Abstract
The assembly of RNA replication complexes on intracellular membranes is an essential step in the life cycle of positive-sense RNA viruses. We have previously shown that Hsp90 chaperone complex activity is essential for efficient Flock House virus (FHV) RNA replication in Drosophila melanogaster S2 cells. To further explore the role of cellular chaperones in viral RNA replication, we used both pharmacologic and genetic approaches to examine the role of the Hsp90 and Hsp70 chaperone systems in FHV RNA replication complex assembly and function in Saccharomyces cerevisiae. In contrast to results with insect cells, yeast deficient in Hsp90 chaperone complex activity showed no significant decrease in FHV RNA replication. However, yeast with a deletion of the Hsp70 cochaperone YDJ1 showed a dramatic reduction in FHV RNA replication that was due in part to reduced viral RNA polymerase accumulation. Furthermore, the absence of YDJ1 did not reduce FHV RNA replication when the viral RNA polymerase and replication complexes were retargeted from the mitochondria to the endoplasmic reticulum. These results identify YDJ1 as an essential membrane-specific host factor for FHV RNA replication complex assembly and function in S. cerevisiae and are consistent with known differences in the role of distinct chaperone complexes in organelle-specific protein targeting between yeast and higher eukaryotes.
Genome replication of positive-sense RNA viruses occurs within membrane-associated macromolecular complexes (5). Although the assembly of these highly active enzymatic complexes in association with intracellular membranes is a critical step in the positive-sense RNA virus life cycle, the mechanisms responsible for viral protein translation, folding, and transport to the appropriate membrane compartment within cells during viral RNA replication complex assembly are poorly understood. The heat shock proteins (Hsps) are a diverse set of molecular chaperones that facilitate cellular protein translation, folding, and trafficking (15). These abundant chaperones also participate in the assembly of membrane-associated protein complexes (47), suggesting that positive-sense RNA viruses may also use cytosolic Hsps as chaperones to assemble viral RNA replication complexes. Consistent with this hypothesis, cellular chaperones have been associated with the replication of numerous positive-sense RNA viruses, including hepatitis C virus (HCV) (42), cucumber necrosis virus (39), brome mosaic virus (BMV) (40), tomato mosaic virus (29), and Sindbis virus (13).
To study the role of cellular chaperones in viral RNA replication complex assembly and function, we used Flock House virus (FHV), a versatile positive-sense RNA virus and well-studied member of the Nodaviridae family (2). The utility of FHV as a model pathogen derives in part from its relatively small genome and robust replication in multiple eukaryotic hosts, including Drosophila melanogaster (14, 26), Caenorhabditis elegans (23), and Saccharomyces cerevisiae (22, 25, 27, 32, 33). The FHV genome is bipartite and consists of two capped but nonpolyadenylated RNA segments (38). The larger 3.1-kb segment, RNA1, encodes protein A, the FHV RNA-dependent RNA polymerase (RdRp), which is essential for the assembly of functional viral RNA replication complexes (1, 2, 18, 22, 25, 33). The smaller 1.4-kb segment, RNA2, encodes the structural capsid protein precursor, which is dispensable for RNA replication but necessary for infectious virion production (2). During viral RNA replication, protein A generates a subgenomic 0.4-kb RNA, RNA3, which is colinear with the 3′ end of RNA1. RNA3 encodes the RNA interference suppressor protein B2 (21), which is required for FHV RNA replication in insects (21), plants (21), and nematodes (23), but not in yeast (33).
FHV RNA replication complexes assemble on the mitochondrial outer membrane in both insect cells (26) and yeast (25), and protein A is sufficient for their appropriate intracellular localization (25). FHV replication complexes are targeted and anchored to the mitochondrial outer membranes in part by an amino-proximal domain in protein A that resembles the transmembrane stop-transfer sequences present in several cellular mitochondrial outer membrane proteins (25, 27). The protein A mitochondrial targeting signal contains no discernible enzymatic function, as fully functional FHV RNA replication complexes are formed when the mitochondrial targeting signal is replaced with a sequence that contains an endoplasmic reticulum (ER)-targeting domain (27). Thus, FHV provides a versatile system to examine the role of both general and membrane-specific host factors in viral RNA replication complex assembly and function.
We have previously shown that the cellular chaperone Hsp90 facilitates the assembly of functional FHV RNA replication complexes in Drosophila S2 cells (8, 19), consistent with the demonstrated role of this abundant cytosolic chaperone in the transport of cellular mitochondrial proteins in higher eukaryotes (47). In this report, we further explore the role of cellular chaperones in FHV RNA replication complex assembly and function using S. cerevisiae as a eukaryotic host. The facile genetics and ready availability of yeast strains with deletions, mutations, or regulated expression of individual genes make S. cerevisiae a useful model host to examine the impact of unique cellular proteins on viral RNA replication (20, 39, 40). We demonstrate that disruption of Hsp90 chaperone complex activity does not significantly impact FHV RNA replication in S. cerevisiae. In contrast, deletion of YDJ1, an Hsp40 family cochaperone required for Hsp70 chaperone complex activity (7), significantly reduced FHV RNA production when replication complexes were targeted to the mitochondrial membrane but had only a marginal effect on ER-targeted FHV RNA replication complex function. These results demonstrate both host- and membrane-specific differences in the role of cellular chaperones in viral RNA replication complex assembly and function.
MATERIALS AND METHODS
Yeast strains, transformation, and culture conditions.
The diploid S. cerevisiae strain BY4743 (MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+ met15Δ0/+ ura3Δ0/ura3Δ0) was used for all experiments to minimize potential confounding effects related to second site mutations more common in haploid deletion strains. Wild-type (wt) BY4743 and diploid strains with homozygous deletions of SBA1, STI1, or YDJ1 were purchased from the American Type Culture Collection (Manassas, VA). Yeast were cultured and transformed as previously described (25), except that the temperature-sensitive phenotype of Δydj1 yeast required growth for 7 days after transformation for satisfactorily large colonies to develop. All experiments were performed at 25°C due to the temperature-sensitive nature of FHV RNA replication and to maintain consistency with published Drosophila studies (8, 19). For induction of Δsba1 and Δsti1 yeast, individual clones were transferred to liquid selective minimal medium containing 2% glucose and grown to stationary phase, washed with sterile distilled water, and resuspended in selective minimal medium with 2% galactose at an optical density at 600 nm (OD600) of 0.1, which was equivalent to approximately 2.5 × 106 cells per ml for both wt and deletion strains. For induction of Δydj1 yeast, cells were grown in liquid selective minimal medium with 2% raffinose, washed in sterile distilled water, and resuspended in selective minimal medium with 2% raffinose plus 2% galactose at an OD600 of 0.1. Unless otherwise stated, experiments were performed with two independently derived clones and results are representative of at least three independent experiments.
Plasmids.
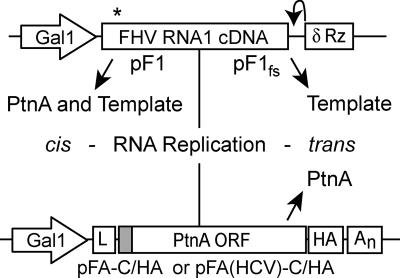
FHV expression plasmids pF1, pF1fs, and pFA-C/HA have been previously described (22, 25, 27, 33). The retargeted protein A expression plasmid pFA(HCV)-C/HA was generated by inserting the NheI/HindIII fragment from pFA-C/HA into pFA-HCV (27). FHV RNA1 expression plasmids pF1 and pF1fs encode galactose-inducible cDNA copies of FHV RNA1 with authentic viral 5′ and 3′ ends that are generated by precise transcription initiation and a hepatitis virus delta ribozyme, respectively, and thus contain the necessary cis elements to serve as replication templates (Fig. 1). In addition, the RNA1 transcribed from pF1 can be translated into protein A and initiate RNA replication in cis, which links protein A accumulation to viral RNA replication (Fig. 1, left). In contrast, the RNA template transcribed from plasmid pF1fs contains an early frameshift-induced stop codon and therefore cannot be translated into protein A. Functional protein A is provided from a second plasmid, pFA-C/HA or pFA(HCV)-C/HA, to initiate RNA replication in trans, where protein A accumulation is independent of viral RNA replication (Fig. 1, right). The protein A expression plasmids pFA-C/HA and pFA(HCV)-C/HA encode a galactose-inducible C-terminally hemagglutinin (HA)-tagged protein A open reading frame flanked by an upstream GAL1 leader sequence and a downstream CYC1 polyadenylation signal sequence. RNAs transcribed from pFA-C/HA and pFA(HCV)-C/HA are efficiently translated into protein A but do not have the necessary cis elements to serve as replication templates.
FIG. 1.
Schematics of FHV replicons. RNA replication in cis (left) is initiated when wild-type FHV RNA1 is transcribed from plasmid pF1 by host RNA polymerase II. Authentic 5′ and 3′ ends are generated by precise transcription initiation and a hepatitis delta ribozyme (δ Rz), respectively, and therefore RNA1 from pF1 can both be translated into protein A (PtnA) and function as an RNA replication template. RNA replication in trans (right) requires that the replication template and PtnA be provided by separate plasmids. The RNA1 template is transcribed from plasmid pF1fs, which contains an early frameshifting mutation (asterisk) that introduces a premature stop codon. Functional protein A is provided in trans from a second plasmid, either pFA-C/HA or pFA(HCV)-C/HA, which generates mRNAs with modified 5′ and 3′ untranslated regions that optimize their translation by introduction of the GAL leader (L) and CYC1 polyadenylation signal (An) but prevent their utilization as replication templates. The shaded box indicates the location of the membrane-targeting signal that is modified in pFA(HCV)-C/HA.
The yeast complementation plasmid pYDJ1 was generously provided by Masayuki Ishikawa (Hokkaido University, Sapporo, Japan) (40), and the plasmids pSC-FLGR and pSC-cGR, which encode the steroid-dependent full-length or constitutively active C-terminally truncated glucocorticoid receptor (GR), respectively, and the steroid-responsive reporter plasmid pSC-GRE-LacZ, were generously provided by Jorgé Iñiguéz (University of Michigan) (17). The plasmid pGAL-LacZ-HA, a galactose-inducible, HA-tagged β-galactosidase expression construct, was generated by inserting a PstI/SalI fragment containing the LacZ gene encoding a C-terminally HA-tagged protein into the PstI/SalI sites of pFA (33).
Antibodies and reagents.
Rabbit polyclonal antibodies against FHV protein A have been described previously (26). Rabbit polyclonal antibodies against the HA epitope tag were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and mouse monoclonal antibodies against porin and 3-phosphoglycerate kinase (PGK) were purchased from Molecular Probes (Eugene, OR). Rabbit polyclonal antibodies against yeast Ydj1p were generously provided by Masayuki Ishikawa (Hokkaido University, Sapporo, Japan). All secondary antibodies for immunoblotting were purchased from Jackson Immunoresearch (West Grove, PA). The Hsp90-specific inhibitor geldanamycin was purchased from Sigma (St. Louis, MO) and stored as a stock solution in dimethyl sulfoxide at −20°C. Deoxycorticosterone was generously provided by Jorgé Iñiguéz and was also stored as a stock solution at −20°C.
Immunoblot and Northern blot analyses.
Total protein was isolated from an equivalent number of yeast cells as previously described (25) and stored at −20°C until analysis. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted as previously described (19). Total RNA was isolated from yeast using hot acidic phenol as previously described (33) and stored at −80°C until analysis. RNA samples were denatured in sample buffer containing 60% formamide and 7% formaldehyde, separated on formaldehyde-1% agarose gels, transferred to ZetaProbe nylon membranes (Bio-Rad, Hercules, CA) by passive capillary transfer overnight, UV cross-linked at 120 mJ per cm2, and probed with either digoxigenin-UTP or [32P]UTP-labeled strand-specific riboprobes as previously described (26). Protein and RNA bands were quantitated by densitometry using either AlphaEaseFC (Alpha Innotech, San Leandro, CA) or ImageQuant TL (Amersham Biosciences, Piscataway, NJ) software.
Glucocorticoid receptor-based Hsp90 activity assay.
Yeast transformed with pSC-GRE-LacZ and pSC-FLGR were inoculated into selective medium in 96-well plates and grown in a moist chamber until saturation, diluted 40-fold into fresh selective test medium containing vehicle, 10 μM deoxycorticosterone, or 10 μM deoxycorticosterone and 10 μM geldanamycin, and incubated overnight at 25°C to induce GR-responsive and Hsp90-dependent β-galactosidase expression. For Hsp90- and steroid-independent GR activity, yeast transformed with pSC-GRE-LacZ and pSC-cGR were cultured in a similar manner but without deoxycorticosterone or geldanamycin. To quantitate reporter gene activity, cells were permeabilized with 120 mM sodium phosphate (pH 7.0), 10 mM potassium chloride, 1 mM magnesium sulfate, 2.5% (wt/vol) 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate, and 20 mM dithiothreitol for 15 min at 37°C and incubated with 0.5 mM chlorophenol red-β-d-galactopyranoside (Roche). Substrate conversion was measured by monitoring the change in OD550 over time, and β-galactosidase activity was normalized to cell density.
Membrane flotation and carbonate extraction.
Thirty OD600 units of yeast were harvested by centrifugation, washed once with distilled water, and converted to spheroplasts with 1,000 U lyticase in 2 ml spheroplasting (SP) buffer (1 M sorbitol, 0.1 M potassium phosphate [pH 7.6]) containing 0.2% β-mercaptoethanol for 20 min at 25°C. Spheroplasts were washed once in SP buffer, resuspended in 0.5 ml lysis buffer (50 mM HEPES [pH 7.5], 50 mM potassium chloride, 5 mM EDTA, 2 mM magnesium chloride, and a 1:100 dilution of yeast protease inhibitor cocktail [Sigma P8215]), stored on ice for 10 min, lysed with 20 strokes of a 2-ml Dounce homogenizer, and centrifuged at 500 × g for 5 min to pellet unlysed cells and nuclei. Clarified lysates were mixed with 50% Nycodenz to a final concentration of 37.5%, and 1.2 ml was loaded under a 3.4-ml 5 to 35% discontinuous Nycodenz gradient in Beckman 13- by 51-mm ultracentrifuge tubes and centrifuged at 100,000 × g in a Beckman MLS 50 rotor for 24 h at 4°C. Equal-volume fractions were collected from both top (low density [LD]) and bottom (high density [HD]) regions of the gradient as previously described (25, 27). For carbonate extraction experiments, LD fractions were pelleted by centrifugation at 17,000 × g for 15 min at 4°C, resuspended in either lysis buffer or in 0.1 M sodium carbonate (pH 11), incubated on ice for 30 min, and centrifuged at 17,000 × g for 15 min at 4°C to separate samples into carbonate-extracted supernatant and pellet fractions. Final samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting as described above.
Statistics.
A two-tailed Student's t test assuming unequal variances was used for all statistical analyses, and a P value of <0.05 was considered statistically significant.
RESULTS
FHV RNA replication in S. cerevisiae is independent of Hsp90 chaperone complex activity.
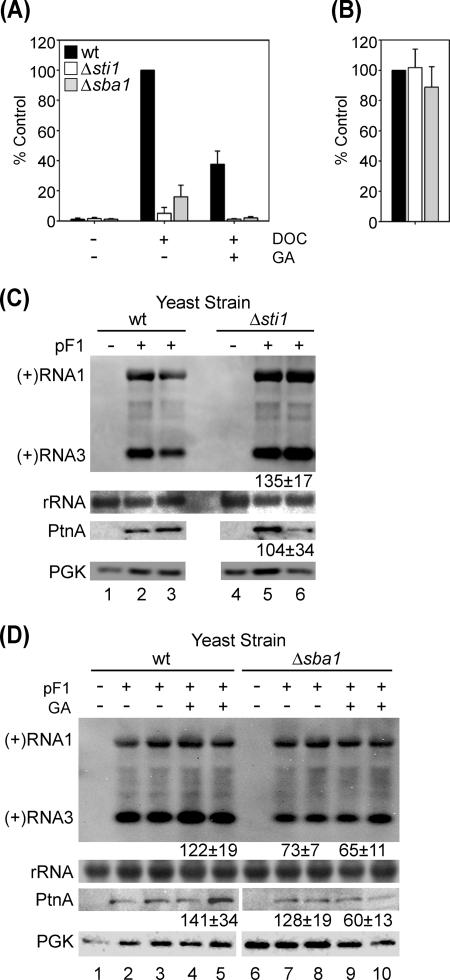
We have previously shown that inhibition of Hsp90 activity suppresses FHV RNA replication in Drosophila S2 cells (19). Since cellular chaperone complexes in yeast and higher eukaryotes are similar in composition but may differ in function (47), we examined the impact of the Hsp90 chaperone complex on FHV RNA replication in S. cerevisiae (Fig. 2). Yeast are less sensitive to Hsp90-specific pharmacologic inhibitors such as geldanamycin due in part to decreased cell permeability (3), and thus we used both pharmacologic and genetic approaches to disrupt S. cerevisiae Hsp90 chaperone complex activity. Yeast express two Hsp90s that are thought to be functionally redundant, which are encoded by the constitutive HSC82 and the inducible HSP82 genes (4). The deletion of both genes is lethal (28), and therefore we used yeast strains with homozygous deletions of the Hsp90 cochaperones STI1 and SBA1. These genes encode the yeast orthologs of Hsp-organizing protein and p23, respectively, and their deletions disrupt Hsp90 chaperone complex activity without significantly altering cell viability (3, 9). To verify the functional chaperone defect in Δsti1 and Δsba1 yeast, we used a GR-based reporter assay (17). The maturation of mammalian steroid receptors into a ligand-ready state is dependent upon a functional Hsp90 chaperone complex (31). Since the S. cerevisiae genome does not encode a GR, we transformed yeast with both a mammalian GR expression plasmid and a glucocorticoid response element-driven β-galactosidase reporter plasmid, induced cells with deoxycorticosterone, and measured reporter gene activity in a β-galactosidase assay (Fig. 2A). In wt BY4743 yeast transformed with the Hsp90-dependent reporter system, 10 μM deoxycorticosterone induced the robust production of β-galactosidase. Consistent with the reduced sensitivity of yeast to the inhibitory activity of pharmacologic Hsp90 inhibitors (3), 10 μM geldanamycin reduced β-galactosidase reporter gene activity by 60% in wt yeast (Fig. 2A), in comparison to the >90% reduction of GR-dependent reporter gene activity in Drosophila S2 cells treated with 5 μM geldanamycin (19). In contrast to results with wt yeast, deletion of STI1 reduced β-galactosidase activity by greater than 90% even in the absence of geldanamycin (Fig. 2A). Similar results were obtained with Δsba1 yeast, where in the absence of geldanamycin, steroid induced β-galactosidase activity was 16% of wt, and this residual activity was reduced to background levels with 10 μM geldanamycin (Fig. 2A). A truncated and constitutively active form of the GR without ligand-binding domains, which is Hsp90 and steroid independent, induced equivalent β-galactosidase activity in wt, Δsti1, and Δsba1 yeast cells (Fig. 2B), indicating that the decrease in deoxycorticosterone-induced reporter activity seen in the absence of STI1 and SBA1 was due to a defect in Hsp90 chaperone complex-dependent GR maturation.
FIG. 2.
FHV RNA replication in S. cerevisiae is independent of Hsp90 chaperone complex activity. (A) Inducible Hsp90-dependent GR activity. Wild-type (black bars), Δsti1 (white bars), and Δsba1 (gray bars) yeast cells transformed with pSC-GRE-LacZ and pSC-FLGR were induced with 10 μM deoxycorticosterone (DOC) in the presence or absence of 10 μM geldanamycin (GA), and β-galactosidase activity was measured in a colorimetric assay. Results are expressed as the percent activity relative to DOC-induced β-galactosidase in wt yeast. (B) Constitutive Hsp90-independent GR activity. Wild-type (black bars), Δsti1 (white bars), and Δsba1 (gray bars) yeast cells transformed with pSC-GRE-LacZ and pSC-cGR were assayed for β-galactosidase activity, and results are expressed as the percent activity relative to wt yeast. (C) FHV RNA replication in Δsti1 yeast. Yeast transformed with the vector control (lanes 1 and 4) or pF1 (lanes 2, 3, 5, and 6) were induced with galactose for 24 h, and total protein and RNA were isolated and analyzed by immunoblotting and Northern blotting, respectively. The positions of FHV genomic and subgenomic positive-sense RNA [(+)RNA1 and (+)RNA3, respectively] are shown on the left. rRNA and PGK are shown as loading controls. (D) FHV RNA replication in Δsba1 yeast. Yeast cells transformed with vector control (lanes 1 and 6) or pF1 (lanes 2 to 5 and 7 to 10) were induced with galactose for 24 h in the presence of vehicle (lanes 1 to 3 and 6 to 8) or 10 μM GA (lanes 4, 5, 9, and 10), and total protein and RNA were isolated and analyzed by immunoblotting and Northern blotting, respectively. Labels and loading controls are as described for panel C. Numbers represent the percent accumulation of (+)RNA3 and protein A (PtnA) in Δsti1 or Δsba1 samples compared to the wt control (lanes 2 and 3). There was no difference in growth between wt, Δsti1, and Δsba1 yeast cells after induction with galactose (data not shown). When cropping was necessary, all panels were from the same exposure of the blot, and all contrast adjustments to the initial image were done prior to cropping.
To examine the impact of Hsp90 activity on FHV RNA replication in yeast, we transformed wt, Δsti1, and Δsba1 yeast with pF1, induced cells with galactose for 24 h, and analyzed FHV protein A and RNA accumulation by immunoblotting and Northern blotting, respectively (Fig. 2C and D). In contrast to the potent inhibitory effect on Hsp90-dependent reporter gene activity (Fig. 2A), the deletion of STI1 did not suppress FHV RNA replication, measured by positive-strand (+)RNA1, (+)RNA3, or protein A accumulation (Fig. 2C, lanes 5 and 6). Although there was an approximate 30% reduction in FHV RNA replication in Δsba1 yeast (Fig. 2D, lanes 7 and 8), this was potentially due to an Hsp90-independent function of SBA1 (11, 12), as the addition of geldanamycin did not further suppress FHV RNA replication in Δsba1 yeast (Fig. 2D, lanes 9 and 10) despite its inhibitory effect on residual GR-dependent reporter gene activity (Fig. 2A). Furthermore, we saw no reduction in FHV RNA replication in wt yeast treated with geldanamycin (Fig. 2D, lanes 4 and 5), despite a 60% reduction in Hsp90 complex chaperone activity (Fig. 2A). Taken together, these results indicated that FHV RNA replication in S. cerevisiae was not dependent upon a functional Hsp90 chaperone complex.
FHV RNA replication in S. cerevisiae is dependent on the Hsp70 cochaperone YDJ1.
One previously described difference in the chaperone activity between yeast and higher eukaryotes is the differential role of Hsp70 and Hsp90 in the delivery of preproteins to the mitochondrial import machinery. In higher eukaryotes, mitochondrial protein targeting and import are facilitated by both the Hsp70 and Hsp90 chaperone complexes, whereas in yeast only the Hsp70 complex is required (47). FHV RNA replication complexes assemble on mitochondrial membranes in both Drosophila S2 cells and yeast (25, 26). Since our results indicated that FHV RNA replication was not dependent upon functional Hsp90 chaperone complex activity in S. cerevisiae, we subsequently examined the role of the Hsp70 chaperone complex. S. cerevisiae encodes at least nine cytosolic isoforms of Hsp70, some of which are thought to have partially overlapping functions (43). The Hsp70 SSA subfamily has been specifically implicated in import into and transport across cellular membranes (6, 10), suggesting that this group of yeast cytosolic Hsp70 chaperones may be particularly relevant to FHV RNA replication complex assembly and function. Deletion of all four SSA genes is lethal (43), and therefore to examine the role of the Hsp70 chaperone complex in FHV RNA replication we used a diploid yeast strain with a homozygous deletion of the Hsp70 cochaperone YDJ1. YDJ1 encodes the yeast ortholog of Hsp40/DnaJ, whose deletion is not lethal but still disrupts Hsp70 chaperone complex activity (7).
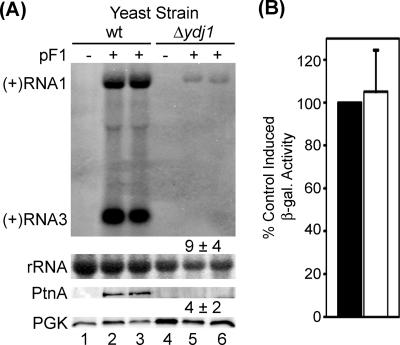
Initial experiments indicated that deletion of YDJ1 in the BY4743 background resulted in a prominent growth defect in selective minimal medium with glucose as the carbon source that produced a doubling time approximately two- to threefold longer than wt yeast and induced a growth arrest in selective minimal medium with galactose as the sole carbon source (data not shown). Thus, to induce FHV replicon expression from the GAL1 promoter-driven plasmid pF1, we initially cultured Δydj1 and wt yeast in selective medium with raffinose and transferred cells to a raffinose-galactose combination to induce replicon expression. To minimize the effects of cell growth on RNA replication, we harvested cells prior to stationary phase for both wt and Δydj1 yeast. Under these growth and induction conditions, deletion of YDJ1 greatly reduced FHV RNA replication in yeast when measured by the accumulation of (+)RNA1, (+)RNA3, or protein A (Fig. 3A). To ensure that the defect in FHV RNA replication was not due to reduced GAL1 promoter activity in the absence of YDJ1, we transformed yeast with the reporter plasmid pGAL-LacZ-HA, induced for 16 h, and assayed for β-galactosidase activity (Fig. 3B). There was no difference between wt and Δydj1 yeast in galactose-induced β-galactosidase activity, indicating that the reduced FHV RNA replication in Δydj1 yeast was not due to a defect in GAL1 promoter activity. Since FHV protein A accumulation and RNA replication are linked processes in cells transformed with pF1 (Fig. 1), we could not determine from these experiments whether the primary defect in Δydj1 yeast was reduced viral RNA polymerase accumulation or replication complex function. Nevertheless, these results suggested that in contrast to the Hsp90 cochaperones STI1 and SBA1, the Hsp70 cochaperone YDJ1 is important for efficient FHV RNA replication in S. cerevisiae.
FIG. 3.
FHV RNA replication is dependent on the Hsp70 cochaperone YDJ1. (A) wt and Δydj1 yeast cells transformed with vector control (lanes 1 and 4) or pF1 (lanes 2, 3, 5, and 6) were induced with galactose for 24 h, and total protein and RNA were isolated and analyzed by immunoblotting and Northern blotting, respectively. Results are presented as described for Fig. 2. Total protein and RNA recovery in Δydj1 yeast cells were increased compared to an equivalent number of wt yeast cells, and therefore sample loading was adjusted and resulted in underloading for Δydj1 RNA samples. Numbers represent the percent accumulation of (+)RNA3 and protein A compared to wt control and normalized to total RNA and protein levels (lanes 2 and 3). (B) GAL1 promoter activity in Δydj1 yeast. wt (black bars) and Δydj1 (white bars) yeast transformed with pGAL-LacZ-HA were induced with galactose for 16 h, and β-galactosidase activity was measured in a colorimetric assay. Results were adjusted to cell number and are expressed as the percent activity relative to wt-induced samples. β-Galactosidase activity in uninduced samples was <1% of wt induced levels.
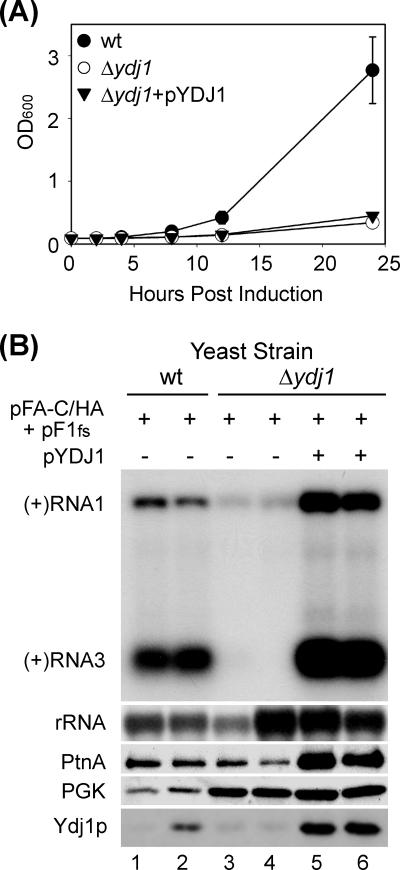
We further explored FHV RNA replication in Δydj1 yeast using the two-plasmid trans replication system depicted in Fig. 1 to separate viral RNA polymerase accumulation from function and complemented Δydj1 mutant yeast with a constitutive exogenous copy of the cochaperone expressed from plasmid pYDJ1 (Fig. 4). We transformed wt, Δydj1, and pYDJ1-complemented Δydj1 yeast with pFA-C/HA and pF1fs, induced with raffinose-galactose and initially examined cell growth for 24 h under induction conditions (Fig. 4A). Proliferation of Δydj1 yeast was markedly reduced compared to wt yeast, with calculated doubling times of 12 h and 5 h, respectively. Surprisingly, this growth defect was not rescued by pYDJ1-mediated complementation, despite Ydj1p overexpression (Fig. 4B, Ydj1p blot lanes 5 and 6), potentially due to unregulated cochaperone expression at 25°C with galactose-containing medium, as the growth defect in Δydj1 yeast was fully complemented by pYDJ1 in dextrose-containing medium at both 25°C and 37°C (data not shown). Nevertheless, the similar growth phenotypes of Δydj1 yeast in the presence and absence of pYDJ1 complementation allowed us to directly examine the impact of the Hsp70 cochaperone on FHV RNA replication without potential confounding effects related to cell growth.
FIG. 4.
FHV RNA replication in trans in Δydj1 yeast. (A) Growth of wt, Δydj1, and complemented (Δydj1+pYDJ1) yeast cells. Yeast cells transformed with pFA-C/HA and pF1fs were induced in selective medium with 2% raffinose plus 2% galactose, and cell density was measured by spectrophotometry (OD600). Note that Δydj1 and the complemented (Δydj1+pYDJ1) yeast cells have the same growth kinetics. The wt yeast were saturated at an OD600 of 4 to 5 in selective medium with 2% raffinose and 2% galactose (data not shown). (B) FHV RNA replication in wt, Δydj1, and complemented Δydj1 yeast cells. Yeast cells transformed with pFA-C/HA and pF1fs were induced for 24 h, and total RNA and protein were isolated from an equal number of cells and analyzed by Northern blotting and immunoblotting, respectively. Note that total RNA and protein accumulation were increased in the Δydj1 strain in both the presence and absence of complementation. We confirmed the genetic deletion of the YDJ1 locus in Δydj1 yeast by PCR (data not shown), and thus the faint bands in lanes 3 and 4 in the Ydj1p blot represent comigrating cellular proteins that cross-reacted with the Ydj1p antiserum, whereas the reduced signal in lane 1 was due to both underloading (note the PGK blot) and replicate variability.
We analyzed FHV RNA replication and protein A accumulation in wt, Δydj1, and pYDJ1-complemented Δydj1 yeast cells transformed with pFA-C/HA and pF1fs at 24 h after galactose induction by quantitative Northern blotting and immunoblotting (Fig. 4B). FHV RNA replication in trans was significantly reduced in Δydj1 compared to wt yeast (Fig. 4B, compare upper blot lanes 1 and 2 to lanes 3 and 4), consistent with cis replication results (Fig. 3). Furthermore, FHV RNA replication was rescued by complementation with pYDJ1 (Fig. 4B, lanes 5 and 6), in contrast to the growth phenotype noted above. When we quantified FHV RNA accumulation normalized to cell number, Δydj1 yeast displayed a >70% reduction in (+)RNA3 levels compared to wt, whereas complementation with pYDJ1 resulted in an increase in (+)RNA3 accumulation to levels greater than in wt yeast (Table 1). Similar results were seen with genomic (+)RNA1 accumulation, although the reduction in Δydj1 yeast was attenuated, likely due to the contribution of replication-independent genomic (+)RNA1 production from host RNA polymerase II-directed transcription. We also conducted in vitro RdRp assays to validate the in vivo results and observed a >80% reduction in FHV RdRp activity in membrane fractions isolated from Δydj1 yeast (data not shown). These results indicated that YDJ1 was necessary for efficient assembly and function of mitochondrion-targeted FHV RNA replication complexes in S. cerevisiae.
TABLE 1.
Quantitative analysis of the effect of YDJ1 on FHV RNA replication in S. cerevisiae
| Target and protein | Relative to no. of cells (% of wt)a
|
Relative to total RNA or protein (% of wt)a
|
||||
|---|---|---|---|---|---|---|
| Δydj1 | Δydj1 + pYDJ1b | Fold increasec | Δydj1 | Δydj1 + pYDJ1b | Fold increasec | |
| Mitochondrion-targeted protein A | ||||||
| (+)RNA1 | 73 ± 26 | 371 ± 63* | 5.1 | 44 ± 14 | 220 ± 40* | 5.0 |
| (+)RNA3 | 27 ± 19 | 319 ± 41* | 11.8 | 16 ± 11 | 188 ± 19* | 11.8 |
| Protein A | 140 ± 11 | 1,130 ± 445 | 8.1 | 32 ± 8 | 208 ± 29* | 6.5 |
| ER-targeted protein A | ||||||
| (+)RNA1 | 143 ± 30 | 314 ± 13* | 2.2 | 112 ± 32 | 245 ± 16* | 2.2 |
| (+)RNA3 | 102 ± 27 | 177 ± 25 | 1.7 | 80 ± 21 | 138 ± 21 | 1.7 |
| Protein A | 245 ± 83 | 513 ± 69 | 2.1 | 62 ± 15 | 160 ± 22* | 2.6 |
Data are the percentage relative to the wild-type control and are means ± standard errors of the means from three independent experiments. Note the complete rescue of protein A, (+)RNA1, and (+)RNA3 expression with pYDJ1 complementation.
An asterisk indicates a P value of <0.05 compared to Δydj1 yeast cells.
The fold increase of pYDJ1-complemented yeast over Δydj1 yeast.
In contrast to FHV RNA accumulation, the deletion of YDJ1 had less of an apparent impact on protein A accumulation (Fig. 4B). When we quantitated immunoblotting results and normalized them to cell number, there was no significant decrease in protein A accumulation in Δydj1 yeast, although complementation with pYDJ1 increased protein A recovery eightfold (Table 1). However, it should be noted that the slow growth phenotype of the Δydj1 mutant and complemented yeast (Fig. 4A) resulted in a three- to fivefold increase in the per cell accumulation of total protein and RNA compared to wt yeast (Fig. 4B, compare the rRNA gel and PGK blot in lanes 1 and 2 to lanes 3 through 6). When we normalized protein accumulation results to total protein loading controls, there was a 68% reduction in protein A accumulation in Δydj1 yeast that was fully complemented by pYDJ1 (Table 1). These results suggested that the FHV RNA replication defect in Δydj1 yeast was due in part to decreased viral polymerase accumulation.
YDJ1 is not essential for efficient ER-targeted FHV RNA replication complex activity.
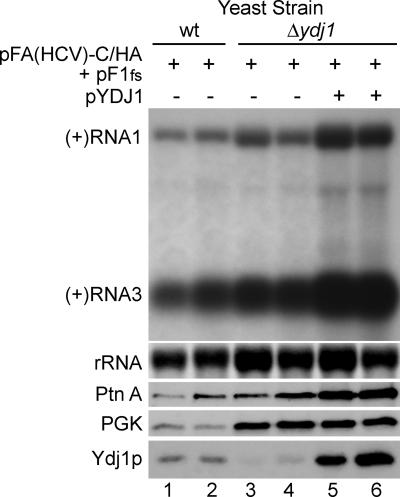
Although cytosolic Hsps are ubiquitous chaperones that influence a wide range of cellular processes, membrane-specific activities for particular chaperones have been described previously (6, 10). To examine whether the role of YDJ1 in FHV RNA replication complex assembly and function was membrane specific, we used a replicon-based system that retargets FHV RNA replication complexes to the ER (27). The wt amino-proximal mitochondrial targeting signal in protein A can be replaced with the ER-targeting signal from the HCV NS5B protein to generate the chimeric protein A expression plasmid pFA(HCV)-C/HA (27). We transformed wt, Δydj1, and pYDJ1-complemented Δydj1 yeast with pFA(HCV)-C/HA and pF1fs, induced with raffinose-galactose for 24 h, and analyzed (+)RNA1, (+)RNA3, and protein A accumulation (Fig. 5 and Table 1). In contrast to results with FHV RNA replication complexes targeted to mitochondria (Fig. 4B), deletion of YDJ1 had a minimal impact on ER-targeted FHV RNA replication complex activity (Fig. 5, lanes 3 and 4). After we normalized results for total protein and RNA levels, ER-targeted FHV protein A accumulation was reduced only 38%, compared to 68% for mitochondrion-targeted protein A in Δydj1 yeast, whereas there was no substantial decrease in (+)RNA1 or (+)RNA3 accumulation (Table 1). These in vivo results were validated by in vitro assays, where membrane fractions from Δydj1 yeast showed in vitro ER-retargeted FHV RdRp activity levels similar to wt yeast (data not shown). Complementation with pYDJ1 did increase (+)RNA1, (+)RNA3, and protein A accumulation (Fig. 5, lanes 3 and 4 compared to lanes 5 and 6), and a quantitative analysis showed an approximate twofold increase in FHV RNA and protein A accumulation with Ydj1p overexpression (Table 1). This is consistent with the finding that overexpression of Ydj1p also increased the accumulation of β-galactosidase expressed from a GAL1 promoter-driven plasmid (data not shown), suggesting that cochaperone overexpression stimulates galactose-inducible promoter activity. These results indicated that in contrast to FHV RNA replication complexes targeted to the mitochondria, those targeted to the ER do not require YDJ1 for efficient assembly and function.
FIG. 5.
YDJ1 is not essential for ER-targeted FHV RNA replication complex activity. wt, Δydj1, and complemented Δydj1 yeast cells transformed with pFA(HCV)-C/HA and pF1fs were induced for 24 h, and total RNA and protein were isolated and analyzed as described for Fig. 4.
Protein A is membrane associated in Δydj1 yeast.
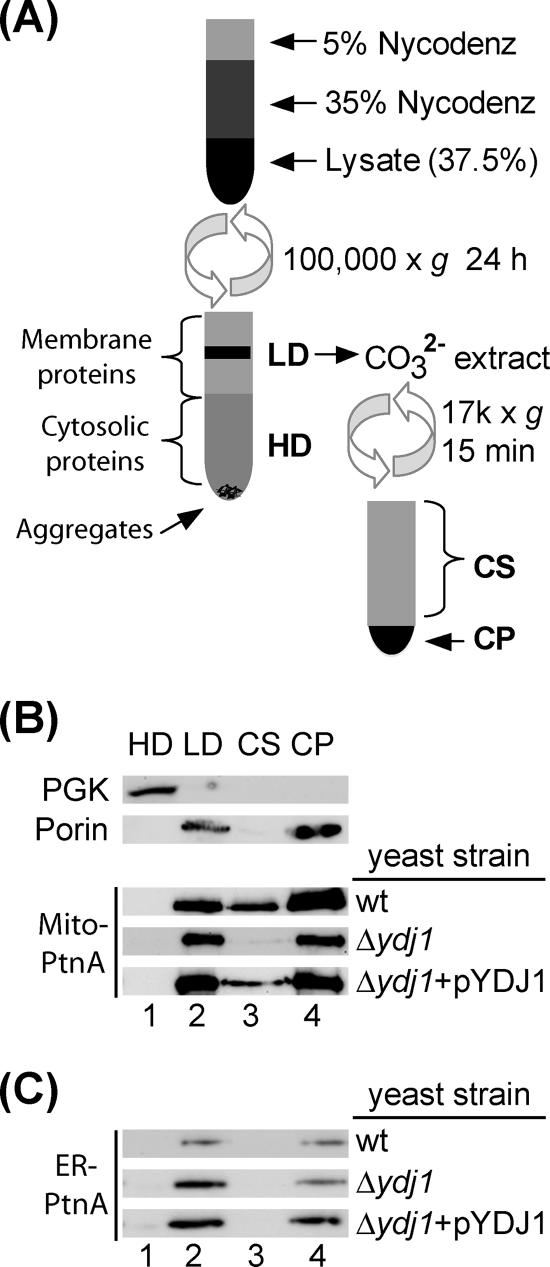
FHV protein A is tightly associated with intracellular membranes in S. cerevisiae (25), and this membrane association is important for complete viral RNA replication complex activity (44, 45). Thus, we examined whether the defect in FHV RNA replication in Δydj1 yeast was due in part to decreased membrane association of the viral polymerase (Fig. 6). Protein A has an amino-proximal transmembrane domain that mediates its membrane association and renders it resistant to alkaline extraction (25). We used this biochemical characteristic in a density-based flotation technique to examine protein A membrane association in Δydj1 yeast (Fig. 6A). We subjected whole-cell lysates to equilibration centrifugation in Nycodenz gradients and isolated LD and HD fractions, which represented membrane-associated and soluble proteins, respectively (25). The LD fraction was further subjected to alkaline extraction and differential centrifugation to separate peripheral from integral membrane proteins. We analyzed fractions by immunoblotting for FHV protein A, the cytosolic enzyme PGK, and the integral membrane protein porin (Fig. 6B). Consistent with previous observations (25, 26), the majority of protein A was recovered in the LD fraction from wt yeast, and neither the deletion of YDJ1 nor its complementation altered this distribution (Fig. 6B, lanes 1 and 2). Furthermore, 84%, 91%, and 81% of membrane-associated protein A was resistant to alkaline extraction in LD fractions from wt, Δydj1, and pYDJ1-complemented strains, respectively (Fig. 6B, lanes 3 and 4). We obtained similar results with ER-retargeted protein A (Fig. 6C). These results indicated that the functional defect in RNA replication complex activity in Δydj1 yeast was not due to grossly altered protein A membrane association.
FIG. 6.
Protein A is tightly membrane associated in Δydj1 yeast. (A) Schematic of membrane flotation protocol. Whole-cell lysates were mixed with Nycodenz to 37.5%, loaded under a discontinuous 5 to 35% Nycodenz gradient, and spun to equilibrium at 100,000 × g. Equal-volume samples were taken from the top LD fraction and bottom HD fraction. LD fractions were further subjected to carbonate extraction at pH 11 for 30 min on ice and subsequently pelleted and separated into supernatant (CS) and pellet (CP) fractions by differential centrifugation. (B) Membrane flotation and carbonate extraction of membranes from wt, Δydj1, and complemented (Δydj1+ pYDJ1) yeast cells transformed with pFA-C/HA and pF1fs. Yeast PGK and porin are cytosolic and membrane proteins, respectively. (C) Membrane flotation and carbonate extraction of membranes from wt, Δydj1, and complemented (Δydj1+ pYDJ1) yeast cells transformed with pFA(HCV)-C/HA and pF1fs.
DISCUSSION
In this report we examined the role of cellular chaperone complexes in FHV RNA replication complex assembly and function in S. cerevisiae. We demonstrated that deletion of YDJ1, an Hsp70 cochaperone, selectively reduced viral RNA replication when replication complexes were targeted to the mitochondria but not those targeted to the ER. The defect in mitochondrion-targeted FHV RNA replication was due in part to decreased viral polymerase accumulation. These results identify YDJ1 as a cellular host factor essential for mitochondrion-targeted FHV RNA replication in yeast. Furthermore, they demonstrate that S. cerevisiae and FHV is a versatile host-pathogen model system to investigate the role of membrane-specific host factors in viral RNA replication complex assembly and function.
The observation that FHV RNA replication in yeast was not significantly reduced in the absence of functional Hsp90 chaperone complex activity (Fig. 2) was in contrast to our results in Drosophila S2 cells, where direct genetic or pharmacologic disruption of Hsp90 function greatly reduced FHV RNA replication (19). One potential explanation for these contradictory results is the previously described differences in chaperone complex activity between yeast and higher eukaryotes with respect to mitochondrial protein targeting (47). The delivery of cellular preproteins to the mitochondrial import machinery is facilitated by both the Hsp70 and Hsp90 chaperone complexes in higher eukaryotes, whereas only the Hsp70 chaperone complex is required in yeast (47). This suggests that FHV uses established cytosolic chaperone pathways to facilitate the synthesis and transport of its RNA polymerase to the appropriate cellular membrane during replication complex assembly. Alternatively, FHV may require Hsp90 activity that is independent of the cochaperones STI1 and SBA1, similar to the direct role of Hsp90 in the maintenance of the cystic fibrosis transmembrane conductance regulator in yeast (46). However, the selective and direct Hsp90 inhibitor geldanamycin had no effect on FHV RNA replication in yeast despite its moderate suppression of Hsp90-dependent GR activity (Fig. 2). Recent work has identified an extensive network of cochaperones used by Hsp70 and Hsp90 in yeast that has raised the possibility of novel chaperone activities (36, 48), and studies are currently under way to examine the contribution of the complete set of known and hypothesized Hsp70 and Hsp90 cochaperones on FHV RNA replication.
The demonstration that the Hsp70 cochaperone YDJ1 was essential for efficient FHV RNA replication is consistent with its previous identification as a host factor necessary for BMV RNA replication (40). However, there are significant differences in the roles that YDJ1 may play in FHV and BMV RNA replication complex assembly and function. The most obvious difference is that BMV normally assembles its viral RNA replication complexes on the ER (34, 35), whereas we found no significant defect in Δydj1 yeast when FHV RNA replication complexes were retargeted to this intracellular membrane compartment (Fig. 5). The ubiquitous nature of Hsp70 chaperone complex activity on numerous cellular processes made the differential effect of YDJ1 deletion on FHV RNA replication complexes targeted to mitochondria or the ER an unanticipated result. One potential explanation for the selective defect in mitochondrion-targeted replication complexes is the observation that ER-retargeted FHV RNA replication complexes demonstrate increased activity via an unknown mechanism (27), and the deletion of YDJ1 may have been insufficient to significantly impact maximal or near-maximal RNA replication complex function. However, Ydj1p overexpression enhanced ER-retargeted FHV RNA replication (Fig. 5), suggesting that maximal replication complex activity was not achieved in wt yeast. Furthermore, ongoing studies in our laboratory have identified yeast deletions that selectively disrupt ER-retargeted FHV RNA replication, including deletions of the Hsp90 cochaperones STI1 and SBA1 (unpublished data). These results suggest that there are distinct membrane-specific chaperone requirements for viral RNA replication complex assembly and function and that the FHV system in S. cerevisiae provides a convenient platform to identify and characterize the molecular mechanisms underlying these requirements.
An additional difference between the role of YDJ1 in FHV and BMV RNA replication is that the Hsp70 cochaperone facilitates assembly of BMV RNA replication complexes by maintenance of viral polymerase cytosolic solubility prior to membrane association but does not influence its accumulation or recruitment to the ER membrane (40). Although we also found normal membrane association of the FHV RNA polymerase in Δydj1 yeast (Fig. 6), a detailed quantitative analysis revealed that Ydj1p facilitated protein A accumulation (Table 1). Furthermore, we found no evidence for FHV protein A aggregation in Δydj1 yeast (Fig. 6), whereas the BMV RNA polymerase forms high-density cytosolic aggregates in the absence of functional Ydj1p (40). However, immunofluorescence microscopy demonstrated that a small fraction of wild-type FHV protein A did not colocalize with mitochondrial markers in Δydj1 yeast (data not shown), suggesting that Ydj1p could influence the cytosolic fate of the FHV RNA polymerase but was not necessary to prevent the formation of insoluble aggregates (Fig. 6). These results indicated that cellular chaperones can have virus-specific effects on positive-strand RNA virus replication, consistent with the limited degree of overlap in host factors identified in genome-wide screens of related positive-strand RNA viruses (20, 30).
There are three general hypotheses to explain the role of cellular chaperones in positive-strand RNA virus replication. The first hypothesis suggests a direct role whereby cellular chaperones physically yet transiently interact with viral RNA replication complex components to facilitate protein folding and stabilization during translation, intracellular targeting, or membrane association. The presence of hydrophobic viral proteins that are required for the association of RNA replication complexes with intracellular membranes (25, 37, 41) may necessitate the use of cytosolic chaperones to prevent premature or inappropriate protein folding prior to membrane association, similar to the role of these chaperones in the maturation of endogenous hydrophobic cellular proteins (15, 16). The second hypothesis also suggests a direct role whereby cellular chaperones constitute an essential component of the functional membrane-associated viral RNA replication complex and may function to stabilize viral proteins or RNA. This hypothesis is consistent with the copurification of Hsp70 chaperones with the membrane-associated replication complexes of cucumber necrosis virus (39), tomato mosaic virus (29), and Sindbis virus (13). Interestingly, Ydj1p has been shown to specifically interact with the SSA family of Hsp70s in yeast (24), and preliminary results suggest that deletion of both SSA1 and SSA2 suppresses FHV RNA replication in S. cerevisiae (S. Weeks and D. Miller, unpublished results), similar to results with cucumber necrosis virus (39). The third hypothesis suggests an indirect role whereby cellular chaperones promote the maturation of particular cellular proteins that subsequently facilitate viral RNA replication complex assembly or function. We are currently using both targeted genetic and global biochemical approaches within the framework of these three hypotheses to further define the role of cellular chaperones in FHV RNA replication.
The three hypotheses outlined above are not mutually exclusive, and there is a high likelihood that individual cellular chaperones may have virus- and cell-specific mechanisms of action. Nevertheless, there is increasing evidence that the use of cellular chaperones may be a common feature during virus replication. Further studies on the roles of these ubiquitous cytosolic proteins on the replication of both closely related and highly divergent pathogens will provide important insight into the elaborate methods that viruses use to complete their life cycles and may identify common targets for the development of novel therapeutics.
Acknowledgments
We thank Kathryn Castorena, Leslie Goo, Bethany Sprader, Allison Simms, and Donna Gschwendt for assistance and Alice Telesnitsky, Mike Imperiale, Malini Raghavan, Anuj Kumar, William Pratt, and all members of the Miller lab for their helpful comments on the research and manuscript. We thank Jorgé Iñiguez-Lluhi, Masayuki Ishikawa, and Elizabeth Craig for generously providing reagents.
This work was funded by National Institutes of Health grant R01-AI062749. S.A.W. was supported by training grant T32-AI007528 and a departmental Willison fellowship.
Footnotes
Published ahead of print on 5 December 2007.
REFERENCES
- 1.Ball, L. A. 1995. Requirements for the self-directed replication of Flock House virus RNA 1. J. Virol. 69720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, L. A., and K. L. Johnson. 1998. Nodaviruses of insects, p. 225-267. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Publishing Corporation, New York, NY.
- 3.Bohen, S. P. 1998. Genetic and biochemical analysis of p23 and ansamycin antibiotics in the function of Hsp90-dependent signaling proteins. Mol. Cell. Biol. 183330-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borkovich, K. A., F. W. Farrelly, D. B. Finkelstein, J. Taulien, and S. Lindquist. 1989. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 93919-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush, G. L., and D. I. Meyer. 1996. The refolding activity of the yeast heat shock proteins Ssa1 and Ssa2 defines their role in protein translocation. J. Cell Biol. 1351229-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caplan, A. J., D. M. Cyr, and M. G. Douglas. 1993. Eukaryotic homologues of Escherichia coli dnaJ: a diverse protein family that functions with HSP70 stress proteins. Mol. Biol. Cell 4555-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castorena, K. M., S. A. Weeks, K. A. Stapleford, A. M. Cadwallader, and D. J. Miller. 2007. A functional heat shock protein 90 chaperone is essential for efficient Flock House virus RNA polymerase synthesis in Drosophila cells. J. Virol. 818412-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, H. C., D. F. Nathan, and S. Lindquist. 1997. In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17318-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshaies, R. J., B. D. Koch, M. Werner-Washburne, E. A. Craig, and R. Schekman. 1988. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332800-805. [DOI] [PubMed] [Google Scholar]
- 11.Freeman, B. C., S. J. Felts, D. O. Toft, and K. R. Yamamoto. 2000. The p23 molecular chaperones act at a late step in intracellular receptor action to differentially affect ligand efficacies. Genes Dev. 14422-434. [PMC free article] [PubMed] [Google Scholar]
- 12.Freeman, B. C., and K. R. Yamamoto. 2002. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 2962232-2235. [DOI] [PubMed] [Google Scholar]
- 13.Frolova, E., R. Gorchakov, N. Garmashova, S. Atasheva, L. A. Vergara, and I. Frolov. 2006. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol. 804122-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher, T. M., and R. R. Rueckert. 1988. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J. Virol. 623399-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 2951852-1858. [DOI] [PubMed] [Google Scholar]
- 16.Hoogenraad, N. J., L. A. Ward, and M. T. Ryan. 2002. Import and assembly of proteins into mitochondria of mammalian cells. Biochim. Biophys. Acta 159297-105. [DOI] [PubMed] [Google Scholar]
- 17.Iniguez-Lluhi, J. A., D. Y. Lou, and K. R. Yamamoto. 1997. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J. Biol. Chem. 2724149-4156. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, K. L., and L. A. Ball. 1999. Induction and maintenance of autonomous Flock House virus RNA1 replication. J. Virol. 737933-7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kampmueller, K. M., and D. J. Miller. 2005. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J. Virol. 796827-6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kushner, D. B., B. D. Lindenbach, V. Z. Grdzelishvili, A. O. Noueiry, S. M. Paul, and P. Ahlquist. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. USA 10015764-15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 2961319-1321. [DOI] [PubMed] [Google Scholar]
- 22.Lindenbach, B. D., J. Y. Sgro, and P. Ahlquist. 2002. Long-distance base pairing in Flock House virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J. Virol. 763905-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, R., M. Maduro, F. Li, H. W. Li, G. Broitman-Maduro, W. X. Li, and S. W. Ding. 2005. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 4361040-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, Z., and D. M. Cyr. 1998. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 27327824-27830. [DOI] [PubMed] [Google Scholar]
- 25.Miller, D. J., and P. Ahlquist. 2002. Flock House virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 769856-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock House virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 7511664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, D. J., M. D. Schwartz, B. T. Dye, and P. Ahlquist. 2003. Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane. J. Virol. 7712193-12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nathan, D. F., M. H. Vos, and S. Lindquist. 1997. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc. Natl. Acad. Sci. USA 9412949-12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishikiori, M., K. Dohi, M. Mori, T. Meshi, S. Naito, and M. Ishikawa. 2006. Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J. Virol. 808459-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panavas, T., E. Serviene, J. Brasher, and P. D. Nagy. 2005. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. USA 1027326-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pratt, W. B., and D. O. Toft. 1997. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 18306-360. [DOI] [PubMed] [Google Scholar]
- 32.Price, B. D., P. Ahlquist, and L. A. Ball. 2002. DNA-directed expression of an animal virus RNA for replication-dependent colony formation in Saccharomyces cerevisiae. J. Virol. 761610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional Flock House virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 7411724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Restrepo-Hartwig, M., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize in the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 708908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Restropo-Hartwig, M., and P. Ahlquist. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 7310303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahi, C., and E. A. Craig. 2007. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. USA 1047163-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt-Mende, J., E. Bieck, T. Hügle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 27644052-44063. [DOI] [PubMed] [Google Scholar]
- 38.Scotti, P. D., S. Dearing, and D. W. Mossop. 1983. Flock House virus: a nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae). Arch. Virol. 75181-189. [DOI] [PubMed] [Google Scholar]
- 39.Serva, S., and P. D. Nagy. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 802162-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomita, Y., T. Mizuno, J. Diez, S. Naito, P. Ahlquist, and M. Ishikawa. 2003. Mutation of host DnaJ homolog inhibits brome mosaic virus negative-strand RNA synthesis. J. Virol. 772990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 27126810-26818. [DOI] [PubMed] [Google Scholar]
- 42.Waxman, L., M. Whitney, B. A. Pollok, L. C. Kuo, and P. L. Darke. 2001. Host cell factor requirement for hepatitis C virus enzyme maturation. Proc. Natl. Acad. Sci. USA 9813931-13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Werner-Washburne, M., D. E. Stone, and E. A. Craig. 1987. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 72568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, S. X., P. Ahlquist, and P. Kaesberg. 1992. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc. Natl. Acad. Sci. USA 8911136-11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, S. X., and P. Kaesberg. 1991. Synthesis of template-sense, single-strand Flock House virus RNA in a cell-free replication system. Virology 183392-396. [DOI] [PubMed] [Google Scholar]
- 46.Youker, R. T., P. Walsh, T. Beilharz, T. Lithgow, and J. L. Brodsky. 2004. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell 154787-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young, J. C., N. J. Hoogenraad, and F. U. Hartl. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 11241-50. [DOI] [PubMed] [Google Scholar]
- 48.Zhao, R., M. Davey, Y. C. Hsu, P. Kaplanek, A. Tong, A. B. Parsons, N. Krogan, G. Cagney, D. Mai, J. Greenblatt, C. Boone, A. Emili, and W. A. Houry. 2005. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120715-727. [DOI] [PubMed] [Google Scholar]