Abstract
A series of Rhodobacter capsulatus AmtB variants were created and assessed for effects on ammonia transport, formation of AmtB-GlnK complexes, and regulation of nitrogenase activity and NifH ADP-ribosylation. Confirming previous reports, H193 and H342 were essential for ammonia transport and the replacement of aspartate 185 with glutamate reduced ammonia transport. Several amino acid residues, F131, D334, and D335, predicted to be critical for AmtB activity, are shown here for the first time by mutational analysis to be essential for transport. Alterations of the C-terminal tail reduced methylamine transport, prevented AmtB-GlnK complex formation, and abolished nitrogenase switch-off and NifH ADP-ribosylation. On the other hand, D185E, with a reduced level of transport, was capable of forming an ammonium-induced complex with GlnK and regulating nitrogenase. This reinforces the notions that ammonia transport is not sufficient for nitrogenase regulation and that formation of an AmtB-GlnK complex is necessary for these processes. However, some transport-incompetent AmtB variants, i.e., F131A, H193A, and H342A, form ammonium-induced complexes with GlnK but fail to properly regulate nitrogenase. These results show that formation of an AmtB-GlnK complex is insufficient in itself for nitrogenase regulation and suggest that partial ammonia transport or occupation of the pore by ammonia is essential for this function.
AmtB of the purple nonsulfur bacterium Rhodobacter capsulatus is a member of the ammonium transporter (Amt)/methylamine permease (Mep) family. Present in bacteria, archaea, and eukaryotes (29), proteins of this family are homotrimers that transport ammonium through the cytoplasmic membrane. The crystal structures of AmtB of Escherichia coli and of Amt-1 of Archaeoglobus fulgidus reveal a cluster of 11 transmembrane helices organized around a hydrophobic pore (1, 18, 42) which has been proposed to act as a channel that allows transport of NH3 (18, 21, 36, 41, 42). In this model, NH4+ binds to residues located inside a periplasmic vestibule, is deprotonated, and is subsequently reprotonated on the cytoplasmic side. However, details of this transport process, including the nature of the transported species (NH4+ or NH3), are still subject to debate (4, 5, 12, 20, 22). Our previous study, showing that nitrogen-fixing cultures of an R. capsulatus amtB mutant strain accumulated extracellular ammonium whereas wild-type cultures did not, supports a role for AmtB in active ammonium uptake (38), and likewise, many studies have shown that a proton gradient seems essential, implying some type of active transport process.
In some organisms, Amt proteins have also been implicated in the posttranslational regulation of nitrogenase (15, 34, 38). Inactivation of amtB of R. capsulatus results in the abolition of NifH ADP-ribosylation-dependent and -independent nitrogenase switch-off (38). A similar effect is obtained when either of the PII proteins, GlnB or GlnK, two soluble trimeric proteins playing a central role in the regulation of nitrogen metabolism, is inactivated (10, 33). Membrane fractionation and pull-down experiments reveal the sequestration of PII proteins by AmtB after the addition of ammonium (9, 15, 17, 33, 34). The crystal structure of the E. coli GlnK-AmtB complex shows the docking of the PII protein via its T loop into the cytoplasmic pore exit of AmtB (7, 13). In Azospirillum brasilense and Rhodospirillum rubrum, sequestration of PII proteins by AmtB is also directly linked to ADP-ribosylation-dependent switch-off of nitrogenase since complex formation is necessary for the capture of DraG, the enzyme responsible for removing the ADP-ribosyl residue from NifH, and reactivating nitrogenase (15, 28, 34). To further characterize the role of R. capsulatus AmtB in ammonia transport and the posttranslational regulation of nitrogenase, variants of AmtB with amino acid substitutions for some highly conserved residues, or alterations of its C terminus, were created and studied. Correlating the results of the analysis of ammonia transport, formation of complexes with GlnK, and nitrogenase regulation by these variant AmtB proteins reveals that the formation of a GlnK-AmtB complex is not sufficient to trigger nitrogenase regulation. Moreover, there is a correspondence between the capacity of AmtB to conduct ammonia and its ability to properly regulate nitrogenase.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and DNA manipulation.
Except where mentioned otherwise, E. coli strains were cultivated in LB medium with the appropriate antibiotics. R. capsulatus strains were maintained in YPS medium (35). For all of the experiments with R. capsulatus, cultures were grown photoheterotrophically to early stationary phase at 30°C in RCV-lactate medium without fixed nitrogen (40). Conjugal transfer of plasmids from E. coli S17.1 to R. capsulatus amtB mutant strain RCAY63 was achieved by filter mating (24). DNA manipulation followed previously described standard procedures (30).
Mutagenesis and construction of plasmids.
C-terminal deletions in R. capsulatus AmtB were obtained by PCR. The complete glnKamtB operon with its ntr-dependent promoter and a deletion of either 81 or 21 nucleotides (nt) at the 3′ end of amtB were amplified by using pAY98 as a template with the appropriate forward and reverse primers containing PstI and EcoRI sites and cloned as approximately 2-kb PstI-EcoRI fragments into the conjugational vector pJB3TC20, resulting in pPLT2 (81-nt deletion) or pPLT28 (21-nt deletion).
Site-directed mutagenesis was achieved by overlap extension PCR. The 5′ part of amtB, a complete copy of glnK, and the ntr promoter of the glnKamtB operon were cloned as a 0.8-kb PstI-SalI fragment from pAY98 into pMECA, resulting in pPLT2III. For each site-directed mutation, amtB without its 5′ part was amplified and modified by using pAY98 as a template and the appropriate forward and reverse primers carrying SalI and EcoRI sites. Following overlap extension PCR, the resulting 1.4-kb SalI-EcoRI fragments were cloned into pPLT2III. Then, the complete glnKamtB operon for each variant was cloned as a 2.4-kb PstI-EcoRI fragment into conjugational vector pJB3TC20, resulting in pPLT6 (D334A D335A), pPLT9 (R146A), pPLT31 (D185E), pPLT41 (H193A), pPLT47 (F131A), and pPLT49 (H342K). The presence of the correct sequence change was verified by sequencing.
pPLT26 contains amtB with a thrombin cleavage site and a histidine tag at its C-terminal end under the control of the T7 promoter. The appropriate PCR product was obtained by using pAY98 as a template and primers containing NdeI and HindIII sites, a histidine tag, and a thrombin cleavage site. The resulting 1.4-kb NdeI-HindIII fragment was cloned into pT7-7. The validity of the construction was verified by sequencing. For expression in R. capsulatus, a 1.38-kb SalI-HindIII fragment of pPLT26 encompassing the histidine-tagged amtB gene was cloned into pBluescript KS−, resulting in pPLT30. Then, the same fragment with EcoRI as the 3′ restriction site instead of HindIII was cloned from pPLT30 into pPLT2III. A 2.3-kb PstI-EcoRI fragment bearing the complete glnKamtB operon with the ntr promoter and the C-terminally histidine-tagged variant of amtB was cloned into pJB3TC20, resulting in pPLT33.
AmtBHis overexpression, purification, and antibody production.
pPLT26 was transformed into C43 for AmtBHis overexpression, and cells were grown as described by Blakey et al. (3). Collection of cells, fractionation, membrane solubilization, and purification (with dodecyl maltoside as a nonionic detergent) of AmtBHis with Talon resin (Clontech, Mississauga, Ontario, Canada) were carried out as previously described (25). The specificity of antibody raised in rabbits against purified AmtBHis was confirmed by the absence of a signal in a Western blot assay of an amtB mutant strain cell extract.
Gel electrophoresis analyses.
Ammonium treatment and subsequent cellular fractionation were carried out as previously described (33). Protein concentration was measured by the Bradford reaction (6). A 5-μg sample of total protein was loaded per well for all sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) experiments. SDS-PAGE and then probing of immunoblots with anti-GlnK antibody (33) were done with 15% polyacrylamide gels, whereas 10% polyacrylamide gels were used when the protein studied was AmtB. After SDS-PAGE, proteins were electrophoretically transferred to polyvinylidene difluoride membranes (Roche, Mississauga, Ontario, Canada) and signals were detected with the ECL enhanced chemiluminescence system (Amersham, Oakville, Ontario, Canada) by using Kodak BioMax XAR film. For quantification, signals were digitized with a Chemi Imager 5500 (Alpha Innotech, San Leandro, CA) and quantified using ImageJ software (NIH, Bethesda, MD). This method was used for quantification of various AmtB proteins and GlnK with reference to a standard curve of signal intensities generated by applying different quantities of purified proteins.
Methylammonium transport, in vivo nitrogenase activity, and NifH ADP-ribosylation.
[14C]methylamine uptake was measured as described previously (27). Radioactivity of the samples was counted with a Beckman LS 6000SC scintillation counter. The procedures used for in vivo nitrogenase activity assay by gas chromatography and determination of NifH ADP-ribosylation status by Western blot assays were previously described (38).
RESULTS
AmtB variants studied.
A number of regions or amino acid residues of AmtB potentially associated with ammonium recruitment and transport or with interaction with GlnK were targeted by mutagenesis (Table 1). The roles proposed for the various amino acid residues indicated in Table 1 were derived from crystallographic studies, mutational analysis, or computational modeling of E. coli AmtB as indicated. Here we use the numbering of R. capsulatus AmtB; the corresponding E. coli residues are indicated in Table 1. The recently published three-dimensional structure of an E. coli AmtB-GlnK complex shows that, in this complex, the cytoplasmic C-terminal tail serves several structural roles, with contacts with the cytoplasmic faces of several helices of the same monomer and with the neighboring subunit such that the last six residues of the C-terminal tail of E. coli AmtB are part of the cytoplasmic pore exit of the adjacent subunit (7). Minor changes in this region of E. coli AmtB reduce methylamine transport rates (31). Both the entire C-terminal cytoplasmic tail (Δ403-430) and the last seven amino acid residues (Δ423-430) of R. capsulatus AmtB were targeted by deletion mutagenesis. H193 and H342 are located inside the pore and presumably serve as proton acceptors or agents of weak stabilization between the substrate and the protein (4, 16, 18, 42). Mutation of these residues abolishes ammonia transport by E. coli AmtB (16). Possible functions of F131 identified by a combination of crystallographic and computational studies are to structurally block the periplasmic side of the pore, rotating to allow the passage of ammonia, and to contribute to the dehydration of ammonium (4, 18, 42). Analysis of X-ray crystallographic structures suggests that D185 plays a structural role in the periplasmic site that recruits ammonium (18, 42). Moreover, molecular dynamic simulations show that D185 could become, upon a switch in the structure of the recruitment vestibule induced by the entrance of ammonium, the ultimate proton acceptor for ammonium (21, 26). The structure of AmtB shows a quasi-twofold symmetry caused by a presumed ancient gene duplication (16); thus, the counterpart of D185 is highly conserved residue D335. Because of similarities in terms of structural roles between D185 and D335, Khademi and Stroud (19) suggested that D335 could also become available for interaction with the substrate after a structural change and thus could be the proton donor for reprotonation of ammonia in the cytoplasmic vestibule. R146, highly conserved in the Amt/Mep family but with no assigned putative function as yet, was also targeted.
TABLE 1.
Putative functions of mutated residues of R. capsulatus AmtB
| R. capsulatus AmtBa variant | Corresponding residue(s) in E. coli AmtBa | Putative function(s) |
|---|---|---|
| F131A | F107 | Blocking periplasmic entrance of pore and/or contributing to dehydration of NH4+ (4, 18, 42) |
| R146A | R122 | None |
| D185E | D160 | Role in periplasmic vestibule recruiting NH4+ (18, 21, 26, 42) |
| H193A | H168 | Proton acceptor or agent of weak stabilization by hydrogen bonding between substrate and AmtB (4, 16, 18, 42) |
| D334A D335A | D309, D310 | Participation in rearrangement of AmtB cytoplasmic face upon GlnK binding (7) and in reprotonation of NH3 after passage through pore (19) |
| H342K | H318 | Proton acceptor or agent of weak stabilization by hydrogen bonding between substrate and AmtB (4, 16, 18, 42) |
| Δ423-430 | Partial C-terminal tail | Part of cytoplasmic pore exit of next subunit (7) |
The numbering shown is with the N-terminal residue of the respective mature AmtB protein as 1. The E. coli AmtB preprotein has been shown to have a signal sequence that is cleaved upon membrane insertion (32). The presumed R. capsulatus mature AmtB protein, after removal of the predicted signal peptide (SignalP 3.0) (2), has an N terminus that is extended by 17 amino acids in comparison with that of E. coli AmtB.
Methylamine transport by AmtB variants.
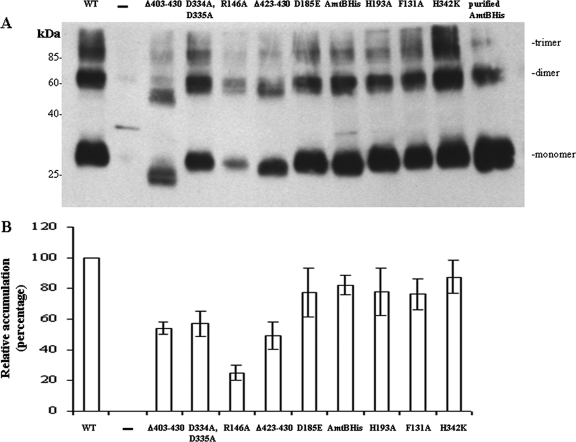
Since GlnK and AmtB form complexes under some conditions, their relative proportion in the cell may be critical. Therefore, the different AmtB variants were expressed as plasmid-borne glnKamtB operons in an amtB mutant strain of R. capsulatus (RCAY63). Quantitative analysis showed that this strategy was successful, with the same relative GlnK/AmtB ratio in cells expressing these proteins from plasmid-borne glnKamtB (4.15 ± 0.7) as the wild-type strain expressing these proteins from glnKamtB on the chromosome (4.2 ± 0.6). Moreover, the total AmtB accumulated when expressed from a plasmid was not much greater (1.5-fold ± 0.4-fold) than that of the wild-type strain with a chromosomally located operon, suggesting that the results obtained could reasonably be extrapolated to the wild-type physiology. Levels of accumulation of AmtB variants were checked by Western blotting of SDS-PAGE gels (Fig. 1A), and membrane localization was determined by fractionation experiments. All of the variants studied, except R146A, were shown to be correctly localized to the membrane (not shown) and appeared to accumulate at significant (50% or greater) levels compared to the wild-type protein (plasmid-borne wild-type glnKamtB) (Fig. 1). The relative level of accumulation of the AmtB R146A variant is only 25% ± 5% (Fig. 1A and B). Apparently, this conserved residue located in the M3-M4 cytoplasmic loop is essential for protein stability. AmtB proteins with C-terminal deletions (Δ403-430, Δ423-430) apparently accumulated at somewhat reduced levels (approximately 50%) (Fig. 1A and B); similar results have been noted in E. coli (9, 31). Except for D334A and D335A (57% ± 8%), all of the other variants (D185E, AmtBHis, H193A, F131A, and H342K) were present at nearly wild-type levels, varying between ∼75% and ∼90% (Fig. 1A and B).
FIG. 1.
Accumulation of AmtB variants. Cultures of an R. capsulatus amtB mutant strain (RCAY63) alone (−) or complemented with a plasmid carrying a glnKamtB operon encoding wild-type AmtB (WT) or various AmtB variants (Δ403-430, D334A D335A, R146A, Δ423-430, D185E, AmtBHis, H193A, F131A, and H342K) were grown under N2-fixing conditions. Whole-cell extracts were subjected to SDS-PAGE (5 μg of total protein loaded per well) and immunoblotting with anti-AmtBHis (A). In the last well, 30 ng of purified AmtBHis was added. Potential monomeric, dimeric, and trimeric forms of AmtB are indicated to the right of the panel. The top of the panel indicates the AmtB variant present. Two independent experiments were done, and the amounts of AmtB variants relative to wild-type AmtB were quantified as described in Materials and Methods (B). As previously noted for E. coli (3, 8, 11), dissociation of AmtB trimers, even under these denaturing conditions, is incomplete.
The transport capabilities of the AmtB variants were assessed by using a methylamine transport assay (38). Complete (Δ403-430) and partial (Δ423-430) C-terminal deletion AmtB proteins are still functional for methylamine transport with, respectively, 26% and 57% of the transport capability of the wild-type protein (Table 2). Addition of a histidine tag at the C-terminal end of R. capsulatus AmtB appears to deregulate methylamine transport with a 1.9-fold greater rate than wild-type AmtB (Table 2). In agreement with results obtained with E. coli AmtB and yeast Mep2 (17, 23), replacement of aspartate 185 with glutamate reduces the methylamine transport rate of R. capsulatus AmtB by ∼50% (Table 2). As in E. coli (16), substitution at either residue H185 or H342 of R. capsulatus AmtB abolishes methylamine transport (Table 2). Here we report the first mutational analysis of F131. In agreement with its predicted critical role, replacement of F131 with alanine abolishes methylamine transport (Table 2). Introducing a D334A D335A double substitution into R. capsulatus AmtB also abolishes methylamine transport (Table 2), suggesting that these two residues, which are highly conserved in the Amt/Mep protein family, are functionally important.
TABLE 2.
Methylamine transport and nitrogenase switch-off in amtB variant strains
| Strain | Relevant AmtB phenotype | CH3NH2+ transporta,b | Nitrogenase switch-offb,c |
|---|---|---|---|
| RCAY63/pAY99 | Wild type | 2.82 ± 0.53 | 0.37 ± 0.09 |
| RCAY63 | AmtB− | 0.50 ± 0.28 | 0.99 ± 0.14 |
| RCAY63/pPLT2 | Δ403-430 | 1.10 ± 0.25 | 0.93 ± 0.09 |
| RCAY63/pPLT6 | D334A, D335A | 0.36 ± 0.50 | 1.01 ± 0.07 |
| RCAY63/pPLT28 | Δ423-430 | 1.82 ± 0.65 | 1.09 ± 0.11 |
| RCAY63/pPLT31 | D185E | 1.73 ± 0.52 | 0.35 ± 0.20 |
| RCAY63/pPLT33 | C-terminal histidine tag | 4.94 ± 0.80 | 1.20 ± 0.15 |
| RCAY63/pPLT41 | H193A | 0.44 ± 0.11 | 0.93 ± 0.15 |
| RCAY63/pPLT47 | F131A | 0.28 ± 0.07 | 0.95 ± 0.09 |
| RCAY63/pPLT49 | H342K | 0.44 ± 0.26 | 0.98 ± 0.16 |
Values are the mean nmol CH3NH2+/mg protein/min ± the standard error of the mean.
Values are from triplicate experiments.
Shown is the mean nitrogenase activity 5 min after addition of NH4Cl to 200 μM divided by nitrogenase activity 5 min before ammonium addition ± the standard error of the mean.
Formation of AmtB-GlnK complexes.
The question arose as to whether AmtB variants showing some transport activity were capable of forming ammonium-induced complexes with GlnK. As well, recent results suggest that AmtB-GlnK interactions are governed by metabolite pools and the modification status of the PII protein (11, 14, 37). In an R. capsulatus amtB mutant strain that is incapable of nitrogenase regulation, sufficient ammonium enters the cell upon its addition to 1 mM to trigger the proper regulation of glutamine synthetase (38) and is sufficient to cause the efficient deuridylylation of GlnK (P.-L. Tremblay and P. C. Hallenbeck, unpublished results). This suggested that AmtB-GlnK complex formation could possibly occur upon ammonium addition to cultures of transport-incompetent AmtB variant strains. Therefore, the ability of the various AmtB variants to form complexes with GlnK upon addition of ammonium was tested by membrane fractionation experiments.
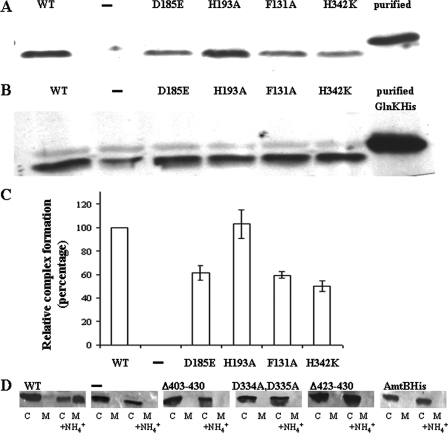
These analyses show that, similar to E. coli (9, 31), complete (Δ403-430) or partial (Δ423-430) deletion of the C-terminal tail completely abolished the membrane sequestration of GlnK (Fig. 2D). Models based on crystallographic studies reveal that the C-terminal region of one subunit of the AmtB homotrimer forms part of the docking bay of the next subunit in which the T loop of the PII protein anchors itself (1, 7, 13). Fractionation experiments with AmtBHis, possessing a C-terminal histidine tag, shows that GlnK is not sequestered to membranes containing this variant after an ammonium shock (Fig. 2D). Since docking of GlnK to AmtB blocks the ammonium transport pore, thereby downregulating ammonia transport, addition of a histidine tag probably physically blocked the formation of a GlnK-AmtB complex (7, 13). As well, GlnK interaction with AmtB D334A D335A was also completely absent (Fig. 2D). Crystallographic studies of free E. coli AmtB versus the AmtB-GlnK complex show a significant change in the conformation of the transporter's cytoplasmic face (7, 18, 42). D334 participates in this rearrangement by forming salt bridge interactions with other conserved residues (7). This could explain the lack of interaction of GlnK with AmtB D334A D335A.
FIG. 2.
Complex formation between AmtB variants and GlnK or GlnB. Cultures of an R. capsulatus amtB mutant strain (RCAY63) alone (−) or complemented with a plasmid carrying a glnKamtB operon encoding wild-type AmtB (WT) or various AmtB variants (D185E, H193A, F131A, and H342K) were grown under N2-fixing conditions. NH4Cl (1 mM) was added 15 min prior to harvest. The membrane fractions (A) and whole-cell extracts (B) of cultures exposed to NH4Cl (1 mM) were subjected to SDS-PAGE (5 μg of total protein loaded per well) and immunoblotting with anti-GlnK antibody. In the last well, 30 ng of purified GlnKHis was added. The loaded variants with which GlnK interacts are indicated above the panels. Two independent experiments were done, and the amount of GlnK complexed with the variant AmtB was quantified relative to wild-type AmtB (C). Additional strains were examined for complex formation between GlnK and AmtB variants by visualizing the ammonium-induced membrane sequestration of GlnK by using fractionation and immunoblotting. Two parallel cultures of each strain were grown under N2-fixing conditions. The strains examined consisted of an R. capsulatus amtB mutant strain (RCAY63) alone (−) or complemented with a plasmid carrying a glnKamtB operon encoding wild-type AmtB (WT) or various AmtB variants (Δ403-430, D334A D335A, Δ423-430, and AmtBHis). NH4Cl (1 mM) was added to one of the parallel cultures 15 min prior to harvest. Cytoplasmic (C) and membrane (M) fractions were subjected to SDS-PAGE (5 μg of total protein loaded per well) and immunoblotting with anti-GlnK. The top of the panel indicates the AmtB variant present (D).
Four of the AmtB variants, H193A, D185E, F131A, and H342K, were capable of forming complexes with GlnK (Fig. 2A and C). The levels of GlnK present in the cell were similar in all of these variants (Fig. 2B). Densitometric analysis showed that, although in all cases substantial amounts were bound, the relative amounts of GlnK in the complexes varied somewhat between the variants. Relative to wild-type AmtB (100%), membrane-bound GlnK in the H193A, D185E, F131A, and H342K AmtB variants was, respectively, 103% ± 12%, 61% ± 6%, 59% ± 3%, and 50% ± 4% (Fig. 2A to C). Interestingly, there did not seem to be a correlation between AmtB transport activity and complex formation with GlnK. H193A, F131A, and H342K, which were negative for transport, all formed complexes with GlnK, as did D185E, which retains substantial transport activity (Table 2).
Nitrogenase switch-off and NifH ADP-ribosylation in AmtB variants.
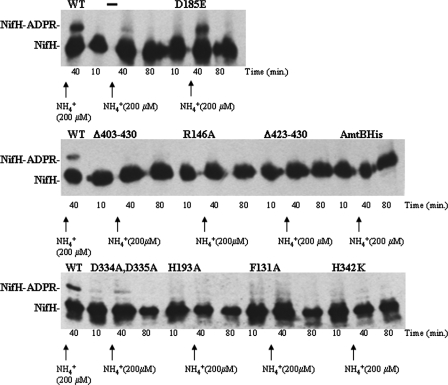
R. capsulatus possesses two different systems implicated in the regulation of nitrogenase activity, one that is linked to ADP-ribosylation of NifH and one that is independent of this covalent modification (39). Both responses can be provoked by the addition of ammonium to the medium, and the relative importance of the two responses varies with the growth conditions (39). However, the formation of an AmtB-GlnK complex plays a key role since an AmtB− strain (38) or a GlnB− strain where deuridylylated GlnK is not sequestered by the membrane (33) is deficient in both responses even though, as previously shown (38), ammonia is entering the cell. Therefore, formation of an AmtB-GlnK complex is necessary for nitrogenase regulation. Thus, it was of interest to examine ammonium-induced switch-off of nitrogenase activity and ADP-ribosylation of NifH in the AmtB variants studied here. Nitrogenase activity was measured by the acetylene reduction method, switch-off was triggered by the addition of 200 μM NH4Cl, and ADP-ribosylation of NifH was monitored electrophoretically (38). With the exception of AmtB D185E, all of the mutants are negative for Mo-nitrogenase switch-off (Table 2) and NifH ADP-ribosylation (Fig. 3). Similar ADP-ribosylation results were obtained with 1 mM ammonium (not shown) and are typical for what is observed with a wild-type strain with glnKamtB on the chromosome (38, 39). Thus, the only AmtB variant capable of forming a complex with GlnK that is competent for proper regulation of nitrogenase switch-off and NifH-ADP ribosylation, D185E, is the variant that retains substantial transport activity.
FIG. 3.
Ammonium-induced ADP-ribosylation of NifH in amtB variant strains. Cultures of R. capsulatus were grown under N2-fixing conditions. Time zero corresponded to the addition of acetylene necessary to start the in vivo nitrogenase activity assay (results presented in Table 1) which was carried out simultaneously with the monitoring of the ADP-ribosylation state of NifH. Where indicated by arrows, NH4Cl was added to 200 μM at 25 min. Culture samples were withdrawn at the indicated times, and the ADP-ribosylation state of NifH of the amtB mutant strain (RCAY63) complemented with the wild-type glnK-amtB operon encoding wild-type AmtB (WT), the uncomplemented amtB mutant strain (−), and the amtB mutant strain complemented with plasmids carrying glnK-amtB operons bearing different mutated amtB genes encoding AmtB variants (D185E, Δ403-430, R146A, Δ423-430, AmtBHis, D334A D335A, H193A, F131A, and H342K) was monitored by immunoblotting.
DISCUSSION
Here we have investigated the role of some key residues in ammonium transport by R. capsulatus AmtB, in AmtB-GlnK complex formation, and in nitrogenase regulation. The different R. capsulatus AmtB variants created and studied here corroborate previous studies with E. coli AmtB and shed new light on the role(s) of some amino acid residues highly conserved in the Amt/Mep family. Since we wished to examine nitrogenase regulation in these variants, it was important to verify experimentally that changing the homologous residues of R. capsulatus AmtB produced effects on methylamine transport and GlnK complex formation similar to what has been observed with the E. coli protein. Absolutely conserved residues H193 and H342, critical for ammonia transport in E. coli (16), are also required for ammonia transport by R. capsulatus AmtB. As previously reported for E. coli (17), changing highly conserved aspartate 185 to a glutamate residue reduces methylamine uptake. Several studies have shown the importance of the C-terminal cytoplasmic tail of E. coli AmtB in optimal ammonia transport (7, 9, 31), and similar results are seen with either a full or a partial deletion of this region in R. capsulatus AmtB. Highly conserved AmtB amino acid residues not previously studied by mutational analysis, F154, D334, and D335, have also been examined here and have now been shown to be indispensable for ammonia transport.
Evidence is accumulating that AmtB-GlnK complexes play a critical role in the posttranslational regulation of nitrogenase in some organisms. A model is emerging in which the AmtB and PII proteins control the activities of the enzymes involved in NifH modification, DraG and DraT, with AmtB and a PII protein being required for the ammonium-induced membrane sequestration of DraG in both A. brasilense (14, 15) and R. rubrum (34). Similar events are likely to occur in R. capsulatus since AmtB is absolutely required for proper nitrogenase regulation (38) and its ammonium-induced membrane sequestration of GlnK appears to be critical in this process (33).
The results obtained with AmtB variants with the C-terminal tail partially or completely deleted or AmtBHis clearly demonstrate that, in spite of various levels of methylamine uptake, without GlnK-AmtB complex formation, there is no posttranslational regulation of nitrogenase. On the other hand, examination of the AmtB F131A, H193A, and H342A variants showed that GlnK sequestration after ammonium shock is not sufficient to ensure nitrogenase switch-off and NifH ADP-ribosylation. The absolute amount of GlnK sequestered to the membrane seems to be relatively unimportant. AmtB D185E appears to have a lower level of membrane-bound GlnK than the wild type, yet it is fully capable of regulating nitrogenase, while AmtB H193A, which has wild-type levels of GlnK sequestration after ammonium shock, is completely incapable of nitrogenase switch-off or NifH ADP-ribosylation.
These results strongly suggest that, in addition to its complex formation with GlnK, partial ammonium transport by AmtB is absolutely required for the posttranslational regulation of nitrogenase in response to an ammonium shock. This raises the questions of how much ammonia progression through the channel is necessary and what the signal transduction pathway is. The results obtained with D185E AmtB suggest that the events in ammonia transport by AmtB associated with nitrogenase regulatory processes occur downstream of NH4+ binding inside the periplasmic vestibule. Regardless of what the details may ultimately prove to be, here we have shown that ammonia transport by AmtB appears to be required for its role as an ammonium sensor in triggering nitrogenase switch-off and NifH ADP-ribosylation in R. capsulatus.
Acknowledgments
This research was supported by a grant from the Natural Sciences and Engineering Council.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Andrade, S. L., A. Dickmanns, R. Ficner, and O. Einsle. 2005. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA 10214994-14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 3.Blakey, D., A. Leech, G. H. Thomas, G. Coutts, K. Findlay, and M. Merrick. 2002. Purification of the Escherichia coli ammonium transporter AmtB reveals a trimeric stoichiometry. Biochem. J. 364527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bostick, D. L., and C. L. Brooks III. 2007. Deprotonation by dehydration: the origin of ammonium sensing in the AmtB channel. PLoS Comput. Biol. 3e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bostick, D. L., and C. L. Brooks III. 2007. On the equivalence point for ammonium (de)protonation during its transport through the AmtB channel. Biophys. J. 92L103-L105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 7.Conroy, M. J., A. Durand, D. Lupo, X. D. Li, P. A. Bullough, F. K. Winkler, and M. Merrick. 2007. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc. Natl. Acad. Sci. USA 1041213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conroy, M. J., S. J. Jamieson, D. Blakey, T. Kaufmann, A. Engel, D. Fotiadis, M. Merrick, and P. A. Bullough. 2004. Electron and atomic force microscopy of the trimeric ammonium transporter AmtB. EMBO Rep. 51153-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coutts, G., G. Thomas, D. Blakey, and M. Merrick. 2002. Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 21536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drepper, T., S. Gross, A. F. Yakunin, P. C. Hallenbeck, B. Masepohl, and W. Klipp. 2003. Role of GlnB and GlnK in ammonium control of both nitrogenase systems in the phototrophic bacterium Rhodobacter capsulatus. Microbiology 1492203-2212. [DOI] [PubMed] [Google Scholar]
- 11.Durand, A., and M. Merrick. 2006. In vitro analysis of the Escherichia coli AmtB-GlnK complex reveals a stoichiometric interaction and sensitivity to ATP and 2-oxoglutarate. J. Biol. Chem. 28129558-29567. [DOI] [PubMed] [Google Scholar]
- 12.Fong, R. N., K. S. Kim, C. Yoshihara, W. B. Inwood, and S. Kustu. 2007. The W148L substitution in the Escherichia coli ammonium channel AmtB increases flux and indicates that the substrate is an ion. Proc. Natl. Acad. Sci. USA 10418706-18711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruswitz, F., J. O'Connell III, and R. M. Stroud. 2007. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 A. Proc. Natl. Acad. Sci. USA 10442-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huergo, L. F., M. Merrick, F. O. Pedrosa, L. S. Chubatsu, L. M. Araujo, and E. M. Souza. 2007. Ternary complex formation between AmtB, GlnZ and the nitrogenase regulatory enzyme DraG reveals a novel facet of nitrogen regulation in bacteria. Mol. Microbiol. 661523-1535. [DOI] [PubMed] [Google Scholar]
- 15.Huergo, L. F., E. M. Souza, M. S. Araujo, F. O. Pedrosa, L. S. Chubatsu, M. B. Steffens, and M. Merrick. 2006. ADP-ribosylation of dinitrogenase reductase in Azospirillum brasilense is regulated by AmtB-dependent membrane sequestration of DraG. Mol. Microbiol. 59326-337. [DOI] [PubMed] [Google Scholar]
- 16.Javelle, A., D. Lupo, L. Zheng, X. D. Li, F. K. Winkler, and M. Merrick. 2006. An unusual twin-His arrangement in the pore of ammonia channels is essential for substrate conductance. J. Biol. Chem. 28139492-39498. [DOI] [PubMed] [Google Scholar]
- 17.Javelle, A., E. Severi, J. Thornton, and M. Merrick. 2004. Ammonium sensing in Escherichia coli: role of the ammonium transporter AmtB and AmtB-GlnK complex formation. J. Biol. Chem. 2798530-8538. [DOI] [PubMed] [Google Scholar]
- 18.Khademi, S., J. O'Connell III, J. Remis, Y. Robles-Colmenares, L. J. Miercke, and R. M. Stroud. 2004. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science 3051587-1594. [DOI] [PubMed] [Google Scholar]
- 19.Khademi, S., and R. M. Stroud. 2006. The Amt/MEP/Rh family: structure of AmtB and the mechanism of ammonia gas conduction. Physiology (Bethesda) 21419-429. [DOI] [PubMed] [Google Scholar]
- 20.Lamoureux, G., M. L. Klein, and S. Berneche. 2007. A stable water chain in the hydrophobic pore of the AmtB ammonium transporter. Biophys. J. 92L82-L84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, Y., Z. Cao, and Y. Mo. 2006. Molecular dynamics simulations on the Escherichia coli ammonia channel protein AmtB: mechanism of ammonia/ammonium transport. J. Am. Chem. Soc. 12810876-10884. [DOI] [PubMed] [Google Scholar]
- 22.Luzhkov, V. B., M. Almlöf, M. Nervall, and J. Aqvist. 2006. Computational study of the binding affinity and selectivity of the bacterial ammonium transporter AmtB. Biochemistry 4510807-10814. [DOI] [PubMed] [Google Scholar]
- 23.Marini, A. M., M. Boeckstaens, F. Benjelloun, B. Cherif-Zahar, and B. Andre. 2006. Structural involvement in substrate recognition of an essential aspartate residue conserved in Mep/Amt and Rh-type ammonium transporters. Curr. Genet. 49364-374. [DOI] [PubMed] [Google Scholar]
- 24.Masepohl, B., W. Klipp, and A. Puhler. 1988. Genetic characterization and sequence analysis of the duplicated nifA/nifB gene region of Rhodobacter capsulatus. Mol. Gen. Genet. 21227-37. [DOI] [PubMed] [Google Scholar]
- 25.Niegowski, D., M. Hedren, P. Nordlund, and S. Eshaghi. 2006. A simple strategy towards membrane protein purification and crystallization. Int. J. Biol. Macromol. 3983-87. [DOI] [PubMed] [Google Scholar]
- 26.Nygaard, T. P., C. Rovira, G. H. Peters, and M. O. Jensen. 2006. Ammonium recruitment and ammonia transport by E. coli ammonia channel AmtB. Biophys. J. 914401-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rapp, B. J., D. C. Landrum, and J. D. Wall. 1986. Methylammonium uptake by Rhodobacter capsulatus. Arch. Microbiol. 146134-141. [Google Scholar]
- 28.Saari, L. L., E. W. Triplett, and P. W. Ludden. 1984. Purification and properties of the activating enzyme for iron protein of nitrogenase from the photosynthetic bacterium Rhodospirillum rubrum. J. Biol. Chem. 25915502-15508. [PubMed] [Google Scholar]
- 29.Saier, M. H., Jr., B. H. Eng, S. Fard, J. Garg, D. A. Haggerty, W. J. Hutchinson, D. L. Jack, E. C. Lai, H. J. Liu, D. P. Nusinew, A. M. Omar, S. S. Pao, I. T. Paulsen, J. A. Quan, M. Sliwinski, T. T. Tseng, S. Wachi, and G. B. Young. 1999. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim. Biophys. Acta 14221-56. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Frisch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Severi, E., A. Javelle, and M. Merrick. 2007. The conserved carboxy-terminal region of the ammonia channel AmtB plays a critical role in channel function. Mol. Membr. Biol. 24161-171. [DOI] [PubMed] [Google Scholar]
- 32.Thornton, J., D. Blakey, E. Scanlon, and M. Merrick. 2006. The ammonia channel protein AmtB from Escherichia coli is a polytopic membrane protein with a cleavable signal peptide. FEMS Microbiol. Lett. 258114-120. [DOI] [PubMed] [Google Scholar]
- 33.Tremblay, P.-L., T. Drepper, B. Masepohl, and P. C. Hallenbeck. 2007. Membrane sequestration of PII proteins and nitrogenase regulation in the photosynthetic bacterium Rhodobacter capsulatus. J. Bacteriol. 1895850-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, H., C. C. Franke, S. Nordlund, and A. Noren. 2005. Reversible membrane association of dinitrogenase reductase activating glycohydrolase in the regulation of nitrogenase activity in Rhodospirillum rubrum; dependence on GlnJ and AmtB1. FEMS Microbiol. Lett. 253273-279. [DOI] [PubMed] [Google Scholar]
- 35.Weaver, P. F., J. D. Wall, and H. Gest. 1975. Characterization of Rhodopseudomonas capsulata. Arch. Microbiol. 105207-216. [DOI] [PubMed] [Google Scholar]
- 36.Winkler, F. K. 2006. Amt/MEP/Rh proteins conduct ammonia. Pfluegers Arch. Eur. J. Physiol. 451701-707. [DOI] [PubMed] [Google Scholar]
- 37.Wolfe, D. M., Y. Zhang, and G. P. Roberts. 2007. Specificity and regulation of interaction between the PII and AmtB1 proteins in Rhodospirillum rubrum. J. Bacteriol. 1896861-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yakunin, A. F., and P. C. Hallenbeck. 2002. AmtB is necessary for NH4+-induced nitrogenase switch-off and ADP-ribosylation in Rhodobacter capsulatus. J. Bacteriol. 1844081-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yakunin, A. F., and P. C. Hallenbeck. 1998. Short-term regulation of nitrogenase activity by NH4+ in Rhodobacter capsulatus: multiple in vivo nitrogenase responses to NH4+ addition. J. Bacteriol. 1806392-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yakunin, A. F., T. V. Laurinavichene, A. A. Tsygankov, and P. C. Hallenbeck. 1999. The presence of ADP-ribosylated Fe protein of nitrogenase in Rhodobacter capsulatus is correlated with cellular nitrogen status. J. Bacteriol. 1811994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, H., Y. Xu, W. Zhu, K. Chen, and H. Jiang. 2007. Detailed mechanism for AmtB conducting NH4+/NH3: molecular dynamics simulations. Biophys. J. 92877-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng, L., D. Kostrewa, S. Berneche, F. K. Winkler, and X. D. Li. 2004. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. USA 10117090-17095. [DOI] [PMC free article] [PubMed] [Google Scholar]