Abstract
Coxsackievirus B3 (CVB3) generates 5′-terminally deleted genomes (TDs) during replication in murine hearts. We show here that CVB3 populations with TDs can also be generated within two to three passages of CVB3 in primary, but not immortalized, cell cultures. Deletions of less than 49 nucleotides increase in size during passage, while 5′ TDs of 49 nucleotides appear to be the maximum deletion size. The cellular environment of contact-inhibited primary cell cultures or the myocardium in vivo is sufficient for the selection of 5′ TDs over undeleted genomes.
The six serotypes of the group B coxsackieviruses (CVB1 to CVB6) (human enteroviruses, Picornaviridae) (18, 22) are small, nonenveloped, positive-strand RNA viruses. The enterovirus RNA genome, which is flanked by 5′ and 3′ nontranslated regions (NTR), is translated upon infection of the host cell (4) and then serves as the template for synthesis of complementary negative-strand RNA, which in turn templates progeny positive-strand genomic RNAs. Human enteroviruses cause acute myocarditis and are etiologically linked as causes of dilated cardiomyopathy, a disease that can proceed to heart failure (2, 3, 11, 20, 22). Enteroviral RNA persistence in adult hearts with myocarditis or dilated cardiomyopathy in the absence of detectable infectious virus has been a common observation (reviewed in references 3, 11, and 12) but remained without adequate explanation until we demonstrated that, during replication in isolated murine cardiomyocytes and in hearts of infected mice, CVB3 can naturally delete sequence information at the genomic 5′ terminus (13). This work showed that, weeks following the inoculation of mice with CVB3, mouse myocardium in which no cytolytic virus was evident was able to transmit virus to cell cultures, which, in turn, showed no cytopathic effects (CPE) despite the detection of CVB3 RNA. Genomic viral RNA lacked native 5′ termini, demonstrating deletions ranging from 7 to 49 nucleotides (nt) from the 5′ genomic terminus; these novel, naturally occurring CVB3 strains with 5′-terminally deleted (TD) viral genomes were termed CVB3TD. The cloning and placement of naturally occurring deletions into an infectious CVB3 cDNA clone revealed that, while the resultant viruses are replication competent, CVB3TD replication is very slow, resulting in no observable CPE on HeLa indicator monolayers. In addition, purified CVB3TD virions encapsidate both positive- and negative-strand RNA in a strand ratio similar to that of total RNA from CVB3TD-infected cell cultures. Because CPE in CVB3TD-infected cell cultures are not measurable, a method was developed to titer infectious CVB3TD using real-time quantitative reverse transcription-PCR (RT-PCR) analysis of the viral RNA content in CVB3TD preparations (13). It was not surprising to note deviations from wild-type enteroviral replication, as the region in the 5′ NTR that is progressively deleted (the cloverleaf RNA structure termed domain I) is also an essential region for viral replication (1, 17, 19). Having generated CVB3TD strains in murine hearts, and having recently shown their naturally occurring existence in myocarditic human heart tissue (N. Chapman, K.-S. Kim, and S. Tracy, submitted for publication), we studied the generation of TD genomes in cell cultures. Toward this end, we compared the impact of primary versus immortalized cell cultures on CVB3 in serial passages to determine whether CVB3TD genomes would arise.
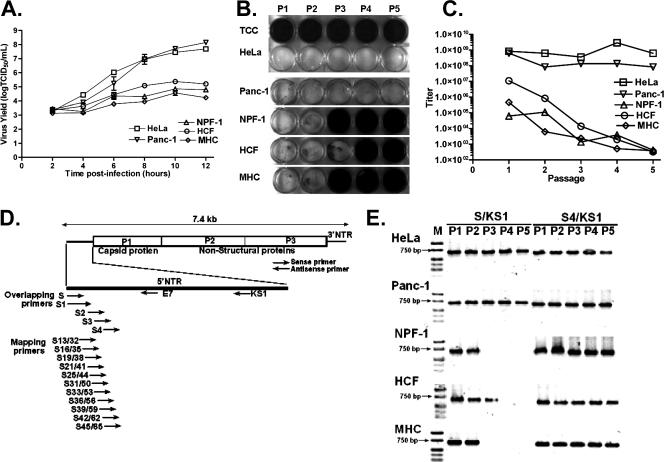
The ability of the cardiovirulent strain CVB3/28 (23) to replicate productively in all cell cultures to be used for this study was confirmed by using single-step growth curves (25). Five cell cultures were employed, two transformed cell lines, HeLa and Panc-1, and three primary cell cultures: NPF-1 (from pancreata of 4- to 6-week-old female nonobese diabetic mice; Taconic Laboratories, Germantown PA), maintained in minimal essential medium; MHC (cultured from the heart of a C3H/HeJ mouse [21]), maintained in Claycomb medium (6); and HCF (human cardiac myocytes; ScienCell Research Laboratories, San Diego CA), maintained in Claycomb medium. Primary cell cultures were used after passages 4 to 15. The replication rate of CVB3 in the Panc-1 (16) and HeLa cell lines was more rapid than in the HCF, MHC, or NPF-1 cell cultures, and virus titers increased 100-fold more than in the primary cell cultures by the end of the experiment (Fig. 1A). The cell cultures were then inoculated with CVB3/28 at a multiplicity of infection (MOI) of 20 50% tissue culture infective doses (TCID50) per cell when cell monolayers were less than 24 h from complete confluence (due to variation in cell size, the number of cells per cm2 varied as a function of the cell culture). Cultures were maintained for 3 days or until lysis was complete; lysis occurred within 24 h in HeLa and Panc-1 cell lines, but little or no CPE was observed in the primary cell cultures during the 3 days of incubation. Sequential passages were carried out by passing one-third of the supernatant volume of the previously frozen/thawed passage onto fresh cell cultures as described previously (13). Each culture supernatant was then assayed by light microscopy on HeLa indicator monolayers for the generation of CPE. HeLa monolayers inoculated with supernatants from MHC and NPF-1 passages 3 to 5 or HCF passages 4 and 5 appeared to be normal, showing no difference from uninfected (control) HeLa cell monolayers (Fig. 1B), while lytic virus was detected in the supernatants of all five passages of CVB3 on HeLa and Panc-1 cell lines. The viral titers (assayed by measuring TCID50 [25]) from HeLa cell monolayers did not decline during passage, but titers declined 1,000- to 10,000-fold in the primary cell culture passages (Fig. 1C), until no CPE and no wild-type virus were observed (TCID50 equivalents in viral RNA for the noncytopathic passages [13]) (Fig. 1B), although viral RNA (presumably CVB3TD) was present (Fig. 1E). The proportion of infected cells in each passage is unknown, and the generation of noncytopathic but replicating virus may occur in only a few cells. However, once established, the infection persists, despite the loss of CPE in primary cell cultures. To demonstrate the presence of CVB3TD, samples of total RNA from infected cell cultures were examined by RT-PCR using progressive overlapping primers inward from the 5′ viral genome terminus (described previously in reference 13) (Fig. 1D). Amplification with primers S4 (nt 45 to 74) and KS1 (nt 667 to 691) detected viral RNA in all passages of the MHC, NPF-1, and HCF cell cultures (Fig. 1E), despite no CPE in some passages (Fig. 1B). Each cell culture in which CPE was observed (Fig. 1B) was positive for the intact 5′-terminal sequence (Fig. 1E), using the S (nt 1 to 20) and KS1 primer set to detect intact 5′ termini. However, in MHC and NPF-1 cells at passage 3 and HCF cells at passage 4 (which showed no CPE [Fig. 1B]), S/KS1 amplimers were not generated (Fig. 1E). These results were consistent with earlier observations in which failure to detect the 5′ termini was due to deletions of the intact 5′-terminal genomic sequence (13). CVB TD were also generated in a similar experiment by passage of two other CVB3 strains, CVB3/CO (24) and CVB3/GA (15 and data not shown), indicating that these observations were not limited to a specific CVB3 strain.
FIG. 1.
CVB3 replicates but does not induce CPE in the indicator HeLa cell monolayers following sequential infection of replication-restricted primary cells. (A) Single-step growth curves of CVB3/28 in primary cell cultures NPF-1, MHC, and HCF and in immortal cell lines HeLa and Panc-1 which were inoculated with an MOI of 20 TCID50 per cell. Error bars show standard deviations. (B) Supernatants of CVB3/28 passaged in primary cell cultures lose the ability to cause CPE in HeLa cell monolayers. P1 to P5, HeLa cells infected with supernatants of passages 1 to 5; TCC, uninfected HeLa cell control. Cell cultures were fixed in acetic acid/acetone and then stained with crystal violet. Titers at each passage were determined either by CPE on HeLa cell monolayers (25) or by real-time quantitative RT-PCR (13). (C) Titers from serial passages shown in panel B are plotted. (D) Relative positions in the CVB3 genome of primers used in this study. (E) CVB3/28 RNAs in primary cell cultures are detected by RT-PCR. Lane M, Hi-Lo DNA marker (Minnesota Molecular, Inc., Minneapolis, MN); lanes P1 to P5, passages 1 to 5. Arrows indicate 750-bp bands.
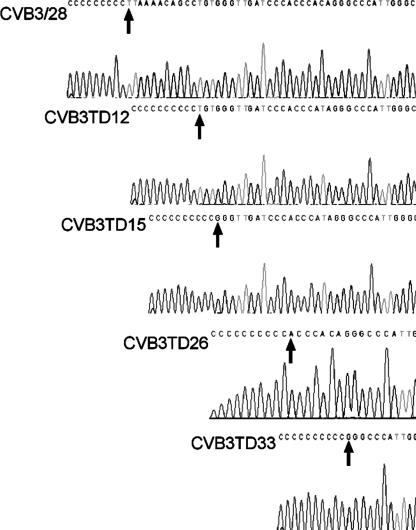
Lowering the MOI to 1 or 10 demonstrated that only passage 1 in HCF, MHC, and NPF-1 cell cultures produced CPE; thereafter, cells showed no CPE and appeared to be uninfected (data not shown). To analyze the deletions by sequence analysis, we chose total RNA isolated from passage 2 in MHC cell cultures infected at an MOI of 1. As previously described (13), cDNA was transcribed with the primer KS1, G10-tailed using terminal deoxynucleotidyl transferase, and then amplified using primers DC-Tail and KS1. Amplified DNA was cloned; we selected 10 plasmids containing 5′ NTR sequences for sequencing using primer E7 (nt 118 to 139) (Fig. 1D). Multiple deletions at the 5′ end were identified (typical sequencing results are shown in Fig. 2). Deletions in the 10 clones included 1 clone each with nt 1 to 11 or nt 1 to 14 deleted and 2 with nt 1 to 25, while 6 of the 10 randomly chosen clones showed deletions of nt 1 to 32. Due to CC and CCC tracts in the 5′ NTR of CVB3/28, it is possible that clones CVB3TD12 and CVB3TD26 represent smaller deletions, but the poly(C)10 tract in the clones makes deletions of 11 and 25 nt most likely. The 5′ end of clone CVB3TD12 (nt 1 to 11 deleted) terminated with UG and the CVB3TD15 (nt 1 to 14) and -33 (nt 1 to 32) RNAs terminated with a GG residue, while CVB3TD26 (nt 1 to 25) terminated with AC; none ended in the canonical enterovirus 5′-terminal dimer 5′-UU (14), again reminiscent of previously described CVB3TD genomes isolated from mouse myocardium (13).
FIG. 2.
5′-Terminal deletions from passage in primary cell culture range from 11 to 32 nt. Sequence analysis of tailed and cloned cDNAs from the second-passage MHC cell culture revealed deletions of various sizes between about 11 and 32 nt. Parental CVB3/28 RNA was similarly assayed for comparison. Arrows indicate the 5′ terminus of each CVB3TD genome. The sequences of the 5′ ends of CVB3TD12, -15, -26, and -33 were determined.
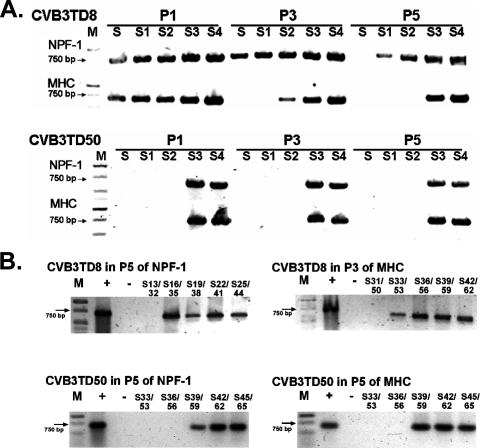
It has yet to be established whether terminal deletions initially occur randomly in different sizes, which then remain as stable populations, or whether shorter deletions enlarge during replication, eventually stabilizing at some approximate size. To date, we have observed deletions of from 7 to 49 nt in the CVB3 genome but none greater than 49 nt (13), which delete most but not all of domain I in the 5′ NTR. We therefore tested the hypothesis that short deletions (exemplified by CVB3TD8, in which nt 1 to 7 are deleted [13]) can enlarge during replication, while the largest deletion observed to date (nt 1 to 49 deleted in CVB3TD50 [13]) remains stable. We inoculated MHC and NPF-1 cell cultures at an MOI of 10 with CVB3TD8 or CVB3TD50 and then sequentially passaged the viruses as described above. Total cell RNA was isolated from each culture, reverse transcribed, and then enzymatically amplified with progressively overlapping primers (13) or with the overlapping mapping primers (Fig. 1D). RT-PCR with the progressively overlapping primers S through S4 indicated that the size of the deletion in the viral genome isolated from CVB3TD8-inoculated cultures had become greater during passage (Fig. 3A). RT-PCR with primers S/KS1 produced amplimers from passages 1 and 3 in the NPF-1 cell cultures, but not from passage 5 in the cultures (Fig. 3A), although CVB3TD8 can be detected by RT-PCR with primers S/KS1. RT-PCR of NPF-1 passage 5 of CVB3TD8 using the mapping primer S13/32 (nt 13 to 32) and KS1 showed no amplification (Fig. 3B), indicating that a deletion of potentially as much as 32 nt had replaced TD8. After three passages of CVB3TD8 in MHC cell cultures, RT-PCR did not detect viral RNA with S1, and by five passages, RT-PCR did not detect viral RNA with S2 (nt 21 to 49) (Fig. 3A). As RT-PCR with the mapping primer S33/53 (nt 33 to 53) detected the viral RNA in MHC passage 3 of CVB3TD8 (Fig. 3B), although the mapping primer S31/50 (nt 31 to 50) did not, the TD8 deletion of nt 1 to 7 had increased within three passages to nearly 50 nt in size. However, RT-PCR of similarly passaged CVB3TD50 showed no change in amplification patterns from passage 1 to 5 in both NPF-1 and MHC cell cultures (Fig. 3A and B). Detection of viral cDNA using the mapping primer S39/59 (nt 39 to 59), but not S36/56 (nt 36 to 56) (Fig. 3B), was consistent with there being little or no increase in the 49-nt deletion of CVB3TD50 through five passages. It appears that TD mutations at their most extreme preserved most of stem-loop d in domain I, a region that may serve as an essential part of the initiation-replication complex in the negative strand of the TD genome. The evolution of the CVB3TD8 genome into a population containing significantly larger genomic 5′-end deletions suggests that further loss of 5′-terminal sequence information is favored once terminal deletions have occurred, while the apparent lack of further evolution of the CVB3TD50 sequence suggests that the remnant of stem-loop d in domain I is the necessary minimum in this structure for viability of the virus.
FIG. 3.
A 5′-terminal deletion of 7 nt evolves to larger deletions in primary cell cultures, but a deletion of 49 nt does not change. Total RNA from passages 1, 3, and 5 of CVB3TD8- or CVB3TD50-inoculated MHC and NPF-1 cell cultures were assayed by RT-PCR analysis using primers covering the 5′ 74 nt of the genome (A) and finer-scale mapping primers (B). Lane M, Hi-Lo DNA marker (Minnesota Molecular, Inc.); P1, P3, P5, passages 1, 3, and 5, respectively; + and −, positive and negative amplification controls, respectively. Lanes are marked by the primer number which indicates the primer's annealing position in the CVB3 genome (GenBank accession no. AY752944). Arrows indicate 750-bp bands.
The results described here demonstrate that the evolution of wild-type enteroviral genomes to the TD form can be studied in cell culture and that the sizes of the TD genomes which arise in cell culture are in line with those characterized from mouse (13) and human heart tissue (N. Chapman, K.-S. Kim, and S. Tracy, submitted), results that validate the cell culture system for further study of this novel enteroviral biology. A recent study (9) that examined a similar, albeit genetically engineered, deletion of the 5′-terminal 32 nt of the CVB3 genome observed no viable progeny virus or genomic RNA replication. This is in contrast to work described here and published previously (13). Because assays for viral translation were limited to 30 min of labeling in transfected cells and slot-blot assays were used to assay for RNA replication (9), it is possible that such approaches were insufficiently sensitive to detect the low levels of virus found in cultures of CVB3TD (which required 24 h of accumulation of viral proteins and RT-PCR for detection of viral RNA [13]). An alternative explanation—that the specific deletion of nt 1 to 32 may itself be lethal—is not supported by our current results, in which we observed CVB3TD genomes occurring naturally in cell culture, with identically sized deletions, or by previous results with similarly sized deletions in mouse hearts (13).
An early report (7) suggested that the production of progeny positive-strand RNA in poliovirus-infected HeLa cells was linked to DNA synthesis and CVB3 replication is more restricted in nonreplicating cells (8). We hypothesize that differential expression levels of specific cellular proteins in primary cells may affect the generation and selection of TD enteroviral genomes, although it is as yet unknown whether specific host factors are in fact limiting in these primary cell cultures. Relevant to this hypothesis is the observation that a host factor known to bind to the 3′ end of the enteroviral negative-strand RNA and believed to play a role in positive-strand RNA replication, nuclear factor hnRNP-C (5), migrates into the cytoplasm during mitosis (10). More-recent work from this group (B. Semler, personal communication) indicates that the cell line SKOV3, an ovarian carcinoma line, expresses low levels of hnRNP-C. During infection with poliovirus, replication of the virus is delayed in these cells, but if hnRNP-C is exogenously supplied through transfection of an expression plasmid, poliovirus titers in the cells increase. Were levels of hnRNP-C to become limiting or significantly lowered in the cytoplasm of cells, such as quiescent or differentiated cells, then TD genomes which lack sequence from the negative-strand 3′ end might well enjoy an evolutionary advantage over wild-type enteroviral RNA, thereby enabling them to become the dominant virus population.
Acknowledgments
K.-S. Kim was supported in part by a grant from the gene delivery/therapy pilot project program (Nebraska Research Initiative) and a fellowship from the American Heart Association. The work was supported by grants from the American Heart Association (N. M. Chapman), the National Institutes of Health, the Juvenile Diabetes Research Foundation, and the American Diabetes Association (S. Tracy). We sincerely thank F. Schiff of the ERACE Foundation, the Stein family, M. Guthrie, and E. Barnett for their generosity in supporting this work in memory of loved ones.
Footnotes
Published ahead of print on 5 December 2007.
REFERENCES
- 1.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63369-380. [DOI] [PubMed] [Google Scholar]
- 2.Andreoletti, L., T. Bourlet, D. Moukassa, L. Rey, D. Hot, Y. Li, V. Lambert, B. Gosselin, J. F. Mosnier, C. Stankowiak, and P. Wattre. 2000. Enteroviruses can persist with or without active viral replication in cardiac tissue of patients with end-stage ischemic or dilated cardiomyopathy. J. Infect. Dis. 1821222-1227. [DOI] [PubMed] [Google Scholar]
- 3.Baboonian, C., M. J. Davies, J. Booth, and W. McKenna. 1997. Coxsackie B viruses and heart disease. Curr. Top. Microbiol. Immunol. 22331-52. [DOI] [PubMed] [Google Scholar]
- 4.Bedard, K. M., and B. L. Semler. 2004. Regulation of picornavirus gene expression. Microbes Infect. 6702-713. [DOI] [PubMed] [Google Scholar]
- 5.Brunner, J. E., J. H. Nguyen, H. H. Roehl, T. V. Ho, K. M. Swiderek, and B. L. Semler. 2005. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 793254-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claycomb, W. C., N. A. Lanson, Jr., B. S. Stallworth, D. B. Egeland, J. B. Delcarpio, A. Bahinski, and N. J. Izzo, Jr. 1998. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA 952979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eremenko, T., A. Benedetto, and P. Volpe. 1972. Virus infection as a function of the host cell life cycle: replication of poliovirus RNA. J. Gen. Virol. 1661-68. [DOI] [PubMed] [Google Scholar]
- 8.Feuer, R., I. Mena, R. Pagarigan, M. K. Slifka, and J. L. Whitton. 2002. Cell cycle status affects coxsackievirus replication, persistence, and reactivation in vitro. J. Virol. 764430-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunziker, I. P., C. T. Cornell, and J. L. Whitton. 2007. Deletions within the 5′UTR of coxsackievirus B3: consequences for virus translation and replication. Virology 360120-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, J. H., K. Y. Paek, K. Choi, T. D. Kim, B. Hahm, K. T. Kim, and S. K. Jang. 2003. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell. Biol. 23708-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, K.-S., K. Hofling, S. D. Carson, N. M. Chapman, and S. Tracy. 2003. The primary viruses of myocarditis. In L. T. Cooper (ed.), Myocarditis: from bench to bedside. Humana Press, Totowa, NJ.
- 12.Kim, K.-S., K. Hofling, N. M. Chapman, and S. Tracy. 2001. The group B coxsackieviruses and myocarditis. Rev. Med. Virol. 11355-368. [DOI] [PubMed] [Google Scholar]
- 13.Kim, K.-S., S. Tracy, W. Tapprich, J. Bailey, C. K. Lee, K. Kim, W. H. Barry, and N. M. Chapman. 2005. 5′-Terminal deletions occur in coxsackievirus B3 during replication in murine hearts and cardiac myocyte cultures and correlate with encapsidation of negative-strand viral RNA. J. Virol. 797024-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klump, W. M., I. Bergmann, B. C. Muller, D. Ameis, and R. Kandolf. 1990. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA: two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J. Virol. 641573-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, C. K., K. Kono, E. Haas, K. S. Kim, K. M. Drescher, N. M. Chapman, and S. Tracy. 2005. Characterization of an infectious cDNA copy of the genome of a naturally occurring, avirulent coxsackievirus B3 clinical isolate. J. Gen. Virol. 86197-210. [DOI] [PubMed] [Google Scholar]
- 16.Lieber, M., J. Mazzetta, W. Nelson-Rees, M. Kaplan, and G. Todaro. 1975. Establishment of a continuous tumor-cell line (panc-1) from a human carcinoma of the exocrine pancreas. Int. J. Cancer 15741-747. [DOI] [PubMed] [Google Scholar]
- 17.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 7510696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In D. M. Knipe et al. (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 19.Sharma, N., B. J. O'Donnell, and J. B. Flanegan. 2005. 3′-Terminal sequence in poliovirus negative-strand templates is the primary cis-acting element required for VPgpUpU-primed positive-strand initiation. J. Virol. 793565-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spotnitz, M. D., and M. Lesch. 2006. Idiopathic dilated cardiomyopathy as a late complication of healed viral (coxsackie B virus) myocarditis: historical analysis, review of the literature, and a postulated unifying hypothesis. Prog. Cardiovasc. Dis. 4942-57. [DOI] [PubMed] [Google Scholar]
- 21.Toraason, M., M. E. Luken, M. Breitenstein, J. A. Krueger, and R. E. Biagini. 1989. Comparative toxicity of allylamine and acrolein in cultured myocytes and fibroblasts from neonatal rat heart. Toxicology 56107-117. [DOI] [PubMed] [Google Scholar]
- 22.Tracy, S., N. Chapman, and B. Mahy. 1997. The coxsackie B viruses. In Current topics in microbiology and immunology, vol. 223. Springer-Verlag, Heidelberg, Germany.
- 23.Tracy, S., K. M. Drescher, N. M. Chapman, K. S. Kim, S. D. Carson, S. Pirruccello, P. H. Lane, J. R. Romero, and J. S. Leser. 2002. Toward testing the hypothesis that group B coxsackieviruses (CVB) trigger insulin-dependent diabetes: inoculating nonobese diabetic mice with CVB markedly lowers diabetes incidence. J. Virol. 7612097-12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tracy, S., K. Hofling, S. Pirruccello, P. H. Lane, S. M. Reyna, and C. J. Gauntt. 2000. Group B coxsackievirus myocarditis and pancreatitis: connection between viral virulence phenotypes in mice. J. Med. Virol. 6270-81. [DOI] [PubMed] [Google Scholar]
- 25.Tu, Z., N. M. Chapman, G. Hufnagel, S. Tracy, J. R. Romero, W. H. Barry, L. Zhao, K. Currey, and B. Shapiro. 1995. The cardiovirulent phenotype of coxsackievirus B3 is determined at a single site in the genomic 5′ nontranslated region. J. Virol. 694607-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]