Abstract
Certain major histocompatibility complex (MHC) class I alleles are strongly associated with control of human immunodeficiency virus and simian immunodeficiency virus (SIV). CD8+ T cells specific for epitopes restricted by these molecules may be particularly effective. Understanding how CD8+ T cells contribute to control of viral replication should yield important insights for vaccine design. We have recently identified an Indian rhesus macaque MHC class I allele, Mamu-B*08, associated with elite control and low plasma viremia after infection with the pathogenic isolate SIVmac239. Here, we infected four Mamu-B*08-positive macaques with SIVmac239 to investigate why some of these macaques control viral replication. Three of the four macaques controlled SIVmac239 replication with plasma virus concentrations below 20,000 viral RNA copies/ml at 20 weeks postinfection; two of four macaques were elite controllers (ECs). Interestingly, two of the four macaques preserved their CD4+ memory T lymphocytes during peak viremia, and all four recovered their CD4+ memory T lymphocytes in the chronic phase of infection. Mamu-B*08-restricted CD8+ T-cell responses dominated the acute phase and accounted for 23.3% to 59.6% of the total SIV-specific immune responses. Additionally, the ECs mounted strong and broad CD8+ T-cell responses against several epitopes in Vif and Nef. Mamu-B*08-specific CD8+ T cells accounted for the majority of mutations in the virus at 18 weeks postinfection. Interestingly, patterns of viral variation in Nef differed between the ECs and the other two macaques. Natural containment of AIDS virus replication in Mamu-B*08-positive macaques may, therefore, be related to a combination of immunodominance and viral escape from CD8+ T-cell responses.
Understanding the immunological and genetic basis of the natural control of AIDS virus replication should assist in human immunodeficiency virus (HIV) vaccine design. Of particular interest are human “elite controllers” (ECs), rare individuals who spontaneously control HIV viremia to extremely low levels (17). Similarly, a limited number of macaques spontaneously control simian immunodeficiency virus (SIV) replication and become ECs (49, 76).
Several lines of evidence suggest that CD8+ T cells play a key role in immune control of immunodeficiency virus replication. The transient in vivo depletion of circulating CD8+ lymphocytes in SIV-infected macaques, including EC macaques, results in dramatic increases in plasma viremia (23, 32, 52, 69). CD8+ T-cell responses also exert selective pressure on replicating viruses, resulting in the emergence of variants that escape immune detection in both HIV (5, 12, 20, 26, 36, 64, 65) and SIV (3, 7, 19, 35, 51, 59, 60) infection. In addition, it is well established that the expression of specific major histocompatibility complex (MHC) class I alleles is associated with reduced plasma viremia and/or slower disease progression in humans (14, 15, 29, 34, 55) and macaques (49, 57, 61, 63, 76, 77). In particular, human long-term nonprogressor and EC cohorts are enriched for HLA-B27 and HLA-B57. Numerous studies have implicated these molecules in the presentation of epitopes that elicit effective HIV-specific CD8+ T-lymphocyte responses (6, 20, 26, 36, 38, 54, 55, 70). This assertion is further supported by studies describing associations between viral escape from the immunodominant HLA-B27-restricted Gag263-272KK10 response and disease progression (8, 20, 26, 36).
Unfortunately, there are many difficulties inherent to studying HIV-infected humans. Viral control appears to be mediated soon after the resolution of acute-phase viremia with the appearance of CD8+ T-cell responses in HIV-infected individuals (11, 41), yet HIV is infrequently diagnosed during primary infection (42, 73). Hence, the study of immune responses initially involved in controlling HIV replication is extremely difficult. An additional complication arises due to the diversity of HIV isolates with which individuals might be infected (24, 27). Therefore, complete immunological monitoring would necessitate the sequencing of acute-phase virus and the synthesis of custom peptide sets matched to the infecting virus for every patient.
AIDS research with nonhuman primates provides an animal model to complement human studies. In particular, SIV-infected ECs also provide examples of successful immune containment of pathogenic immunodeficiency virus replication. However, unlike in human studies, researchers working with macaques have direct control over key variables such as virus strain, host genotype, and route of infection. Inoculum variability is eliminated because macaques can be infected with a clonal viral stock, e.g., SIVmac239, enabling complete and accurate tracking of early immune responses by use of corresponding peptides in ex vivo immunological assays. Moreover, the timing of immune responses after infection, the associated viral sequence evolution, and plasma virus concentrations may be closely monitored. Hence, the immunology and pathogenesis of acute infection can be more readily studied in macaques than in humans and may aid in our understanding of the correlates of immune protection.
Previously, we assembled a cohort of 192 Indian rhesus macaques, all infected with SIVmac239 (49, 76). Fourteen of these animals were considered ECs and controlled virus replication to fewer than 1,000 viral RNA (vRNA) copies/ml in the chronic phase of SIV infection. Nine of the 14 ECs expressed Mamu-B*17 (76). In addition to Mamu-B*17, we recently discovered that Mamu-B*08 is enriched in EC cohorts and associated with reduced chronic-phase plasma virus concentrations (49). Over 50% of Mamu-B*08-positive macaques become ECs. Interestingly, a preliminary binding motif for Mamu-B*08 appears comparable to that of HLA-B27, an allele associated with elite control in humans (48). The similarity between Mamu-B*08 and HLA-B27 makes SIVmac239-infected, Mamu-B*08-positive macaques an ideal system for modeling human ECs.
While not all Mamu-B*08- or Mamu-B*17-positive macaques control viral replication (49, 75, 76), we and others have previously shown that control of SIV replication is not due to genes linked to Mamu-B*17 (75) or to polymorphisms in several host genes, including CCR5, CXCR6, GPR15, RANTES, interleukin-10 (IL-10), APOBEC3G, tumor necrosis factor alpha, and TSG101 (72). Rather, CD8+ T cells appear to play a critical role in the natural containment of SIV replication (23, 61).
Here, we investigated early immune responses and viral evolution in four Mamu-B*08-positive Indian rhesus macaques during primary SIVmac239 infection. From previous studies (49), we estimated that two of the four macaques would control viral replication. We hypothesized that this control would be a function of the Mamu-B*08-specific CD8+ T-cell responses and viral escape. We found that containment of SIV replication began before 10 weeks postinfection in three of the four Mamu-B*08-positive macaques. During peak viremia, the CD4+ memory T-cell subset decreased in only two of the four Mamu-B*08-positive macaques. However, the CD4+ memory T-cell numbers later rebounded in these two animals, and CD4+ memory T cells were then maintained in the chronic phase of infection in all four macaques. While robust CD8+ immune responses were detected in all four animals, macaques with the broadest Mamu-B*08-restricted immune responses were the most successful at viral containment. Mutations within Mamu-B*08 epitopes were detected in all four SIVmac239-infected macaques. Interestingly, different patterns of viral variation in Nef distinguished the two ECs from the non-EC macaques.
MATERIALS AND METHODS
Animals and viruses.
Indian rhesus macaques (Macaca mulatta) were genotyped for the MHC class I alleles Mamu-A*01, -A*02, -A*08, -A*11, -B*01, -B*03, -B*04, -B*08, -B*17, and -B*29 by using PCR amplification with sequence-specific primers as previously described (33, 49). While all four macaques expressed Mamu-B*08, none of the four macaques expressed Mamu-B*17. Macaques were infected intravenously with 100 50% tissue culture infective doses of the pathogenic molecular clone SIVmac239 (37) (GenBank accession no. M33262). SIV-infected animals were maintained at the National Primate Research Center (University of Wisconsin—Madison, Madison, WI) and cared for according to the regulations and guidelines of the University of Wisconsin Institutional Animal Care and Use Committee.
Quantification of vRNA in plasma.
The plasma virus concentrations of SIVmac239-infected Indian rhesus macaques were monitored by quantitative reverse transcription-PCR (QRT-PCR) as previously described (23, 47). Briefly, vRNA was isolated from EDTA-anticoagulated plasma, reverse transcribed, and detected using a one-step QRT-PCR kit (Invitrogen, Carlsbad, CA). The QRT-PCR was performed under the previously described conditions on a LightCycler 1.2 (Roche, Indianapolis, IN). Each QRT-PCR assay was performed with an internal standard curve prepared by 10-fold serial dilutions of a synthetic SIV gag transcript. The copy number for each sample was then determined by interpolation onto the standard curve, using LightCycler software version 4.0. Under normal conditions, the threshold for detection in this assay is 30 vRNA eq/ml.
Quantification of circulating CD4+ T lymphocytes.
The absolute counts of CD4+ T cells and CD4+ memory T cells/μl of blood were determined using a two-platform method. Briefly, the frequency of CD3+ CD4+ (CD4+ T cell) or CD3+ CD4+ CD95+ (CD4+ memory T cell) cells within the lymphocyte population was determined by flow cytometry using anti-human CD4 PerCP (clone SK3; BD Biosciences, San Jose, CA), anti-human CD95 fluorescein isothiocyanate (FITC) (clone DX2; BD Biosciences), and anti-human CD3 allophycocyanin (clone SP34-2; BD Biosciences) monoclonal antibodies. Data were acquired on a FACSCalibur (BD Biosciences) flow cytometer and analyzed by FlowJo software version 8.4.5 (Tree Star, Inc., Ashland, OR). Lymphocyte counts per μl of blood were obtained from complete blood count analysis performed on a Pentra 60C+ Hematology analyzer (ABX Diagnostics, Irvine, CA). The CD4+ T-cell or CD4+ memory T-cell count/μl of blood was then determined by multiplying the lymphocyte count/μl blood by the frequency of each cell subset within the lymphocyte gate.
IFN-γ ELISPOT assay.
Enzyme-linked immunospot (ELISPOT) assays were performed as previously described (47). Briefly, peripheral blood mononuclear cells (PBMC) were isolated from EDTA-anticoagulated blood by using Ficoll-Paque PLUS (GE Healthcare Bio-Sciences, Uppsala, Sweden) and density centrifugation. A total of 1 × 105 PBMC were used per well in precoated ELISpotPLUS kits (MABTECH Inc, Mariemont, OH) according to the manufacturer's instructions for the detection of gamma interferon (IFN-γ)-secreting cells. All tests were performed in duplicate using individual peptides at 10 μM or peptide pools (10 15-mer peptides overlapping by 11 amino acids) at 1 μM. Fifteen-mer peptides were provided by the NIH AIDS Research and Reference Reagent Program (Germantown, MD). Peptides fewer than 15 amino acids in length that contained epitopes restricted by Mamu-A*01 (2, 4), Mamu-A*02 (46, 67, 71), and Mamu-B*08 (48) were synthesized at the University of Wisconsin Biotechnology Center (Madison, WI). The Mamu-B*08-restricted Env epitope Env573-581KL9 has not been previously described (J. T. Loffredo et al., unpublished data). The positive control, concanavalin A (Sigma, St. Louis, MO), was used at a final concentration of 5 μg/ml. The negative-control wells were devoid of any stimulation. The 96-well plates were incubated for 12 to 18 h at 37°C in 5% CO2.
CD8+ cell-depleted ELISPOT assays were incorporated to identify CD4+ T-cell responses. In these cases, freshly isolated or cryopreserved PBMC were depleted of CD8+ cells by using a CD8 microbead kit for nonhuman primates (Miltenyi, Auburn, CA) along with LS columns (Miltenyi) according to the manufacturer's protocols. Labeled cells were removed by magnetic separation, and the remaining cells were used as described above in IFN-γ ELISPOT assays. Fluorescence-activated cell sorting analysis using CD3 FITC (clone SP34-2; BD Biosciences), CD8 PerCP (clone SK1; BD Biosciences), and CD4 allophycocyanin (clone SK3; BD Biosciences) cell surface markers confirmed that the CD8+ cell depletions removed >99% of the CD8+ lymphocytes.
Wells were imaged and counted with AID EliSpot reader version 3.4.0 or 4.0 (AID, Strassberg, Germany) and analyzed as previously described (46, 47). A response was considered positive if the mean number of spot-forming cells (SFC) from the duplicate sample wells exceeded the background level (mean of wells without peptide stimulation) plus 2 standard deviations. Background levels were subtracted from each well, and assay results are shown as numbers of SFC per 1 × 106 PBMC. Responses of <50 SFC per 1 × 106 PBMC were not considered positive.
MHC class I tetramer and surface staining.
Ex vivo MHC class I tetramer stains were performed on freshly isolated or cryopreserved PBMC as previously described (46). MHC class I tetramers were constructed with minor modifications as previously described (31, 46). Cryopreserved PBMC were thawed at 37°C and washed twice in R10 (RPMI 1640 medium [HyClone, Logan, UT] supplemented with 10% fetal calf serum [HyClone], 2 mM l-glutamine [HyClone], and 1× antibiotic-antimycotic solution [HyClone]) before staining.
Briefly, ∼5 × 105 cells were stained with 5 μl of 0.1 mg/ml tetramer stocks, 3 μl of CD3 FITC (clone SP34-2; BD Biosciences), and 5 μl of CD8 PerCP (clone SK1; BD Biosciences) in ∼100 μl of R10. After the cells were fixed in 1% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA), approximately 0.5 × 105 to 1 × 105 lymphocyte-gated events were acquired on a FACSCalibur (BD Biosciences) and analyzed using FlowJo 8.4.5 (TreeStar, Inc.). Percentages represent the numbers of lymphocyte-gated events that were CD3+ CD8+ and MHC class I tetramer positive. The threshold of detection in these assays was 0.02% CD3+ CD8+ MHC class I tetramer-positive gated lymphocytes.
ICS assay.
IL-2 and IFN-γ intracellular cytokine staining (ICS) assays were performed on freshly isolated PBMC as previously described (23, 71). Briefly, each test contained ∼5 × 105 PBMC. Peptide pools were used at a concentration of 10 μM. SIV peptide pools each contained 10 15-mer peptides overlapping by 11 amino acids. Fifteen-mer peptides were provided by the NIH AIDS Research and Reference Reagent Program (Germantown, MD). Approximately 0.5 × 105 to 1 × 105 lymphocyte-gated events were acquired on a FACSCalibur (BD Biosciences) and analyzed using FlowJo 8.4.5 (TreeStar, Inc.). All values were normalized by subtracting the background staining level (negative control of PBMC in media without stimulation).
Sequencing of plasma vRNA.
Viral sequencing was performed based on methods previously described (23, 60). Briefly, vRNA was extracted from plasma by using a Qiagen MinElute kit (Valencia, CA). We used a Qiagen One Step RT-PCR kit to amplify overlapping regions between approximately 300 and 800 nucleotides in length that spanned the entire SIVmac239 genome. The RT-PCR conditions for all amplicons were as follows: 50°C for 30 min; 95°C for 15 min; 45 cycles of 94°C for 30 s, 53°C for 1 min, and 72°C for 150 s; and 68°C for 20 min. Cycling ramp rates were 2°C per second. The amplified cDNA was purified using a Qiagen PCR purification kit. Plasmids containing cloned sequences were purified using a QIAprep Spin Miniprep kit (Qiagen). Both strands of each amplicon were sequenced on a 3730 DNA analyzer (Applied Biosystems, Foster City, CA). Sequences were assembled using Aligner version 1.6.3 (CodonCode, Deadham, MA). DNA sequences were conceptually translated and aligned to the wild-type SIVmac239 sequence with the MacVector 9.0 trial version (Accelrys, Burlington, MA).
Statistical analysis.
Statistical analyses consisted of two sample t tests for CD4+ memory T-cell (CD3+ CD4+ CD95+ gated lymphocytes) and total CD4+ T-cell (CD3+ CD4+ gated lymphocytes) counts in Mamu-B*08-positive and Mamu-B*08-negative SIVmac239-infected macaques at seven different time points. The time points were categorized into seven intervals, as 0, 2, 6, 8, 10, 12, and 18 (or 28) weeks postinfection. Mean values for comparison between Mamu-B*08-positive (n = 4) and Mamu-B*08-negative (n = 4) macaques at each of the time points were obtained using the TTEST procedure in version 9.1 of SAS (Cary, NC).
The Z-score test was employed to compare the viral loads observed in each of the four Mamu-B*08-positive macaques against the means of the progressor (n = 175) and EC (n = 10) cohorts at 12 different time points. None of the SIVmac239-infected macaques in the progressor and EC cohorts expressed Mamu-B*08. The viral load data from the 187 animals within these two cohorts were obtained from a prior investigation (49). The time points were defined here as the numbers of weeks, between 0 and 20, after SIVmac239 infection, subdivided into 12 intervals. The goal was to ascertain whether the viral loads in Mamu-B*08-positive macaques were significantly different from the mean of the EC cohort or significantly lower than the mean of the progressor cohort.
Before performing inferential testing, we checked for key underlying assumptions of the t tests and Z-score tests (i.e., normality of residuals and homoscedasticity). The failure of the data to support these assumptions led us to transform the data for both CD4 counts and viral loads via the natural logarithm to improve conformity to the assumptions. To circumvent the assumption of normality of residuals, we employed one-way nonparametric analysis using an exact Wilcoxon rank sum test with the continuity correction for the CD4+ T-cell counts only to further verify our results. Because statistical significance and direction of effects were the same across these analytic approaches, we based our interpretations on results from t tests only.
Nucleotide sequence accession numbers.
The SIV genome sequences from 18 weeks postinfection were given the following GenBank accession numbers: EU280803 (r91003-SIVmm239), EU280805 (r01027-SIVmm239), EU280804 (r00032-SIVmm239), and EU280806 (r02019-SIVmm239).
RESULTS
Mamu-B*08-positive, SIV-infected macaques control SIVmac239 viral replication and recover their CD4+ memory T lymphocytes after the acute phase of infection.
We infected four Mamu-B*08-positive Indian rhesus macaques in an attempt to understand the dynamics of early infection and the way that the majority of macaques expressing this protective allele control viral replication. After challenging the macaques intravenously with the cloned viral isolate SIVmac239, we followed plasma viral concentrations, peripheral blood CD4+ counts (including total and memory subsets), antigen-specific responses (CD8+ and CD4+), and viral evolution during the first 20 weeks after SIV infection.
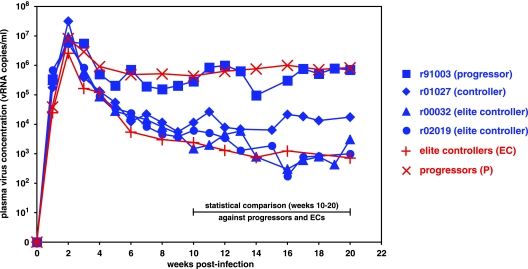
Three of the four macaques controlled their viral set points of SIVmac239 replication below 20,000 vRNA copies/ml, and two of these had viral set points of ∼1,000 vRNA copies/ml (Fig. 1). Only one macaque, animal r91003 (Mamu-A*01 positive and -B*08 positive), displayed plasma virus concentrations no different from those seen in 175 SIVmac239-infected macaques that progressed to AIDS (∼500,000 vRNA copies/ml) and was thus termed a progressor. In contrast, animal r01027 (Mamu-A*01-positive and -B*08-positive) had a viral set point of <20,000 vRNA copies/ml and was termed a slow progressor/controller. Controller macaques maintain viral set points >10-fold lower than our previously studied SIV-infected macaque cohort (49). Macaques r00032 (Mamu-A*02 positive and -B*08 positive) and r02019 (Mamu-B*08 positive) controlled replication of this pathogenic SIVmac239 isolate to ∼1,000 vRNA copies/ml and were classified as ECs. When comparing the plasma virus concentrations of the three Mamu-B*08-positive macaques that controlled SIV viremia with those of 175 macaques that progressed to AIDS, we found that the EC macaques r00032 and r02019 had viral set points significantly below those of 175 macaques that progressed to AIDS (P = 0.0066 and P = 0.0076, respectively). The difference between the viral set point of controller r01027 and those of the progressor macaques was near statistical significance (P = 0.0578). We also found that the plasma virus concentrations in r01027, r00032, and r02019 first began to differentiate themselves from those in the progressors between weeks 6 and 8 postinfection and trended toward significance as early as 4 weeks postinfection (data not shown). These data further suggest that the initial immunopathogenic events of immunodeficiency virus infection are crucial in controlling viral replication.
FIG. 1.
Three of four Mamu-B*08-positive Indian rhesus macaques control pathogenic SIVmac239 viral replication. Longitudinal SIVmac239 plasma virus concentrations were plotted for four Mamu-B*08-positive macaques (blue lines) and the geometric mean of viral loads from 10 Mamu-B*08-negative ECs and 175 Mamu-B*08-negative animals that progressed to AIDS (red lines) from a previous study (49). SIV-infected macaques r00032 (Mamu-A*02 positive and Mamu-B*08 positive) and r02019 (Mamu-B*08 positive) were considered ECs, with viral set points of ∼1 × 103 vRNA copies/ml. These two macaques had significantly lower viral set points (weeks 10 to 20 postinfection) than the macaques that progressed to AIDS (P = 0.0066 for r00032 and P = 0.0076 for r02019). Animal r01027 (Mamu-A*01 positive and Mamu-B*08 positive) is considered a “controller,” with viremia >1 log lower than typical SIV replication. The viral set point of r01027 was near statistical significance compared to those of the macaques that progressed to AIDS (P = 0.0578). None of these three macaques had P values significantly different from that for the viral set point of the EC cohort. One of the four macaques did not control SIVmac239 replication to <20,000 vRNA copies/ml after 10 weeks postinfection. Animal r91003 (Mamu-A*01 positive and Mamu-B*08 positive) exhibited typical viremia at ≥10 weeks postinfection that was not statistically different from that of the macaques that progressed to AIDS.
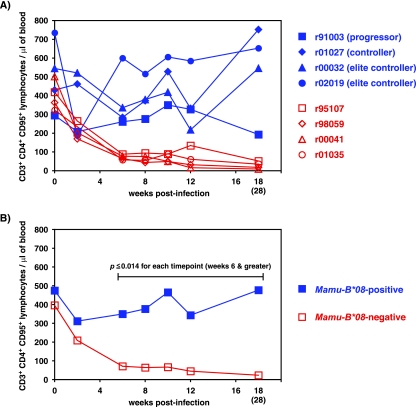
We also measured concentrations of circulating CD4+ T cells in our four SIV-infected Mamu-B*08-positive macaques. High levels of acute-phase immunodeficiency virus replication leads to the destruction of the CD4+ memory T-cell compartment (44, 53), thereby crippling the immune system during the critical stage of HIV/SIV infection. As expected, four Mamu-B*08-negative macaques (r95107, r98059, r00041, and r01035) that progressed to AIDS lost their CD4+ memory T cells (defined as CD3+ CD4+ CD95+ lymphocytes) during the acute phase of infection and never recovered their CD4+ memory T lymphocytes (Fig. 2). Two Mamu-B*08-negative animals, r98059 and r00041, were infected with SIVmac239 at the same time as the four Mamu-B*08-positive macaques, while r95107 and r01035 were two Mamu-B*08-negative historical controls. By comparison, only two of the four Mamu-B*08-positive macaques experienced an acute-phase loss in their CD4+ memory T-cell levels during peak viremia. CD4+ memory T-cell levels rebounded in all four macaques (Fig. 2). For each time point between weeks 6 and 18 postinfection, the Mamu-B*08-positive macaques preserved a significantly higher number of CD4+ memory T cells than the four Mamu-B*08-negative macaques (P ≤ 0.014). Interestingly, the Mamu-B*08-positive progressor macaque r91003 began to show a decline in its CD4+ memory T-cell count only at 18 weeks postinfection, perhaps due to high plasma virus concentrations. The total CD4+ T-cell counts (defined as CD3+ CD4+ lymphocytes) were not statistically different between the groups of Mamu-B*08-positive and -B*08-negative macaques (data not shown).
FIG. 2.
CD4+ memory T cells are maintained in Mamu-B*08-positive macaques infected with SIVmac239. (A) Absolute counts of CD4+ memory T cells were obtained from the four SIVmac239-infected, Mamu-B*08-positive macaques (blue lines) in addition to the four SIVmac239-infected, Mamu-B*08-negative macaques (red lines). Mamu-B*08-negative macaques r98059 and r00041 were infected along with the four Mamu-B*08-positive macaques. CD4+ memory T-cell counts from two additional Mamu-B*08-negative, -B*17-negative macaques (animals r95107 and r01035) were acquired from cryopreserved PBMC. Archived PBMC were not available at week 2 from r01035 or at week 18 from r01035 and r00041. In place of the week 18 time point, absolute counts of CD4+ memory T cells were acquired from the closest available time point (week 28), indicated in parentheses. (B) Geometric means of absolute counts of CD4+ memory T cells. The blue line represents the four Mamu-B*08-positive macaques, and the red line represents the four Mamu-B*08-negative macaques. By use of log-transformed arithmetic means, statistically significant differences were found between the absolute CD4+ memory T-cell counts of the Mamu-B*08-positive and -B*08-negative groups at the following time points: week 6 (P = 0.001), week 8 (P < 0.001), week 10 (P < 0.001), week 12 (P = 0.014), and week 18 (or 28) (P = 0.001).
Dominance of Mamu-B*08-restricted CD8+ T-cell responses during the acute phase of SIVmac239 infection.
We initially examined the entire repertoire of SIV-specific immune responses by using ex vivo IFN-γ ELISPOT with 15-mer peptides that overlapped by 11 amino acids for the entire viral proteome (Fig. 3). These assays also included eight minimal optimal epitopes restricted by Mamu-B*08 (48) as well as known epitopes and other peptides that bind to Mamu-A*01 (2, 4) and Mamu-A*02 (46, 67, 71). In addition, we performed MHC class I tetramer staining on the immunodominant epitopes restricted by Mamu-A*01 (Tat28-35SL8 and Gag181-189CM9) and Mamu-A*02 (Gag71-79GY9 and Nef159-167YY9) during primary infection in macaques that expressed these alleles (data not shown). Since CD8+ T-cell responses usually peak at 3 weeks postinfection (66), we chose this time point to extensively investigate the possible role of SIV-specific CD8+ T cells in disease protection or progression during the acute phase.
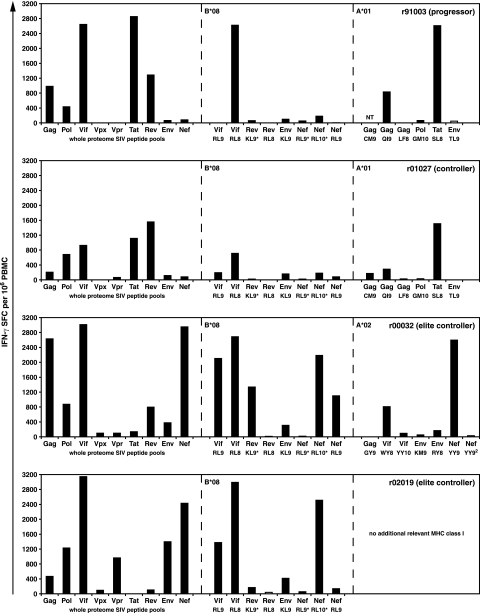
FIG. 3.
Ex vivo whole PBMC IFN-γ ELISPOT using peptides spanning the entire SIVmac239 proteome and the relevant MHC class I-restricted minimal optimal CD8+ T-cell epitopes at 3 weeks postinfection. Eighty-one peptide pools (10 15-mer peptides overlapping by 11 amino acids) were tested in IFN-γ ELISPOT assays spanning the complete SIVmac239 proteome. Total responses for each protein were calculated by adding the mean values of the individual peptide pools for each SIV protein. Individual minimal optimal peptides were included to detect responses restricted by Mamu-A*01 and Mamu-A*02. While the Mamu-A*01-restricted Gag181-189CM9 epitope was not tested (NT) in r91003, we can verify that a response to this epitope was detected at 3 weeks postinfection by the Gag161-211(E) peptide pool that contains the Gag181-189CM9 sequence (692 SFC/106 PBMC). MHC class I tetramer staining in r91003 confirms the presence of Gag181-189CM9-specific CD8+ T cells at this time point (see text). Two Mamu-A*02-restricted CD8+ T-cell epitopes are labeled as Nef YY9. Nef YY9 refers to the epitope at positions 159 to 167 (YTSGPGIRY), while Nef YY92 refers to the epitope at positions 221 to 229 (YTYEAYVRY). Minimal optimal peptides were used for the following Mamu-B*08-restricted epitopes: Vif123-131RL9, Vif172-179RL8, Rev44-51RL8, Env573-581KL9, and Nef246-254RL9. Mamu-B*08 epitopes annotated with an asterisk were represented with peptides slightly larger than the minimal optimal as these responses were in the process of being fine mapped (Rev12-20KL9* represents two overlapping 15-mer peptides at positions 5 to 23, Nef8-16RL9* represents a 10-mer peptide at positions 7 to 16, and Nef137-146RL10* represents an 11-mer peptide at positions 136 to 146). Background levels were subtracted from each well. Mean responses of <50 SFC per 1 × 106 cells (white bars) were not considered positive.
CD8+ T-cell responses against the majority of the SIV proteins were detected in all four SIV-infected macaques. While these macaques all expressed Mamu-B*08, their immune responses should also be influenced by other known and unknown MHC class I alleles that they expressed. As expected, the progressor macaque r91003 (also Mamu-A*01 positive) made robust immune responses against the immunodominant Mamu-A*01-restricted CD8+ T-cell epitopes Gag181-189CM9 and Tat28-35SL8 (Fig. 3 and data not shown). These CD8+ T-cell responses peaked at 3 weeks postinfection, with ∼0.39% (Gag181-189CM9) and ∼2.24% (Tat28-35SL8) of CD3+ CD8+ lymphocytes staining with the relevant tetramer. Aside from these two Mamu-A*01-restricted responses, along with the Mamu-A*01-restricted response against Gag254-262QI9, the majority of the CD8+ T-cell response generated by r91003 targeted a single Mamu-B*08-restricted epitope in Vif and an epitope of unknown MHC class I restriction in Rev. Progressor macaque r91003 made no substantial CD8+ T-cell responses against Nef, although three known Mamu-B*08-restricted CD8+ T-cell epitopes are located in this protein (48).
The controller macaque r01027 (also expressing Mamu-A*01) mounted SIV-specific responses of a similar magnitude against the Mamu-A*01-restricted CD8+ T-cell epitopes Tat28-35SL8 and Gag181-189CM9 (Fig. 3 and data not shown). MHC class I tetramer staining showed that the CD8+ T-cell response against Gag181-189CM9 peaked at 2 weeks postinfection (∼0.77%), while the Tat28-35SL8-specific CD8+ T cells reached ∼1.95% at 3 weeks postinfection. Along with a robust CD8+ T-cell response against Vif172-179RL8, several Mamu-B*08-restricted responses of lower magnitude (<200 SFC/106 PBMC) were detected in r01027 (Fig. 3). A sizeable CD8+ T-cell response of unknown MHC restriction to Rev was also identified in this macaque.
At 3 weeks postinfection, the two EC macaques r00032 and r02019 mounted total SIV-specific immune responses near or exceeding 10,000 SFC/106 PBMC (Fig. 4). These included several robust CD8+ T-cell responses to both the Vif and Nef epitopes restricted by Mamu-B*08 (Fig. 3 and 4). Additionally, animal r00032 (also Mamu-A*02 positive) made a response against the immunodominant Mamu-A*02-restricted CD8+ T-cell epitope Nef159-167YY9 (∼2.4% at 2 and 3 weeks postinfection) (data not shown). Notably, we did not detect a response to Gag71-79GY9, which is frequently codominant with Nef159-167YY9 in Mamu-A*02-positive animals (46, 71). However, two responses, of >500 SFC/106 PBMC and of unknown MHC class I restriction, were directed against Gag [Gag241-291(G) and Gag361-411(J) peptide pools] in EC r00032 (Fig. 3). While EC macaque r02019 did not express any other characterized MHC class I alleles that restrict known minimal optimal epitopes, a substantial response(s) of unknown restriction against Vpr was identified.
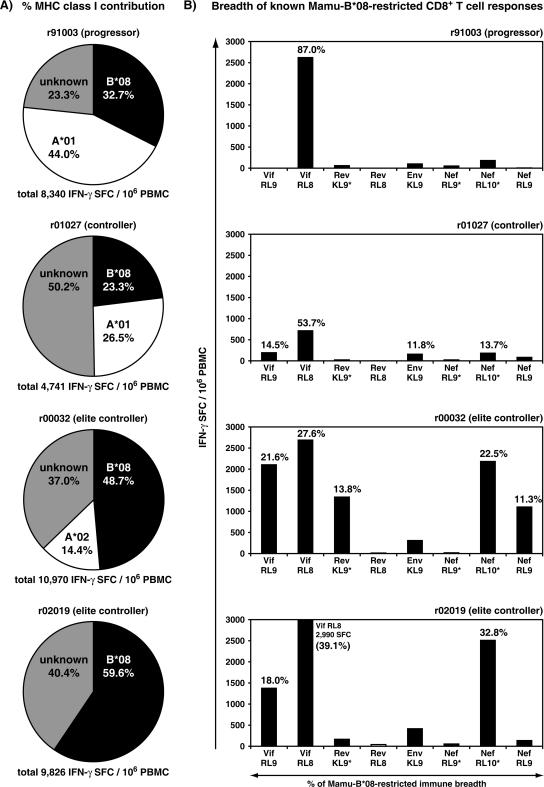
FIG. 4.
Involvement of Mamu-B*08-restricted CD8+ T-cell responses in the overall CD8+ T-cell-mediated immune response against SIVmac239 at 3 weeks postinfection. (A) Mamu-B*08-restricted CD8+ T cells make a major contribution to the total SIV-specific immune response during the acute phase of infection. The contributions of known responses are shown for Mamu-A*01, Mamu-A*02, and Mamu-B*08 by attributing peptide pools to particular MHC class I molecules based on the locations of known SIV epitopes in these pools. (B) Mamu-B*08-restricted CD8+ T cells recognize a broader epitope repertoire in EC macaques early in SIV infection. CD8+ T-cell responses accounting for >10% of the total Mamu-B*08-restricted immune response are indicated above the appropriate bar. Minimal optimal peptides were used for the following Mamu-B*08-restricted epitopes: Vif123-131RL9, Vif172-179RL8, Rev44-51RL8, Env573-581KL9, and Nef246-254RL9. Mamu-B*08 epitopes annotated with an asterisk were represented with peptides slightly larger than the minimal optimal as these responses were in the process of being fine mapped (Rev12-20KL9* represents two overlapping 15-mer peptides positions 5 to 23, Nef8-16RL9* represents a 10-mer peptide at positions 7 to 16, and Nef137-146RL10* represents an 11-mer peptide at positions 136 to 146). Data for both panel A and panel B were derived from ex vivo IFN-γ ELISPOT assays at 3 weeks postinfection (Fig. 3). Background levels were subtracted from each well. Mean responses of <50 SFC per 1 × 106 cells (white bars) were not considered positive.
Using the week 3 ELISPOT pool response data along with responses to known minimal optimal epitopes and MHC class I tetramer stains, we defined the proportion of the total SIV-specific immune responses restricted by Mamu-B*08 and other MHC class I molecules (Fig. 4). Mamu-A*01-positive macaques mount two immunodominant, acute-phase CD8+ T-cell responses, Gag181-189CM9 and Tat28-35SL8 (3, 61), that typically account for >50% of the total SIV-specific CD8+ responses (56). However, the normally dominant Mamu-A*01-restricted immune responses contributed only a slightly larger percentage of the total SIV-specific immune response than the Mamu-B*08-restricted immune responses in progressor macaque r91003 and controller r01027. Mamu-A*01-restricted CD8+ T cells accounted for 44% of the total SIV-specific immune response in r91003 and 26.5% of the total SIV-specific immune response in r01027, compared to 32.7% and 23.3%, respectively, for Mamu-B*08-restricted CD8+ T cells (Fig. 4A). Animal r00032 expressed both Mamu-A*02 and Mamu-B*08. While Mamu-A*02-restricted responses accounted for 14.4% of the SIV-specific response, Mamu-B*08-restricted responses accounted for 48.7% of the total number of SIV-specific CD8+ T cells. Macaque r02019 did not express any other characterized MHC class I alleles aside from Mamu-B*08, and Mamu-B*08-restricted CD8+ T cells accounted for 59.6% of its total SIV-specific response.
Despite the presence of as many as 12 MHC class I alleles expressed in a given macaque (13, 16, 62), Mamu-B*08-restricted CD8+ T cells accounted for a large proportion (23.3 to 59.6%) of the total SIV-specific response. Furthermore, the contribution of Mamu-B*08-restricted SIV-specific immune responses is likely an underestimate because, unlike for Mamu-A*01 (2, 4) and Mamu-A*02 (46), a comprehensive peptide binding motif and epitope mapping study has not yet been completed for Mamu-B*08.
Increased breadth of Mamu-B*08-restricted CD8+ T-cell responses against epitopes in Vif and Nef was associated with elite control.
Despite the fact that all four macaques made strong Mamu-B*08-restricted CD8+ T-cell responses, only two animals were ECs, controlling viral replication to approximately 1,000 vRNA copies/ml (49, 76). We next analyzed the breadth of the Mamu-B*08-restricted immune responses to examine the possible role that multiple epitope-specific, Mamu-B*08-restricted CD8+ T-cell responses might play in the control of SIV replication. At 3 weeks postinfection, the progressor macaque r91003 made a narrowly focused immunodominant response against Vif172-179RL8 that constituted 87% of the Mamu-B*08-restricted CD8+ T cells detected in this macaque (Fig. 4B). Controller macaque r01027 also made an immunodominant response to this same epitope (53.7%, Vif172-179RL8). However, three other Mamu-B*08-restricted CD8+ T-cell responses (Vif123-131RL9, Env573-581KL9, and Nef137-146RL10) each accounted for >10% of the Mamu-B*08-restricted immune breadth in r01027. By contrast, the EC macaques r00032 and r02019 made robust CD8+ T-cell responses to several of the eight Mamu-B*08-bound peptides, none of which accounted for >50% of the Mamu-B*08-restricted CD8+ T-cell response. At 3 weeks postinfection, five of the eight Mamu-B*08-restricted responses were at >1,000 SFC/106 PBMC for the EC r00032, while the other EC r02019 had three responses exceeding this magnitude (Fig. 4B).
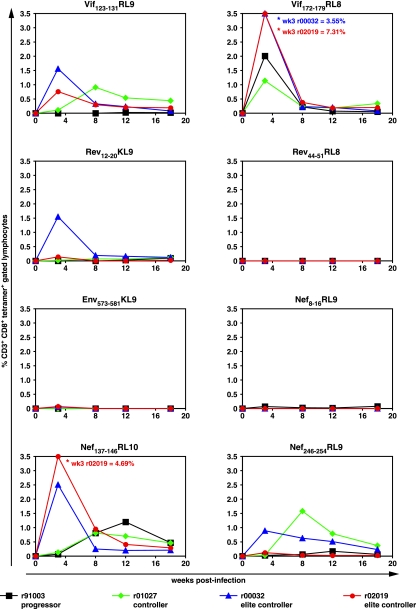
To study changes in the Mamu-B*08-restricted epitope breadth over time, we performed MHC class I tetramer stains on cryopreserved PBMC during the acute to early chronic phase of SIV infection for all eight Mamu-B*08-restricted CD8+ T-cell epitopes (Fig. 5). We found that the MHC class I tetramer stains largely agreed with the magnitude of the IFN-γ ELISPOT responses at 3 weeks postinfection (Fig. 4) and subsequent time points (data not shown). At 3 weeks postinfection, ECs r00032 and r02019 typically displayed the largest percentages of SIV-specific CD8+ T cells for each of the eight Mamu-B*08-restricted CD8+ T-cell responses. These two ECs targeted three epitopes, two in Vif (RL9 and RL8) and one in Nef (RL10), with massive CD8+ T-cell responses (0.76 to 7.31%) (Fig. 5). Controller macaque r01027 displayed several low-level responses aside from its immunodominant Vif172-179RL8 that were maintained over time. In contrast, progressor r91003 mounted a Mamu-B*08-restricted immune response focused primarily against the Vif172-179RL8 epitope at 3 weeks postinfection, although a sizeable response against Nef137-146RL10 was detected later after infection (Fig. 5). Interestingly, only r91003 failed to make a substantial response against the Vif123-131RL9 epitope during any of the time points, potentially implicating this response in Mamu-B*08-restricted control. Not surprisingly, only low-level frequencies for the majority of the Mamu-B*08-restricted CD8+ T cells were found in the three macaques controlling viremia at 18 weeks postinfection. This diminution of their robust acute-phase responses was likely due to small amounts of antigen stimulation due to low SIV plasma virus concentrations after primary infection.
FIG. 5.
Comparison of Mamu-B*08-restricted CD8+ T-cell responses detected by MHC class I tetramers during the first 18 weeks postinfection. Cryopreserved PBMC from the four Mamu-B*08-positive macaques (black, r91003; green, r01027; blue, r00032; and red, r02019) were thawed and stained at the indicated time points for the eight Mamu-B*08-restricted epitopes. Results of MHC class I tetramer stains are shown here as percentages of CD3+ CD8+ MHC class I tetramer-positive gated lymphocytes. The threshold of detection in these assays was 0.02% CD3+ CD8+ MHC class I tetramer-positive gated lymphocytes.
Stronger and broad CD4+ T-cell responses in the Mamu-B*08-positive EC macaques.
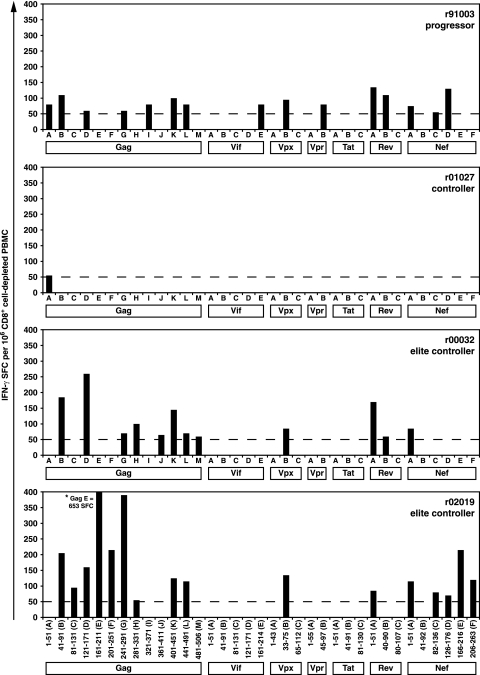
Several previous studies have shown an association between strong and broad CD4+ T-cell responses and control of HIV/SIV replication (10, 23, 25, 28, 50, 68). CD4+ T cells may also play a role in the prevention of CD8+ T-cell exhaustion (45). Therefore, we investigated the involvement of CD4-mediated immune responses in the four Mamu-B*08-positive macaques. At 6 weeks postinfection, we performed ex vivo IL-2 and IFN-γ ICS assays to determine whether responses to peptide pools detected in the PBMC IFN-γ ELISPOTs at 3 weeks postinfection were mediated by CD4+ or CD8+ T cells. Progressor r91003 and controller r01027 did not make any appreciable CD4+ responses at this time point (data not shown). In contrast, EC macaque r00032 made three responses directed against the Gag121-171(D), Rev1-51(A), and Nef1-51(A) pools, while EC macaque r02019 responded to the Gag241-291(G) pool. All of these responses were at or below 0.1% of the level for CD4+ lymphocytes (data not shown).
We next determined whether CD4+ T-cell responses broadened after resolution of primary viremia, as a durable, long-lasting control may also be aided by CD4+ T-cell responses (45). IFN-γ ELISPOT assays were performed on PBMC depleted of CD8+ cells at 20 weeks after SIV infection (Fig. 6). Controller r01027 made only one detectable CD4+ T-cell response to a peptide pool in Gag. Surprisingly, despite high levels of SIV viremia, the number of CD4+ T-cell responses made by progressor r91003 was comparable to those made by the EC macaques r00032 and r02019. However, all of the CD4+ T-cell responses detected in r91003 were low frequency, with between 50 and 150 SFC/106 CD8+ cell-depleted PBMC. In comparison, the ECs exhibited stronger CD4+ immune responses. EC r00032 mounted three CD4+ T-cell responses, while EC r02019 had six CD4+ T-cell responses at >150 SFC/106 CD8+ cell-depleted PBMC. Almost half of the CD4+ T-cell responses generated by r91003, r00032, and r02019 were against Gag, a protein that is frequently recognized by CD4+ T cells in successful vaccinees and ECs (10, 23, 25, 28, 68, 74). A number of the low-level responses (<100 SFC/106 CD8+ cell-depleted PBMC) from these three macaques were also found in frozen IFN-γ CD8+ cell-depleted PBMC ELISPOT assays between weeks 14 and 26 postinfection (data not shown).
FIG. 6.
Detection of CD4+ T-cell responses at 20 weeks postinfection by ex vivo CD8+ cell-depleted IFN-γ ELISPOT. Thirty-five peptide pools, with 10 15-mer peptides overlapping by 11 amino acids, spanning the entire SIVmac239 proteome (except for Pol and Env due to limited availability of PBMC) were tested in IFN-γ ELISPOT assays on PBMC depleted of CD8+ cells. Each column represents the number of SFC per 106 CD8+ cell-depleted PBMC directed against a single peptide pool at 20 weeks postinfection. Only positive responses are indicated. Background levels were subtracted from each well. Mean responses of <50 SFC per 1 × 106 cells (dashed line) were not considered positive.
Early viral variation in several Mamu-B*08-restricted CD8+ T-cell epitopes.
It has been shown that CD8+ T cells exert enormous selective pressures and can lead immunodeficiency virus escape as early as 4 weeks postinfection, (3, 5, 7, 8, 12, 19, 20, 26, 35, 36, 51, 59, 60, 64, 65). We therefore investigated whether viral escape was a factor in determining whether a Mamu-B*08-positive macaque develops high viral loads or controls SIV replication. Sequencing the entire SIVmac239 genome from plasma virus at 18 weeks postinfection revealed only a few substitutions in the clonal SIVmac239 virus (see Fig. S1 in the supplemental material). In total, mutations resulting in 33 amino acid substitutions (17 complete amino acid substitutions and 16 incomplete replacements) were found in plasma virus from the four SIV-infected macaques (Table 1). A deletion in the RNA that encodes the gp120 Env protein was also found in progressor macaque r91003 (positions 418 to 424) (see Fig. S1 in the supplemental material). Of these 33 amino acid substitutions, 7 were previously documented to be the result of suboptimal nucleotides that routinely mutate in the majority of Indian rhesus macaques in vivo, likely increasing the fitness of SIVmac239 (1). An additional two substitutions located in Env also do not appear to be associated with MHC class I expression (Table 1). Of the remaining 24 substitutions, 16 were within defined MHC class I epitopes, while 8 cannot be accounted for at this time.
TABLE 1.
Amino acid replacements in the circulating plasma virus of the four Mamu-B*08-positive macaques at 18 weeks after SIVmac239 infection
| Type of substitution | No. of amino acid replacements
|
||
|---|---|---|---|
| Complete | Mixed base | Total | |
| MHC class I associated | |||
| Mamu-B*08a | 6 | 4 | 10 |
| Mamu-A*01b | 2e | 4e | 6 |
| Mamu-A*02c | 0 | 0 | 0 |
| Not MHC associated | |||
| Suboptimal | 6 | 1 | 7 |
| Otherd | 2 | 0 | 2 |
| Unknown | 1 | 7 | 8 |
| Total | 17 | 16 | 33 |
Expressed by r91003 (progressor), r01027 (controller), r00032 (EC), and r02019 (EC).
Expressed by r91003 (progressor) and r01027 (controller).
Expressed by r00032 (EC).
Substitutions at position 67 in Env were previously detected in 33 of 35 SIV-infected macaques regardless of MHC class I genotype (60).
This number includes concomitant changes in the overlapping open reading frame for Vpr (see Fig. S1C in the supplemental material).
Mamu-B*08-restricted CD8+ T cells were responsible for selecting the majority of the viral variation in the replicating plasma virus at 18 weeks postinfection. We found 10 amino acid replacements in or near Mamu-B*08-restricted epitopes (Table 1; also see Fig. S1 in the supplemental material). Six of these were the result of complete nucleotide substitutions, while four were identified as mixed-base substitutions. Surprisingly, viral variation was observed in four of the eight known Mamu-B*08-restricted CD8+ T-cell epitopes (Vif123-131RL9, Vif172-179RL8, Nef136-147RL10, and Nef246-254RL9), with at least one mutation detected in each of the four macaques at 18 weeks postinfection.
Selective pressures exerted by Mamu-A*01-restricted CD8+ T cells accounted for six amino acid replacements found at 18 weeks postinfection (Table 1; also see Fig. S1 in the supplemental material). Escape mutations were detected in both Mamu-A*01-positive macaques (r91003 and r01027) in the Tat28-35SL8 epitope, consistent with the rapid evolution of this sequence (3, 59). Three concomitant changes in the overlapping open reading frame for Vpr were also identified. The only other substitution that was likely due to Mamu-A*01-restricted CD8+ T cells was in the Env726-735ST10 epitope in controller macaque r01027. No variation was identified in Mamu-A*02-restricted epitopes in EC macaque r00032.
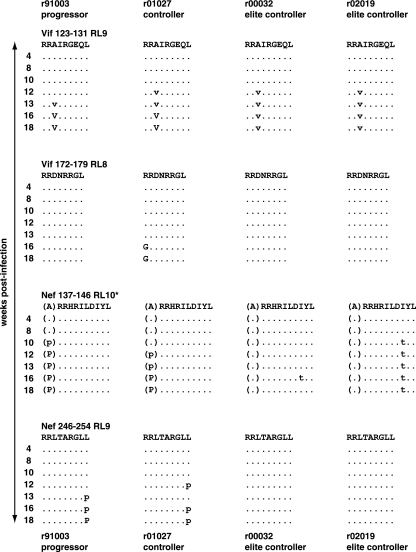
We further defined the viral ontogeny in these four Mamu-B*08-restricted epitopes (Vif123-131RL9, Vif172-179RL8, Nef136-147RL10, and Nef246-254RL9) by population sequencing of replicating plasma virus at seven time points between weeks 4 and 18 postinfection (Fig. 7). Epitope mutations were detected as early as 10 weeks after SIV infection in Nef136-147RL10 in the EC r02019. The majority of the viral variants detected at 18 weeks postinfection appeared between weeks 12 and 13 postinfection. Nonetheless, we saw viral variation in four of the eight Mamu-B*08-restricted epitopes, indicating selective pressure mediated by Mamu-B*08-restricted CD8+ T cells on multiple epitopes. A majority of these mutations were observed previously in the chronic phase, and selection was correlated with expression of Mamu-B*08 (48).
FIG. 7.
Mutations in Mamu-B*08-restricted CD8+ T-cell epitopes occurred as early as 10 weeks postinfection. The ontogeny of substitutions in Vif123-131RL9, Vif172-179RL8, Nef137-146RL10, and Nef245-254RL9 was followed by sequencing of plasma virus between 4 and 18 weeks after SIVmac239 infection. The Nef137-146RL10 epitope is annotated with an asterisk to signify that the amino acid residue immediately N-terminal to the epitope, alanine (A), is also displayed. Viral variation at this position is associated with Mamu-B*08 expression (48). Amino acids identical to the wild-type sequence are shown as dots. Complete amino acid replacements are shown in uppercase; sites of mixed-base heterogeneity are shown in lowercase.
Viral variation in the Mamu-B*08-restricted CD8+ T-cell epitopes appeared to be more pronounced in the progressor (r91003) and controller (r01027) macaques (Fig. 7). The Vif123-131RL9 epitope showed the same pattern of viral variation (alanine to valine) in all four macaques. By contrast, mutations in the second Vif epitope, Vif172-179RL8, were detected only in the controller r01027. The Nef136-146RL10 epitopes displayed two patterns of viral variation. The first pattern was seen in the two ECs and involved an isoleucine (I)-to-threonine (T) change at position eight of the epitope. The second pattern was found in the other two animals and involved a change of alanine (A) to proline (P) in the amino acid N-terminal to Nef136-146RL10 that may alter antigen processing (48). Interestingly, a similar A-to-P substitution immediately N-terminal of an HLA-B57-restricted HIV epitope in Gag is known to alter antigen processing and to abrogate recognition (18). The second Nef epitope, Nef246-254RL9, accumulated substitutions only in progressor r91003 and controller r01027. In both cases, there was a leucine (L)-to-proline (P) substitution at the C terminus of the epitope.
DISCUSSION
Previous investigations have linked particular MHC class I alleles to control of HIV replication (14, 15, 29, 34, 55), yet understanding how CD8+ T cells restricted by these protective alleles contribute to viral control remains a mystery. Recently, we identified an association between the Indian rhesus macaque MHC class I allele Mamu-B*08 and control of SIVmac239 replication (49). While identifying CD8+ T-cell responses restricted by this allele in EC macaques, we also discovered that the preliminary peptide binding motif of Mamu-B*08 appears to be similar to the peptide binding motif of HLA-B27 (48), an MHC class I allele associated with control of HIV replication in humans. The high percentage of Mamu-B*08-positive macaques that become ECs (∼50%) and the functional similarity of Mamu-B*08 to HLA-B27 make these MHC class I-defined macaques ideal for modeling human ECs. We therefore studied the immunopathogenic events of acute-phase SIV infection in four Mamu-B*08-positive macaques in an attempt to further understand CD8-mediated viral control in ECs.
In this observational study, we show that three of four Mamu-B*08-positive macaques controlled replication of the pathogenic SIVmac239 isolate (Fig. 1). Two of these macaques (r00032 and r02019) were ECs with plasma viral concentrations of approximately 1,000 vRNA copies/ml at 20 weeks postinfection. The third macaque (r01027) showed some measure of control of this highly pathogenic virus, with a plasma viral concentration of <20,000 vRNA copies/ml at 20 weeks postinfection. Only macaque r91003 had plasma viremia similar to the viral set points in the majority of animals that progressed to AIDS (∼5 × 105 vRNA copies/ml).
We then investigated how the majority of Mamu-B*08-positive macaques controlled viral replication. Recent experiments have demonstrated that the rapid depletion of the CD4+ memory T cells during the acute phase of HIV/SIV infection might be an important factor contributing to disease progression (44, 53). Interestingly, while the CD4+ memory T cells in the PBMC were depleted during primary SIV infection in two of the four Mamu-B*08-positive macaques, all four of the Mamu-B*08-positive macaques recovered their CD4+ memory T cells by the chronic phase of infection (Fig. 2). By contrast, four Mamu-B*08-negative macaques infected with SIVmac239 experienced acute-phase loss of their CD4+ memory T-cell subset and none of the Mamu-B*08-negative macaques recovered this important CD4+ T-cell subset. At 18 weeks postinfection, the progressor macaque (r91003) started to show signs of CD4+ memory T-cell depletion, likely related to the high plasma virus concentrations in this animal.
Mamu-B*08-restricted CD8+ T-cell responses contributed substantially to the acute phase of SIVmac239 infection (Fig. 4). However, the pattern of immunodominance varied from animal to animal, and breadth seemed to correlate with successful control of immunodeficiency virus replication. At 3 weeks postinfection, progressor r91003 focused its Mamu-B*08-restricted CD8+ T-cell responses primarily on only one of the eight mapped Mamu-B*08 CD8+ T-cell epitopes, Vif172-179 RL8. Vif172-179 RL8-specific cells accounted for 87% of the Mamu-B*08-restricted CD8+ T cells detected in this animal. By contrast, the two ECs (r00032 and r02019) divided their robust CD8+ T-cell responses among several Mamu-B*08-restricted epitopes (Fig. 3 to 5). Controller macaque r01027 also made several Mamu-B*08-restricted CD8+ T-cell responses at 3 weeks postinfection, but these were of a much lower magnitude than those of the ECs. Indeed, both EC macaques targeted at least the two epitopes in Vif (Vif123-131RL9 and Vif172-179RL8) and one epitope in Nef (Nef137-146RL10) with high-frequency CD8+ T-cell responses. Thus, high-frequency CD8+ T-cell responses against epitopes in Vif and Nef appeared to correlate with a successful disease outcome.
The Vif123-131RL9 response may be especially crucial in the control of viral replication. Vif123-131RL9-specific CD8+ T cells were generated to a sizeable frequency only in the three macaques that controlled viral replication (Fig. 3 to 5). Interestingly, we have recently shown that two SIVmac239Δnef-vaccinated Mamu-B*08-positive macaques controlled replication of a pathogenic heterologous challenge with SIVsmE660 (M. R. Reynolds et al., unpublished data). No replication of the challenge virus was seen in one of the two Mamu-B*08-positive macaques, whereas the other had a peak of 14,000 vRNA copies/ml at 2 weeks postchallenge. In the macaque that experienced replication of the SIVsmmE660 challenge virus, we detected only two vaccine-induced anamnestic CD8+ T-cell responses. Both of these were restricted by Mamu-B*08 responses (Vif123-131RL9 and Env573-581KL9). During the acute phase after SIVsmE660 challenge, the Env573-581KL9-specific CD8+ T cells expanded to a frequency of ∼1%. However, the Vif123-131RL9-specific CD8+ T-cell response was immunodominant, peaking at 4.84% of the level for CD3+ CD8+ lymphocytes in the peripheral blood at 3 weeks postchallenge, implicating this response in the control of viral replication.
We then examined whether escape from Mamu-B*08-restricted responses could account for progression or control of SIVmac239 replication. It has been previously shown that escape mutations could lead to loss of viral control in SIV-infected macaques (7, 35). Also, viral escape from the immunodominant HIV-specific response against the HLA-B27-restricted epitope Gag263-272KK10 has been associated with loss of control of viral replication (8, 20, 26, 36). We sequenced replicating plasma virus at 18 weeks postinfection and found that the majority of the amino acid replacements at this time were selected for by Mamu-B*08-restricted CD8+ T-cell responses (Table 1). By comparison, viral variation was not detected in any of the Mamu-A*02-restricted epitopes, and amino acid replacements were present in only two Mamu-A*01-restricted epitopes (Tat28-35SL8 and Env726-735ST10) (see Fig. S1 in the supplemental material). Four of the eight Mamu-B*08-restricted epitopes (Vif123-131RL9, Vif172-179RL8, Nef136-147RL10, and Nef246-254RL9) exhibited viral variation consistent with mutations identified in a previous study (48). These results may suggest that Mamu-B*08-restricted CD8+ T cells exert more selective pressure than other MHC class I-restricted responses during the critical early phase of infection. Alternatively, the mutations observed in these epitope sequences could occur in regions of the viral genome that are not under strong evolutionary constraints. Our group and others have previously shown that fitness costs may play a role in determining the rate at which escape mutations accumulate and revert in vivo (21, 22, 43).
We also followed the ontogeny of mutations by population sequencing these four Mamu-B*08-restricted epitopes at various time points. We discovered the first evidence of viral variation in circulating plasma virus at 10 weeks postinfection and mutations within several Mamu-B*08-restricted epitopes by 13 weeks postinfection (Fig. 7). Interestingly, the patterns of amino acid substitutions in the Nef137-146RL10 and Nef246-254RL9 epitopes differentiated the four Mamu-B*08-positive macaques into two separate groups, the ECs and the non-ECs. It will be intriguing to see if viral control is lost through the accumulation of additional mutations as the three macaques that control SIV replication progress further into the chronic phase of infection.
Similar to those in HLA-B27 and HLA-B57-positive individuals (6, 9, 38), the acute-phase CD8+ T-cell responses in Mamu-B*08-positive macaques were dominated by Mamu-B*08-specific CD8+ T-cell responses (Fig. 3 and Fig. 4). Surprisingly, Mamu-B*08 appeared to reduce the dominant influence of Mamu-A*01 on CD8+ T-cell responses. Previously, such immunodomination was described to occur in macaques expressing both Mamu-A*01 and Mamu-A*02 during the acute phase of SIVmac251 infection (58). Typically, in Mamu-A*01-positive macaques, the Mamu-A*01-restricted Tat28-35SL8- and Gag181-189CM9-specific CD8+ T cells account for >50% of the acute-phase SIV-specific immune responses (56). However, for both r91003 and r01027, the Mamu-A*01-restricted contribution was below 50%. Responses restricted by Mamu-A*02 in r00032 also appeared to be diminished in this Mamu-B*08-positive macaque compared to those in Mamu-A*02-positive macaques that do not express Mamu-B*08. It should be emphasized, however, that these data are derived from only three animals and that larger cohorts of Mamu-B*08-positive macaques expressing other MHC class I molecules should be studied to clarify this issue.
HIV disease progression in HLA-B27-positive humans appears to be associated with viral escape from the immunodominant Gag263-272KK10 response during the chronic phase of infection (8, 20, 26, 36). However, additional HIV-specific CD8+ T-cell responses may be involved in the initial control of viremia. Because immune responses in HIV-infected individuals are normally characterized using IFN-γ ELISPOT with consensus peptides, often after primary infection, definition of the entire breadth of the HLA-B27-restricted CD8+ T-cell responses may be incomplete. While preliminary, our current findings appear to indicate that the breadth of CD8+ T-cell responses restricted by protective MHC class I alleles, rather than a single immunodominant response, may be important in determining the control of replication of the AIDS virus.
Surprisingly, while Mamu-B*08 appears to bind similar peptides to HLA-B27, an immunodominant CD8+ T-cell response directed against Gag has not been identified for Mamu-B*08. Rather, Mamu-B*08 restricts robust CD8+ T-cell responses largely from Vif and Nef. Moreover, we are currently attempting to identify all of the SIV epitopes restricted by Mamu-B*08. At this time, we have identified only a single Gag-specific CD8+ T-cell response, and it is both low frequency and recognized in only a few Mamu-B*08-positive macaques (J. T. Loffredo et al., unpublished data). Hence, while CD8+ T cells targeting Gag have been shown to be extremely effective in controlling immunodeficiency virus replication (7, 8, 20, 26, 35, 36, 39, 51, 70), it may be possible for individuals to control viremia by directing responses against other proteins as well. Mamu-B*08-positive, SIV-infected macaques may offer an intriguing model for studying such a mechanism. Interestingly, both HLA-B27 and Mamu-B*08 present many epitopes containing two N-terminal basic amino acids. These peptides are relatively resistant to peptidase activity, and thus, those peptides may be more stable, and this may result in more efficient MHC class I antigen presentation (30).
Approximately 50% of Mamu-B*08-positive macaques become ECs, controlling replication of the pathogenic SIVmac239 isolate to <1,000 vRNA copies/ml (49). Given that the chronic-phase viral set points in 175 macaques that progressed to AIDS were ∼500,000 vRNA copies/ml, and approximately two-thirds of macaques die by 1 year postinfection (40), this level of control is remarkable. However, not all Mamu-B*08-positive macaques become ECs after SIVmac239 infection, and the immune system's role in successful viral containment remains difficult to define. Understanding how some of these macaques become progressors or controllers might give us key insights into how to make an effective HIV vaccine. Here, we provide evidence from a small yet provocative study that the immunodominance and breadth of CD8+ T-cell responses restricted by protective MHC class I alleles may facilitate the development of elite control. Further experiments are needed to explore this intriguing idea, especially given the fundamental implications that this might have for future vaccines.
Supplementary Material
Acknowledgments
We thank the MHC Genotyping Core at the WNPRC (William Rehrauer, Chrystal Glidden, Gretta Borchardt, and Debi Fisk) for genotyping our Indian rhesus macaques. We also gratefully acknowledge Gnankang Napoé, Emma Gostick, and David A. Price for assistance with the construction of MHC class I tetramers, Taeko Soma for QPCR, Kim Weisgrau for technical assistance, and Alex Blasky for assistance with DNA sequencing. Laura Valentine and Matthew Reynolds provided helpful discussions. We also thank the Virology, Genetics, Immunology, and Animal core laboratories as well as Research Support Services at the National Primate Research Center, University of Wisconsin—Madison (WNPRC) for technical assistance. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: complete SIVmac239 peptide sets (15-mer peptides overlapping by 11 amino acids) of Gag (item no. 6204), Vif (item no. 6205), Tat (item no. 6207), Pol (item no. 6443), Rev (item no. 6448), Vpr (item no. 6449), Vpx (item no. 6450), Env (item no. 6883), and full-length Nef (item no. 8762).
This research was supported by National Institutes of Health (NIH) contract HHSN266200400088C and NIH grants R01 AI049120, R01 AI052056, R24 RR015371, and R24 RR016038 to D.I.W. as well as R21 AI068586 to T.C.F. Additionally, this publication was made possible in part by grant number P51 RR000167 from the National Center for Research Resources (NCRR), a component of the NIH, awarded to the WNPRC. This project has also been funded in part by a grant from the Japan Health Sciences Foundation to D.I.W. This work was conducted in part at a facility constructed with support from Research Facilities Improvement grant numbers RR15459-01 and RR020141-01 (WNPRC).
This publication's contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCRR or the NIH.
Footnotes
Published ahead of print on 5 December 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alexander, L., L. Denekamp, S. Czajak, and R. C. Desrosiers. 2001. Suboptimal nucleotides in the infectious, pathogenic simian immunodeficiency virus clone SIVmac239. J. Virol. 754019-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. M., B. R. Mothe, J. Sidney, P. Jing, J. L. Dzuris, M. E. Liebl, T. U. Vogel, D. H. O'Connor, X. Wang, M. C. Wussow, J. A. Thomson, J. D. Altman, D. I. Watkins, and A. Sette. 2001. CD8+ lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule Mamu-A*01: implications for vaccine design and testing. J. Virol. 75738-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407386-390. [DOI] [PubMed] [Google Scholar]
- 4.Allen, T. M., J. Sidney, M. F. del Guercio, R. L. Glickman, G. L. Lensmeyer, D. A. Wiebe, R. DeMars, C. D. Pauza, R. P. Johnson, A. Sette, and D. I. Watkins. 1998. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J. Immunol. 1606062-6071. [PubMed] [Google Scholar]
- 5.Allen, T. M., M. Altfeld, S. C. Geer, E. T. Kalife, C. Moore, K. M. O'sullivan, I. Desouza, M. E. Feeney, R. L. Eldridge, E. L. Maier, D. E. Kaufmann, M. P. Lahaie, L. Reyor, G. Tanzi, M. N. Johnston, C. Brander, R. Draenert, J. K. Rockstroh, H. Jessen, E. S. Rosenberg, S. A. Mallal, and B. D. Walker. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J. Virol. 7913239-13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLoS Med. 3e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415335-339. [DOI] [PubMed] [Google Scholar]
- 8.Betts, M. R., B. Exley, D. A. Price, A. Bansal, Z. T. Camacho, V. Teaberry, S. M. West, D. R. Ambrozak, G. Tomaras, M. Roederer, J. M. Kilby, J. Tartaglia, R. Belshe, F. Gao, D. C. Douek, K. J. Weinhold, R. A. Koup, P. Goepfert, and G. Ferrari. 2005. Characterization of functional and phenotypic changes in anti-Gag vaccine-induced T cell responses and their role in protection after HIV-1 infection. Proc. Natl. Acad. Sci. USA 1024512-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bihl, F., N. Frahm, L. Di Giammarino, J. Sidney, M. John, K. Yusim, T. Woodberry, K. Sango, H. S. Hewitt, L. Henry, C. H. Linde, J. V. Chisholm III, T. M. Zaman, E. Pae, S. Mallal, B. D. Walker, A. Sette, B. T. Korber, D. Heckerman, and C. Brander. 2006. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J. Immunol. 1764094-4101. [DOI] [PubMed] [Google Scholar]
- 10.Boaz, M. J., A. Waters, S. Murad, P. J. Easterbrook, and A. Vyakarnam. 2002. Presence of HIV-1 Gag-specific IFN-gamma+IL-2+ and CD28+IL-2+ CD4 T cell responses is associated with nonprogression in HIV-1 infection. J. Immunol. 1696376-6385. [DOI] [PubMed] [Google Scholar]
- 11.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 686103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3205-211. [DOI] [PubMed] [Google Scholar]
- 13.Boyson, J. E., C. Shufflebotham, L. F. Cadavid, J. A. Urvater, L. A. Knapp, A. L. Hughes, and D. I. Watkins. 1996. The MHC class I genes of the rhesus monkey. Different evolutionary histories of MHC class I and II genes in primates. J. Immunol. 1564656-4665. [PubMed] [Google Scholar]
- 14.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 2831748-1752. [DOI] [PubMed] [Google Scholar]
- 15.Carrington, M., and S. J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54535-551. [DOI] [PubMed] [Google Scholar]
- 16.Daza-Vamenta, R., G. Glusman, L. Rowen, B. Guthrie, and D. E. Geraghty. 2004. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 141501-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deeks, S. G., and B. D. Walker. 2007. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27406-416. [DOI] [PubMed] [Google Scholar]
- 18.Draenert, R., S. Le Gall, K. J. Pfafferott, A. J. Leslie, P. Chetty, C. Brander, E. C. Holmes, S. C. Chang, M. E. Feeney, M. M. Addo, L. Ruiz, D. Ramduth, P. Jeena, M. Altfeld, S. Thomas, Y. Tang, C. L. Verrill, C. Dixon, J. G. Prado, P. Kiepiela, J. Martinez-Picado, B. D. Walker, and P. J. Goulder. 2004. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 199905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 51270-1276. [DOI] [PubMed] [Google Scholar]
- 20.Feeney, M. E., Y. Tang, K. A. Roosevelt, A. J. Leslie, K. McIntosh, N. Karthas, B. D. Walker, and P. J. Goulder. 2004. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 788927-8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedrich, T. C., E. J. Dodds, L. J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, D. T. Evans, R. C. Desrosiers, B. R. Mothe, J. Sidney, A. Sette, K. Kunstman, S. Wolinsky, M. Piatak, J. Lifson, A. L. Hughes, N. Wilson, D. H. O'Connor, and D. I. Watkins. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10275-281. [DOI] [PubMed] [Google Scholar]
- 22.Friedrich, T. C., C. A. Frye, L. J. Yant, D. H. O'Connor, N. A. Kriewaldt, M. Benson, L. Vojnov, E. J. Dodds, C. Cullen, R. Rudersdorf, A. L. Hughes, N. Wilson, and D. I. Watkins. 2004. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J. Virol. 782581-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedrich, T. C., L. E. Valentine, L. J. Yant, E. G. Rakasz, S. M. Piaskowski, J. R. Furlott, K. L. Weisgrau, B. Burwitz, G. E. May, E. J. Leon, T. Soma, G. Napoe, S. V. Capuano III, N. A. Wilson, and D. I. Watkins. 2007. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 813465-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 2962354-2360. [DOI] [PubMed] [Google Scholar]
- 25.Giraldo-Vela, J. P., R. Rudersdorf, C. Chung, Y. Qi, L. T. Wallace, B. Bimber, G. J. Borchardt, D. L. Fisk, C. E. Glidden, J. T. Loffredo, S. M. Piaskowski, J. R. Furlott, J. P. Morales-Martinez, N. A. Wilson, W. M. Rehrauer, J. D. Lifson, M. Carrington, and D. I. Watkins. 2007. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of SIV-infected rhesus macaque elite controllers. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 26.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3212-217. [DOI] [PubMed] [Google Scholar]
- 27.Goulder, P. J., and D. I. Watkins. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4630-640. [DOI] [PubMed] [Google Scholar]
- 28.Hel, Z., W. P. Tsai, E. Tryniszewska, J. Nacsa, P. D. Markham, M. G. Lewis, G. N. Pavlakis, B. K. Felber, J. Tartaglia, and G. Franchini. 2006. Improved vaccine protection from simian AIDS by the addition of nonstructural simian immunodeficiency virus genes. J. Immunol. 17685-96. [DOI] [PubMed] [Google Scholar]
- 29.Hendel, H., S. Caillat-Zucman, H. Lebuanec, M. Carrington, S. O'Brien, J. M. Andrieu, F. Schachter, D. Zagury, J. Rappaport, C. Winkler, G. W. Nelson, and J. F. Zagury. 1999. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J. Immunol. 1626942-6946. [PubMed] [Google Scholar]
- 30.Herberts, C. A., J. J. Neijssen, J. de Haan, L. Janssen, J. W. Drijfhout, E. A. Reits, and J. J. Neefjes. 2006. Cutting edge: HLA-B27 acquires many N-terminal dibasic peptides: coupling cytosolic peptide stability to antigen presentation. J. Immunol. 1762697-2701. [DOI] [PubMed] [Google Scholar]
- 31.Hutchinson, S. L., L. Wooldridge, S. Tafuro, B. Laugel, M. Glick, J. M. Boulter, B. K. Jakobsen, D. A. Price, and A. K. Sewell. 2003. The CD8 T cell coreceptor exhibits disproportionate biological activity at extremely low binding affinities. J. Biol. Chem. 27824285-24293. [DOI] [PubMed] [Google Scholar]
- 32.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaizu, M., G. J. Borchardt, C. E. Glidden, D. L. Fisk, J. T. Loffredo, D. I. Watkins, and W. M. Rehrauer. 2007. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8(+) T cell epitopes. Immunogenetics 59693-703. [DOI] [PubMed] [Google Scholar]
- 34.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2405-411. [DOI] [PubMed] [Google Scholar]
- 35.Kawada, M., H. Igarashi, A. Takeda, T. Tsukamoto, H. Yamamoto, S. Dohki, M. Takiguchi, and T. Matano. 2006. Involvement of multiple epitope-specific cytotoxic T-lymphocyte responses in vaccine-based control of simian immunodeficiency virus replication in rhesus macaques. J. Virol. 801949-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kestler, H., T. Kodama, D. Ringler, M. Marthas, N. Pedersen, A. Lackner, D. Regier, P. Sehgal, M. Daniel, N. King, et al. 1990. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science 2481109-1112. [DOI] [PubMed] [Google Scholar]
- 38.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432769-775. [DOI] [PubMed] [Google Scholar]
- 39.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 1346-53. [DOI] [PubMed] [Google Scholar]
- 40.King, N. W., L. V. Chalifoux, D. J. Ringler, M. S. Wyand, P. K. Sehgal, M. D. Daniel, N. L. Letvin, R. C. Desrosiers, B. J. Blake, and R. D. Hunt. 1990. Comparative biology of natural and experimental SIVmac infection in macaque monkeys: a review. J. Med. Primatol. 19109-118. [PubMed] [Google Scholar]
- 41.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 684650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo, A. M., J. S. Haukoos, M. D. Witt, M. L. Babaie, and R. J. Lewis. 2005. Recognition of undiagnosed HIV infection: an evaluation of missed opportunities in a predominantly urban minority population. AIDS Patient Care STDS 19239-246. [DOI] [PubMed] [Google Scholar]
- 43.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. S. John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10282-289. [DOI] [PubMed] [Google Scholar]
- 44.Li, Q., L. Duan, J. D. Estes, Z. M. Ma, T. Rourke, Y. Wang, C. Reilly, J. Carlis, C. J. Miller, and A. T. Haase. 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 4341148-1152. [DOI] [PubMed] [Google Scholar]
- 45.Lichterfeld, M., D. E. Kaufmann, X. G. Yu, S. K. Mui, M. M. Addo, M. N. Johnston, D. Cohen, G. K. Robbins, E. Pae, G. Alter, A. Wurcel, D. Stone, E. S. Rosenberg, B. D. Walker, and M. Altfeld. 2004. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J. Exp. Med. 200701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loffredo, J. T., J. Sidney, C. Wojewoda, E. Dodds, M. R. Reynolds, G. Napoe, B. R. Mothe, D. H. O'Connor, N. A. Wilson, D. I. Watkins, and A. Sette. 2004. Identification of seventeen new simian immunodeficiency virus-derived CD8+ T cell epitopes restricted by the high frequency molecule, Mamu-A*02, and potential escape from CTL recognition. J. Immunol. 1735064-5076. [DOI] [PubMed] [Google Scholar]
- 47.Loffredo, J. T., B. J. Burwitz, E. G. Rakasz, S. P. Spencer, J. J. Stephany, J. P. Vela, S. R. Martin, J. Reed, S. M. Piaskowski, J. Furlott, K. L. Weisgrau, D. S. Rodrigues, T. Soma, G. Napoe, T. C. Friedrich, N. A. Wilson, E. G. Kallas, and D. I. Watkins. 2007. The antiviral efficacy of simian immunodeficiency virus-specific CD8+ T cells is unrelated to epitope specificity and is abrogated by viral escape. J. Virol. 812624-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loffredo, J. T., T. C. Friedrich, E. J. Leon, J. J. Stephany, D. S. Rodrigues, S. P. Spencer, A. T. Bean, D. R. Beal, B. J. Burwitz, R. A. Rudersdorf, L. T. Wallace, S. M. Piaskowski, G. E. May, J. Sidney, E. Gostick, N. A. Wilson, D. A. Price, E. G. Kallas, H. Piontkivska, A. L. Hughes, A. Sette, and D. I. Watkins. 2007. CD8+ T cells from SIV elite controller macaques recognize Mamu-B*08-bound epitopes and select for widespread viral variation. PLoS ONE 2e1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loffredo, J. T., J. Maxwell, Y. Qi, C. E. Glidden, G. J. Borchardt, T. Soma, A. T. Bean, D. R. Beal, N. A. Wilson, W. M. Rehrauer, J. D. Lifson, M. Carrington, and D. I. Watkins. 2007. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 818827-8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lun, W. H., A. Takeda, H. Nakamura, M. Kano, K. Mori, T. Sata, Y. Nagai, and T. Matano. 2004. Loss of virus-specific CD4(+) T cells with increases in viral loads in the chronic phase after vaccine-based partial control of primary simian immunodeficiency virus replication in macaques. J. Gen. Virol. 851955-1963. [DOI] [PubMed] [Google Scholar]
- 51.Matano, T., M. Kobayashi, H. Igarashi, A. Takeda, H. Nakamura, M. Kano, C. Sugimoto, K. Mori, A. Iida, T. Hirata, M. Hasegawa, T. Yuasa, M. Miyazawa, Y. Takahashi, M. Yasunami, A. Kimura, D. H. O'Connor, D. I. Watkins, and Y. Nagai. 2004. Cytotoxic T lymphocyte-based control of simian immunodeficiency virus replication in a preclinical AIDS vaccine trial. J. Exp. Med. 1991709-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matano, T., R. Shibata, C. Siemon, M. Connors, H. C. Lane, and M. A. Martin. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 72164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mattapallil, J. J., D. C. Douek, B. Hill, Y. Nishimura, M. Martin, and M. Roederer. 2005. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 4341093-1097. [DOI] [PubMed] [Google Scholar]
- 54.Migueles, S. A., A. C. Laborico, W. L. Shupert, M. S. Sabbaghian, R. Rabin, C. W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, and M. Connors. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 31061-1068. [DOI] [PubMed] [Google Scholar]
- 55.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 972709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mothe, B. R., H. Horton, D. K. Carter, T. M. Allen, M. E. Liebl, P. Skinner, T. U. Vogel, S. Fuenger, K. Vielhuber, W. Rehrauer, N. Wilson, G. Franchini, J. D. Altman, A. Haase, L. J. Picker, D. B. Allison, and D. I. Watkins. 2002. Dominance of CD8 responses specific for epitopes bound by a single major histocompatibility complex class I molecule during the acute phase of viral infection. J. Virol. 76875-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muhl, T., M. Krawczak, P. Ten Haaft, G. Hunsmann, and U. Sauermann. 2002. MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 1693438-3446. [DOI] [PubMed] [Google Scholar]
- 58.Newberg, M. H., K. J. McEvers, D. A. Gorgone, M. A. Lifton, S. H. Baumeister, R. S. Veazey, J. E. Schmitz, and N. L. Letvin. 2006. Immunodomination in the evolution of dominant epitope-specific CD8+ T lymphocyte responses in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 176319-328. [DOI] [PubMed] [Google Scholar]
- 59.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8493-499. [DOI] [PubMed] [Google Scholar]
- 60.O'Connor, D. H., A. B. McDermott, K. C. Krebs, E. J. Dodds, J. E. Miller, E. J. Gonzalez, T. J. Jacoby, L. Yant, H. Piontkivska, R. Pantophlet, D. R. Burton, W. M. Rehrauer, N. Wilson, A. L. Hughes, and D. I. Watkins. 2004. A dominant role for CD8+-T-lymphocyte selection in simian immunodeficiency virus sequence variation. J. Virol. 7814012-14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Connor, D. H., B. R. Mothe, J. T. Weinfurter, S. Fuenger, W. M. Rehrauer, P. Jing, R. R. Rudersdorf, M. E. Liebl, K. Krebs, J. Vasquez, E. Dodds, J. Loffredo, S. Martin, A. B. McDermott, T. M. Allen, C. Wang, G. G. Doxiadis, D. C. Montefiori, A. Hughes, D. R. Burton, D. B. Allison, S. M. Wolinsky, R. Bontrop, L. J. Picker, and D. I. Watkins. 2003. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J. Virol. 779029-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Otting, N., C. M. Heijmans, R. C. Noort, N. G. de Groot, G. G. Doxiadis, J. J. van Rood, D. I. Watkins, and R. E. Bontrop. 2005. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc. Natl. Acad. Sci. USA 1021626-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pal, R., D. Venzon, N. L. Letvin, S. Santra, D. C. Montefiori, N. R. Miller, E. Tryniszewska, M. G. Lewis, T. C. VanCott, V. Hirsch, R. Woodward, A. Gibson, M. Grace, E. Dobratz, P. D. Markham, Z. Hel, J. Nacsa, M. Klein, J. Tartaglia, and G. Franchini. 2002. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J. Virol. 76292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, and A. J. McMichael. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354453-459. [DOI] [PubMed] [Google Scholar]
- 65.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 941890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reynolds, M. R., E. Rakasz, P. J. Skinner, C. White, K. Abel, Z. M. Ma, L. Compton, G. Napoe, N. Wilson, C. J. Miller, A. Haase, and D. I. Watkins. 2005. CD8+ T-lymphocyte response to major immunodominant epitopes after vaginal exposure to simian immunodeficiency virus: too late and too little. J. Virol. 799228-9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinson, S., W. A. Charini, M. H. Newberg, M. J. Kuroda, C. I. Lord, and N. L. Letvin. 2001. A commonly recognized simian immunodeficiency virus Nef epitope presented to cytotoxic T lymphocytes of Indian-origin rhesus monkeys by the prevalent major histocompatibility complex class I allele Mamu-A*02. J. Virol. 7510179-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rosenberg, E. S., J. M. Billingsley, A. M. Caliendo, S. L. Boswell, P. E. Sax, S. A. Kalams, and B. D. Walker. 1997. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 2781447-1450. [DOI] [PubMed] [Google Scholar]
- 69.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283857-860. [DOI] [PubMed] [Google Scholar]
- 70.Streeck, H., M. Lichterfeld, G. Alter, A. Meier, N. Teigen, B. Yassine-Diab, H. K. Sidhu, S. Little, A. Kelleher, J. P. Routy, E. S. Rosenberg, R. P. Sekaly, B. D. Walker, and M. Altfeld. 2007. Recognition of a defined region within p24 Gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J. Virol. 817725-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vogel, T. U., T. C. Friedrich, D. H. O'Connor, W. Rehrauer, E. J. Dodds, H. Hickman, W. Hildebrand, J. Sidney, A. Sette, A. Hughes, H. Horton, K. Vielhuber, R. Rudersdorf, I. P. De Souza, M. R. Reynolds, T. M. Allen, N. Wilson, and D. I. Watkins. 2002. Escape in one of two cytotoxic T-lymphocyte epitopes bound by a high-frequency major histocompatibility complex class I molecule, Mamu-A*02: a paradigm for virus evolution and persistence? J. Virol. 7611623-11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weiler, A., G. E. May, Y. Qi, N. Wilson, and D. I. Watkins. 2006. Polymorphisms in eight host genes associated with control of HIV replication do not mediate elite control of viral replication in SIV-infected Indian rhesus macaques. Immunogenetics 581003-1009. [DOI] [PubMed] [Google Scholar]
- 73.Weintrob, A. C., J. Giner, P. Menezes, E. Patrick, D. K. J. Benjamin, J. Lennox, C. D. Pilcher, J. J. Eron, and C. B. Hicks. 2003. Infrequent diagnosis of primary human immunodeficiency virus infection: missed opportunities in acute care settings. Arch. Intern. Med. 1632097-2100. [DOI] [PubMed] [Google Scholar]
- 74.Wilson, N. A., J. Reed, G. S. Napoe, S. Piaskowski, A. Szymanski, J. Furlott, E. J. Gonzalez, L. J. Yant, N. J. Maness, G. E. May, T. Soma, M. R. Reynolds, E. Rakasz, R. Rudersdorf, A. B. McDermott, D. H. O'Connor, T. C. Friedrich, D. B. Allison, A. Patki, L. J. Picker, D. R. Burton, J. Lin, L. Huang, D. Patel, G. Heindecker, J. Fan, M. Citron, M. Horton, F. Wang, X. Liang, J. W. Shiver, D. R. Casimiro, and D. I. Watkins. 2006. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 805875-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wojcechowskyj, J. A., L. J. Yant, R. W. Wiseman, S. L. O'Connor, and D. H. O'Connor. 2007. Control of simian immunodeficiency virus SIVmac239 is not predicted by inheritance of Mamu-B*17-containing haplotypes. J. Virol. 81406-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yant, L. J., T. C. Friedrich, R. C. Johnson, G. E. May, N. J. Maness, A. M. Enz, J. D. Lifson, D. H. O'Connor, M. Carrington, and D. I. Watkins. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 805074-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, Z. Q., T. M. Fu, D. R. Casimiro, M. E. Davies, X. Liang, W. A. Schleif, L. Handt, L. Tussey, M. Chen, A. Tang, K. A. Wilson, W. L. Trigona, D. C. Freed, C. Y. Tan, M. Horton, E. A. Emini, and J. W. Shiver. 2002. Mamu-A*01 allele-mediated attenuation of disease progression in simian-human immunodeficiency virus infection. J. Virol. 7612845-12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.