Abstract
Human endogenous retroviruses (HERVs) account for up to 9% of the human genome and include more than 800 elements related to betaretroviruses. While mouse mammary tumor virus (MMTV) is the accepted etiological agent of mammary tumors in mice, the role of retroviral elements in human breast cancer remains elusive. Here, we performed a comprehensive microarray-based analysis of overall retroviral transcriptional activities in 46 mammary gland tissue specimens representing pairs of nonmalignant and tumor samples from 23 patients. An analysis of nonmalignant tissue samples revealed a distinct, mammary gland-specific HERV expression profile that consists of 18 constitutively active HERV taxa. For corresponding tumor samples, a general trend toward lower levels of HERV transcription was observed, suggesting common regulatory mechanisms. In various subsets of patients, however, increased transcript levels of single class I HERV families (HERV-T, HERV-E, and HERV-F) and several class II families, including HML-6, were detected. An analysis of transcribed HML-6 sequences revealed either the activation of some or the increased activity of several proviral loci. No evidence for MMTV or human MMTV-like virus transcripts was found, indicating that transcriptionally active, MMTV analogous, exogenous viruses were not present in the breast cancer samples analyzed.
Great efforts have been invested in searching for the etiology of human breast cancer, a malignancy accounting for one-fifth of all female cancers worldwide. Although many studies have identified several risk factors, such as age, diet, hormonal balance, and genetic predisposition, a clear underlying cause for the disease, especially for sporadic cases of breast cancer, remains unknown. The current data suggest that breast cancer most likely is a multifactorial disease encompassing many different causes and factors (2, 30). Moreover, it has been suggested that an infectious agent contributes to the development of human breast cancer (16, 45). Of note, a novel human retrovirus (xenotropic murine leukemia virus) has recently been associated with human prostate cancer (7, 48).
Since type B mouse mammary tumor virus (MMTV) is the major etiological agent of mammary gland neoplasia in laboratory mice, researchers have searched extensively for a related human retrovirus that could be responsible for human breast cancer. The existence of such a virus, although postulated for many years, has not been conclusively demonstrated, although a long line of indirect evidence for it exists. This evidence is reflected by reports on the expression of type B envelope glycoprotein (gp52) (32) and the occurrence of virus-like particles in breast cancer biopsy specimens (8), in milk (38), and in cultures of breast cancer-derived cell lines (20, 40) as well as the detection of antibodies directed against gp52 in breast cancer patients (52). However, supporting observations have been confounded by a failure to continually observe virus particles in human tumors and by numerous controversial reports. Moreover, the presence of endogenous MMTV-related sequences in the human genome (1, 4, 36, 37, 46, 47) and their ubiquitous transcriptional activities in normal human tissues, including mammary gland tissue (31, 33, 42, 54), has complicated a systematic investigation.
Human endogenous retroviruses (HERVs) are natural components of the human genome and are considered remnants of ancient germ line infections by exogenous retroviruses that have been genetically fixed and transmitted in a Mendelian fashion (for a review, see references 27 and 44). During evolution, these elements were amplified and spread throughout the genome by repeated events of retrotransposition and/or reinfection. The human genome sequencing project revealed that 8 to 9% of the human genome is of retroviral origin (23). Around 826 of these elements (class II HERVs) are betaretrovirus-like and therefore distantly related to exogenous MMTV (33).
Although the majority of HERVs are noninfectious, replication-defective retroviral fossils, at least some members of each HERV family were found to still be transcriptionally active (12, 33, 42, 43). Furthermore, tissue-specific HERV expression profiles could be established for all human tissues investigated so far, confirming that HERVs are permanent components of the human transcriptome (13, 42). In a few studies, a prevalence of HERV transcripts, in particular class II elements, such as members of the HML-2 family, was reported for breast cancer tissues and cell lines (5, 50, 51).
Recently, several reports described a novel human MMTV-like virus (HMLV) in human breast cancer that shares at least 95% sequence identity with MMTV, maintains open reading frames, has replicative potential, and is considered to be of exogenous origin (6, 9, 11, 26, 49). However, the limited reproducibility of original molecular observations has heated a highly controversial debate about the positive association of HMLV and human breast cancer (3, 28, 29, 53, 54).
To test a possible association of endogenous or exogenous MMTV-related retroviruses and human breast cancer, we compared the transcriptional activities of HERVs, including MMTV-related HERV-K elements (HML subgroups) and putative human MMTV-like elements (HMLV). Here, we report on transcriptional signatures found in 46 mammary gland tissue specimens representing pairs of nonmalignant and tumor samples derived from 23 patients (one pair per patient), by means of a powerful microarray-based test system.
MATERIALS AND METHODS
Patient samples.
Pairs of human mammary tumor and corresponding nonmalignant breast tissue samples (n = 46) derived from the same patients (n = 23) were investigated. Tissue specimens were obtained from the Department of Pathology, Medical Faculty Mannheim, University of Heidelberg (Mannheim, Germany) after the breast surgeries of 23 female German patients. Informed consent was obtained from all patients according to the Declaration of Helsinki. As shown in Table S1 in the supplemental material, all tumors were unilateral invasive ductal carcinomas (grade I, n = 1; grade II, n = 11; and grade III, n = 11). The median age of the patients was 63.0 years (range, 41 to 87). The mean tumor size was 36 mm (range, 21 to 85 mm). Patient samples corresponding to tumor cell-rich and adjacent tumor cell-free tissue areas were prepared immediately upon tumor surgery and consecutively snap-frozen in liquid nitrogen. Tissues were kept at −80°C until RNA extraction.
RNA preparation.
Total RNA was extracted from ground tissue samples according to a guanidinium isothiocyanate-cesium chloride ultracentrifugation protocol (13) and dissolved in diethylpyrocarbonate-treated distilled water. Subsequently, mRNA was purified using Dynabeads paramagnetic particles as described by the manufacturer (Dynal, Hamburg, Germany). To remove genomic DNA contamination, all mRNA samples were treated with 100 U/μg RNase-free DNase (Roche Diagnostics GmbH, Mannheim, Germany) in 100 mM sodium acetate (pH 5.0) and 5 mM MgSO4. Subsequently, 25 ng of each mRNA preparation was tested by PCR with mixed oligonucleotide primers (MOP), omitting the reverse transcription (RT) step. Only mRNA preparations negative for amplification products were used for subsequent reverse transcription and MOP multiplex PCR.
RT-PCR and microarray experiments.
Reverse transcription of mRNA, hybridization probe synthesis, and labeling by MOP PCR as well as DNA chip preparation, hybridization, and postprocessing of retrovirus-specific microarrays were performed as described previously (13, 42). To evaluate the discriminatory power of the HERV chip, 25 pmol of synthetic Cy3-labeled oligonucleotides complementary to the capture probes (Fig. 1) was used in the standardized experimental setting.
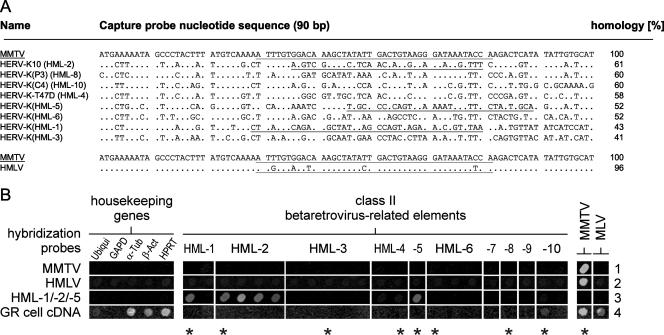
FIG. 1.
Alignment of human MMTV-like (HML) DNA chip capture probes and demonstration of microarray discriminatory power. (A) Aligned nucleotide sequences of MMTV, HMLV, and endogenous HERV-K-related proviral elements (HML taxa) correspond to a 90-bp stretch of genomic DNA flanked by sequences encoding two highly conserved amino acid motifs (VLPQG and YV/MDDI/V/L, not included) commonly found in retroviral pol(RT) genes. See references 13 and 42 for the origins and classifications of HERV chip capture probes and amplification primers. Synthetic oligonucleotide probes, complementary to HERV-K(HML-1), HERV-K10 (HML-2 family), HERV-K(HML-5), MMTV, and HMLV capture probes used for optimization of the discriminatory power of the microarray assay, are underlined. (B) Specific hybridization signals were obtained employing Cy3-modified synthetic hybridization probes specific for MMTV (row 1), HMLV (row 2), and HML-1, HERV-K10 (HML-2 family), and HML-5 (row 3). Sensitive detection of MMTV was demonstrated by analyzing 100 ng of total RNA derived from murine GR cells in a standardized retrovirus DNA chip assay (row 4). Asterisks denote capture probe sequences aligned in panel A. Ubiqui, ubiquitin; GAPD, glyceraldehyde 3-phosphate dehydrogenase; α-Tub, alpha tubulin; β-Act, beta actin.
Microarray evaluation and HERV signal quantification.
Hybridized microarrays were scanned using an Affymetrix GMS 418 scanner (laser power setting, 100%; gain setting, 50%), and the resulting images (16-bit TIFF files) were subjected to densitometric analysis using ImaGene 4.0 software (BioDiscovery, Inc., Los Angeles, CA). Densitometric data were used for signal quantification. To discriminate positive signals from background, an arbitrary cutoff value of 4,000 relative signal intensity units was used as in the study by Frank and coworkers (13). This cutoff value corresponds to twofold background intensity values of the respective chip and proved to be in good agreement with the optical appearance of raw images when observed on a color-calibrated monitor in a darkened room. To account for the influence of mRNA quality, HERV signals were normalized to RNA levels for the hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene, a housekeeping gene showing the most consistent transcript levels. Thus, microarray-based relative HERV transcript levels were expressed as a ratio of HERV to HPRT.
Quantification of HERV transcripts by QRT-PCR.
For the amplification of pol(RT) sequences, HERV subgroup-specific pol primers for RT244(HML-3) (forward primer, 5′-GCT GAG CAA GAC TGT GAA TG-3′, and reverse primer, 5′-CAT ACG ACT GGC AAA TTG TG-3′), HERV-FRD (see reference 42), and HERV-K-T47D (forward primer, 5′-GTC GCT CAG GCT ACA TGC-3′, and reverse primer, 5′-AGT GAA CGA TGT AAC ATC GG-3′) were employed. In general, HERV-specific primers for LightCycler real-time RT-PCR were designed in such a way that for each HERV, except HML-6 (the primers of which were adopted from reference 33), one primer matched the capture probe sequences used in the corresponding microarray experiments, whereas a second primer was located 100 to 150 bp upstream of the first primer (see reference 42). For normalization, the housekeeping gene for beta-glucuronidase (GUS) was included in the analysis, as described previously, using previously published amplification primers (14). For verification purposes, the gene for HPRT was also used as an additional housekeeping gene (for primer sequences, see reference 42) in a set of representative samples (patients 13, 16, 19, and 20). As reference sequences, standard plasmids containing cloned pol(RT) fragments of HERV-K-T47D (accession no. AF020092; nucleotides [nt] 3762 to 6542), HERV-FRD (accession no. U27240; nt 1 to 2873), and HERV-K(HML-6) (accession no. AC092674.2; nt 26836 to 26982) as well as plasmids containing HPRT (accession no. BT019350.1; nt 32 to 292) and GUS (accession no. NM_000181.2; nt 1705 to 1909 [14]) gene fragments were used. The plasmid containing a HERV-K(HML-3) sequence (accession no. AH013886 [33]) was obtained from J. Blomberg (University of Uppsala, Sweden). Quantitative RT-PCR (QRT-PCR) was performed using 2 μl of 1:3 cDNA sample dilutions and primers at 0.5 μM each in LightCycler FastStart DNA MasterPLUS SYBR green I ready-to-use hot-start PCR mix containing Taq DNA polymerase, reaction buffer, deoxynucleoside triphosphates (dTTP was replaced by dUTP), SYBR green I dye, and MgCl2 (Roche Diagnostics GmbH, Mannheim, Germany). Amplification was performed using a 10-min denaturation step at 95°C, followed by 45 cycles of 1 s at 95°C, 5 s at 50°C, and 12 s at 72°C. Furthermore, extensive standardization of PCRs was performed initially through melting curve analysis of respective amplicons in order to minimize primer pair formation (data not shown). Relative quantification of HERV pol transcription was performed using LightCycler software (version 3.5; Roche Molecular Biochemicals). Data obtained from triplicate analyses were expressed as gene ratios for paired patient samples (malignant versus nonmalignant breast tissues).
Amplification, cloning, and sequence analysis of HML-6 transcripts.
For the amplification of HML-6 transcripts, PCR was performed using 4 μl of undiluted cDNA from both nonmalignant and tumor tissues. Primers HML6RTfw (33), HMLRTrv3 (5′-AAC AAT AGT TCC TAA GTA TTA GTA TGG-3′), and HML6RTc16 (5′-AAC AAT AGT GCC TAA GTA CTG GTA TC-3′) were employed at concentrations of 1.0, 0.96, and 0.04 μM, respectively. The 50-μl PCR mix contained 1× reaction buffer, 1.5 mM MgCl2, deoxynucleoside triphosphates at 0.2 mM each, and 2.5 U Taq DNA polymerase (Invitrogen, Inc., Carlsbad, CA). Reaction conditions comprised an initial denaturation at 94°C for 5 min; 35 cycles at 94°C for 45 s, annealing at 51.4°C for 45 s, and elongation at 72°C for 30 s, followed by a final elongation step at 72°C for 10 min. HML-6-specific pol PCR products were purified (QIAquick PCR purification kit, Qiagen, Hilden, Germany) and cloned into the pCR2.1-TOPO vector (TOPO TA cloning kit, Invitrogen), and TOP10F′ bacterial cells were chemically transformed with ligation products. Plasmid DNA was isolated from insert-containing colonies according to the manufacturer's protocol (QuickLyse Miniprep kit; Qiagen). Subsequently, cloned HML-6 cDNAs were analyzed by sequencing (Institut für Immunologie und Genetik, Kaiserslautern, Germany) and mapped to their respective genomic loci. For the latter procedure, we used the BLAT tool at the Human Genome Browser database (19) with cloned cDNA sequences as probes to search the March 2006 version of the human genome. Straightforward assignment of HML-6 cDNAs to proviral loci was feasible because of about 9% sequence divergence between the various HML-6 loci in the human genome.
RESULTS
This investigation was initiated to establish an overall retroviral expression profile for the human mammary gland and to evaluate the possible implications of differentially active retroviral elements in human breast cancer. Our rationales were, first, to identify constitutively active HERVs (i.e., HERVs which are active in all mammary gland samples analyzed and thus constitute a typical tissue-specific HERV transcription pattern (13) and, second, to compare incidence and transcript levels of differentially expressed proviruses for pairs of malignant and normal tissue samples from the 23 patients (one pair per patient).
Tissue blocks from all patients were macroscopically prepared to minimize the amount of normal cells contained in tumor tissue samples and to make sure that normal tissue samples were free of contaminating tumor cells. To examine the transcriptome of the human mammary gland for retroviral sequences, we employed a previously established, retrovirus-specific microarray that allows simultaneous detection and identification of a wide variety of HERV elements as well as human and mammalian exogenous retroviruses (41). The microarray consists of 52 representative HERV pol(RT)-derived sequences from 18 major HERV families, all of which include at least one full-length provirus in the human genome (27), and 11 pol(RT)-derived sequences from mammalian exogenous retroviruses, including MMTV. The DNA chip has been successfully used to analyze the transcriptional HERV activity in various human tissues and to comprehensively profile retrovirus expression in brain samples from patients with schizophrenia and bipolar disorders (13, 41, 42).
Discriminatory power of the retrovirus-specific microarray for betaretroviruses.
To assure the best performance of the DNA chip assay in terms of detection, identification, and discrimination of endogenous HERV-K(HML) sequences as well as exogenous MMTV and the putative human MMTV-like virus HMLV, DNA sequence alignments were performed with corresponding capture probes (Fig. 1A). The sequence identity of MMTV to representative members of HERV-K HML subgroups ranges from 41% to 61%, whereas the putative HMLV provirus described by Liu and coworkers (26) is more closely related to MMTV (96% sequence identity, five mismatches) in the corresponding reverse transcriptase region. Based on calculated sequence similarities, a set of pilot chip hybridization experiments was carried out to identify hybridization conditions for optimal discrimination (Fig. 1B). As hybridization probes, a standardized panel of five synthetic Cy3-labeled oligonucleotides corresponding to the pol(RT) regions of MMTV, HMLV, HERV-K10 (HML-2 family), HERV-K(HML-1), and HERV-K(HML-5) was used. Hybridization with MMTV- and HMLV-specific oligonucleotide probes resulted in one single hybridization signal each (Fig. 1B, rows 1 and 2, respectively). Therefore, the MMTV-specific capture probe can be used to detect HMLV sequences in biological samples. Furthermore, hybridizations were sufficiently specific to avoid cross-hybridization between HMLV and endogenous HERV-K(HML)-related capture probes. HERV-K10 (HML-2 family), HERV-K(HML-1), and HERV-K(HML-5)-specific control oligonucleotides also resulted in the expected signals (Fig. 1B, row 3), illustrating the specificity of our assay. The high sensitivity of the microarray was demonstrated previously in dilution experiments using pig endogenous retrovirus DNA. As few as 25 copies of pig endogenous retrovirus DNA could be detected in 100 ng human cDNA in a standardized assay (41). High sensitivity was further demonstrated by the detection of MLV-reverse transcriptase nucleic acid contamination introduced by commercial enzyme preparations (13).
To test for the ability of the MOP cocktail to amplify MMTV/HMLV sequences, chip hybridization experiments were performed using cDNA samples derived from murine GR cells harboring a productive strain of MMTV (17). As shown in Fig. 1B, row 4, the microarray clearly detected MMTV infection. Moreover, MLV transcripts that are a characteristic feature of all cells of murine origin were observed in GR cells. Interestingly, mouse GR cells seem to contain HML-10-related transcripts, as revealed by the occurrence of a signal at the respective position on the chip. This result may be due to transcribed IAPEz sequences that are detected as best matches when the mouse genome sequence is searched with a representative HML-10 sequence (18). Except for GAPD (glyceraldehyde-3-phosphate dehydrogenase), hybridization signals of housekeeping genes point to the close phylogenetic relationship of these genes in mice and humans.
HERV activity signature of human mammary gland tissue.
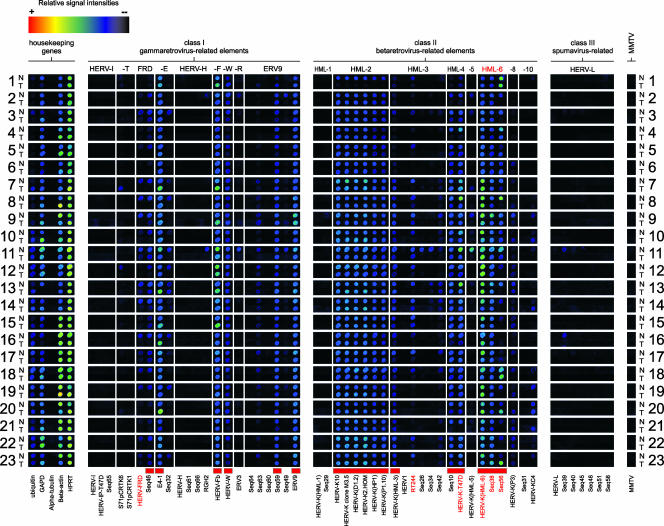
Paired mRNA samples extracted from nonmalignant and tumor cells were tested in triplicate microarray hybridizations according to a standardized chip hybridization protocol (15). A digitally processed alignment of a representative data set for all patient samples is shown in Fig. 2. A panel of housekeeping genes served as an internal control for RNA quality. Of these, the gene for HPRT showed the most consistent transcriptional levels, indicating comparable RNA quality for all 46 RNA samples. All nonmalignant samples showed a mammary gland-specific HERV transcription signature clearly distinct from that found in other human tissues (13, 42). As expected from the results of previous studies (41, 42), not all HERV taxa appear equally active in mammary gland tissue. Differential transcriptional activity was observed, ranging from HERV taxa with ubiquitous or frequent activity to HERV taxa with rare or no detectable expression. HERV taxa were clustered into three groups corresponding to elements with (i) ubiquitous (incidence was 22 or 23 out of 23 samples), (ii) differential (incidence was 2 to 21 of 23 samples), and (iii) no or rare transcriptional activity (incidence was 0 or 1 of 23 samples). HERV taxa with ubiquitously detectable transcripts are considered core components of the human mammary gland transcriptome. These HERV sequences are marked in Fig. 2 and 3.
FIG. 2.
HERV transcriptional activity in human breast cancer. A total of 46 samples representing pairs of normal (N) and malignant (T) mammary gland tissue specimens from the same patient (n = 23) were compared by microarray hybridization. A housekeeping gene panel served as an internal control. Cy3-labeled DNA hybridization probes were generated by RT-PCR on mRNA according to the standardized protocol (41). HERV elements representing the mammary gland-specific HERV activity profile are indicated by red boxes (bottom line). QRT-PCR was performed for a subset of four differentially active HERV elements (HERV-FRD, RT244, HERV-K-T47D, and Seq56), shown in red letters. For detailed information about the identity of targets and capture probes see references 13 and 42. Albeit false color mapping was used for image visualization, the printed image may not properly display signal gradation, especially for weak signals.
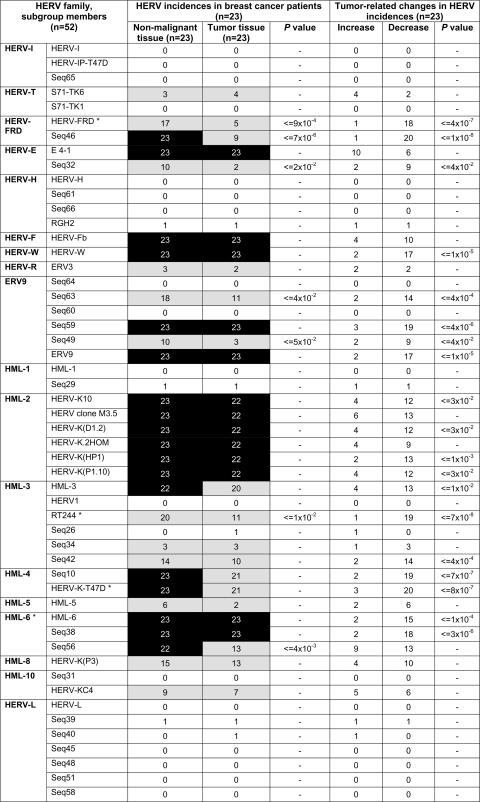
FIG. 3.
Qualitative analysis of HERV transcription in pairs of mammary gland tissue specimens corresponding to nonmalignant and tumor cells of 23 breast cancer patients. For the determination of HERV incidences, only those HERVs with signal intensities above a defined cutoff value (4,000 relative units) were evaluated. Constitutively active HERVs are marked by black boxes (incidence, 22 or 23 out of 23), and differentially active (2 to 21 out of 23) and inactive (0 or 1 out of 23) HERV elements are depicted by gray and white boxes, respectively. Tumor-related changes in HERV incidences were defined by normalizing particular HERV transcript levels with their corresponding HPRT transcript levels, whereby only alterations greater than 10% were judged as changes. Asterisks denote HERV elements that were subjected to QRT-PCR. −, the P value was not significant after Fisher's exact test.
The retroviral core activity signature is composed of 18 proviruses from nine families of both class I and II HERV elements. Ubiquitous class I gammaretroviral elements are HERV-FRD (Seq46), HERV-E (E4-1), HERV-F (HERV-Fb), HERV-W, two ERV9 elements (Seq59 and ERV9), and members of the class II betaretrovirus elements, including all six HML-2 elements [HERV-K10, HERV-K clone M3.5, HERV-K(D1.2), HERV-K2.HOM, HERV-K(HP1), and HERV-K(P1.10)], HML-3 [HERV-K(HML-3)], HML-4 (HERV-K-T47D, Seq10), and all three HML-6 elements [HERV-K(HML-6), Seq38, and Seq56]. Spumavirus-related class III elements are not constitutively active in the mammary glands. In addition, differentially active HERVs (2 to 21 out of 23 samples) were identified in nonmalignant tissue samples (Fig. 3). These transcripts originate from class I (HERV-T, HERV-FRD, HERV-E, HERV-R, and ERV9) and class II families (HML-3, -5, -8, and -10). Human exogenous retroviruses (human immunodeficiency virus types 1 and 2, HTLV-1 and -2, and human foamy virus) were not detected (data not shown).
Searching for MMTV homologous sequences.
In contrast to the results of several prodromal test hybridization experiments employing synthetic MMTV- and HMLV-specific hybridization probes and cDNA from murine GR cells as controls (Fig. 1B, rows 1, 2, and 4), we did not detect MMTV-specific signals in any of the nonmalignant and tumor tissue samples (Fig. 2). Likewise, both DNA and RNA samples derived from the human mammary carcinoma cell line T47D, which was described as HMLV env positive (6, 49), turned out to be negative for MMTV/HMLV pol transcripts and proviral DNA in our assays (data not shown).
Comparative analysis of nonmalignant and tumor tissue profiles.
In order to search for tumor-related HERV elements, we compared the expression profiles of pairs of nonmalignant and tumor tissue samples (Fig. 2). Remarkably, none of the HERVs lacking pol transcription in nonmalignant mammary gland tissue became transcriptionally activated in the corresponding tumor. A comparison of HERV incidences revealed that the expression profiles of tumor samples were generally similar to those of nonmalignant tissue samples. As shown in Fig. 3, constitutively active HERV taxa (HERV-FRD, HERV-E, HERV-F, HERV-W, ERV9, HML-2, HML-3, HML-4, and HML-6) that make up the characteristic signature are represented equally in both tissues except for HERV-FRD (Seq46) and HML-6 (Seq56) elements, which display significantly lower HERV incidences of transcriptional activity in tumor tissues. A similar distribution in normal and tumor tissue was also observed for the variably transcribed HERVs, confirming individual differences in HERV expression patterns and thereby underlining the importance of pairwise comparisons.
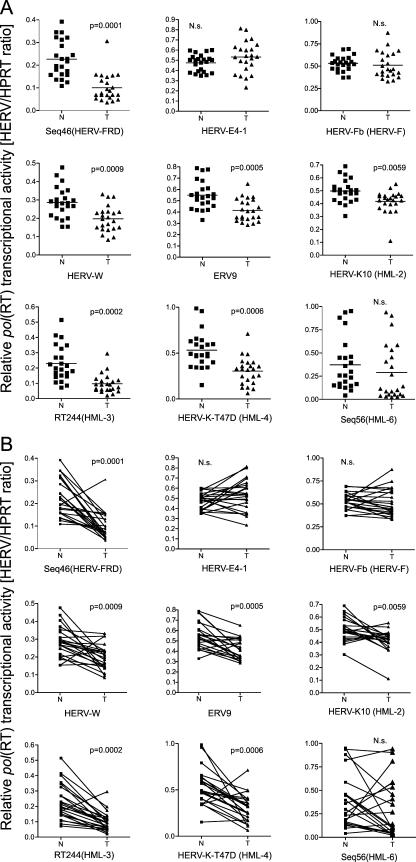
Quantitative analysis of overall HERV transcript levels based on microarray densitometry revealed significantly lower transcript levels in tumors compared to nonmalignant mammary gland tissue. As a representative set of data, scatter plots (representing nonmalignant versus tumor tissue samples) for nine HERV taxa of the characteristic core expression pattern are shown in Fig. 4. Irrespective of the general trend, all active HERVs (n = 30) (Fig. 3) displayed, at least in some cases (median, 2; range, 0 to 10), an increase of transcripts in the tumor (Fig. 3 and 4B). Of note, while the number of downregulated HERVs (median, 17; range, 5 to 24) in tumors from most patients exceeded the number of upregulated HERVs (median, 3; range, 0 to 14), there was a reverse trend in one patient (patient 11 [data not shown]). Interestingly, this patient represents one of two patients with an invasive mixed medullary type carcinoma (see Table S1 in the supplemental material).
FIG. 4.
Quantitative analysis of HERV transcript levels based on microarray densitometry was performed for nine HERV elements, representing core components of the human mammary gland HERV transcriptome, and is illustrated by group (A) and pairwise comparisons (B). Based on mean values of triplicate DNA chip hybridizations, representative relative abundancies of HERV transcripts in each sample pair (N indicates normal tissue, and T indicates tumor tissue) were normalized by respective HPRT transcript levels. P values are given for median value comparisons of N and T groups using the Wilcoxon signed-rank test. N.s., not significant.
QRT-PCR on HERV activity.
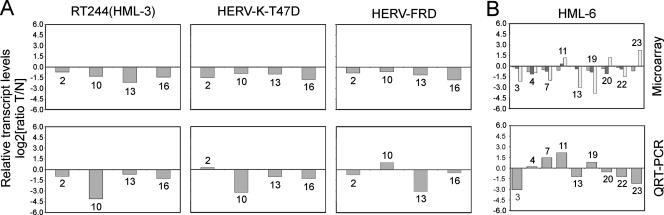
Interesting candidate HERVs, as revealed by microarray analysis, were further quantified by QRT-PCR. In this respect, we focused on three HERV taxa that were among those with the most significant transcriptional decreases in tumor tissue (Fig. 3 and 4), represented by element RT244 (HML-3 family), HERV-K-T47D (HML-4 family), and HERV-FRD. QRT-PCR was performed on paired tissue samples of a representative set of patients (n = 4; patients 2, 10, 13, and 16) (Fig. 5A) by employing HERV subgroup-specific pol primers for RT244(HML-3), HERV-FRD, and HERV-K-T47D that generated specific PCR products when genomic DNA was used as the test target (data not shown). Notably, the HERV subgroup-specific QRT-PCR amplicons overlap with the corresponding microarray capture probe sequences (see Materials and Methods). Comparing microarray and QRT-PCR data, we observed a general concordance for all three HERVs, i.e., downregulation of HERVs in tumor tissue, except for HERV-FRD, which showed an inverse trend in a single patient (patient 10).
FIG. 5.
(A) Tissue type-specific (malignant versus nonmalignant) transcript level alterations of RT244(HML-3), HERV-K-T47D (HML-4 family), and HERV-FRD derived from DNA chip densitometry (upper panels) were compared with QRT-PCR data using subgroup-specific amplification primers (lower panels). (B) Analogous analysis of three HML-6 members [HERV-K(HML-6), Seq38, and Seq56]. Microarray analysis (upper panel) and QRT-PCR, using primers specific for the HML-6 family, according to the method of Muradrasoli and coworkers (33) (lower panel) was performed. Numbers above and below the bars correspond to identification numbers of analyzed patient samples. All ratio calculations (tumor [T] versus normal [N]) are based on values normalized to their respective housekeeping gene (HK6) levels [(HERV/HK6)T/(HERV/HK6)N] that were subsequently log2 transformed. Microarray and QRT-PCR data are based on triplicate experiments as outlined in Materials and Methods.
Although exhibiting a lower incidence in tumors compared to that in nonmalignant tissues (Fig. 3), Seq56 (HML-6 family) indicates increased transcription in a remarkably high number of tumors (i.e., in nearly 40% of patients) (Fig. 4B). Since it was not possible to obtain functional subgroup-specific primers for Seq56 (data not shown), recently published degenerate primers specific for the HML-6 family were used (33). In contrast to primers used for QRT-PCR analysis of RT244(HML-3), HERV-FRD, and HERV-K-T47D, both HML-6 primers are located upstream from the microarray capture probe sequence. Interestingly, three out of nine analyzed patients (Fig. 5B) showed increased transcriptional levels in tumor tissue (patients 7, 11, and 19). When we compared QRT-PCR with microarray data (Fig. 5) for the three HML-6 taxa, HERV-K(HML-6), Seq38, and Seq56, each revealed concordant results for five (patients 3, 13, 20, 22, and 23), five (patients 1, 11, 13, 20, and 22), and four (patients 1, 11, 13, and 22) patients, respectively. Compared to the results for RT244(HML-3), HERV-FRD, and HERV-K-T47D, lower concordance between microarray and QRT-PCR data might be explained by differences in primer specificity, thus amplifying preferentially different HML-6 targets. Furthermore, specific capture probes used for the microarray allow for discrimination between several HML-6 subtypes, whereas primers used for QRT-PCR cover transcripts of nearly the whole HML-6 family (33). Nevertheless, in most cases, QRT-PCR data show the same tendency as do data from the densitometric microarray analysis, suggesting that a comparison of paired samples from the same patient in our microarray system might allow for semiquantitative evaluation. The same tendency was also observed when the gene for HPRT was used instead of the gene for GUS as a housekeeping gene in a set of representative samples (data not shown).
Differential transcription of HML-6 in normal and tumor tissues.
In order to characterize in more detail HML-6 transcripts in nonmalignant and tumor mammary gland tissue specimens, PCR was performed with the primer HML6RTfw (33), already used in QRT-PCR analysis, in combination with two specifically designed reverse primers (HML6RTrv3 and HML6RTc16). Based on sequence comparisons, those primers are able to amplify a PCR product from at least 20 HML-6 loci in the human genome, among them all three HML-6 subgroups represented as capture probes on the HERV chip (HML-6, Seq38, and Seq56). Sequence analysis of cloned HML-6 PCR products obtained from genomic DNA demonstrated the specificity of PCR primers as well as the amplification of a number of HML-6 loci (data not shown). We sequenced, in total, 86 HML-6 cDNAs derived from nonmalignant and tumor tissues from patients 7 and 11 and assigned their sequences to individual HML-6 loci in the human genome. While most cDNA sequences displayed 100% or close to 100% sequence identity with a genomic HML-6 locus, 11 cDNA sequences were excluded from further analysis because of significantly lower best hits, ranging between 93.6% and 98%. Those sequences were most likely recombination products between different transcripts/cDNAs that arose during RT-PCR (10). We thus further analyzed 75 sequences in total (between 16 and 21 cDNAs per sample). Our analysis identified transcripts from six different HML-6 loci located on human chromosomes 5, 11, 12, 19, and 20 (Table 1). For each sample, a locus on chromosome 19 (HML6-c19A) (Table 2) was cloned most frequently, indicating higher transcriptional activity for that locus than for other active HML-6 loci. When comparing normal versus tumor tissue from patient 11, we found cDNAs from two HML-6 loci in tumor tissue (HML6-c12 and HML6-c19B) but not in normal tissue. For patient 7, transcripts from two HML-6 loci were found only in normal tissue (HML6-c11 and HML6-c20), whereas one locus was found only in tumor tissue (HML6-c19B).
TABLE 1.
Localization of transcriptionally active HML-6 proviruses in the human genome sequencea
| Provirus | Chromosomal localizationb | Chromosome bandc |
|---|---|---|
| HML6-c5d | chr 5:70907221-70907558 | 5q13.2 (+) |
| HML6-c11 | chr 11:7880587-7880910 | 11p15.4 (−) |
| HML6-c12 | chr 12:110744034-110744371 | 12q24.12 (−) |
| HML6-c19A | chr 19:57003399-57003736 | 19q13.33 (−) |
| HML6-c19B | chr 19:9483047-9483385 | 19q13.2 (−) |
| HML6-c20 | chr 20:25327489-25327823 | 20p11.21 (−) |
Localization data are as given by the Human Genome Browser (March 2006 version). chr, chromosome.
Chromosomal localization of HML-6 proviral portions amplified by RT-PCR.
Cytogenetic localization of HML-6 proviruses. The orientations of proviruses on the chromosomes are given in parentheses.
Best hits of probe sequences to that locus include an identical hit to a chromosome 5 alternate assembly of the SMN1 gene region (chr5_h2_hap1:1532284-1532621 [http://www.ncbi.nlm.nih.gov/genome/guide/human/release_notes.html]).
TABLE 2.
Relative cloning frequencies of cDNAs from various HML-6 proviruses from different patient samplesa
| Provirus | Frequency for patient sample
|
|||
|---|---|---|---|---|
| 7N | 7T | 11N | 11T | |
| HML6-c5 | 9.52 | 15 | 11.11 | 25 |
| HML6-c11 | 9.52 | 5.56 | 18.75 | |
| HML6-c12 | 6.25 | |||
| HML6-c19A | 76.19 | 80 | 83.33 | 31.25 |
| HML6-c19B | 5 | 12.50 | ||
| HML6-c20 | 4.76 | 6.25 | ||
| Absolute no. of analyzed cDNAs | 21 | 20 | 18 | 16 |
Provirus names are as detailed in Table 1. Normal and tumor tissues from patients 7 and 11 are designated “N” and “T,” respectively. Numbers indicate relative cloning frequencies (given as percentages) of the cDNA of a particular provirus. Absolute numbers of analyzed cDNAs per patient sample are given in the bottom row.
DISCUSSION
Identification of a characteristic HERV transcription profile in human mammary gland.
Analysis of HERV transcription patterns in nonmalignant mammary gland tissues revealed a core transcription signature that consists of members of class I families HERV-FRD, HERV-E, HERV-F, HERV-W, and ERV9 and class II families HML-2, -3, -4, and -6 and is in accordance with previously published data (42). A further important outcome of this study is the definition of variably active HERVs, which include elements of class I and II HERVs (Fig. 3), that may reflect individual epigenetic differences in the HERV methylation status or chromatin structure (21, 24) and in the availability of cellular transcription factors (35, 39). Such individual variability appears to be a typical feature of HERV transcription and has also been shown, for example, for the human brain (13). These data must be taken into account in view of identifying altered HERV activity in mammary carcinomas.
Breast cancer-related alterations in HERV activity.
The paramount aim of this study was to explore alterations in HERV transcriptional activity in mammary carcinomas, especially the activation of HERVs that have also been postulated for other malignant diseases (reviewed in reference 34). Potentially pathogenic HERVs, dormant in the human genome, may be activated by exogenous triggers, such as radiation, chemical mutagens, or other forms of cellular stress. However, as for HERV families completely lacking detectable transcriptional activity in nonmalignant mammary gland tissue, our data provide no evidence for the onset of the expression of these HERVs in tumor tissues (Fig. 2 and 3). Irrespective of the lack of transcriptional activation of silent HERV elements, tumor-related differences in incidence and transcript levels of HERVs could provide the molecular basis to illuminate the potential role of HERVs in human breast cancer or their usefulness as markers of malignancy.
The comparison of HERV profiles derived from corresponding nonmalignant and tumor tissue samples revealed a tumor-related HERV profile that broadly reflects the observed retroviral core profile of the nonmalignant mammary gland. The use of paired samples enabled us to overcome problems from interindividual variability in HERV activity described above and to state differences most likely due to the malignant status of the analyzed cells. Unanticipated, generally lower activity was confirmed in tumor tissue for most HERV taxa appearing as conspicuous changes in both incidences and transcript levels (Fig. 3 and 4). Since the relative HERV transcription shown in Fig. 4 represents HERV-to-HPRT ratios, one could argue that observed lower HERV transcript levels might be due to enhanced HPRT expression in tumors. However, this argument seems unlikely as the mean HPRT signal intensities (Fig. 2, housekeeping genes column) are similar within malignant and nonmalignant tissues (data not shown). Our finding is also in line with other studies showing that many HERV elements, including HERV-K(HML) subgroups, are active in a tissue type-specific, concerted fashion in normal breast tissue (43, 54). Although these studies did not use paired tumor/control tissue samples, the high number of samples from both diagnostic groups allowed for statistical analysis of incidences and expression levels. Yin and coworkers (54) used a semiquantitative PCR technique to compare the expressions of five HML families (HML-1, -2, -3, -5, and -6) in 60 breast cancer tissue specimens and 58 nonmalignant control tissue specimens. Stauffer and coworkers (43) analyzed the digital expression patterns of HERV-W, -H, and -E and HML-2 families in normal and cancerous tissues by searching human expressed sequence tag databases. Both approaches revealed largely unchanged or reduced HERV transcription levels in malignant tissue compared to the levels in nonmalignant breast tissue.
The observed overall decrease of HERV activity in breast carcinomas appears to be an epiphenomenon of tumor development and/or progression rather than a causative event. On the molecular level, tumor-related altered transcriptional activity may be explained by the fact that retroviral long-terminal repeats carry essentially very similar regulatory sequences as cellular promoters and therefore obey basal rules of the transcriptional machinery of the “host” cell. Malignant transformation, phenotypically apparent by alterations in proliferation, differentiation, and cell-cell interaction is associated with changes in the transcriptional milieu, and HERVs may be transcriptionally activated or shut down in response to differences in chromatin modeling, methylation status, and the presence of transcription factors. Transcriptional changes of a multitude of cellular genes, including up- and downregulation of transcription factors and other regulatory proteins have been monitored in different stages of breast tumor development (22, 25). Therefore, differences observed in our experiments may reflect changes in the transcriptional machinery in breast cancer cells and may result from the malignant transformation, but do not necessarily point to HERVs as putative etiological agents of malignancy.
Upregulation of distinct HERVs in a remarkable percentage of patients.
While most HERVs show decreased transcript levels in tumor tissue, we found enhanced activity of some distinct HERVs in more than 20% of analyzed patients. In a few previous studies, some single HERV-K elements were reported to be expressed predominantly in breast carcinomas or breast cancer cell lines. Among those are members of the HERV-K10 subgroup (5), HERV-K102, and HERV-K109 (50, 51), all belonging to the HML-2 family. However, we note that these studies used different retroviral target sequences (gag and env), were restricted to a rather small number of HERV subgroups (HERV-R, HERV-E, and HML-2), and in particular, used rather small numbers of unpaired nonmalignant and malignant tissues from different patients, thus representing a heterogeneous set of samples.
In our investigation, conspicuous HERV sequences were S71-TK6 (HERV-T family), E4-1 (HERV-E family), HERV-Fb (HERV-F family), HML-2 [all family members except HERV-K(HP1)], HML-3 (HML-3 family), HERV-K(P3) (HML-8 family), HERV-KC4 (HML-10 family), and Seq56 (HML-6 family) because of their upregulation in various subsets of analyzed patients (Fig. 3). These HERVs may represent promising candidates for future efforts toward identifying potential clinical correlates to tumor staging and prognosis.
Seq56 is of particular interest, since there is a striking increase of transcript levels in about 40% of tumors relative to corresponding nonmalignant tissues. Seq56 is identical in sequence to an HML-6 locus on human chromosome 16 (localization, 16:34611961-34612050). Transcripts from the particular chromosome 16 locus were not identified in our study, although the PCR primers employed, in principle, could also amplify cDNAs from that locus. It therefore seems that this locus is, at least, not expressed at significant levels. Further sequence comparison shows that other HML-6 transcripts, several of them identified in this study, could probably also be detected by Seq56 capture probes. It is therefore possible that the upregulation of transcripts from HML-6 loci other than the chromosome 16 locus increased Seq56 microarray signal intensities in tumors compared to the intensities in normal tissues. It is still unclear whether the upregulation of specific HML-6 proviral loci, as reflected by our sequencing data, is due to low copy numbers of tumor-specific HML-6 cDNA clones (Table 2). Increased expression levels for HML-6 sequences, as observed by microarray analysis, could be explained either by upregulated activities of a few specific HML-6 loci or by the upregulation of several HML-6 loci due to more general regulatory differences between normal and tumor tissues. Interestingly, unusually high expression of an HML-6 transcript was also reported for a multifocal breast carcinoma by Yin and coworkers (54). In this study, the expression levels of HML families were investigated in 60 breast cancer tissue samples and compared to those of nonmalignant control tissue samples (see above). Although this specific sequence (HML-6.2BC1) cannot be detected by our microarray because it lacks the pol region used for our assay (55), these data substantiate members of the HML-6 family as a distinctive feature in human breast cancer.
No evidence for exogenous retroviruses in human breast cancer.
Our study furthermore contributes to the ongoing debate on the role of exogenous retroviral sequences in breast cancer. Despite the high discriminatory power and sensitivity of our PCR-based microarray test (Fig. 1), we could not detect HMLV-related pol sequences in breast cancer samples. This result could be due either to the downregulation of HMLV transcription at the time of sample preparation or to a complete absence of HMLV in the examined tissue (28, 29, 53). Besides a lack of HMLV in patient samples, we also could not detect HMLV RNA or DNA in T47D cells (data not shown) that have been described as HMLV positive (6, 49), which points to a lack of HMLV in the T47D cells used in our experiments. Furthermore, microarray analyses of patient samples revealed no activity of other exogenous retroviruses, such as MLV-related xenotropic retroviruses (7, 48).
Conclusions.
We established a consistent HERV expression profile specific for the human mammary gland that comprises constitutively and differentially active HERVs. However, our study challenges the existence of exogenous MMTV-related retroviruses in breast cancer. The obtained retroviral expression profiles confirm several previous findings about the biology of HERVs and add more elements to the list of potential agents that may contribute to the pathogenesis of malignant diseases or represent novel disease markers. This investigation expands the current knowledge of HERVs in cancer tissue and should provide support for the design of future studies aimed at the relevance of mobile genetic elements in human carcinogenesis.
Supplementary Material
Acknowledgments
This work was financially supported by the Medizinische Fakultät Mannheim der Universität Heidelberg. Jens Mayer was supported by funds from Deutsche Forschungsgemeinschaft and HOMFOR.
We thank Jonas Blomberg (Section of Virology, Department of Medical Sciences, Uppsala University, Uppsala, Sweden) for providing HERV standard plasmids. We are grateful to Ulf Krause for assistance in mRNA preparation. We thank Alex D. Greenwood (Old Dominion University, Norfolk, VA) for critically reading the manuscript.
Footnotes
Published ahead of print on 12 December 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Andersson, M. L., M. Lindeskog, P. Medstrand, B. Westley, F. May, and J. Blomberg. 1999. Diversity of human endogenous retrovirus class II-like sequences. J. Gen. Virol. 80255-260. [DOI] [PubMed] [Google Scholar]
- 2.Antoniou, A. C., and D. F. Easton. 2006. Models of genetic susceptibility to breast cancer. Oncogene 255898-5905. [DOI] [PubMed] [Google Scholar]
- 3.Bindra, A., S. Muradrasoli, R. Kisekka, H. Nordgren, F. Warnberg, and J. Blomberg. 2007. Search for DNA of exogenous mouse mammary tumor virus-related virus in human breast cancer samples. J. Gen. Virol. 881806-1809. [DOI] [PubMed] [Google Scholar]
- 4.Burmeister, T., A. D. Ebert, W. Pritze, C. Loddenkemper, S. Schwartz, and E. Thiel. 2004. Insertional polymorphisms of endogenous HERV-K113 and HERV-K115 retroviruses in breast cancer patients and age-matched controls. AIDS Res. Hum. Retrovir. 201223-1229. [DOI] [PubMed] [Google Scholar]
- 5.Ejthadi, H. D., J. H. Martin, J. Junying, D. A. Roden, M. Lahiri, P. Warren, P. G. Murray, and P. N. Nelson. 2005. A novel multiplex RT-PCR system detects human endogenous retrovirus-K in breast cancer. Arch. Virol. 150177-184. [DOI] [PubMed] [Google Scholar]
- 6.Etkind, P., J. Du, A. Khan, J. Pillitteri, and P. H. Wiernik. 2000. Mouse mammary tumor virus-like ENV gene sequences in human breast tumors and in a lymphoma of a breast cancer patient. Clin. Cancer Res. 61273-1278. [PubMed] [Google Scholar]
- 7.Fan, H. 2007. A new human retrovirus associated with prostate cancer. Proc. Natl. Acad. Sci. USA 1041449-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feller, W. F., and H. C. Chopra. 1968. A small virus-like particle observed in human breast cancer by means of electron microscopy. J. Natl. Cancer Inst. 401359-1373. [PubMed] [Google Scholar]
- 9.Fernandez-Cobo, M., S. M. Melana, J. F. Holland, and B. G. Pogo. 2006. Transcription profile of a human breast cancer cell line expressing MMTV-like sequences. Infect. Agents Cancer 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flockerzi, A., J. Maydt, O. Frank, A. Ruggieri, E. Maldener, W. Seifarth, P. Medstrand, T. Lengauer, A. Meyerhans, C. Leib-Mösch, E. Meese, and J. Mayer. 2007. Expression pattern analysis of transcribed HERV sequences is complicated by ex vivo recombination. Retrovirology 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford, C. E., M. Faedo, and W. D. Rawlinson. 2004. Mouse mammary tumor virus-like RNA transcripts and DNA are found in affected cells of human breast cancer. Clin. Cancer Res. 107284-7289. [DOI] [PubMed] [Google Scholar]
- 12.Forsman, A., Z. Yun, L. Hu, D. Uzhameckis, P. Jern, and J. Blomberg. 2005. Development of broadly targeted human endogenous gammaretroviral pol-based real time PCRs quantitation of RNA expression in human tissues. J. Virol. Methods 12916-30. [DOI] [PubMed] [Google Scholar]
- 13.Frank, O., M. Giehl, C. Zheng, R. Hehlmann, C. Leib-Mösch, and W. Seifarth. 2005. Human endogenous retrovirus expression profiles in samples from brains of patients with schizophrenia and bipolar disorders. J. Virol. 7910890-10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank, O., B. Brors, A. Fabarius, L. Li, M. Haak, S. Merk, U. Schwindel, C. Zheng, M. C. Müller, N. Gretz, R. Hehlmann, A. Hochhaus, and W. Seifarth. 2006. Gene expression signature of primary imatinib-resistant chronic myeloid leukemia patients. Leukemia 201400-1407. [DOI] [PubMed] [Google Scholar]
- 15.Frank, O., L. Jones-Brando, C. Leib-Mösch, R. Yolken, and W. Seifarth. 2006. Altered transcriptional activity of human endogenous retroviruses in neuroepithelial cells after infection with Toxoplasma gondii. J. Infect. Dis. 1941447-1449. [DOI] [PubMed] [Google Scholar]
- 16.Glaser, S. L., J. L. Hsu, and M. L. Gulley. 2004. Epstein-Barr virus and breast cancer: state of the evidence for viral carcinogenesis. Cancer Epidemiol. Biomarkers Prev. 13688-697. [PubMed] [Google Scholar]
- 17.Indik, S., W. H. Günzburg, B. Salmons, and F. Rouault. 2005. Mouse mammary tumor virus infects human cells. Cancer Res. 656651-6659. [DOI] [PubMed] [Google Scholar]
- 18.Jurka, J., V. V. Kapitonov, A. Pavlicek, P. Klonowski, O. Kohany, and J. Walichiewicz. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110462-467. [DOI] [PubMed] [Google Scholar]
- 19.Kent, W. J., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, and D. Haussler. 2002. The human genome browser at UCSC. Genome Res. 12996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keydar, I., T. Ohno, R. Nayak, R. Sweet, F. Simoni, F. Weiss, S. Karby, R. Mesa-Tejada, and S. Spiegelman. 1984. Properties of retrovirus-like particles produced by a human breast carcinoma cell line: immunological relationship with mouse mammary tumor virus proteins. Proc. Natl. Acad. Sci. USA 814188-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khodosevich, K., Y. Lebedev, and E. D. Sverdlov. 2004. Large-scale determination of the methylation status of retrotransposons in different tissues using a methylation tags approach. Nucleic Acids Res. 32e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein, C. A., S. Seidl, K. Petat-Dutter, S. Offner, J. B. Geigl, O. Schmidt-Kittler, N. Wendler, B. Passlick, R. M. Huber, G. Schlimok, P. A. Baeuerle, and G. Riethmüller. 2002. Combined transcriptome and genome analysis of single micrometastatic cells. Nat. Biotechnol. 20387-392. [DOI] [PubMed] [Google Scholar]
- 23.Lander, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409860-921. [DOI] [PubMed] [Google Scholar]
- 24.Lavie, L., M. Kitova, E. Maldener, E. Meese, and J. Mayer. 2005. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 79876-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Y., J. Pan, J. L. Li, J. H. Lee, C. Tunkey, K. Saraf, J. C. Garbe, M. Z. Whitley, S. A. Jelinsky, M. R. Stampfer, and S. A. Haney. 2007. Transcriptional changes associated with breast cancer occur as normal human mammary epithelial cells overcome senescence barriers and become immortalized. Mol. Cancer 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, B., Y. Wang, S. M. Melana, I. Pelisson, V. Najfeld, J. F. Holland, and B. G. Pogo. 2001. Identification of a proviral structure in human breast cancer. Cancer Res. 611754-1759. [PubMed] [Google Scholar]
- 27.Mager, D. L., and P. Medstrand. 2003. Retroviral repeat sequences, p. 57-63. In D. Cooper (ed.), Nature encyclopedia of the human genome. Nature Publishing Group, London, United Kingdom.
- 28.Mant, C., and J. Cason. 2004. A human murine mammary tumour virus-like agent is an unconvincing aetiological agent for human breast cancer. Rev. Med. Virol. 14169-177. [DOI] [PubMed] [Google Scholar]
- 29.Mant, C., C. Gillett, C. D'Arrigo, and J. Cason. 2004. Human murine mammary tumour virus-like agents are genetically distinct from endogenous retroviruses and are not detectable in breast cancer cell lines or biopsies. Virology 318393-404. [DOI] [PubMed] [Google Scholar]
- 30.McPherson, K., C. M. Steel, and J. M. Dixon. 2000. ABC of breast diseases. Breast cancer—epidemiology, risk factors, and genetics. BMJ 321624-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medstrand, P., and J. Blomberg. 1993. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J. Virol. 676778-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mesa-Tejada, R., I. Keydar, M. Ramanarayanan, T. Ohno, C. Fenoglio, and S. Spiegelman. 1978. Detection in human breast carcinomas of an antigen immunologically related to a group-specific antigen of mouse mammary tumor virus. Proc. Natl. Acad. Sci. USA 751529-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muradrasoli, S., A. Forsman, L. Hu, V. Blikstad, and J. Blomberg. 2006. Development of real-time PCRs for detection and quantitation of human MMTV-like (HML) sequences HML expression in human tissues. J. Virol. Methods 13683-92. [DOI] [PubMed] [Google Scholar]
- 34.Nelson, P. N., P. R. Carnegie, J. Martin, E. H. Davari, P. Hooley, D. Roden, S. Rowland-Jones, P. Warren, J. Astley, and P. G. Murray. 2003. Demystified… Human endogenous retroviruses. Mol. Pathol. 5611-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okahara, G., S. Matsubara, T. Oda, J. Sugimoto, Y. Jinno, and F. Kanaya. 2004. Expression analyses of human endogenous retroviruses (HERVs): tissue-specific and developmental stage-dependent expression of HERVs. Genomics 84982-990. [DOI] [PubMed] [Google Scholar]
- 36.Ono, M., T. Yasunaga, T. Miyata, and H. Ushikubo. 1986. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J. Virol. 60589-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reus, K., J. Mayer, M. Sauter, H. Zischler, N. Müller-Lantzsch, and E. Meese. 2001. HERV-K(OLD): ancestor sequences of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 758917-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkar, N. H., and D. H. Moore. 1972. On the possibility of a human breast cancer virus. Nature 236103-106. [DOI] [PubMed] [Google Scholar]
- 39.Schön, U., W. Seifarth, C. Baust, C. Hohenadl, V. Erfle, and C. Leib-Mösch. 2001. Cell type-specific expression and promoter activity of human endogenous retroviral long terminal repeats. Virology 279280-291. [DOI] [PubMed] [Google Scholar]
- 40.Seifarth, W., C. Baust, A. Murr, H. Skladny, F. Krieg-Schneider, J. Blusch, T. Werner, R. Hehlmann, and C. Leib-Mösch. 1998. Proviral structure, chromosomal location, and expression of HERV-K-T47D, a novel human endogenous retrovirus derived from T47D particles. J. Virol. 728384-8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seifarth, W., B. Spiess, U. Zeilfelder, C. Speth, R. Hehlmann, and C. Leib-Mösch. 2003. Assessment of retroviral activity using a universal retrovirus chip. J. Virol. Methods 11279-91. [DOI] [PubMed] [Google Scholar]
- 42.Seifarth, W., O. Frank, U. Zeilfelder, B. Spiess, A. D. Greenwood, R. Hehlmann, and C. Leib-Mösch. 2005. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 79341-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stauffer, Y., G. Theiler, P. Sperisen, Y. Lebedev, and C. V. Jongeneel. 2004. Digital expression profiles of human endogenous retroviral families in normal and cancerous tissues. Cancer Immun. 42. [PubMed] [Google Scholar]
- 44.Sverdlov, E. D. 2005. Retroviruses and primate genome evolution. Landes Bioscience, Georgetown, TX.
- 45.Szabo, S., A. M. Haislip, and R. F. Garry. 2005. Of mice, cats, and men: is human breast cancer a zoonosis? Microsc. Res. Tech. 68197-208. [DOI] [PubMed] [Google Scholar]
- 46.Tönjes, R. R., F. Czauderna, and R. Kurth. 1999. Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J. Virol. 739187-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tristem, M. 2000. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J. Virol. 743715-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urisman, A., R. J. Molinaro, N. Fischer, S. J. Plummer, G. Casey, E. A. Klein, K. Malathi, C. Magi-Galluzzi, R. R. Tubbs, D. Ganem, R. H. Silverman, and J. L. DeRisi. 2006. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Wang, Y., V. Go, J. F. Holland, S. M. Melana, and B. G. Pogo. 1998. Expression of mouse mammary tumor virus-like env gene sequences in human breast cancer. Clin. Cancer Res. 42565-2568. [PubMed] [Google Scholar]
- 50.Wang-Johanning, F., A. R. Frost, G. L. Johanning, M. B. Khazaeli, A. F. LoBuglio, D. R. Shaw, and T. V. Strong. 2001. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin. Cancer Res. 71553-1560. [PubMed] [Google Scholar]
- 51.Wang-Johanning, F., A. R. Frost, B. Jian, L. Epp, D. W. Lu, and G. L. Johanning. 2003. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene 221528-1535. [DOI] [PubMed] [Google Scholar]
- 52.Witkin, S. S., N. H. Sarkar, R. A. Good, and N. K. Day. 1980. An enzyme-linked immunoassay for the detection of antibodies to the mouse mammary tumor virus: application to human breast cancer. J. Immunol. Methods 3285-91. [DOI] [PubMed] [Google Scholar]
- 53.Witt, A., B. Hartmann, E. Marton, R. Zeillinger, M. Schreiber, and E. Kubista. 2003. The mouse mammary tumor virus-like env gene sequence is not detectable in breast cancer tissue of Austrian patients. Oncol. Rep. 101025-1029. [PubMed] [Google Scholar]
- 54.Yin, H., P. Medstrand, M. L. Andersson, A. Borg, H. Olsson, and J. Blomberg. 1997. Transcription of human endogenous retroviral sequences related to mouse mammary tumor virus in human breast and placenta: similar pattern in most malignant and nonmalignant breast tissues. AIDS Res. Hum. Retrovir. 13507-516. [DOI] [PubMed] [Google Scholar]
- 55.Yin, H., P. Medstrand, A. Kristofferson, U. Dietrich, P. Aman, and J. Blomberg. 1999. Characterization of human MMTV-like (HML) elements similar to a sequence that was highly expressed in a human breast cancer: further definition of the HML-6 group. Virology 25622-35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.