Abstract
Hepatitis C virus (HCV) infection is a common cause of chronic hepatitis and is currently treated with alpha interferon (IFN-α)-based therapies. However, the underlying mechanism of IFN-α therapy remains to be elucidated. To identify the cellular proteins that mediate the antiviral effects of IFN-α, we created a HEK293-based cell culture system to inducibly express individual interferon-stimulated genes (ISGs) and determined their antiviral effects against HCV. By screening 29 ISGs that are induced in Huh7 cells by IFN-α and/or up-regulated in HCV-infected livers, we discovered that viperin, ISG20, and double-stranded RNA-dependent protein kinase (PKR) noncytolytically inhibited the replication of HCV replicons. Mechanistically, inhibition of HCV replication by ISG20 and PKR depends on their 3′-5′ exonuclease and protein kinase activities, respectively. Moreover, our work, for the first time, provides strong evidence suggesting that viperin is a putative radical S-adenosyl-l-methionine (SAM) enzyme. In addition to demonstrating that the antiviral activity of viperin depends on its radical SAM domain, which contains conserved motifs to coordinate [4Fe-4S] cluster and cofactor SAM and is essential for its enzymatic activity, mutagenesis studies also revealed that viperin requires an aromatic amino acid residue at its C terminus for proper antiviral function. Furthermore, although the N-terminal 70 amino acid residues of viperin are not absolutely required, deletion of this region significantly compromises its antiviral activity against HCV. Our findings suggest that viperin represents a novel antiviral pathway that works together with other antiviral proteins, such as ISG20 and PKR, to mediate the IFN response against HCV infection.
Hepatitis C virus (HCV) is the sole member of the genus Hepacivirus in the family Flaviviridae (43). It establishes persistent infections in the vast majority of infected individuals and is the only known positive-stranded RNA virus that causes persistent life-long infections in humans. Currently, HCV chronically infects more than 170 million people worldwide. Although the initial infection is largely asymptomatic, prolonged infection carries a high risk of chronic hepatitis, cirrhosis, and primary hepatocellular carcinoma (2).
Although it has been elegantly demonstrated that HCV can evade the host cellular innate defense response through proteolytic cleavage of RIG-I/MDA5 adaptor protein MAVS and Toll-like receptor 3 adaptor protein TRIF (7, 22, 25, 42, 44, 48, 69), microarray studies performed with liver samples obtained from HCV transiently infected chimpanzees and chronically infected humans revealed that the induction of interferon (IFN)-stimulated genes (ISGs) in HCV-infected livers is a hallmark of the virus infection (5, 6, 33, 39, 58, 61). These discoveries suggest that the HCV-infected liver is a constant battlefield between the virus and host innate immunity defense systems, and thus IFN-mediated innate responses induced by HCV may play an important role in shaping the pathogenesis and clinical outcomes of HCV infection (55). Moreover, alpha interferon (IFN-α) and ribavirin combination therapy is the current standard therapeutic regimen for chronic hepatitis C and can lead to sustained virological response in only 40 to 50% of treated patients (46, 50). The parameters determining the success or failure of the antiviral therapy are not understood, and their identification represents a major challenge in HCV biology (17). Accordingly, elucidation of the mechanism by which IFN-α controls HCV replication represents an important step toward understanding the pathobiology of HCV infection and molecular basis of IFN treatment of chronic hepatitis C.
IFN-α treatment of cells alters the expression of hundreds of genes (15, 32, 45). So far, the nature of the IFN-induced cellular proteins that specifically target viral components and, hence, are responsible for the inhibition of HCV replication has not been completely defined (29). Previous studies suggested that overexpression of known IFN-α-inducible genes, such as ISG p56 and viperin, but not MxA, partially inhibited HCV replication in HCV replicon-containing Huh7 cells (23, 33, 47, 64). Additionally, HCV replicons were found to replicate more efficiently in double-stranded RNA-dependent protein kinase (PKR)-deficient mouse embryonic fibroblasts (MEFs), and in Huh7 cells, in which adenosine deaminase acts on RNA 1 (ADAR 1), gene expression was reduced by small interfering RNA (siRNA) transfection (11, 62). These findings suggest that those ISGs might play a role in controlling HCV replication and mediating IFN-induced antiviral effects against HCV.
To gain a better understanding of the molecular mechanism by which IFN-α controls HCV infection, we attempted to identify the IFN-induced cellular proteins that mediate the antiviral response of the cytokine. In the studies presented herein, we took advantage of a commercially available human embryonic kidney (HEK293)-derived cell line, FLP-IN T Rex (Invitrogen), for inducible ISG expression and replication of HCV subgenomic replicons in HEK293 cells to determine the antiviral effects of individual ISGs. Among 29 ISGs tested (see Table 1, below), we found that induction of PKR, ISG20, and viperin expression in HEK293 cells inhibited HCV replication in a noncytopathic fashion. Mechanistic studies revealed that inhibition of HCV replication by PKR, ISG20, and viperin depends on their protein kinase, 3′-5′ exonuclease, and putative radical S-adenosyl-l-methionine (SAM) enzymatic activities, respectively. Hence, our work suggests that the IFN response against HCV is mediated by at least three distinct cellular antiviral pathways.
TABLE 1.
Induction of ISGs in Huh7 cells by IFN-α and in HCV-infected human and chimpanzee livers
| ISG | GenBank accession no. | Reference(s) reporting induction by:
|
Effect on HCV in 293 cellsa | |
|---|---|---|---|---|
| IFN-α in Huh7 cells | HCV-infected livers | |||
| ISG56 | 001548 | 32, 39, 45, 49, 53 | 5, 6, 33, 39, 58, 61 | − |
| GBP1 | 002053 | 32, 39, 45 | 6, 35, 58 | − |
| ISG9-27 | 003641 | 32, 45, 49, 53 | 6, 35, 61 | − |
| 1-8D | 006435 | 53 | 6 | − |
| 1-8U | 021034 | 53 | − | |
| MTAP44 | D28915 | 32, 53 | 5, 6, 33 | − |
| ADAR1 | 001111 | 6, 35, 39 | − | |
| ISG20 | 002201 | 39, 45 | 6, 39 | + |
| Viperin | AF442151 | 33, 39, 49 | 6, 33, 61 | + |
| PKR | AH008429 | 32, 45, 53 | + | |
| ISG15 | 005101 | 32, 39, 45 | 5, 6, 33, 39, 58, 61 | − |
| OAS1 | 016816 | 32, 39, 45 | 5, 6, 33, 39, 61 | − |
| OAS1 | 002534 | 32, 39, 45, 53 | 5, 6, 39, 61 | − |
| OAS-L | 003733 | 32, 39, 45 | 6, 35, 61 | − |
| OAS-L | 198213 | 32, 39, 45 | 6, 35, 61 | − |
| PLSCR1 | 021105 | 39, 45, 53 | 6, 39 | − |
| MAPK8 | 002750 | 32 | − | |
| SAMHD1 | 015474 | 32 | 33, 39 | − |
| IFI44 | 006417 | 39, 45, 49 | 6, 61 | − |
| IFI44L | 006820 | 39 | 39 | − |
| UBE2L6 | 004223 | 32, 45 | 6, 39, 61 | − |
| USP18 | 017414 | 32, 39, 45 | 6, 39 | − |
| BST2 | 004335 | 45, 53 | 35 | − |
| FLJ20637 | AK000644 | 53 | 6, 61 | − |
| FLJ38348 | AK095667 | 53 | 6 | − |
| STAF50 | X82200 | 39, 45 | 5, 6, 33, 39, 58 | − |
| ISG12 | BN000227 | 39 | 6, 39 | − |
| FLJ20035 | AK000042 | 45 | 6 | − |
| C1ORF9 | DQ053381 | 45 | − | |
+ indicates the ISG inhibits the replication of HCV replicons.
MATERIALS AND METHODS
Cell culture.
Huh7 cells were cultured in a complete Dulbecco's modified Eagle's medium (DMEM) that includes DMEM supplemented with 10% fetal bovine serum (FBS), penicillin G, streptomycin, nonessential amino acids, and l-glutamine. The HEK293-derived cell line FLP-IN T Rex (Invitrogen) was maintained in complete DMEM containing 10 μg/ml blasticidin and 5 μg/ml zeocin. HCV subgenomic replicon-containing Huh7 (GS4.1) and HeLa (SL1) cell lines were described previously (28, 73). HEK293HCVrep was derived from a single G418-resistant clone of SL1 total RNA-transfected FLP/IN T Rex cells. All three replicon-containing cell lines were cultured with complete DMEM containing 500 μg/ml G418.
cDNA cloning and plasmid construction.
To obtain cDNA clones of human IFN-α-stimulated genes, Huh7 cells were treated with 500 IU/ml of IFN-α for 6 h, and total cellular RNA was extracted with TRIzol reagent (Invitrogen). First-strand cDNA was made with an oligo(dT)12-18 primer and SuperScript III DNA polymerase (Invitrogen) by following the manufacturer's directions. Full-length cDNA of each of 29 individual ISGs with an N-terminal FLAG tag (or C-terminal FLAG and V5 tags) was amplified by PCR (primer sequences are available upon request). The purified PCR fragments were digested with restriction enzymes Afl II and NotI and cloned into a pcDNA5/FRT/ΔCAT vector that was derived from pCDNA5/FRT/CAT (Invitrogen) by removing the chloramphenicol acetyltransferase (CAT)-coding sequence with ApaI and XhoI digestion and self-ligation. The identity of each cDNA clone was verified by nucleotide sequence analysis. FLAG-tagged viperin, ISG20, and PKR point mutants were constructed by overlap extension PCR. Briefly, two separate PCRs were performed to amplify two overlapping fragments of the coding region of the target molecule using four primers. The point mutations were introduced by the two middle primers. The final PCR products were cloned into the vector pcDNA5/FRT/ΔCAT. N-terminal FLAG-tagged N- or C-terminally truncated viperin mutants were constructed by cloning the PCR products, amplified with a pair of primers bracketing the desired regions of viperin cDNA, into the pcDNA5/FRT/ΔCAT vector. All resulting DNA clones were sequenced to verify the desired mutation(s).
Establishment of stable cell lines that inducibly express individual ISGs.
FLP-IN T Rex (Invitrogen) cells were cotransfected with a pcDNA5/FRT/ΔCAT-derived individual ISG expression plasmid and pOG44 (Invitrogen) at a molar ratio of 1:1. Two days after transfection, cells were trypsinized and reseeded at less than 25% confluence. The ISG cDNA-integrated cells were selected with 250 μg/ml hygromycin and 5 μg/ml blasticidin. Two weeks later, separate colonies appeared, and the pool of such cells was expanded to generate cell lines that express ISG proteins upon the addition of tetracycline into the culture medium. To assure tight control of ISG expression by tetracycline, the ISG expression cell lines were selected and maintained in DMEM supplemented with 10% certified tetracycline-free FBS (HyClone). Expression of the desired ISG by each cell line was confirmed by Western blot analysis with an antibody against FLAG or V5 epitope tag.
RNA electroporation, colony formation efficiency assay, and establishment of the HCV replicon cell line that inducibly expresses ISG.
To determine the effects of each ISG on HCV replication, the individual ISG-expressing cell lines established above were transfected with total cellular RNA extracted from SL1 cells. G418-resistant colonies were selected with G418 in the presence and absence of 1 μg/ml tetracycline. Briefly, subconfluent ISG-expressing cells maintained in tetracycline-free medium were trypsinized and washed once with complete DMEM and twice with serum-free DMEM-F-12 medium. Cells were then resuspended in serum-free DMEM-F-12 at a concentration of 1 × 107 cells/ml. Ten micrograms of SL1 total cellular RNA was mixed with 200 μl of cells in a 2-mm gap cuvette (BTX) and immediately pulsed with the parameters described previously (28). The pulsed cells were left at room temperature for 10 min and then diluted into 10 ml DMEM-10% tetracycline-free FBS and plated into 100-mm-diameter dishes. Forty-eight hours after plating, medium was changed with complete DMEM containing 500 μg/ml G418 with or without 1 μg/ml tetracycline. Two to 3 weeks later, G418-resistant colonies were picked from one of the plates selected without tetracycline and expanded into HCV replicon-containing cell lines that inducibly express the ISG, and the remaining plates were fixed with 10% formaldehyde and stained with 1% crystal violet in 50% ethanol to facilitate colony counting.
RNA extraction and Northern blot hybridization.
Total cellular RNA was extracted with TRIzol reagent (Invitrogen) by following the manufacturer's directions. Ten micrograms of total RNA was fractionated on a 1% agarose gel containing 2.2 M formaldehyde and transferred onto nylon membranes. Membranes were hybridized with riboprobes specific for plus-stranded HCV replicon RNA and β-actin mRNA under the conditions described previously (28).
Western blot assay.
Cell monolayers were washed once with phosphate-buffered saline buffer and lysed with 1× Laemmli buffer. A fraction of cell lysate was separated on sodium dodecyl sulfate-12% polyacrylamide gels and electrophoretically transferred onto a polyvinylidene difluoride membrane (Bio-Rad). Membranes were blocked with phosphate-buffered saline containing 5% nonfat dry milk and probed with antibodies against FLAG tag (catalog no. 9272; Sigma), V5 epitope tag (Invitrogen), HCV NS5A (a gift of Chen Liu, Florida State University, Jacksonville), and β-actin (Chemicon International). Bound antibodies were revealed by horseradish peroxidase-labeled secondary antibodies and visualized with an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech) according to the protocol of the manufacturer.
RESULTS
HCV replicons efficiently replicate in FLP-IN T Rex cells.
Although HCV has been considered primarily a hepatotropic virus, the subgenomic replicons derived from both HCV genotypes 1b and 2a have been demonstrated to replicate in a variety of nonhepatocyte-derived cell lines, such as HeLa, HEK293, and murine MEFs, following introduction of the replicons into the cells by electroporation (1, 11, 36, 73). As observed in human hepatoma Huh7 cells, the replication of HCV replicons in those cell lines can be efficiently inhibited by IFN-α (1, 11, 30).
In previous studies, the antiviral effects of individual IFN-induced cellular genes were usually assayed by two complementary methods: the overexpression or ablation of an individual ISG by transient transfection of ISG cDNA, or siRNA targeting the ISG mRNA (62, 64). Because in those transient-transfection experiments only a part of the cell population was transfected and expressed the transgene, the antiviral effects observed under those conditions may have been underestimated. For example, if only 50% of HCV replicon-containing cells were transfected, a maximal onefold change of HCV RNA and protein levels could be possibly detected by Northern blot hybridization and Western blot assays. Moreover, the antiviral effects of IFN-α are quite possibly mediated by multiple antiviral pathways (70) and, hence, ablation of a single ISG's function by siRNA could potentially be compensated by other antiviral pathways. To circumvent these problems, we intended to establish HCV replicon-containing cell lines that inducibly expressed individual ISGs, thereby allowing more careful evaluation of the effects on HCV replication.
In searching for potential cell lines that could support efficient replication of HCV replicons and also be conveniently engineered for inducible expression of ISGs, we speculated that a HEK293-derived cell line, FLP-IN T Rex (Invitrogen), might be a good candidate for this purpose (10). The genome of this cell line contains stable integrations of a single FLP recombination target (FRT) site and a gene that expresses a TET repressor. Cotransfection of the cells with a FRT site-containing plasmid (pcDNA5/FRT/ISG) that encodes ISG and plasmid pOG44 that expresses Flp IN recombinase results in the integration of ISG cDNA through the FRT site with its expression under the control of TET-on promoter.
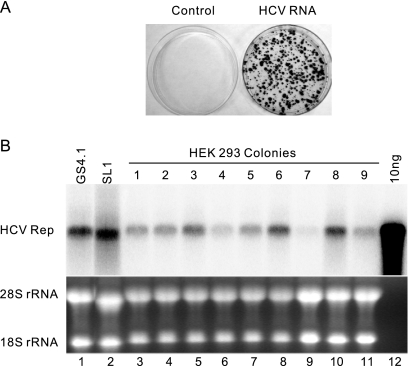
To ensure that FLP-IN T Rex cells can support the replication of HCV subgenomic replicons, the cells were transfected with total RNA extracted from parental HeLa cells or a HeLa cell-derived cell line that replicates HCV genotype 1b (Con 1 strain) subgenomic replicons (SL1) (73). The transfected cells were selected with G418. After 2 to 3 weeks, G418-resistant colonies were observed only in SL1 but not parental HeLa cellular RNA-transfected plates. The colony formation efficiency was 300 to 1,000 colonies per 10 μg of total SL1 RNA (Fig. 1A), which is at least 10-fold higher than that observed with HEK293 cells transfected with Huh7-derived HCV replicon RNA, as reported by Ali and colleagues (1). Individual colonies were picked and expanded into cell lines (HEK293HCVrep). HCV RNA replication and protein expression in those cell lines were confirmed by Northern and Western blot assays (Fig. 1B and data not shown). The levels of HCV RNA in HEK293HCVrep cell lines were similar to those observed in HCV replicon-containing Huh7 (GS4.1) (30) and HeLa (SL1) cells. In agreement with the previous report (1), we demonstrated that IFN-α efficiently inhibited HCV replication in HEK293HCVrep cell lines, and the 50% inhibitory concentration under the conditions that cells were treated with the cytokine for 3 days was 2 IU/ml (data not shown).
FIG. 1.
HCV replicons efficiently replicate in FLP-IN T Rex-derived cells. (A) Cell colony formation conferred by SL1 cell-derived HCV replicon replication. FLP-IN T Rex cells were electroporated with total RNA extracted from parental HeLa cells (left) and SL1 cells (right) and selected with medium containing 500 μg/ml of G418 for 3 weeks. Cell foci were stained with crystal violet, and a photograph is shown. (B) GS4.1 and SL1 are Huh7- and HeLa-derived cell lines expressing HCV subgenomic replicons, respectively. Ten micrograms of total RNA isolated from GS4.1, SL1, and nine HEK293HCVrep (lane 3 to 11) cell lines that were established from G418-resistant cell colonies of FLP-IN T Rex cells transfected with SL1-derived RNA was analyzed by Northern blot analysis with an [α-32P]UTP-labeled riboprobe that is complementary to the plus strand of the HCV NS3-coding region. In vitro-transcribed HCV replicon RNA (10 ng; lane 12) served as a molecular weight marker (M) and hybridization control. rRNAs served as loading controls.
Establishment of HEK293 cell lines that inducibly express individual IFN-α-induced cellular proteins.
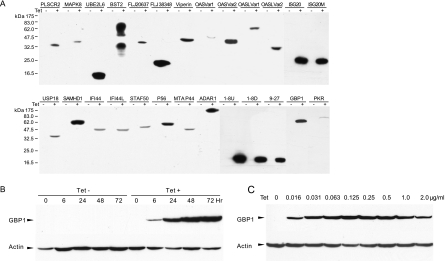
To determine the effects of ISGs on HCV replicon replication, we first established FLP-IN T Rex-derived cell lines that inducibly expressed individual ISGs. Twenty-nine ISGs were chosen for this study, based on their ability to be induced by IFN-α in Huh7 cells and/or up-regulated in HCV chronically infected human and chimpanzee livers (Table 1). Stable cell lines that inducibly express each of the 29 ISGs were established, as described in Materials and Methods. The resulting cell lines were designated with a suffix identifying the specific ISG they contain, such as FLP-IN/p56, where p56 is the ISG. Expression of the desired N-terminal FLAG-tagged ISG proteins by each cell line was confirmed in a Western blot assay (Fig. 2A). The expression profile of ISG proteins upon induction by tetracycline was characterized in detail by using the FLP-IN/GBP1 cell line as an example. As shown in Fig. 2B, the expression of GTP binding protein 1 (GBP1) was undetectable in the cells maintained in tetracycline-free medium. Upon the addition of the antibiotic, GBP1 was induced quickly and became detectable at 6 h after induction. The steady-state level of GBP1 was reached at 48 h after induction. The results presented in Fig. 2C demonstrate that GBP1 expression in this cell line is tightly controlled by tetracycline and can be efficiently induced by as little as 0.016 μg/ml of the antibiotic. Because many lots of commercial FBS are contaminated with a low concentration of tetracycline, all cell lines used in this study were cultured with certified tetracycline-free FBS to ensure the tight control of ISG expression.
FIG. 2.
Characterization of ISG expression following tetracycline induction in FLP-IN/ISG cell lines. (A) Establishment of stable cell lines that inducibly express individual ISG proteins. FLP-IN T Rex cell lines that inducibly express individual ISGs were established as described in Materials and Methods. The cells were cultured in the absence or presence of tetracycline for 48 h and then harvested. The levels of N-terminal FLAG-tagged ISG protein expression in cell lysates were determined by Western blot analysis with a monoclonal antibody against FLAG tag. (B) Kinetics of ISG induction. A stable cell line that expresses GBP1 (FLP-IN/GBP1) was cultured in the absence or presence of 1 μg/ml tetracycline, and cells were harvested at the indicated time after the addition of the antibiotic. The levels of GBP1 protein expression in cell lysates were determined by Western blot analysis with a monoclonal antibody against FLAG epitope tag (Sigma). (C) Dose response of tetracycline. FLP-IN/GBP1 cells were cultured in the absence and presence of indicated concentrations of tetracycline for 48 h, and cells were then harvested. The levels of GBP1 protein expression in cell lysates were determined by Western blot analysis. β-Actin served as a loading control and was detected by using a monoclonal antibody against human β-actin.
Effects of ISG expression on G418-resistant colony formation conferred by HCV subgenomic RNA transfection.
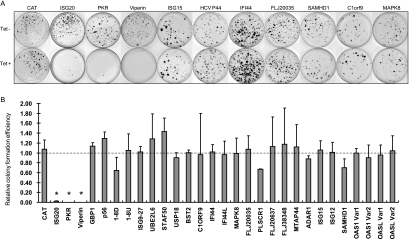
To investigate the effects of ISG expression on HCV replication, the individual ISG-expressing cell lines (FLP-IN/ISG) described above were transfected with total cellular RNA extracted from SL1 cells. HCV replicon-replicating cells were selected with 500 μg/ml of G418 in the presence or absence of 1 μg/ml tetracycline. Two to 3 weeks after the selection, G418-resistant cell colonies became visible and were stained with crystal violet to facilitate colony counting. Photographs of the representative plates from SL1 RNA-transfected FLP-IN/ISG cell lines and a cell line that inducibly expresses CAT, serving as a negative control, are presented in Fig. 3A. The relative colony formation efficiency (RCFE) was expressed as a ratio of the colony number obtained in cells selected with G418 in the presence of tetracycline over that obtained from cells that were selected with G418 in the absence of tetracycline. Thus, the value of RCFE should be 1 if the ISG does not inhibit HCV replication or <1 if the ISG inhibits HCV replication. Average RCFE values obtained from three plates for each of 30 cell lines are presented in Fig. 3B.
FIG. 3.
Effects of ISG expression on HCV replicon-dependent colony formation. (A) Effects of ISG expression on colony formation. CAT- and ISG-expressing FLP-IN T Rex stable cell lines were electroporated with total RNA extracted from SL1 cells and selected with G418 in the absence or presence of 1 μg/ml tetracycline for 2 to 3 weeks as described in Materials and Methods. Cell foci were stained with crystal violet and photographed. A pair of the representative plates from each of the 10 ISG- and control protein CAT-expressing cell lines that were cultured in the absence (upper panel) or presence (lower panel) of tetracycline is presented. (B) Effects of ISG expression on efficiency of HCV replicating cell colony formation. The numbers of cell foci were counted from three plates cultured in either the absence or presence of tetracycline from each of the ISG- and CAT-expressing cell lines. The RCFE was expressed and plotted as a ratio of the cell foci number obtained from cells that were selected in the presence of tetracycline over that obtained from cells that were cultured in the absence of the antibiotic. *, P < 0.01.
The results showed that while the expression of a control protein, CAT, as well as the majority of ISG proteins did not significantly affect the efficiency of HCV replicon-replicating cell colony formation, induction of viperin expression completely prevented colony formation. Furthermore, colony formation efficiency was decreased approximately 50- and 500-fold in cells in which expression of ISG20 and PKR was induced, respectively.
Effects of viperin, ISG20, and PKR expression on cell growth.
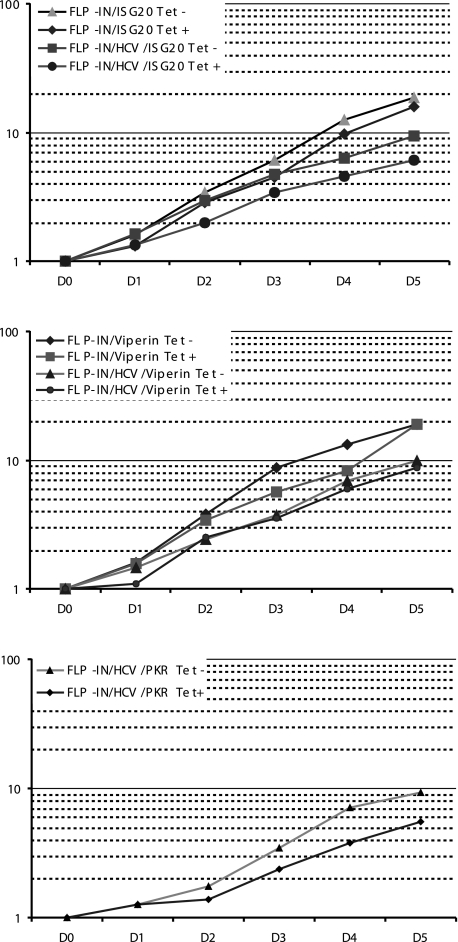
The reduction of colony formation efficiency in cultured cells associated with expression of viperin, ISG20, or PKR could be due to either the inhibition of HCV replication or cell death induced by the expression of the ISG proteins. To distinguish between those two possibilities, individual G418-resistant cell colonies were picked from plates selected with G418 in the absence of tetracycline and expanded into cell lines designated as FLP-IN/HCV/ISG (such as FLP-IN/HCV/viperin). Effects of each of the three ISGs on the growth of both parental FLP IN/ISG and FLP IN/HCV/ISG cells were examined by culturing the cells in the absence or presence of tetracycline for 5 days, respectively. Growth of the cells was monitored daily by cell counting, and average rates of cell growth related to cell number seeded were plotted (Fig. 4). These results clearly demonstrated that, although the growth of HCV replicon-containing cells was generally slower than in cells that did not harbor replicons, expression of viperin or ISG20 in either HCV replicon-containing or replicon-free cells did not significantly affect cell growth. Consistent with previous reports (26), expression of PKR in HCV replicon-containing cells modestly inhibited cell growth, but apparent cell death was not observed in a trypan blue exclusion staining assay (data not shown). Furthermore, when the three ISG-expressing cell lines were cultured in the presence of tetracycline and maintained through three passages for 3 weeks, all three cell lines remained viable. Hence, the results imply that the reduction of colony formation efficiency by expression of the three ISGs is most possibly due to their ability to inhibit HCV replication, rather than direct cytotoxicity.
FIG. 4.
Effects of viperin, ISG20, and PKR on cell growth. Indicated cell lines were seeded in six-well plates at a density of 1 × 105 cells per well in medium with or without 1 μg/ml tetracycline. Three wells of cells from each of the cell lines cultured with or without tetracycline were trypsinized at 1, 2, 3, 4, and 5 days after seeding. Average rates of cell growth related to cell numbers seeded were plotted.
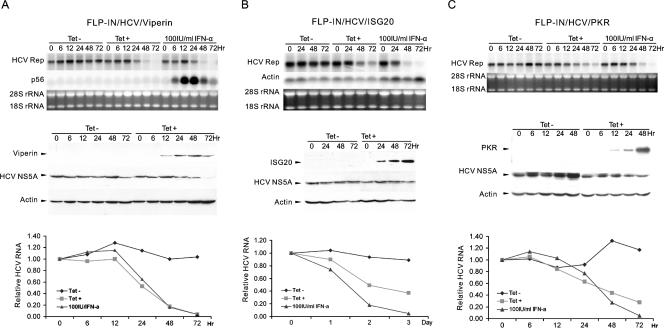
Induction of viperin, ISG20, or PKR potently inhibits HCV replication.
To directly determine the effect of ISG expression on HCV replication, three HCV replicon-containing cell lines (FLP-IN/HCV/ISG) that express viperin, ISG20, or PKR were cultured in the absence or presence of tetracycline for 3 days. Cells were harvested at the indicated time points. Total cellular RNA was extracted, and the levels of intracellular HCV RNA were determined by Northern blot hybridization (Fig. 5A, B, and C, upper panels). Expression of the desired ISGs and levels of HCV NS5A protein in the cell lines were determined by Western blot analysis (Fig. 5A, B, and C, middle panels). In agreement with the results obtained from the colony formation assay presented above, expression of viperin (Fig. 5A), ISG20 (Fig. 5B), or PKR (Fig. 5C) significantly reduced the intracellular levels of HCV RNA in a time-dependent manner. HCV protein NS5A expression was also reduced following the induction of viperin but was less profoundly affected in ISG20- and PKR-expressing cell lines (Fig. 5A, B, and C, middle panels). Among those three ISGs, viperin appeared to be the most potent inhibitor of HCV replication. As shown in Fig. 5A, the rate of the reduction of HCV RNA following viperin induction was similar to that achieved with 100 IU/ml IFN-α treatment (Fig. 5A, lower panel). In addition, consistent with the results obtained from the colony formation assay, expression of CAT and the other ISG proteins did not affect the levels of HCV RNA in HEK293 cells (data not shown).
FIG. 5.
Effects of viperin, ISG20, and PKR expression on replication of HCV replicons in HEK293 cells. FLP-IN/HCV/viperin (A), FLP-IN/HCV/ISG20 (B), and FLP-IN/HCV/PKR (C) cell lines were cultured in the absence or presence of 1 μg/ml tetracycline or treated with 100 IU/ml IFN-α in the absence of the antibiotic for 3 days. Cells were harvested at the indicated time points. Ten micrograms of total RNA was analyzed by Northern blot hybridizations with an [α-32P]UTP-labeled riboprobe that is complementary to the plus strand of the HCV NS3-coding region. rRNA and β-actin mRNA served as loading controls (upper panels of A, B, and C). Expression of viperin, ISG20, and PKR and levels of HCV NS5A in the cell lysates were determined by Western blot analyses with antibodies against FLAG tag or HCV NS5A. β-Actin served as a loading control (middle panels of A, B, and C). The amounts of HCV RNA were quantified with the help of a Quantity-One PhosphorImager (Bio-Rad), and the values were plotted as the fraction (percent) of the values obtained with pretreated cells (lower panels of A, B, and C).
Inhibition of HCV replication by PKR depends on its protein kinase activity.
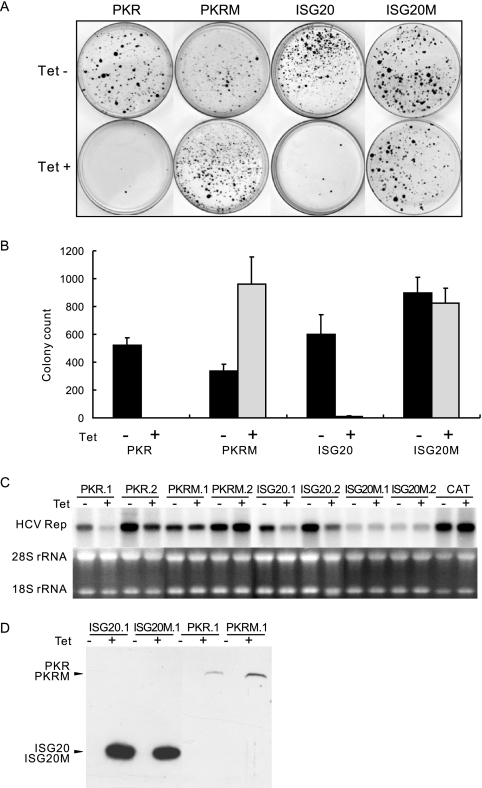
PKR is a double-stranded RNA-dependent protein kinase. The best-studied target of PKR is the alpha subunit of eukaryotic translation initiation factor 2 (eIF-2α). Upon its activation by double-stranded RNA, PKR phosphorylates eIF-2α on Ser51 and consequently causes a general reduction in protein translation (16, 66). To determine if the inhibitory effect of PKR on HCV replication depends on its protein kinase activity, the conserved lysine residue in the ATP binding pocket of the enzyme was replaced with an arginine residue to yield a dominant-negative form of a PKR variant, PKRK296R (13, 14). A cell line that inducibly expresses PKRK296R was established and designated FLP-IN/PKRM. The cells were transfected with total cellular RNA extracted from SL1 cells, and HCV replicon-replicating cells were selected with 500 μg/ml of G418 in the presence or absence of 1 μg/ml tetracycline. As expected, there were few colonies formed under conditions where wild-type PKR was induced. But, interestingly, the colony formation efficiency was increased approximately threefold under conditions where PKRK296R expression was induced (Fig. 6A and B). This result suggests that PKRK296R regulates endogenous PKR function in a dominant-negative fashion and, thereby, enhances the replication of HCV replicons.
FIG. 6.
Requirements of the enzymatic activities of PKR and ISG20 on their antiviral effects against HCV. (A) Stable FLP-IN T Rex-derived cells lines that inducibly express N-terminal FLAG-tagged wild-type and enzymatically inactive mutant PKR (PKRM) and ISG20 (ISG20M) were electroporated with total RNA extracted from SL1 cells and selected with G418 in the absence or presence of 1 μg/ml tetracycline for 3 weeks as described in Materials and Methods. Cell foci were stained with crystal violet and photographed. A pair of the representative plates from each of cell lines that were cultured in the absence (upper panel) or presence (lower panel) of tetracycline is presented. (B) Averages and standard derivations of cell foci numbers obtained from three plates of each cell line under the indicated selection conditions were plotted. (C) Two independent HCV replicon-containing cell lines that inducibly express wild-type and mutant PKR or ISG20 were cultured in the absence or presence of 1 μg/ml tetracycline for 3 days. Cells were then harvested, and 10 μg of total RNA was analyzed by Northern blot hybridizations with an [α-32P]UTP-labeled riboprobe that is complementary to the plus strand of the HCV NS3-coding region. rRNA served as a loading control. (D) Inducible expression of wild-type and mutant PKR and ISG20 proteins in the stable cell lines was determined by Western blot analysis with a monoclonal antibody against FLAG tag.
To further examine the effects of endogenous PKR on HCV replication, individual G418-resistant colonies were picked from plates selected in the absence of tetracycline and expanded into cell lines. The cells were then cultured in the absence or presence of tetracycline for 3 days. Levels of intracellular HCV RNA were determined by Northern blot hybridization. Consistent with the results presented above, expression of wild-type PKR significantly reduced the levels of HCV RNA, but ironically, the levels of HCV RNA in cells expressing PKRK296R were not affected upon the addition of tetracycline (Fig. 6C).
That the induction of the dominant-negative form of PKR only promotes colony formation, but fails to increase HCV RNA levels in cells supporting persistent HCV RNA replication, implies that constitutively expressed endogenous PKR may control the establishment of HCV replication in newly infected (or transfected) cells but does not play a significant role in controlling HCV replication once a persistent infection is fully established. This interpretation is consistent with our previous observation that levels of cellular eIF-2α phosphorylation are not increased in the majority of HCV replicon-containing Huh7 and HeLa cell lines examined (30).
Inhibition of HCV replication by ISG20 depends on its 3′-to-5′ exonuclease activity.
ISG20 is a 3′-to-5′ exonuclease with a strong preference for single-stranded RNA and minor activity toward single-stranded DNA (51). According to the bioinformatic analysis and recently published crystal structural data, ISG20 belongs to the DEDD superfamily of 3′-to-5′ exonucleases that is defined by four conserved acidic residues, three aspartates (D) and one glutamate (E), distributed among three separate sequence motifs (Exo I to III) (19, 21, 34, 74). Previous studies demonstrated that replacement of Asp96 in Exo II with a glycine inactivates the enzymatic activity of ISG20 (19). As shown in Fig. 6A, B, and C, compared with wild-type ISG20, the ISG20D96G mutant (ISG20M) completely lost the antiviral effects against HCV, suggesting that the 3′-to-5′ exonuclease activity of ISG20 is essential to inhibit HCV replication. Interestingly, as shown in Fig. 5B (upper panel), despite the significant reduction of HCV RNA levels in wild-type ISG20-expressing cells, the level of β-actin mRNA in the same cells was not apparently affected, indicating that ISG20 selectively attacks viral RNA but not cellular mRNA. Further studies on the molecular mechanism of the substrate selection of ISG20 exonuclease are clearly warranted.
Viperin is a putative radical SAM enzyme.
The results presented above and work published previously indicate that viperin is an important mediator of the interferon response against HCV and some other viruses (33, 54, 67). Thus far, the molecular mechanism by which viperin inhibits virus infection remains elusive. An interesting possibility is that viperin might induce the expression of other antiviral proteins, such as interferons, and indirectly inhibit virus replication. However, the results presented in Fig. 5A (upper panel) indicate that while IFN-α treatment of cells efficiently induces ISG p56 expression, induction of viperin does not induce the expression of this ISG. Furthermore, culture of HCV replicon-containing cells, such as HEK293HCVrep and GS4.1, with medium supplemented with an equal volume of culture fluid harvested from FLP-IN/HCV/viperin cells cultured in the presence of tetracycline did not affect HCV replication (data not shown). These results suggest that the antiviral effects of viperin are not mediated by the induction of interferon and/or other antiviral cytokines.
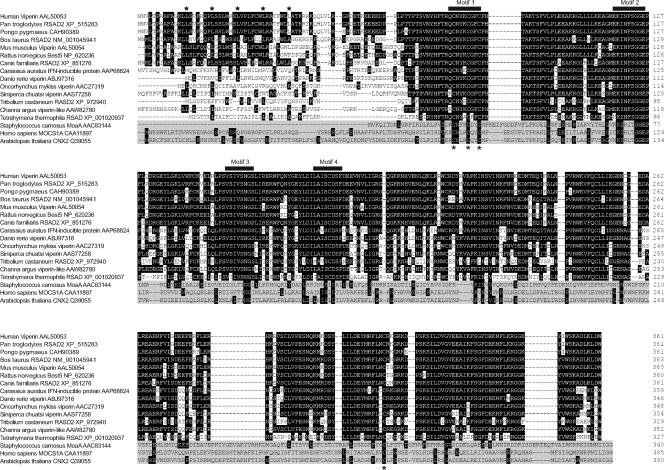
Interestingly, the sequence alignment analysis by Boudinot and colleagues revealed that viperin contains conserved motifs found in enzymes required for the synthesis of molybdopterin (Mao A), heme D1 (NIRJ), and PQQ (PQQIII) (8). Those enzymes were recently categorized into a family of more than 600 putatively related enzymes, denoted as radical SAM enzymes (40, 59, 65). As with viperin, the precise chemical reactions catalyzed by the vast majority of radical SAM enzymes remain to be discovered (24). Based on the catalytic mechanisms revealed from studies on a few radical SAM enzymes (4, 41), they all share one common feature, an unconventional [4Fe-4S] cluster coordinated by three rather than four closely spaced cysteine residues in a CxxxCxxC motif (Fig. 7). Furthermore, radical SAM enzymes require their cofactor or cosubstrate SAM to be placed in the immediate vicinity of the [4Fe-4S] cluster to directly coordinate the cluster and allow electron transfer from one to the other. Hence, the conserved motif I is essential for the enzymatic activity and is considered to be one of the sequence signatures of radical SAM enzymes (18, 38, 40, 41).
FIG. 7.
Multiple alignments of amino acid sequences of viperins from 14 animal species and three representative radical SAM enzymes. The names of the species and enzymes and the GenBank accession numbers of their proteins are indicated on the left. The putative leucine zip and four conserved motifs identified in radical SAM enzymes are highlighted. The sequences of the three radical SAM enzymes are shaded.
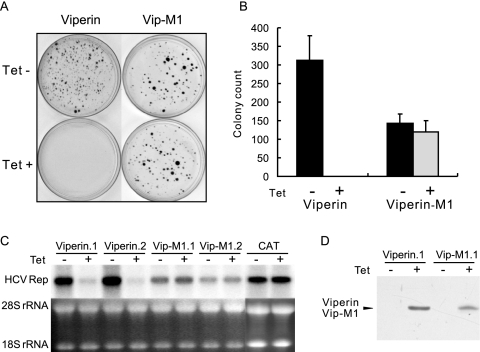
To determine if this motif is essential for the antiviral activity of viperin, the three cysteine residues in the conserved motif I of viperin protein were replaced with three alanines to yield a mutant viperin, viperin M1. As shown in Fig. 8, consistent with the results presented above, expression of wild-type viperin completely prevented HCV replicon-containing cell colony formation and inhibited HCV RNA replication, but the mutant protein completely lost antiviral effects against HCV (Fig. 8A, B, and C). Proper expression of wild-type and mutant viperin proteins was confirmed in a Western blot assay (Fig. 8D). These results strongly suggest that viperin is a radical SAM enzyme and its antiviral activity depends on its enzymatic activity. To our knowledge, this is the first experimental demonstration of a possible enzymatic function of viperin.
FIG. 8.
Viperin is a putative radical SAM enzyme. (A) Stable FLP-IN T Rex-derived cell lines that inducibly express N-terminal FLAG-tagged wild-type viperin and the mutant viperin M1 were electroporated with total RNA extracted from SL1 cells and selected with G418 in the absence or presence of 1 μg/ml tetracycline for 3 weeks as described in Materials and Methods. Cell foci were stained with crystal violet and photographed. A pair of the representative plates from each cell line that were cultured in the absence (upper panel) or presence (lower panel) of tetracycline is presented. (B) Averages and standard derivations of cell foci numbers obtained from three plates of each cell line under the indicated selection conditions were plotted. (C) HCV replicon-containing cell lines that inducibly express wild-type and M1 mutant viperin were cultured in the absence or presence of 1 μg/ml tetracycline for 3 days. Cells were then harvested, and 10 μg of total RNA was analyzed by Northern blot hybridizations with an [α-32P]UTP-labeled riboprobe that is complementary to the plus strand of the HCV NS3-coding region. rRNA served as a loading control. (D) Inducible expression of wild-type and M1 mutant viperin in the stable cell lines was determined by Western blot analysis with a monoclonal antibody against FLAG tag.
Structural and function analysis of viperin protein.
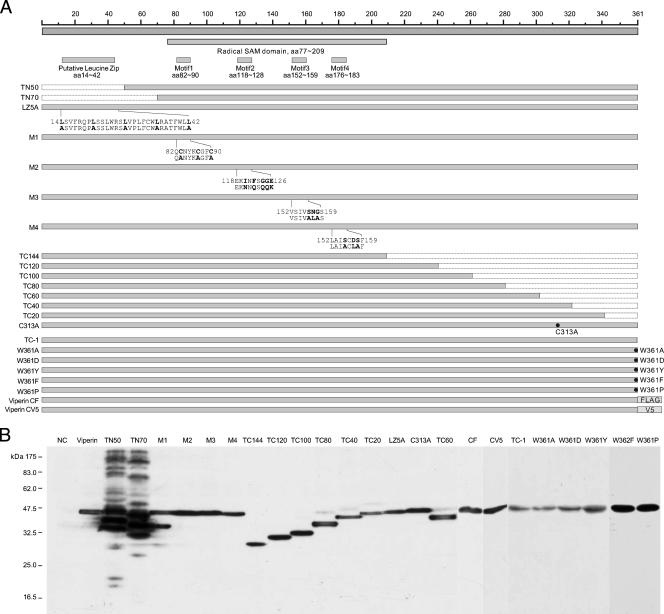
Protein database searching revealed that viperin proteins are encoded by vertebrates ranging from fish to human. Multiple sequence alignments of viperins from 14 species indicate that, with the exception of the amino-terminal 77 amino acid residues, these sequences are highly conserved (Fig. 7). The four conserved motifs (motifs I to IV) common to radical SAM enzymes are conserved in viperins of all species examined. Based on the sequence alignments, the structure of viperins can be designated as have an N-terminal variable domain (amino acids [aa] 1 to 76), followed by a radical SAM domain (aa 77 to 209) in the middle and a C-terminal conserved domain (aa 210 to 361) (Fig. 7 and 9A). In the N-terminal variable region, a putative transmembrane domain spanning approximately the first 50 amino acid residues is predicted by the TMpred server (http://www.ch.embnet.org/software/TMPRED_form.html). In addition, four leucine-6x-leucine repeats (leucine zip motif) are found in human, chimpanzee, and mouse viperins, but not in viperins encoded by fish and several other lower species (Fig. 7).
FIG. 9.
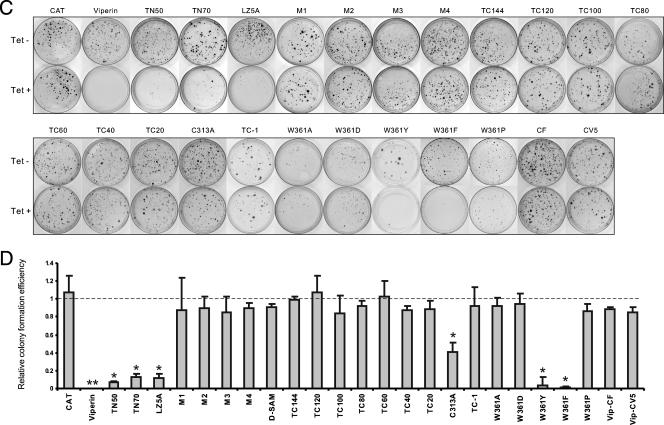
Structural and functional analysis of viperin. (A) Schematic representation of the structures of viperin and its mutants. The top panels show the overall structural organization of viperin. The positions of the putative leucine zip motif, radical SAM domain, and four conserved motifs identified in radical SAM enzymes are indicated. The lower panels highlight the nature of the mutations in 23 viperin mutants, which include sequential deletions from the N terminal (TN50 and TN70) and C terminal (TC1 to TC144), point mutations in the four conserved motifs of radical SAM enzymes (M1 to M4), a leucine zip motif (LZ5), and Cys313 (C313A) and Trp361 at the C terminus (W361A, W361D, W361Y, W361F, and W361P). The nature of the point mutations is highlighted in bold underneath the sequences. In addition, two mutant viperins were constructed by adding FLAG or V5 epitope tag at the C terminus (CF and CV5). (B) Inducible expression of mutant viperins in FLP-IN T Rex cells. Cells were transfected with plasmids expressing individual mutant viperins and cultured in the presence of 1 μg/ml of tetracycline for 2 days, and the levels of FLAG-tagged viperin and its mutants in cell lysates were determined by Western blot analysis with a monoclonal antibody against FLAG tag. C-terminal V5-tagged viperin was detected with an antibody against V5 tag. (C) Stable cell lines that inducibly express CAT or wild-type and mutant viperins were electroporated with total RNA extracted from SL1 cells and selected with G418 in the absence or presence of 1 μg/ml tetracycline for 3 weeks as described in Materials and Methods. Cell foci were stained with crystal violet and photographed. A pair of the representative plates from each of 25 cell lines that were cultured in the absence (upper panel) or presence (lower panel) of tetracycline is presented. (D) The numbers of cell foci were counted from three plates from each of the cell lines cultured under both selection conditions. RCFE is defined in the legend of Fig. 3. *, P < 0.05; **, P < 0.01.
To determine the structural and functional relationship of viperin, plasmids that express mutant viperins with (i) N-terminal and C-terminal sequential deletions, (ii) mutations of three other conserved motifs (motifs II to IV) in the radical SAM domain which are also essential for binding of the [4Fe-4S] cluster and SAM, (iii) mutations of the putative leucine zip motif in the N-terminal variable region, and (iv) a point mutation of the conserved Cys313 (C313A) were constructed (Fig. 9A). The plasmids were transfected into FLP-IN T Rex cells and cultured in medium containing 1 μg/ml of tetracycline for 2 days. The expression of the desired mutant viperins was confirmed by Western blot analysis (Fig. 9B). Stable cell lines expressing each of the individual mutant viperins were established as described above and were electroporated with total RNA extracted from SL1 cells, and G418-resistant colonies were selected in the absence and presence of tetracycline, respectively.
The results (Fig. 9C and D) of the colony formation assay revealed the following observations. First, in addition to motif I, all three other conserved motifs in the SAM radical domain of viperin are essential for its antiviral effects. Second, despite the weaker antiviral effects in comparison with the wild-type viperin, mutant viperins harboring deletions of the N-terminal 50 or 70 amino acid residues still significantly reduced the efficiency of HCV replicon-containing cell colony formation. Similarly, the antiviral effect of a mutant viperin containing leucine-to-alanine mutations in the putative leucine zip motif (LZ5A) was also significantly reduced in comparison with the wild-type viperin. As shown in Fig. 9B, deletions of the N-terminal variable region of viperin resulted in the accumulation of mutant proteins in cells to a higher level and formation of high-molecular-weight aggregates, suggesting that this region, possibly the putative transmembrane domain and also the leucine zip structure, might be important for the proper folding and/or membrane association of the protein. Third, residue Cys313 is conserved among viperins of all species examined (Fig. 7). Interestingly, the colony formation efficiency was only slightly reduced (onefold) in cells that expressed C313A mutant viperin, indicating that the residue is important for the antiviral effects of the protein. Finally, the results obtained from a series of sequential C-terminal-truncated viperins revealed that the deletion of only one amino acid residue (Trp361, designated TC-1) from the C terminus resulted in complete loss of the antiviral effects of viperin, indicating that Trp361 at the C terminus, which is absolutely conserved in viperins examined from all the species (Fig. 7), is required for its antiviral function.
To further investigate the structural requirement of the C terminus, Trp361 was mutated into an alanine, aspartate, tyrosine, phenylalanine, or a proline residue to yield mutants W361A, W361D, W361Y, W361F, and W361P, respectively. The colony formation assay showed that although W361A, W361D, and W361P mutants did not inhibit HCV replication, the W361Y and W361F mutants significantly reduced the colony formation efficiency, but to a lesser extent, than that observed with the wild-type viperin (Fig. 9C and D). Furthermore, addition of epitope tags (FLAG and V5) at the C terminus of viperin to block its authentic C terminus resulted in a complete ablation of its antiviral activity (Fig. 9C and D). These results indicate that functional viperin requires an amino acid residue containing an aromatic ring at its C terminus.
DISCUSSION
Interferons are a family of multifunctional cytokines characterized by their ability to interfere with virus infection through inducing the expression of tens or even hundreds of IFN-stimulated genes (56, 60). In our efforts toward the identification of IFN-induced cellular genes that mediate the antiviral effects of IFN-α against HCV, we found that expression of three IFN-α-induced cellular proteins, PKR, ISG20, and viperin, noncytopathically inhibits the replication of HCV subgenomic replicons in HEK293-derived cells. The inhibitory effects of p56 and ADAR1 on HCV replicon replication had been suggested by previous studies (62, 64) but could not be observed under our experimental conditions. The reason for this discrepancy is currently not known. Interestingly, a recent report showed that ADAR1 can actually enhance replication of vesicular stomatitis virus (VSV) by interacting with PKR and thereby inhibiting its kinase activity (52).
PKR is one of the best-studied components of host cellular innate immunity to virus infections (3). In addition to being an eIF-2α kinase that regulates viral and cellular protein translation upon activation by double-stranded RNA and other factors, PKR also has a role in signal transduction and transcriptional control through the NF-κB pathway and in the modulation of cell growth, differentiation, apoptosis, and oncogenic transformation (26). While they are generally in agreement with the observations made for HCV genotype 2a (JFH1) replicon-containing MEFs that PKR inhibits HCV replication (11), our results further demonstrate that constitutively expressed endogenous PKR plays a role in controlling the initiation of HCV infection but does not affect HCV replication in replicon cells in which persistent viral replication is fully established. Interestingly, induction of PKR expression in replicon-containing cells does inhibit HCV replication. Furthermore, our results demonstrate that inhibition of HCV replication by PKR depends on its protein kinase activity and, therefore, it is most possibly the phosphorylation of eIF-2α that consequentially inhibits the translation initiation of HCV RNA (64).
ISG20 is a type I IFN-inducible 3′-5′ exonuclease that preferentially cleaves single-stranded RNA (51). It has been shown that this exonuclease is a component of the nuclear PML body (21, 27). Previous studies demonstrated that ISG20 expression confers resistance to the infection of various RNA viruses, including VSV and human immunodeficiency virus, but not DNA viruses, such as adenoviruses (19, 20, 68). Our work, for the first time, demonstrates that ISG20 also inhibits the replication of HCV. The prior studies, as well as ours, clearly demonstrate that the antiviral effects of ISG20 depend on its 3′-5′ exonuclease activity, suggesting that ISG20 may directly degrade viral RNA as observed with another type I IFN-activated cellular endonuclease, RNase L (68). If this is the case, a legitimate question is how ISG20 selectively targets viral but not cellular RNA, as demonstrated in this study. One possibility is that ISG20 forms a complex with other cellular proteins that guide ISG20 to viral RNA, essentially as observed with the zinc finger antiviral protein, which forms a complex with the cellular exosome and recruits the latter to retroviral and alphaviral RNA (31). This hypothesis will be tested in our future studies.
Viperin was initially identified as a human cytomegalovirus- and IFN-γ-inducible protein in fibroblasts (71, 72). Since then, it has been shown that the protein can be induced by the infection of many other viruses, such as VSV, dengue virus, yellow fever virus, human polyomavirus JC, and HCV, in cultured cells and in vivo (9, 12, 33, 37, 63). Detailed analyses of the viperin promoter sequence and transcription regulation mechanism reveal that viperin gene expression is activated by both Toll-like receptor 3-dependent and RIG-I-dependent pathways in a type I IFN signaling-dependent manner. The key factor that regulates viperin promoter activity is the ISGF3 complex, but not IRF3 (57). Those results are consistent with the observations suggesting that viperin is primarily a type I IFN-inducible gene.
The rapid and robust induction of viperin gene expression by a range of different viruses and its evolutionary conservation in vertebrates suggest that viperin is an important component of innate immunity against virus infections. Indeed, besides the potent antiviral effects against HCV demonstrated in this report, viperin has been shown to inhibit the infection of a few other DNA and RNA viruses, such as human cytomegalovirus (12), human immunodeficiency virus (54), Sindbis virus (67), and VSV (J.-T. Guo, unpublished observation). An important question that remains to be answered is how viperin inhibits such a diverse group of viruses. It is unlikely that viperin directly binds to viral proteins, thereby interfering with their functions. While our work rules out the possibility that viperin may indirectly inhibit HCV replication by inducing antiviral cytokines, we have obtained strong evidence to suggest that viperin is a radical SAM enzyme and that its antiviral function depends on its putative enzymatic activity. It has been shown that the radical SAM enzymes catalyze the biosynthesis of diverse types of molecules. Hence, a possible antiviral mechanism of viperin could be that the enzyme catalyzes the synthesis of a molecule that either directly inhibits viral replication or activates a latent cellular antiviral protein or pathway to indirectly limit viral replication. If this is indeed the case, identification of the molecule(s) synthesized by viperin might lead to the development of novel antivirals that activate the viperin pathway to control the infection of HCV and many other viruses but would be devoid of the side effects of IFN-α therapy.
In addition to demonstrating that the radical SAM domain, which contains essential motifs to coordinate the [4Fe-4S] cluster and cofactor SAM, is essential for the antiviral activity of viperin, our structural and functional analyses also revealed important roles of both N-terminal variable and C-terminal conserved domains of viperin in its antiviral function. Previous studies suggested that the heterogeneity of the SAM radical protein superfamily is most pronounced in the C-terminal region, which is responsible for binding of substrates and auxiliary cofactors. Our mutagenesis studies revealed that viperin requires an aromatic amino acid residue at its C terminus for proper antiviral function, suggesting that the C terminus of viperin might be involved in substrate recognition and/or interaction with partner proteins or cofactors that are essential for the enzymatic activity. Furthermore, the N-terminal region of viperin, which is highly variable among viperins from different species, contains a putative transmembrane domain and a leucine zip motif (Fig. 7), suggesting a role for this region in proper membrane association and/or subcellular localization of viperin (12). Our results indicate that although this region is not absolutely required for the antiviral function of viperin, deletion of this region or mutation of the putative leucine zip motif significantly reduces its antiviral activity. Further investigations toward understanding the role of the N-terminal variable region in protein folding, membrane association, and the relationship with antiviral activity of viperin are under way.
It is worthy of note that the FLP-IN T Rex cell-based ISG-inducible expression assay developed in this study has proven to be a valuable tool for dissecting IFN′s antiviral mechanism against HCV and should also be useful in the future for elucidating the molecular mechanisms by which IFN-mediated innate immunity controls the infection of other viruses. However, it is obvious that certain ISGs might work together and require other IFN-induced factors for their antiviral function and, thus, would not be identified as antiviral proteins by the single-ISG expression assay described in this report. Hence, the results obtained from this study should represent a minimum repertoire of IFN-induced antiviral pathways that limit HCV replication.
In conclusion, the work described here reports that inhibition of HCV replication by IFN-α is possibly mediated by at least three distinct antiviral pathways exemplified by viperin, ISG20, and PKR. Further studies toward understanding the role of those pathways and their underlying mechanisms in the innate host defense against HCV should provide valuable insight into HCV pathobiology and lead to the development of novel antivirals for the treatment of chronic hepatitis C.
Acknowledgments
We thank Andrea Cuconati and Pamela Norton for critical reading of the manuscript and Gang Chen for statistical analysis.
This work was supported by a grant from the National Institutes of Health (AI061441) and by the Hepatitis B Foundation through an appropriation of the Commonwealth of Pennsylvania.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Ali, S., C. Pellerin, D. Lamarre, and G. Kukolj. 2004. Hepatitis C virus subgenomic replicons in the human embryonic kidney 293 cell line. J. Virol. 78491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 2017-35. [DOI] [PubMed] [Google Scholar]
- 3.Barber, G. N. 2005. The dsRNA-dependent protein kinase, PKR and cell death. Cell Death Differ. 12563-570. [DOI] [PubMed] [Google Scholar]
- 4.Berkovitch, F., Y. Nicolet, J. T. Wan, J. T. Jarrett, and C. L. Drennan. 2004. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 30376-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 757059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bigger, C. B., B. Guerra, K. M. Brasky, G. Hubbard, M. R. Beard, B. A. Luxon, S. M. Lemon, and R. E. Lanford. 2004. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J. Virol. 7813779-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binder, M., G. Kochs, R. Bartenschlager, and V. Lohmann. 2007. Hepatitis C virus escape from the interferon regulatory factor 3 pathway by a passive and active evasion strategy. Hepatology 461365-1374. [DOI] [PubMed] [Google Scholar]
- 8.Boudinot, P., P. Massin, M. Blanco, S. Riffault, and A. Benmansour. 1999. vig-1, a new fish gene induced by the rhabdovirus glycoprotein, has a virus-induced homologue in humans and shares conserved motifs with the MoaA family. J. Virol. 731846-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boudinot, P., S. Riffault, S. Salhi, C. Carrat, C. Sedlik, N. Mahmoudi, B. Charley, and A. Benmansour. 2000. Vesicular stomatitis virus and pseudorabies virus induce a vig1/cig5 homologue in mouse dendritic cells via different pathways. J. Gen. Virol. 812675-2682. [DOI] [PubMed] [Google Scholar]
- 10.Chang, J., S. O. Gudima, C. Tarn, X. Nie, and J. M. Taylor. 2005. Development of a novel system to study hepatitis delta virus genome replication. J. Virol. 798182-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, K. S., Z. Cai, C. Zhang, G. C. Sen, B. R. Williams, and G. Luo. 2006. Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities. J. Virol. 807364-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin, K. C., and P. Cresswell. 2001. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 9815125-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Addario, M., A. Roulston, M. A. Wainberg, and J. Hiscott. 1990. Coordinate enhancement of cytokine gene expression in human immunodeficiency virus type 1-infected promonocytic cells. J. Virol. 646080-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Der, S. D., and A. S. Lau. 1995. Involvement of the double-stranded-RNA-dependent kinase PKR in interferon expression and interferon-mediated antiviral activity. Proc. Natl. Acad. Sci. USA 928841-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 9515623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dey, M., C. Cao, A. C. Dar, T. Tamura, K. Ozato, F. Sicheri, and T. E. Dever. 2005. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2α substrate recognition. Cell 122901-913. [DOI] [PubMed] [Google Scholar]
- 17.Di Bisceglie, A. M., and J. H. Hoofnagle. 2002. Optimal therapy of hepatitis C. Hepatology 36S121-S127. [DOI] [PubMed] [Google Scholar]
- 18.Duin, E. C., M. E. Lafferty, B. R. Crouse, R. M. Allen, I. Sanyal, D. H. Flint, and M. K. Johnson. 1997. [2Fe-2S] to [4Fe-4S] cluster conversion in Escherichia coli biotin synthase. Biochemistry 3611811-11820. [DOI] [PubMed] [Google Scholar]
- 19.Espert, L., G. Degols, C. Gongora, D. Blondel, B. R. Williams, R. H. Silverman, and N. Mechti. 2003. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J. Biol. Chem. 27816151-16158. [DOI] [PubMed] [Google Scholar]
- 20.Espert, L., G. Degols, Y. L. Lin, T. Vincent, M. Benkirane, and N. Mechti. 2005. Interferon-induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J. Gen. Virol. 862221-2229. [DOI] [PubMed] [Google Scholar]
- 21.Espert, L., P. Eldin, C. Gongora, B. Bayard, F. Harper, M. K. Chelbi-Alix, E. Bertrand, G. Degols, and N. Mechti. 2006. The exonuclease ISG20 mainly localizes in the nucleolus and the Cajal (coiled) bodies and is associated with nuclear SMN protein-containing complexes. J. Cell Biochem. 981320-1333. [DOI] [PubMed] [Google Scholar]
- 22.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 3001145-1148. [DOI] [PubMed] [Google Scholar]
- 23.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82723-733. [DOI] [PubMed] [Google Scholar]
- 24.Frey, P. A., and S. J. Booker. 2001. Radical mechanisms of S-adenosylmethionine-dependent enzymes. Adv. Protein Chem. 581-45. [DOI] [PubMed] [Google Scholar]
- 25.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436939-945. [DOI] [PubMed] [Google Scholar]
- 26.Garcia, M. A., J. Gil, I. Ventoso, S. Guerra, E. Domingo, C. Rivas, and M. Esteban. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 701032-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gongora, C., G. David, L. Pintard, C. Tissot, T. D. Hua, A. Dejean, and N. Mechti. 1997. Molecular cloning of a new interferon-induced PML nuclear body-associated protein. J. Biol. Chem. 27219457-19463. [DOI] [PubMed] [Google Scholar]
- 28.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 758516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo, J. T., J. A. Sohn, Q. Zhu, and C. Seeger. 2004. Mechanism of the interferon alpha response against hepatitis C virus replicons. Virology 32571-81. [DOI] [PubMed] [Google Scholar]
- 30.Guo, J. T., Q. Zhu, and C. Seeger. 2003. Cytopathic and noncytopathic interferon responses in cells expressing hepatitis C virus subgenomic replicons. J. Virol. 7710769-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo, X., J. Ma, J. Sun, and G. Gao. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. USA 104151-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi, J., R. Stoyanova, and C. Seeger. 2005. The transcriptome of HCV replicon expressing cell lines in the presence of alpha interferon. Virology 335264-275. [DOI] [PubMed] [Google Scholar]
- 33.Helbig, K. J., D. T. Lau, L. Semendric, H. A. Harley, and M. R. Beard. 2005. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology 42702-710. [DOI] [PubMed] [Google Scholar]
- 34.Horio, T., M. Murai, T. Inoue, T. Hamasaki, T. Tanaka, and T. Ohgi. 2004. Crystal structure of human ISG20, an interferon-induced antiviral ribonuclease. FEBS Lett. 577111-116. [DOI] [PubMed] [Google Scholar]
- 35.Ji, X., R. Cheung, S. Cooper, Q. Li, H. B. Greenberg, and X. S. He. 2003. Interferon alfa regulated gene expression in patients initiating interferon treatment for chronic hepatitis C. Hepatology 37610-621. [DOI] [PubMed] [Google Scholar]
- 36.Kato, T., T. Date, M. Miyamoto, Z. Zhao, M. Mizokami, and T. Wakita. 2005. Nonhepatic cell lines HeLa and 293 support efficient replication of the hepatitis C virus genotype 2a subgenomic replicon. J. Virol. 79592-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khaiboullina, S. F., A. A. Rizvanov, M. R. Holbrook, and S. St. Jeor. 2005. Yellow fever virus strains Asibi and 17D-204 infect human umbilical cord endothelial cells and induce novel changes in gene expression. Virology 342167-176. [DOI] [PubMed] [Google Scholar]
- 38.Kozbial, P. Z., and A. R. Mushegian. 2005. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanford, R. E., B. Guerra, H. Lee, D. Chavez, K. M. Brasky, and C. B. Bigger. 2006. Genomic response to interferon-alpha in chimpanzees: implications of rapid downregulation for hepatitis C kinetics. Hepatology 43961-972. [DOI] [PubMed] [Google Scholar]
- 40.Layer, G., D. W. Heinz, D. Jahn, and W. D. Schubert. 2004. Structure and function of radical SAM enzymes. Curr. Opin. Chem. Biol. 8468-476. [DOI] [PubMed] [Google Scholar]
- 41.Layer, G., J. Moser, D. W. Heinz, D. Jahn, and W. D. Schubert. 2003. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical SAM enzymes. EMBO J. 226214-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 1022992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindenbach, B. D., and C. M. Rice. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436933-938. [DOI] [PubMed] [Google Scholar]
- 44.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 1036001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcello, T., A. Grakoui, G. Barba-Spaeth, E. S. Machlin, S. V. Kotenko, M. R. MacDonald, and C. M. Rice. 2006. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology 1311887-1898. [DOI] [PubMed] [Google Scholar]
- 46.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 3391485-1492. [DOI] [PubMed] [Google Scholar]
- 47.Melen, K., P. Keskinen, A. Lehtonen, and I. Julkunen. 2000. Interferon-induced gene expression and signaling in human hepatoma cell lines. J. Hepatol. 33764-772. [DOI] [PubMed] [Google Scholar]
- 48.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 4371167-1172. [DOI] [PubMed] [Google Scholar]
- 49.Naka, K., K. Takemoto, K. Abe, H. Dansako, M. Ikeda, K. Shimotohno, and N. Kato. 2005. Interferon resistance of hepatitis C virus replicon-harbouring cells is caused by functional disruption of type I interferon receptors. J. Gen. Virol. 862787-2792. [DOI] [PubMed] [Google Scholar]
- 50.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282103-107. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen, L. H., L. Espert, N. Mechti, and D. M. Wilson III. 2001. The human interferon- and estrogen-regulated ISG20/HEM45 gene product degrades single-stranded RNA and DNA in vitro. Biochemistry 407174-7179. [DOI] [PubMed] [Google Scholar]
- 52.Nie, Y., G. L. Hammond, and J. H. Yang. 2007. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J. Virol. 81917-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radaeva, S., B. Jaruga, F. Hong, W. H. Kim, S. Fan, H. Cai, S. Strom, Y. Liu, O. El-Assal, and B. Gao. 2002. Interferon-alpha activates multiple STAT signals and down-regulates c-Met in primary human hepatocytes. Gastroenterology 1221020-1034. [DOI] [PubMed] [Google Scholar]
- 54.Rivieccio, M. A., H. S. Suh, Y. Zhao, M. L. Zhao, K. C. Chin, S. C. Lee, and C. F. Brosnan. 2006. TLR3 ligation activates an antiviral response in human fetal astrocytes: a role for viperin/cig5. J. Immunol. 1774735-4741. [DOI] [PubMed] [Google Scholar]
- 55.Seeger, C. 2005. Salient molecular features of hepatitis C virus revealed. Trends Microbiol. 13528-534. [DOI] [PubMed] [Google Scholar]
- 56.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55255-281. [DOI] [PubMed] [Google Scholar]
- 57.Severa, M., E. M. Coccia, and K. A. Fitzgerald. 2006. Toll-like receptor-dependent and -independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J. Biol. Chem. 28126188-26195. [DOI] [PubMed] [Google Scholar]
- 58.Smith, M. W., Z. N. Yue, M. J. Korth, H. A. Do, L. Boix, N. Fausto, J. Bruix, R. L. Carithers, Jr., and M. G. Katze. 2003. Hepatitis C virus and liver disease: global transcriptional profiling and identification of potential markers. Hepatology 381458-1467. [DOI] [PubMed] [Google Scholar]
- 59.Sofia, H. J., G. Chen, B. G. Hetzler, J. F. Reyes-Spindola, and N. E. Miller. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 291097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67227-264. [DOI] [PubMed] [Google Scholar]
- 61.Su, A. I., J. P. Pezacki, L. Wodicka, A. D. Brideau, L. Supekova, R. Thimme, S. Wieland, J. Bukh, R. H. Purcell, P. G. Schultz, and F. V. Chisari. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 9915669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor, D. R., M. Puig, M. E. Darnell, K. Mihalik, and S. M. Feinstone. 2005. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J. Virol. 796291-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verma, S., K. Ziegler, P. Ananthula, J. K. Co, R. J. Frisque, R. Yanagihara, and V. R. Nerurkar. 2006. JC virus induces altered patterns of cellular gene expression: interferon-inducible genes as major transcriptional targets. Virology 345457-467. [DOI] [PubMed] [Google Scholar]
- 64.Wang, C., J. Pflugheber, R. Sumpter, Jr., D. L. Sodora, D. Hui, G. C. Sen, and M. Gale, Jr. 2003. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J. Virol. 773898-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, S. C., and P. A. Frey. 2007. S-Adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem. Sci. 32101-110. [DOI] [PubMed] [Google Scholar]
- 66.Williams, B. R. 2001. Signal integration via PKR. Sci. STKE 2001RE2. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, Y., C. W. Burke, K. D. Ryman, and W. B. Klimstra. 2007. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 811246-11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou, A., B. A. Hassel, and R. H. Silverman. 1993. Expression cloning of 2-5A-dependent RNase: a uniquely regulated mediator of interferon action. Cell 72753-765. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, A., R. J. Molinaro, K. Malathi, and R. H. Silverman. 2005. Mapping of the human RNase L promoter and expression in cancer and normal cells. J. Interferon Cytokine Res. 25595-603. [DOI] [PubMed] [Google Scholar]
- 70.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258435-440. [DOI] [PubMed] [Google Scholar]
- 71.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 9514470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 9413985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu, Q., J. T. Guo, and C. Seeger. 2003. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 779204-9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuo, Y., and M. P. Deutscher. 2001. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 291017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]