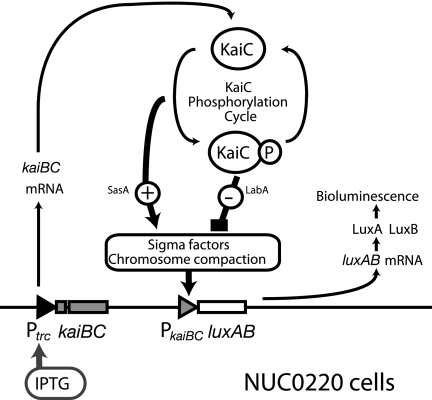
FIG. 6.
Regulatory structure of PkaiBC transcription in NUC0220 cells. Regulation of gene expression in NUC0220 cells is illustrated. PkaiBC was regulated by KaiC phosphorylation cycle through positive and negative pathways. Possible mediators from KaiC to PkaiBC were illustrated. Note that the activity of the trc promoter that regulates kaiBC transcription is primarily controlled by IPTG concentration in a dose-dependent manner and is only slightly affected by KaiC. Thus, the regulatory structure kaiBC expression in NUC0220 can be assumed to be an open loop. See the text for further explanation.
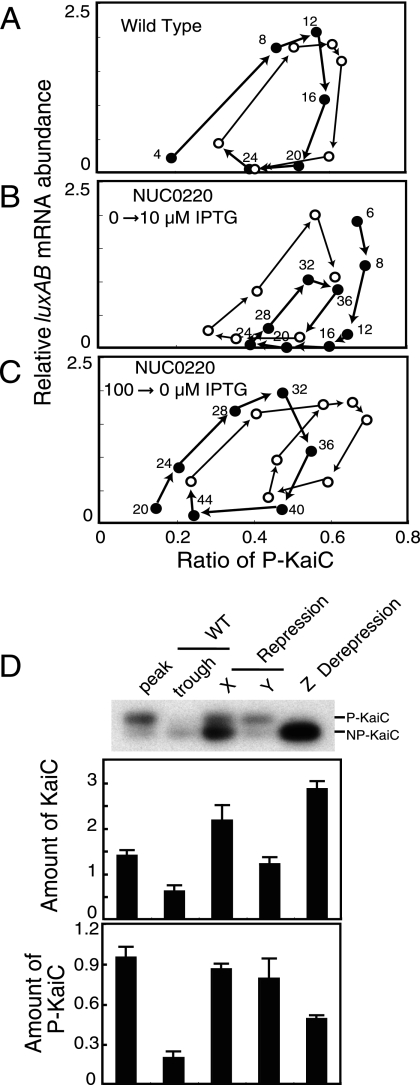
To analyze the temporal relationship between two rhythmic parameters, luxAB mRNA levels and the KaiC phosphorylation ratio (Fig. 3 and 4), we plotted the former parameter against the latter. At first, as a reference, we plotted the relationship for wild-type cells (see Fig. S2 in the supplemental material). As shown in Fig. 5A, the abundance of luxAB mRNA increased proportionally with the KaiC phosphorylation ratio. Apparently, increasing PkaiBC activity was regulated by this ratio. On the other hand, decreasing promoter activity did not directly correlate with the ratio. We then plotted time profiles of the kaiBC induction (Fig. 3) and its elimination experiments (Fig. 4). As shown in Fig. 5B and C, correlation between PkaiBC activation and the phosphorylation ratio was also observed in experiments with NUC0220. We plotted other parameters, such as luxAB mRNA against the KaiC levels, phosphorylated-KaiC (P-KaiC) levels, and unphosphorylated-KaiC levels to try to find consistent relationships with PkaiBC activation. However, no parameters other than the KaiC phosphorylation ratio showed a positive correlation (data not shown). Therefore, the KaiC phosphorylation ratio is likely a major determinant in the activation of PkaiBC.
FIG. 5.
Correlation of kaiBC promoter activity with the KaiC phosphorylation ratio (A to C) and the amount of KaiC at repression or derepression of PkaiBC (D). (A) Positive correlation between activation of the kaiBC promoter and the KaiC phosphorylation ratio. The relationships between luxAB mRNA accumulation and KaiC phosphorylation in wild type (A; see Fig. S2 in the supplemental material), NUC0220 after the addition of 10 μM IPTG (B; see Fig. 3), and NUC0220 after the removal of IPTG (C; see Fig. 4) were examined. The data shown in Fig. S2 in the supplemental material and Fig. 3 and 4 were replotted on a graph with the KaiC phosphorylation ratio on the x axis and the luxAB mRNA amount on the y axis. The first half of the trace is plotted with closed circles, and the second half is plotted by open circles. The number in the figure indicates a time point in each experiment. (D) Quantitative estimation of KaiC required for the repression and derepression of kaiBC promoter activity. KaiC accumulation at the peak (CT16) and trough (CT4) in the wild type and at three time points—“X,” “Y” (Fig. 2 and 3; for repression), and “Z” (Fig. 4; for derepression)—in NUC0220 were quantitatively reexamined by Western blot analysis. The upper panel shows the total amount of KaiC, and the lower panel shows the amount of P-KaiC. The average level of KaiC in the wild type was standardized to 1. The amount of P-KaiC was calculated from the relative KaiC amount and the phosphorylation ratio. Bars represent the means ± the SD from three independent experiments.
Monitoring derepression of PkaiBC after the removal of 100 μM IPTG (Fig. 4C) suggested intracellular KaiC levels that permit robust circadian rhythmicity. By ignoring weak rhythmicity, the intracellular KaiC level was monotonically decreased after removal. Concurrent monitoring during this process (Fig. 4D and E) observed robust rhythms of luxAB mRNA levels and KaiC phosphorylation in only a limited range of KaiC levels (0.5- to 3-fold the average KaiC level in the wild type). In fact, the dynamic range of KaiC concentration that allows in vitro KaiC phosphorylation rhythms is also 0.3 to 3 (M. Nakajima et al., unpublished data). KaiC homeostasis in Synechococcus cells is apparently adjusted to be in this range to permit the KaiC phosphorylation rhythms. Restoration of circadian rhythms by Ptrc-kaiBC (14, 26) or PpurF-kaiBC (2) would also be attained by induction of KaiC in a similar range.
What parameter contributes to the repression of PkaiBC activity? We first assumed that the repression of PkaiBC activity is mainly controlled by the total amount of KaiC. In NUC0220 cells, KaiC thresholds for the repression of PkaiBC could be estimated at times “X” (in Fig. 2) and “Y” (in Fig. 3), whereas the KaiC level for derepression could be estimated at time “Z” (in Fig. 4). We then quantitatively reexamined these levels of KaiC (Fig. 5D) and compared them to the highest and lowest KaiC levels in wild-type cells. The amounts of KaiC for repression at times “X” and “Y” were 2.2- and 1.2-fold the average KaiC accumulation in wild-type cells, respectively. However, the amount of KaiC for derepression was ∼3-fold that in wild-type cells, which is much higher than the range of KaiC fluctuation in rhythmic cells. Therefore, the amount of KaiC is unlikely to be a major factor in repression of PkaiBC under rhythmic conditions.
Alternatively, we hypothesized that the levels of phosphorylated KaC might be the primary parameter for PkaiBC repression because overexpression of a mutant version of KaiC that cannot be phosphorylated did not show acute repression of PkaiBC activity (16). P-KaiC amounts can be calculated from the relative KaiC amount and phosphorylation ratio (Fig. 5D). Interestingly, the amount of P-KaiC required for repression was ca. 0.8 to 1.0 under two IPTG concentrations. Moreover, the P-KaiC level for derepression (ca. 0.5) was lower than that for repression. These results are compatible with the model that repression of PkaiBC activity is mainly regulated by the amount of P-KaiC, because the lower threshold for derepression allows cyclic regulation of PkaiBC by this parameter. In addition, note that regulation by P-KaiC would be more robust than that by KaiC accumulation because the two parameters change in the same phase in wild-type cells (Fig. 5D, WT).
What molecular machinery can mediate transfer of information regarding KaiC states to PkaiBC activity? The SasA-RpaA two-component system (20) is one candidate. It has been reported that phosphotransfer from the histidine kinase (SasA) to the cognate receiver (RpaA) changed as the circadian state of KaiC phosphorylation in vitro. As shown in Fig. 5A to C, the phase of PkaiBC activation is accompanied by an elevation of the KaiC phosphorylation ratio, and phosphotransfer activity in vitro was also coupled to elevation of the KaiC phosphorylation rhythm. This correlation strongly supports the model that an elevation in KaiC phosphorylation state activates PkaiBC through the SasA-RpaA pathway. Based on analyses as described above, we illustrated the regulation of PkaiBC by KaiC in NUC0220 cells (Fig. 6). During the phosphorylation phase, the SasA-RpaA pathway was driven by the KaiC phosphorylation reaction to activate PkaiBC. For this pathway to function with the kinetics we observed here, an integrative process of the activation signal (such as accumulation of sigma factors or changes in chromosome compaction) would be included in the regulatory process. The activation of PkaiBC would then be lost at the peak of KaiC phosphorylation because phosphotransfer to SasA-RpaA pathway would be terminated by the shift of the phosphorylation reaction to dephosphorylation.
We recently identified a novel gene, labA, that encodes a component necessary for KaiC-dependent repression of gene expression in cyanobacteria (21). LabA is therefore a candidate mediator of P-KaiC repression of PkaiBC. When the maximum phosphorylation of KaiC was attained, active KaiC-KaiA association was switched to stable KaiC-KaiB association (8) and, simultaneously, the autokinase activity of KaiC was switched to autophosphatase activity (17). This change in KaiC state would facilitate the repression of PkaiBC by P-KaiC.
Supplementary Material
Acknowledgments
We thank Keiko Imai for providing plasmid pNS2KmTΔHincII-Ptrc; Yoichi Nakahira and Hideo Iwasaki for helpful advice in constructing recombinant DNA; and Yohko Kitayama, Yasuhito Taniguchi, and Michinori Mutsuda for helpful comments on this work.
This research was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (15GS0308 to T.K. and T.O.) and the Japan Society for the Promotion of Science (17370088 to T.O.). Y.M. is supported by a Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science (no. 18006440).
Footnotes
Published ahead of print on 28 December 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bustos, S. A., and S. S. Golden. 1991. Expression of the psbDII gene in Synechococcus sp. strain PCC 7942 requires sequences downstream of the transcription start site. J. Bacteriol. 1737525-7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ditty, J. L., S. R. Canales, B. E. Anderson, S. B. Williams, and S. S. Golden. 2005. Stability of the Synechococcus elongatus PCC 7942 circadian clock under directed anti-phase expression of the kai genes. Microbiology 1512605-2613. [DOI] [PubMed] [Google Scholar]
- 3.Dunlap, J. C., J. J. Loros, and P. J. DeCoursey (ed.). 2004. Chronobiology: biological timekeeping. Sinauer, Sunderland, MA.
- 4.Hardin, P. E., J. C. Hall, and M. Rosbash. 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343536-540. [DOI] [PubMed] [Google Scholar]
- 5.Imai, K., T. Nishiwaki, T. Kondo, and H. Iwasaki. 2004. Circadian rhythms in the synthesis and degradation of a master clock protein KaiC in cyanobacteria. J. Biol. Chem. 27936534-36539. [DOI] [PubMed] [Google Scholar]
- 6.Ishiura, M., S. Kutsuna, S. Aoki, H. Iwasaki, C. R. Andersson, A. Tanabe, S. S. Golden, C. H. Johnson, and T. Kondo. 1998. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 2811519-1523. [DOI] [PubMed] [Google Scholar]
- 7.Iwasaki, H., T. Nishiwaki, Y. Kitayama, M. Nakajima, and T. Kondo. 2002. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. USA 9915788-15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kageyama, H., T. Nishiwaki, M. Nakajima, H. Iwasaki, T. Oyama, and T. Kondo. 2006. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol. Cell 23161-171. [DOI] [PubMed] [Google Scholar]
- 9.Kitayama, Y., H. Iwasaki, T. Nishiwaki, and T. Kondo. 2003. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 222127-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitayama, Y., T. Kondo, Y. Nakahira, H. Nishimura, Y. Ohmiya, and T. Oyama. 2004. An in vivo dual-reporter system of cyanobacteria using two railroad-worm luciferases with different color emissions. Plant Cell Physiol. 45109-113. [DOI] [PubMed] [Google Scholar]
- 11.Liu, Y., N. F. Tsinoremas, C. H. Johnson, N. V. Lebedeva, S. S. Golden, M. Ishiura, and T. Kondo. 1995. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 91469-1478. [DOI] [PubMed] [Google Scholar]
- 12.Liu, Y., S. S. Golden, T. Kondo, M. Ishiura, and C. H. Johnson. 1995. Bacterial luciferase as a reporter of circadian gene expression in cyanobacteria. J. Bacteriol. 1772080-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merrow, M. W., N. Y. Garceau, and J. C. Dunlap. 1997. Dissection of a circadian oscillation into discrete domains. Proc. Natl. Acad. Sci. USA 943877-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakahira, Y., M. Katayama, H. Miyashita, S. Kutsuna, H. Iwasaki, T. Oyama, and T. Kondo. 2004. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proc. Natl. Acad. Sci. USA 101881-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakajima, M., K. Imai, H. Ito, T. Nishiwaki, Y. Murayama, H. Iwasaki, T. Oyama, and T. Kondo. 2005. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308414-415. [DOI] [PubMed] [Google Scholar]
- 16.Nishiwaki, T., Y. Satomi, M. Nakajima, C. Lee, R. Kiyohara, H. Kageyama, Y. Kitayama, M. Temamoto, A. Yamaguchi, A. Hijikata, M. Go, H. Iwasaki, T. Takao, and T. Kondo. 2004. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc. Natl. Acad. Sci. USA 10113927-13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishiwaki, T., Y. Satomi, Y. Kitayama, K. Terauchi, R. Kiyohara, T. Takao, and T. Kondo. 2007. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 264029-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pittendrigh, C. S. 1993. Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 5516-54. [DOI] [PubMed] [Google Scholar]
- 19.Rust, M. J., J. S. Markson, W. S. Lane, D. S. Fisher, and E. K. O'Shea. 2007. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science 318809-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takai, N., M. Nakajima, T. Oyama, R. Kito, C. Sugita, M. Sugita, T. Kondo, and H. Iwasaki. 2006. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. USA 10312109-12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniguchi, Y., M. Katayama, R. Ito, N. Takai, T. Kondo, and T. Oyama. 2007. labA: a novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes Dev. 2160-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terauchi, K., Y. Kitayama, T. Nishiwaki, K. Miwa, Y. Murayama, T. Oyama, and T. Kondo. 2007. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. USA 10416377-16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomita, J., M. Nakajima, T. Kondo, and H. Iwasaki. 2005. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307251-254. [DOI] [PubMed] [Google Scholar]
- 24.Williams, S. B., I. Vakonakis, S. S. Golden, and A. C. LiWang. 2002. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc. Natl. Acad. Sci. USA 9915357-15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu, Y., T. Mori, and C. H. Johnson. 2000. Circadian clock-protein expression in cyanobacteria: rhythms and phase setting. EMBO J. 193349-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu, Y., T. Mori, and C. H. Johnson. 2003. Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 222117-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.